Introduction
Breast cancer is the most common type of cancer
affecting women worldwide, with ~280,000 cases diagnosed annually
(1). Breast cancer is classified as
luminal A or B, human epidermal growth factor receptor 2
(HER2)-positive or -negative, and triple-negative or -positive
according to the expression of hormone receptors and HER2 (2). Triple-negative breast cancer (TNBC)
tumors lack expression of estrogen receptor (ER), progesterone
receptor (PR) and HER2, have relatively poor outcomes, and cannot
be treated with endocrine or HER2-targeted therapies (3).
MicroRNAs (miRNAs/miRs) constitute a large family of
small non-coding RNAs comprised of 20–22 nucleotides that regulate
target gene expression, mainly at the post-transcriptional level
(4). Dysregulated miRNAs are
involved in a broad spectrum of cellular processes in TNBC,
exerting tumor-promoting or -suppressing effects depending on the
cellular targets involved in tumor initiation, promotion, malignant
conversion and metastasis (5).
Notably, miRNAs have attracted considerable attention for their
regulatory involvement in the initiation, progression and
metastasis of breast cancer, and aberrant miRNA expression profiles
have been reported in breast cancer (6).
Stanniocalcin 1 (STC1), which was first identified
in bony fish, is a disulfide-bound glycoprotein hormone involved in
plasma calcium and phosphate homeostasis (7). Previous studies have reported that
STC1 functions as an oncogene in various types of cancer; for
example, STC1 has been reported to promote metastasis in ovarian
cancer and cancer-associated fibroblast-derived STC1 can promote
stemness in hepatocellular carcinoma (8,9).
Notably, STC1 promotes the growth and metastasis of breast cancer
tumors (10,11). However, studies on STC1 function in
TNBC are lacking. Therefore, understanding oncogenes and their
regulators might be critical to developing potential antitumor
therapeutics.
The Cancer Genome Atlas (TCGA) database comprises a
collection of clinical data, DNA/RNA sequences and DNA methylation
profiles of at least 500 cases of 20 different tumor types
(12). The Gene Expression Omnibus
(GEO) repository contains publicly available microarray,
next-generation sequencing and other forms of high-throughput
functional genomic data (https://www.ncbi.nlm.nih.gov/geo) (13). The present study aimed to identify
differential miRNA expression patterns in samples from patients
with breast cancer using miRNA sequence expression profiles
downloaded from TCGA and GEO. The present study identified
hsa-miR-606 as a differentially expressed miRNA and investigated
its tumor-suppressing role in TNBC cells. In addition, the targets
of miR-606 and its effects on tumor growth in a mouse xenograft
model were investigated, and its effects on metastasis were
assessed in a mouse tail vein injection model.
Materials and methods
Cells, animals and reagents
The human TNBC cell lines HS 578T, BT20, MDA-MB-231
and BT549 were obtained from the Korean Cell Line Bank; Korean Cell
Line Research Foundation, and have been authenticated in the past 3
years using a short tandem repeat analysis profile. The cells were
cultured in Dulbecco's Modified Eagle Medium (DMEM; 1X; Welgene,
Inc.) supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc.) and penicillin-streptomycin (PS; 100 U/ml;
Gibco; Thermo Fisher Scientific, Inc.). The human breast cancer
cell lines T47D, SK-BR-3 and MCF7 were also obtained from the
Korean Cell Line Bank; Korean Cell Line Research Foundation, and
have been authenticated in the past 3 years using a short tandem
repeat analysis profile. These cells were cultured in Roswell Park
Memorial Institute 1640 (1X; Welgene, Inc.) supplemented with 10%
fetal bovine serum and penicillin-streptomycin (100 U/ml). The
human breast cell line MCF10A was obtained from the American Type
Culture Collection, and was cultured in DMEM/Nutrient Mixture F12
(F12) (1X; Welgene, Inc.) supplemented with 10% fetal bovine serum
and penicillin-streptomycin (100 U/ml). The cells were maintained
at 5% CO2 and 37°C in a humidified incubator, and were
mycoplasma free.
Animal experimental procedures were reviewed and
approved by the CHA University Animal Care and Use Committee
(approval no. 210152; Seongnam, South Korea). A total of 32 female
BALB/c nu/nu mice (age, 4 weeks, weight, 18–20 g) were purchased
from Orient Bio, and were maintained at 23±1°C and 50±10% humidity
under a 12-h light/dark cycle. Food and water were provided ad
libitum under specific pathogen-free conditions.
G-Fectin and Lipofectamine® 3000
transfection reagents were purchased from Genolution, Inc. and
Invitrogen; Thermo Fisher Scientific, Inc., respectively.
Acquisition of miRNA expression
data
To investigate the expression of miRNAs in patients
with breast cancer, a microarray dataset (GSE118782) was obtained
from the publicly available GEO database (National Center for
Biotechnology Information; http://www.ncbi.nlm.nih.gov/geo). The GSE118782
dataset includes RNA extracted from the plasma of 30 patients with
breast cancer and 10 healthy control women; the levels of small
noncoding RNA were quantitated using Affymetrix microarrays
(14).
Cell transfection with RNA
oligonucleotides
The miR-606 mimics and miR-606 inhibitor
(anti-miR-606) were synthesized by Genolution, Inc. as an RNA
duplex or as a 2′-O-methyl-modified oligoribonucleotide single
strand with the following sequences: miR-606 mimics,
5′-AAACUACUGAAAAUCAAAGAU-3′ and anti-miR-606,
5′-AUCUUUGAUUUUCAGUAGUUU-3′. The sequences were obtained from the
miRBase database (version 22.1: http://www.mirbase.org). The sequences for the control
miRNA and control anti-miRNA (Genolution, Inc.) are:
5′-CCUCGUGCCGUUCCAUCAGGUAGUU-3′ or 5′-CAGUACUUUUGUGUAGUACAA-3′,
respectively.
For transfection with miRNAs, MDA-MB-231 and BT549
cells at 30% confluence were transfected with the miRNAs at a final
concentration of 40 nM for 48 h at 37°C using G-Fectin according to
the manufacturer's instructions.
Tumor xenograft and lung metastasis
experiments
To establish a tumor xenograft model, 5-week-old
female BALB/c nu/nu mice were subcutaneously injected with
MDA-MB-231 cells (107 cells in 100 µl PBS). Once the
size of the tumor reached 150 mm3, miRNA mimics were
injected intratumorally, according to the manufacturer's protocol.
Briefly, 10 µg miRNA mimics and 1.2 µl in vivo-jetPEI
(Polyplus-transfection SA) were mixed at a volume of 50 µl,
incubated for 15 min at room temperature and then injected into
mice. To assess tumor size, the small diameter (SD) and large
diameter (LD) of the tumor were measured twice a week using a
Vernier caliper, and the tumor volume (V) was calculated using the
equation: V=(SD2 × LD) × (π/6). At the endpoint, the
mice were sacrificed in a CO2 euthanasia chamber with a
fill rate of 30% vol/min and the euthanasia of the mice was
confirmed by a lack of breathing and faded eye color. The tumors
were then collected for RT-qPCR and IHC analysis. In addition, to
establish a lung metastasis model, MDA-MB-231 cells were
transfected with miRNA mimics, resuspended at 4×105
cells in 200 µl PBS, and injected into the tail vein of mice after
48 h. Subsequently, the mice were sacrificed in a CO2
euthanasia chamber with a fill rate of 30% vol/min at 6 weeks
post-injection, and their lungs were collected. The dissected lungs
were fixed in 3.7% paraformaldehyde for 10 min at 25°C and stained
with Bouin's solution for 5 min at 25°C, after which the nodules
formed in the lungs were counted.
Reverse-transcription quantitative
(RT-q)PCR analysis
Total RNA was isolated from cultured cell lines
(e.g., MCF10A, T47D, SK-BR-3, MCF7, HS 578T, BT20, MDA-MB-231 and
BT549) or dissected xenograft tumor tissue using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. To quantify mRNA or miRNA expression,
total RNA (1 µg) was reverse transcribed into cDNA using Maxime RT
PreMix (oligo dT Primer; Intron Biotechnology, Inc.) or an HB miR
Multi Assay Kit SYSTEM I (HeimBiotek, Inc.), respectively. The RT
conditions of mRNA or miRNA were 45°C for 1 h and at 95°C for 5 min
or 37°C for 1 h and 95°C for 5 min, respectively. Additionally,
qPCR for mRNA or miRNA quantification was performed using AMPIGENE
qPCR Green Mix Lo-ROX (Enzo Life Sciences, Inc.) or an HB miR Multi
Assay Kit SYSTEM I (HeimBiotek, Inc.), respectively, in accordance
with the manufacturers' instructions. The qPCR thermocycling
conditions for detection of mRNA or miRNA expression levels were
Denaturation at 95°C for 2 min, followed by 40 cycles at 95°C for 5
sec and 60°C for 20 sec or denaturation at 95°C for 15 min,
followed by 40 cycles at 95°C for 5 sec and 60°C for 40 sec,
respectively. The mRNA expression levels were normalized to those
of GAPDH, and the miRNA expression levels were normalized to those
of RNU6B. The mRNA or miRNA expression levels were calculated using
the 2−ΔΔCq method (15).
The sequences of the specific primers were as follows: Activin A
receptor, type I (ACVR1), forward 5′-ACGTGGAGTATGGCACTATC-3′,
reverse 5′-CGACACACTCCAACAGTGTA-3′; DCLRE1A, forward
5′-GTGGAGATGGTATTCAGCAG-3′, reverse 5′-CAGAAGACCATCAGGACACT-3′;
STC1, forward 5′-GACACAGTCAGCACAATCAG-3′, reverse
5′-GAGGAGGACTTTCAGCTTCT-3′; GAPDH, forward
5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′,
RNU6B, forward 5′-CTCGCTTCGGCAGCACA-3′, reverse
5′-AACGCTTCACGAATTTGCGT-3′, and miR-606, forward
5′-AAACTACTGAAAATCAAA-3′, reverse 5′-GATACAAGTGCCTGACCACT-3′.
Cell counting assay
MDA-MB-231 and BT 549 cells were seeded in 6-well
plates at 30% confluence and transfected with miRNA mimics. A total
of 24, 48, 72 and 96 h post-transfection, cells were stained with
trypan blue (Gibco; Thermo Fisher Scientific, Inc.) at 25°C for 5
min and were counted using a hemocytometer (NanoEntek, Inc.) and an
Olympus CKX52 inverted light microscope (×100 magnification;
Olympus Corporation).
Colony formation assay
MDA-MB-231 and BT549 cells were seeded in 60-mm
dishes at a density of 500 cells/dish 48 h post-transfection with
miRNA mimics. Thereafter, the dishes were incubated at 37°C in a
humidified incubator containing 5% CO2 for 2 weeks, and
the medium was replaced with fresh medium every 4 days.
Subsequently, the cells were rinsed once with PBS, fixed with 4%
formaldehyde at 25°C for 10 min, washed in distilled water once and
stained with 0.05% crystal violet solution at 25°C for 30 min,
after which the colonies (>50 cells/colony) were counted.
Annexin V/PI apoptosis assay
MDA-MB-231 and BT549 cells were transfected with
miRNA mimics in 6-well plates at 30% confluence. The apoptotic
population in transfected cells was estimated by Annexin-FITC/PI
double staining using the Annexin V-FITC Apoptosis Detection Kit I
(BD Biosciences), according to the manufacturer's protocols. The
stained cells were analyzed using a CytoFLEX Flow Cytometer
(Beckman Coulter, Inc.) and CytExpert software version 2.4 (Beckman
Coulter, Inc.).
Western blotting
MDA-MB-231 and BT549 cells were transfected with
miRNA mimics using G-Fectin. Subsequently, the cells were collected
in PBS in an Eppendorf tube and lysed with PRO-PREP™ Protein
Extraction Solution (Intron Biotechnology, Inc.) to harvest the
proteins. Subsequently, the protein concentrations were measured
using the Pierce Bicinchoninic Acid Protein Assay Kit (Thermo
Fisher Scientific, Inc.). Thereafter, 20 µg proteins were separated
by SDS-PAGE on 8–12% gels at 100 V for 2 h and transferred onto
PVDF membranes at 95 V for 2 h. After transfer, the membranes were
blocked with 5% skim milk in Tris-buffered saline (TBS)-Tween 20
(TBST; Intron Biotechnology, Inc.) at room temperature for 1 h and
incubated with specific primary antibodies in blocking solution
overnight at 4°C. The specific primary antibodies were as follows:
Anti-proliferating cell nuclear antigen (PCNA; cat. no. sc-25280,
1:1,000), anti-BAX (cat. no. sc-23959, 1:1,000), anti-fibronectin
(cat. no. sc-8422, 1:1,000), anti-E-cadherin (cat. no. sc-8426,
1:1,000), anti-β-actin (cat. no. sc-47778, 1:1,000), and anti-STC1
(cat. no. sc-293435, 1:1,000) (all from Santa Cruz Biotechnology,
Inc.); anti-cleaved-poly ADP ribose polymerase (PARP; cat. no.
9541, 1:1,000) and anti-cleaved-caspase 3 (cat. no. 9664, 1:1,000)
(both from Cell Signaling Technology, Inc.); anti-DNA cross-link
repair 1A (DCLRE1A; cat. no. A303-747A, 1:1,000; Bethyl
Laboratories, Inc.; Thermo Fisher Scientific, Inc.); and
anti-N-cadherin (cat. no. 610920, 1:1,000; BD Biosciences). The
membranes were then washed three times with TBST every 10 min and
incubated with HRP-conjugated anti-mouse IgG or anti-rabbit IgG
secondary antibodies (cat. nos. A1012S and A1013S, 1:2,000; ACE
BioLabs) and in TBST at room temperature for 2 h, followed by
detection using a clear western ECL substrate (Bio-Rad
Laboratories, Inc.).
Sphere formation assay
For sphere formation, cells were cultured in cancer
stem cell (CSC)-like cell growth medium, which consisted of
DMEM/F12 serum-free medium supplemented with 2% B-27 (Gibco; Thermo
Fisher Scientific, Inc.), 20 ng/ml Recombinant Human EGF (Gibco;
Thermo Fisher Scientific, Inc.), and 20 ng/ml Recombinant Human
FGFb (Gibco; Thermo Fisher Scientific, Inc.). The cells were seeded
at 3,000 cells/well in 6-well ultra-low cluster plates (Corning
Life Sciences) using CSC-like cell medium, cultured until the
sphere size reached 50 µm, and transfected with the miRNA mimics at
a final concentration of 100 nM using G-Fectin at 37°C. A total of
4 days post-transfection, images of the spheres were captured using
an Olympus CKX52 inverted light microscope (×40 magnification), and
the number and diameter of spheres were measured.
Wound healing assay
A wound healing assay was used to measure cell
migration ability. Briefly, MDA-MB-231 and BT549 cells were
transfected with miRNA mimics at ~100% confluence in 6-well plates.
To create a straight wound, cell monolayers were scratched with a
pipette tip and incubated with fresh serum-free medium. Images were
captured at 0, 24 and 48 h after wounding using an Olympus CKX52
inverted light microscope (×100 magnification), and the wound area
was measured using ImageJ 1.52a (National Institutes of
Health).
Transwell assay
MDA-MB-231 and BT549 cells were transfected with
miRNA mimics. For the invasion and migration assays, the cells were
suspended in serum-free DMEM and seeded at 5×104
cells/well into the upper chamber of a Transwell plate (8.0-µm pore
size; Corning Life Sciences), with or without Matrigel coating,
respectively. For Matrigel coating, the upper chambers were
incubated with Matrigel at 25°C for 30 min. The lower chambers were
filled with DMEM supplemented with 10% FBS and 1% PS. After
incubation for 60–72 h at 37°C, the migrated and invaded cells on
the lower surface of the filters were fixed for 3 min at 25°C in
100% methanol, stained for 5 min at 25°C in 0.05% crystal violet
solution and then washed three times for 1 min in distilled water.
Stained cells were counted using an Olympus CKX52 microscope (×200
magnification) in three randomly selected fields.
Selection of miR-606 putative target
genes
Putative miR-606 target genes were predicted using
the miRNA target prediction programs DIANA-MICROT version 4
(https://dianalab.e-ce.uth.gr/html/universe/index.php?r=microtv4),
TargetScan release 8.0 (https://www.targetscan.org), and miRDB version 6.0
(http://www.mirdb.org). The common genes from all
three algorithms were selected to obtain specific candidate
genes.
Luciferase reporter assay
The possible binding sites of miR-606 in the human
STC1 3′-UTR were predicted using the miRNA target prediction
program TargetScan (https://www.targetscan.org). The four 3′-UTR fragments
containing the putative miR-606 binding site (STC1 3′-UTR-1, STC1
3′-UTR-2, STC1 3′-UTR-3, and STC1 3′-UTR-4) were cloned into a
pGL3UC vector (provided by V.N. Kim, Seoul National University,
Seoul, South Korea) by Macrogen, Inc. MDA-MB-231 and BT549 cells
were seeded in 24-well plates (5×104 cells/well) and
incubated for 24 h, after which the cells were co-transfected with
the reporter plasmid or empty pGL3UC (100 ng), Renilla
plasmid (100 ng) and miR-606 (40 nM) for 48 h at 37°C using
Lipofectamine 3000. A total of 48 h post-transfection, the
transfected cells were lysed using 1X Passive Lysis Buffer (Promega
Corporation) to obtain the release of the firefly and
Renilla luciferase reporter enzymes into the cell lysate,
and then the luciferase activity of the lysed cells was measured
using a Dual-Luciferase Reporter Assay System (Promega Corporation)
according to the manufacturer's protocol. The relative firefly
luciferase activity was normalized to Renilla luciferase
activity.
ELISA
STC1 protein concentrations were determined in the
supernatants of MDA-MB-231 and BT549 cells transfected with miRNA
mimics using a Human Stanniocalcin 1/STC ELISA Kit (Abcam, cat. no.
ab213829). The cell culture supernatant was obtained via
centrifugation of cells at 400 × g for 10 min at 4°C. STC1
concentration was measured using an Epoch Microplate
Spectrophotometer (BioTek Instruments, Inc.) according to the
manufacturer's protocol.
IHC analysis
For the IHC analysis of resected tumor tissues, a
ready-to-use IHC/ICC kit (cat. no. K405-50; BioVision Inc.) was
used according to the manufacturer's protocol. Briefly, tumor
tissues were fixed with 3.7% paraformaldehyde for 24 h at 25°C and
embedded in paraffin. The paraffin-embedded tumor tissue sections
(5 µm) were then deparaffinized with xylene, rehydrated gradually
in ethanol and microwaved for antigen retrieval in Antigen
Retrieval Buffer (cat. no. ab93678; Abcam). Endogenous peroxidase
activity was blocked in 3% hydrogen peroxide for 30 min at 25°C and
sections were then incubated with blocking buffer (BioVision, Inc.)
for 15 min at 25°C. Thereafter, the slides were washed with PBS and
incubated with specific primary antibodies overnight at 4°C. The
following primary antibodies were used: Anti-STC1 (cat. no.
LS-B14899, 1:200; LifeSpan BioScience), anti-PCNA (1:2,000),
anti-BCL-2 (cat. no. sc-7382, 1:100; Santa Cruz Biotechnology,
Inc.) and anti-BAX (1:100). Next, the slides were incubated with
horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG
polyclonal antibodies (cat. no. K405-50-4; BioVision Inc.) for 20
min at 25°C, washed with PBS and were subsequently developed by
staining with 3,3′-diaminobenzidine for 10 min at 25°C. For H&E
staining, tumor or lung tissue was fixed with 3.7% paraformaldehyde
for 24 h at 25°C and embedded in paraffin. The paraffin-embedded
tumor or lung tissue sections (5 µm) were then deparaffinized with
xylene and rehydrated gradually in ethanol. The sections were
incubated with hematoxylin solution (cat. no. 03971;
MilliporeSigma) for 5 min, washed twice with distilled water,
incubated with eosin solution (cat. no. 318906; MilliporeSigma) for
3 min, and then washed with ethanol. Images were captured using the
ZEISS Axioscan 7 microscope in brightfield scanning mode (Zeiss
GmbH).
Overall survival (OS) analysis
To examine the association between OS and miR-606 or
STC1 expression, survival analysis was performed using the K-M
plotter database (www.kmplot.com; accessed on August 4, 2022) (16). The parameters were set in the Breast
cancer miRNA database of the K-M plotter program as follows: i)
Select a dataset, TCGA; ii) gene symbol, hsa-miR-606; iii) divided
patients by auto-selecting the best cutoff; iv) survival, OS; v)
molecular subtype, TNBC. The parameters were set in the Breast
cancer mRNA database of the K-M plotter program as follows: i) gene
symbol, STC1; ii) divided patients by auto-selecting the best
cutoff; iii) survival, OS; iv) ER status, PR status, HER2 status,
negative; v) subtype, basal.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism Software 8 (Dotmatics). Comparisons between groups
were evaluated using the one-way analysis of variance with Tukey's
post hoc test or unpaired Student's t-test. Data are presented as
the mean ± standard error of the mean. For in vitro analyses,
experiments were performed in triplicate. P<0.05 was considered
to indicate a statistically significant difference.
Results
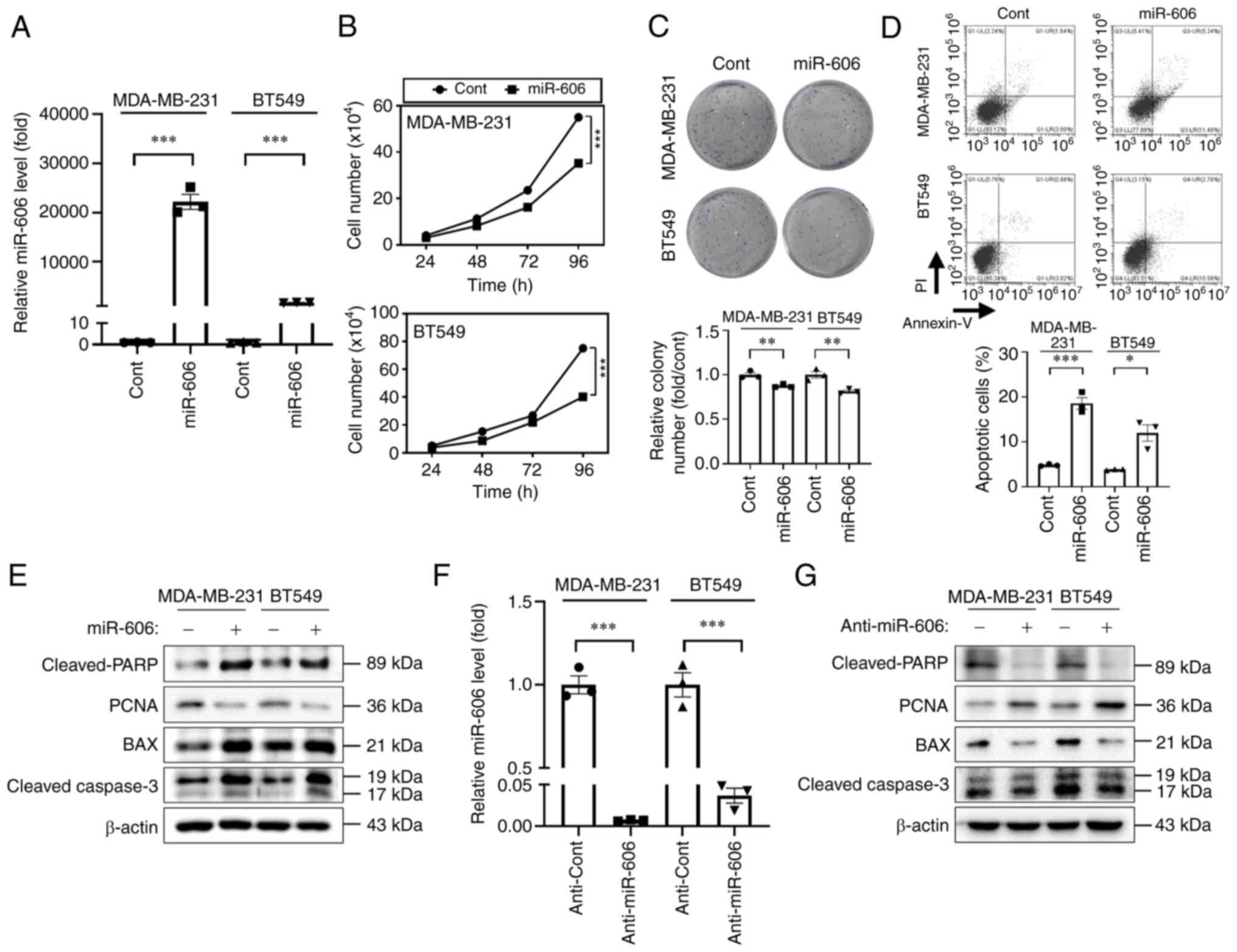
miR-606 induces the apoptosis of TNBC
cells
The present study first investigated the miRNAs that
could regulate breast cancer cell proliferation. The expression
profiles of human miRNAs from a GEO dataset (GSE118782) revealed
that miR-606 was downregulated in the plasma of patients with
breast cancer (Fig. S1A).
Furthermore, miR-606 expression levels were lower in TNBC cells
(i.e., HS 578T, BT20, MDA-MB-231 and BT549) compared with those in
normal breast cells or in cell lines of other breast cancer
subtypes (i.e., MCF10A, T47D, SK-BR-3 and MCF7) (Fig. S1B). To determine the functional
roles of miR-606 in tumor growth inhibition, the TNBC cell lines
MDA-MB-231 and BT549 with the lowest miR-606 expression (Fig. S1B) were transfected with miR-606
mimics. Transfection efficacy was confirmed using RT-qPCR analysis,
which showed that miR-606 expression levels were significantly
increased after transfection for 48 h compared with after
transfection for 0 h with miR-606 mimics (Fig. S2). In addition, as shown in
Fig. 1A, the expression levels of
miR-606 were significantly increased in cells transfected with
miR-606 mimics compared with in those transfected with the control
miRNA for 48 h. The proliferation-inhibiting activity of miR-606
was confirmed by counting the number of cells at different time
points post-transfection; miR-606 overexpression significantly
suppressed the proliferation of MDA-MB-231 and BT549 cells
(Fig. 1B). Colony formation was
also reduced following miR-606 mimics transfection (Fig. 1C). Moreover, transfection with
miR-606 mimics significantly induced the early + late apoptosis of
MDA-MB-231 and BT549 cells (Fig.
1D), and miR-606-induced apoptosis was accompanied by an
increase in the expression levels of cleaved-PARP, BAX and
cleaved-caspase 3, and a decrease in the expression levels of the
proliferative protein PCNA (Fig.
1E). To investigate the effect of anti-miR-606 on MDA-MB-231
and BT549 TNBC cell lines, RT-qPCR analysis was performed
post-transfection. The results revealed a significant reduction in
miR-606 expression levels after anti-miR-606 transfection (Fig. 1F). By contrast, anti-miR-606
significantly inhibited the expression levels of apoptosis markers
and induced the expression of the proliferation marker in
MDA-MB-231 and BT549 cells (Fig.
1F). Collectively, these results suggested that miR-606 induces
TNBC cell apoptosis.
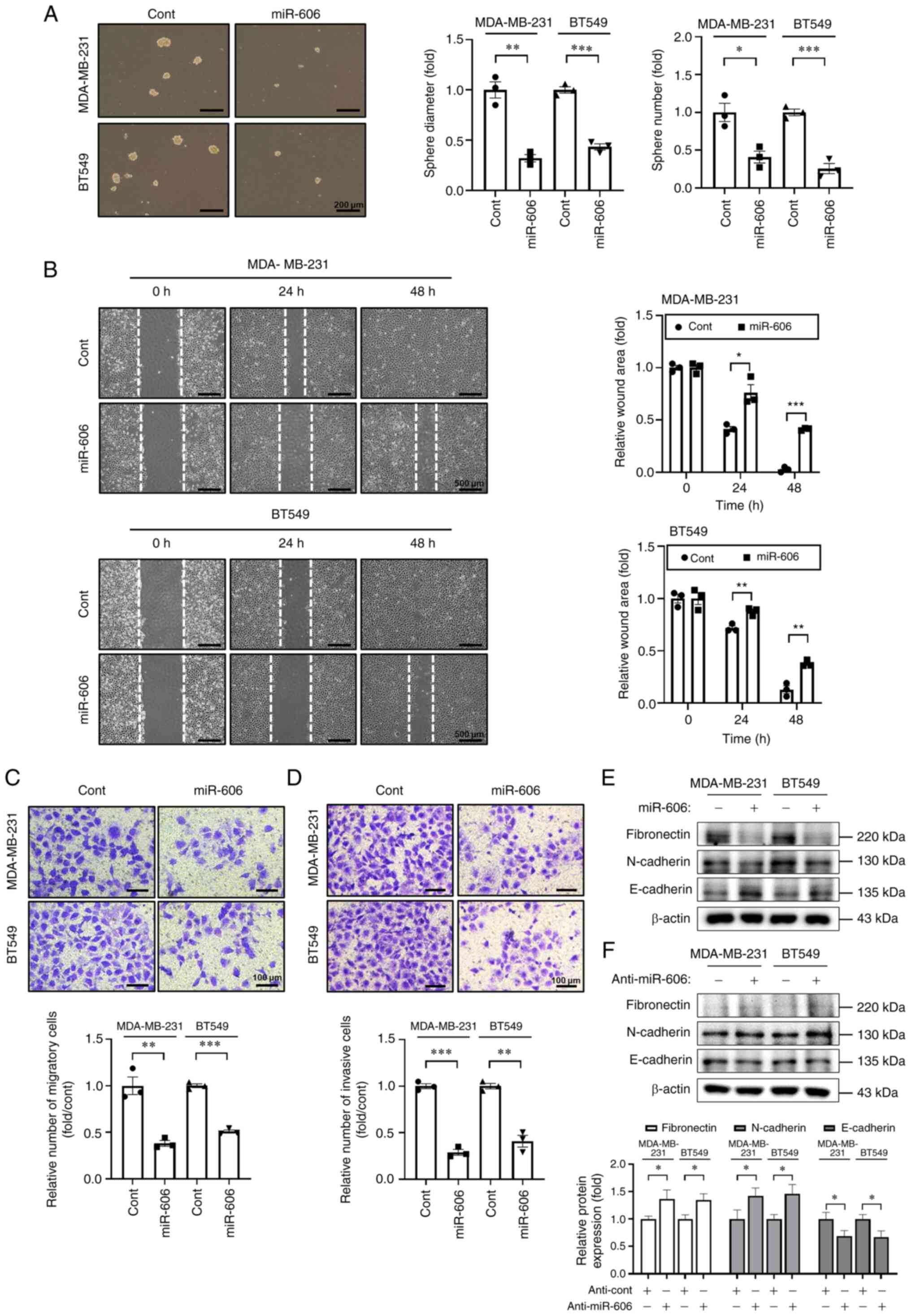
miR-606 suppresses the sphere
formation, migration and invasion of TNBC cells
Subsequently, the effects of miR-606 on various
tumorigenesis-associated characteristics of TNBC cells were
evaluated. It was first determined whether miR-606 influenced the
preservation of the CSC-like phenotype of TNBC cells. To enrich
CSC-like cells, the cells were cultured under serum-free
conditions, and the self-renewal capacity of TNBC cells was
investigated. The tumor sphere-forming ability of MDA-MB-231 and
BT549 cells in CSC-like cell growth medium was estimated 4 days
after transfection with miR-606 mimics or control miRNA. It was
observed that the size and number of tumor spheres were
significantly decreased in cells transfected with miR-606 mimics
(Fig. 2A).
Aggressive and metastatic cancer cells are
characterized by enhanced cell migration and invasion. Therefore,
wound healing assay, Transwell assay and western blotting were
performed to determine the effects of miR-606 on the migratory and
invasive abilities of TNBC cells. The results showed that
transfection with miR-606 mimics significantly suppressed wound
healing in MDA-MB-231 and BT549 cells (Fig. 2B). Furthermore, transfection with
miR-606 mimics significantly decreased the migratory and invasive
abilities of MDA-MB-231 and BT549 cells (Fig. 2C and D). Moreover, in the miR-606
mimics-transfected cells, the expression levels of the mesenchymal
cell markers fibronectin and N-cadherin were downregulated, whereas
those of the epithelial cell marker E-cadherin were upregulated
(Fig. 2E). By contrast, in the
anti-miR-606-transfected cells, the expression levels of
mesenchymal cell markers were increased and those of the epithelial
cell marker were decreased (Fig.
2F). Taken together, these results indicated that miR-606 may
inhibit the tumorigenesis-associated characteristics of TNBC
cells.
miR-606 directly targets STC1
The miRNA target prediction algorithms miRDB,
TargetScan and DIANA were used to predict the target genes of
miR-606; several candidate targets were identified (Fig. 3A). Three common genes were predicted
by all three algorithms: ACVR1, DCLRE1A and STC1. Therefore, the
mRNA and protein expression levels of these three target candidates
were evaluated after miR-606 mimics transfection in MDA-MB-231 and
BT549 cells. In both cell lines, the mRNA expression levels of
DCLRE1A and STC1 were significantly downregulated in miR-606
mimics-transfected cells; however, the mRNA expression levels of
ACVR1 were not significantly downregulated in miR-606
mimics-transfected MDA-MB-231 cells (Fig. 3B). Moreover, in both cell lines, the
protein expression levels of STC1 were markedly reduced by miR-606
overexpression, whereas those of DCLRE1A were not altered (Fig. 3C). Additionally, it was revealed
that the expression levels of STC1 were higher in TNBC cells
compared with those in normal breast cells or in cell lines of
other breast cancer subtypes (Fig.
S3A), and the mRNA expression levels of STC1 were downregulated
or upregulated in TNBC cell lines following miR-606 mimics or
anti-miR-606 transfection, respectively (Fig. S3B and C). Similarly, the secreted
STC1 protein levels in the supernatants of miR-606
mimics-transfected MDA-MB-231 and BT549 cells were significantly
reduced compared with those in the supernatant of control
miRNA-transfected MDA-MB-231 and BT549 cells (Fig. 3D). Furthermore, the protein
expression levels of STC1 were increased in
anti-miR-606-transfected MDA-MB-231 and BT549 cells (Fig. 3E). Therefore, STC1 was further
investigated using a reporter assay to determine whether it is a
direct target of miR-606. Four putative miR-606-binding sites,
complementary to the seed region of miR-606, were identified in the
3′-UTR of STC1. Therefore, the putative miR-606-binding sequence
was cloned into a modified pGL3 reporter vector, pGL3UC (Fig. 3F), and the luciferase reporter
activity was measured 48 h post-co-transfection of MDA-MB-231 and
BT549 cells with the reporter construct and miR-606 mimics.
Co-transfection with miR-606 mimics and STC1 3′UTR-1 or −4
significantly reduced the relative luciferase activity compared
with that in cells transfected with miR-606 mimics and empty pGL3UC
vector (Fig. 3G). Collectively,
these results indicated that STC1 is a direct target of
miR-606.
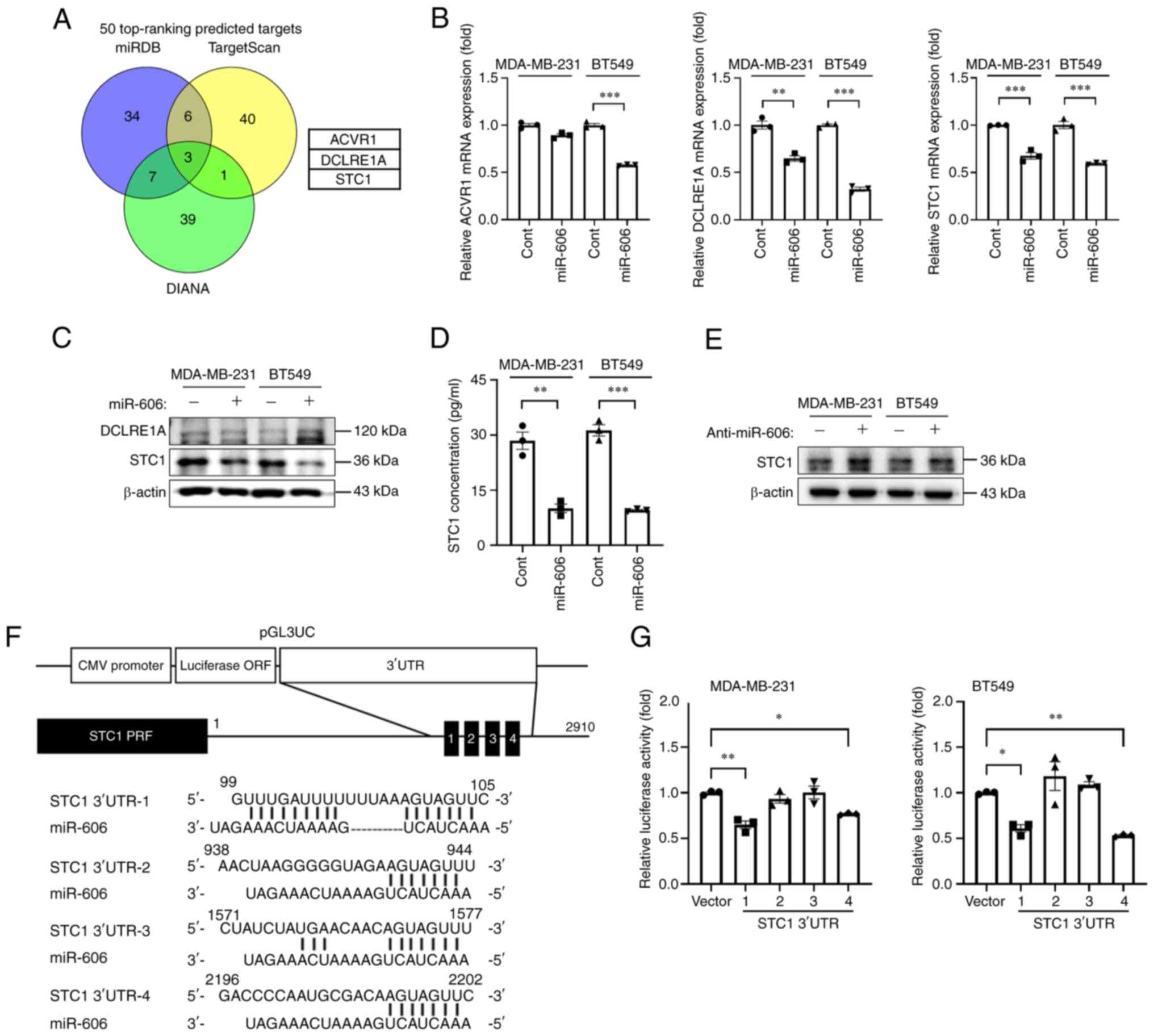 | Figure 3.STC1 is a direct target gene of
miR-606. (A) Venn diagram of the putative target genes of miR-606,
determined using three miRNA target prediction programs. (B)
Reverse transcription-quantitative PCR analysis of the relative
expression levels of three putative target genes (ACVR1, DCLRE1A
and STC1) in TNBC cells transfected with miR-606 mimics. (C)
Western blot analysis of the protein expression levels of two
putative target genes (DCLRE1A and STC1) in TNBC cells transfected
with miR-606 mimics. (D) Secreted protein levels of STC1 in the
supernatant of TNBC cells transfected with miR-606 mimics. (E)
Protein expression levels of the target gene, STC1, in TNBC cells
transfected with anti-miR-606. (F) Construction of the vector
containing four putative miR-606 binding sites in the human STC1
3′-UTR. (G) Luciferase activity of TNBC cells co-transfected with
the four indicated STC1 3′-UTR vectors and miR-606 mimics.
Luciferase activity was measured 48 h post-co-transfection. Data
are presented as the mean ± SEM. Data were analyzed using (B and D)
Student's t-test and (G) one-way ANOVA. *P<0.05, **P<0.005
and ***P<0.001. TNBC, triple-negative breast cancer; ACVR1,
activin A receptor, type 1; DCLRE1A, DNA cross-link repair 1A;
STC1, Stanniocalcin 1; Cont, control; miR, microRNA. |
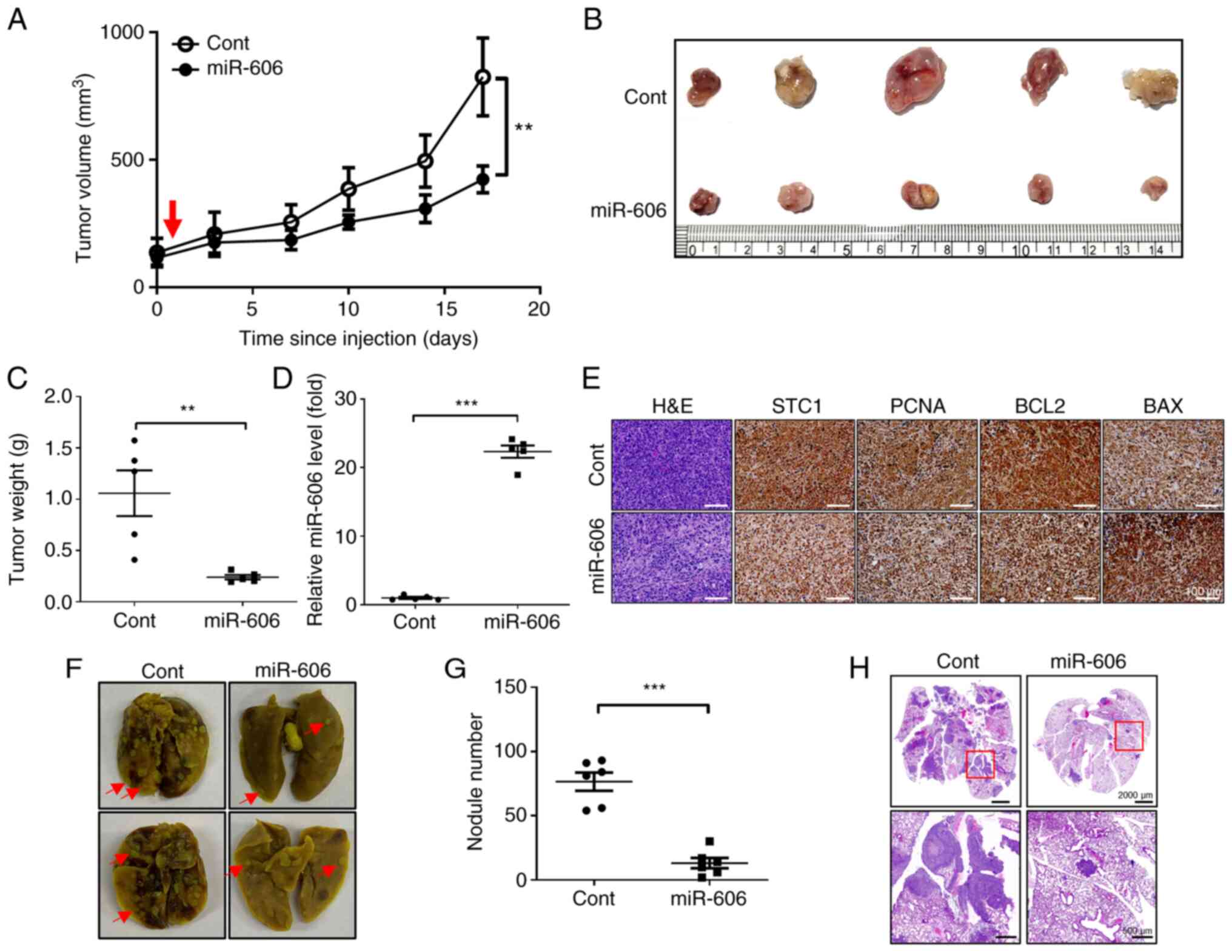
miR-606 inhibits tumor growth and
metastasis in a mouse xenograft model and a mouse model of lung
metastasis
To confirm the tumor-suppressive effects of miR-606
in vivo, the changes in xenograft tumor volume were
investigated after miR-606 mimics transfection. Tumors established
using MDA-MB-231 cells were injected with miR-606 mimics or control
miRNA alongside an in vivo transfection reagent. The results
revealed that the tumor growth rate was significantly suppressed in
mice injected with miR-606 mimics compared with that in mice
injected with control miRNA (Fig.
4A). Additionally, the endpoint tumor weights of miR-606
mimics-injected mice were significantly lower than those of control
miRNA-injected mice (Fig. 4B and
C). The expression levels of miR-606 were also increased in the
dissected tumors of mice injected with miR-606 mimics (Fig. 4D). Moreover, IHC analysis and
hematoxylin and eosin staining revealed that intratumoral
injections of miR-606 mimics decreased the number of tumor cells
and the expression levels of the target proteins STC1, PCNA and
BCL-2, but increased BAX expression (Fig. 4E).
To determine whether miR-606 inhibits metastasis
in vivo, the number of metastatic lung nodules was detected
following tail vein injection of breast cancer cell lines in a
mouse model. Mouse models of lung metastasis were established by
injecting control miRNA- or miR-606 mimics-transfected MDA-MB-231
cells into the tail veins of mice whose lungs were resected after 6
weeks. After lung resection, the number of metastatic nodules in
the lungs of mice injected with miR-606 mimics-transfected
MDA-MB-231 cells were significantly decreased compared with that in
the lungs of mice injected with control miRNA-transfected
MDA-MB-231 cells (Fig. 4F and G).
In addition, hematoxylin and eosin staining revealed a reduction in
metastatic loci in the lungs of mice injected with miR-606
mimics-transfected MDA-MB-231 cells (Fig. 4H). Taken together, these results
indicated that miR-606 functions as a tumor and metastasis
suppressor in vivo.
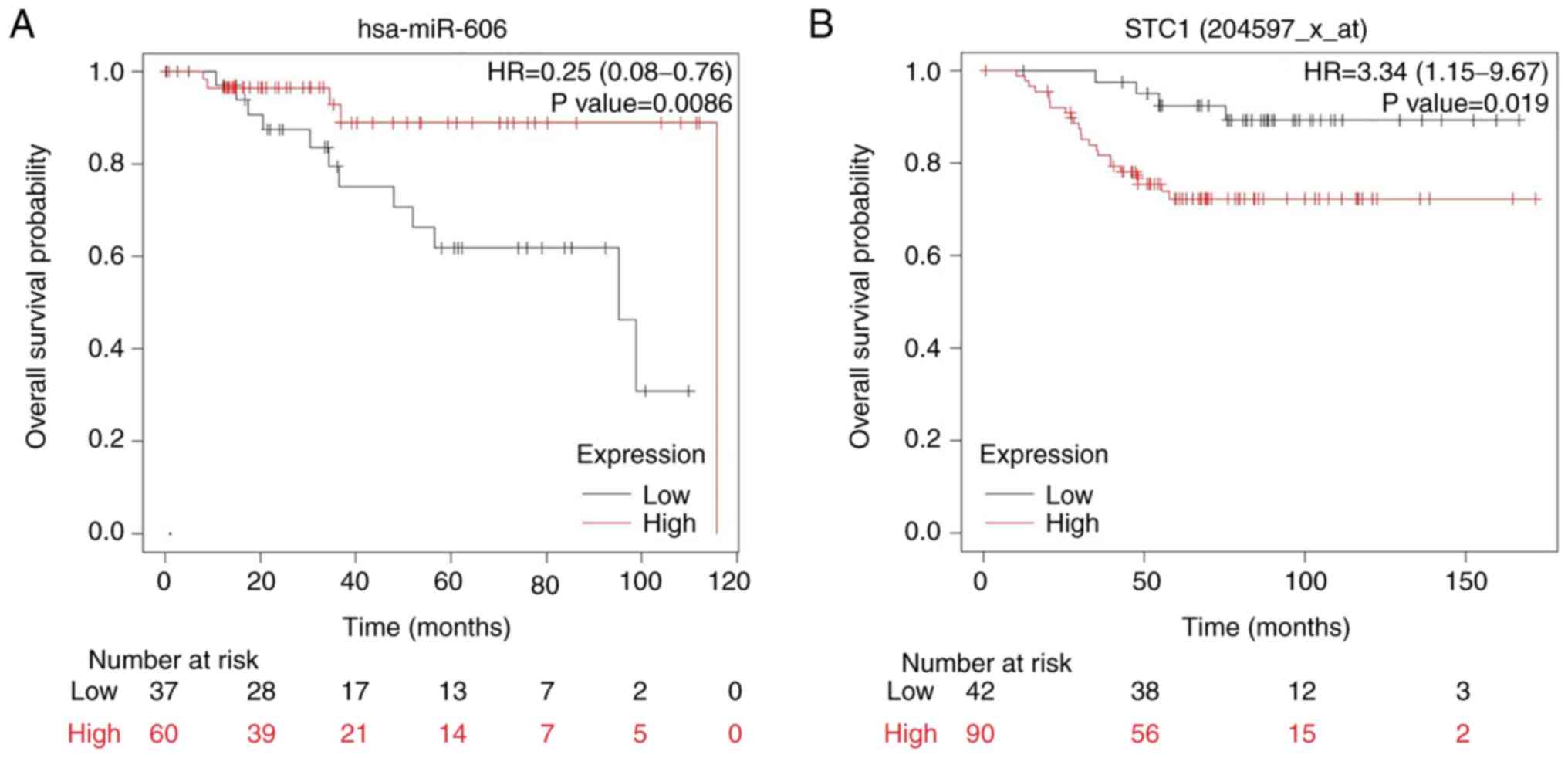
miR-606 and STC1 expression levels are
associated with the OS of patients with TNBC
To identify the association between miR-606 and the
OS of patients with TNBC, the expression profiles of 97 patients
with breast cancer were analyzed from TCGA database using the K-M
plotter tool. The patients were divided into high- and low-miR-606
expression groups, comprising 60 and 37 patients, respectively.
Kaplan-Meier survival analysis revealed that patients with low
miR-606 expression had a significantly lower OS than those with
high miR-606 expression (Fig.
5A).
Furthermore, to determine the association between
STC1 and the OS of patients with TNBC, the STC1 expression profiles
of 132 patients with breast cancer were analyzed from TCGA database
using the K-M plotter tool. The patients were divided into high-
and low-STC1 expression groups, comprising 90 and 42 patients,
respectively. Kaplan-Meier survival analysis revealed that patients
with high STC1 expression had a significantly lower OS than those
with low STC1 expression (Fig. 5B).
Collectively, these results suggested that miR-606 and STC1 may act
as prognostic indicators of the OS of patients with TNBC.
Discussion
TNBC is an aggressive subtype of breast cancer with
a high rate of recurrence and distant metastasis (17). Owing to the absence of ER, PR and
HER2 in TNBC, currently available hormones or targeted therapies
are ineffective in treating TNBC (18). Therefore, new prognostic and
therapeutic biomarkers are urgently needed for the treatment of
TNBC. Notably, miRNA-based therapeutics have emerged as a novel
strategy for cancer treatment, since miRNAs regulate pathways
associated with tumor progression and migration, and their
dysregulation has been linked to various types of cancer (19,20).
The potential of miRNAs as novel biomarkers has been demonstrated
in various cancers, including breast cancer (21). However, the functions of several
miRNAs, including the novel miR-606, remain unknown. In the present
study, miR-606 was identified as a potential tumor suppressor in
TNBC and STC1 was revealed to be a direct target gene of
miR-606.
STC1, which was first identified in bony fish, is a
56-kDa disulfide-bound glycoprotein hormone involved in plasma
calcium and phosphate homeostasis (7). Mammalian STC1 is expressed in various
tissues (18) and is involved in
biological processes, including mesenchymal-epithelial transition,
intramembranous and endochondral bone formation, and metabolic
rate, other than calcium and phosphate homeostasis (22–24).
The expression of STC-1 is altered during various developmental,
physiological and pathological processes, including pregnancy,
lactation, angiogenesis and apoptosis (25–27).
In addition, several studies have reported the relationship between
STC1 and various types of cancer. For example, STC1 overexpression
has been shown to induce apoptosis in cervical cancer (23) and to mediate metastasis in
colorectal cancer (28,29). Moreover, in hepatocellular
carcinoma, higher STC1 expression has been associated with smaller
tumor size (30).
The role of STC1 in breast cancer is complex and
multifaceted (31). STC1 is
associated with invasion and metastasis in TNBC cells, and high
STC1 expression is associated with poor prognosis in patients with
TNBC (10,32). Additionally, STC1 has been
implicated in breast cancer chemoresistance (31). By contrast, in hormone
receptor-positive breast cancer, high STC1 expression levels have
been reported to be associated with a favorable prognosis (33). Moreover, high STC1 expression is
associated with a relatively favorable prognosis in the early
postoperative period, but is associated with an increased risk of
breast cancer relapse in later stages (34). An association between STC1 and tumor
suppression has also been established in other studies. For
example, STC1 has been shown to be associated with BRCA1, a tumor
suppressor gene in breast cancer (24). Additionally, STC1 can influence
components of the tumor microenvironment, such as cancer-associated
fibroblasts (35).
In the present study, miR-606 was identified as a
miRNA with tumor-suppressive properties in TNBC. The results of the
present study demonstrated that miR-606 could effectively target
and inhibit the activity of the STC1 gene, thus exhibiting
anticancer effects both in vitro and in vivo.
Furthermore, to validate the findings of the current study, an
analysis was conducted using publicly available databases. This
analysis confirmed that in patients with TNBC elevated STC1
expression levels and reduced miR-606 levels resulted in shorter OS
rates. Notably, the present clinical analysis had a limitation, as
it did not explore inverse relationship between miR-606 and STC1
within the same group of patients.
In conclusion, the findings of the present study
revealed that miR-606 may inhibit TNBC cell proliferation, stem
cell-like ability, migration and invasion by targeting STC1.
Consequently, it could act as a tumor and metastasis suppressor,
suggesting its potential in the development of anticancer miRNA
therapeutics, particularly for TNBC.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Ms. Gyurim Lee, Ms.
Yeongji Kim, Mr. Seungchan An and Ms. Hanna Kim (Orthopedic Surgery
Lab of CHA University) for their helpful comments, suggestions and
assistance with animal experiments.
Funding
This work was supported by a National Research Foundation of
Korea (NRF) grant funded by the Korean government (MSIT) (grant
nos. RS-2023-00210067, 2022R1A2C2005916 and RS-2023-00245268).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The datasets generated and/or analyzed during the current
study are available in the GSE118782 repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118782.
Authors' contributions
IK and SL made substantial contributions to
conception and design, and contributed to manuscript review,
funding acquisition, investigation and methodology. SC was involved
in experiments, acquisition of the data, interpretation of the data
and writing of the original draft. HJA was involved in experiments,
acquisition of the data, interpretation of the data, writing of the
original draft and project administration. HJY and MJS were
involved in experiments and formal analysis. JO, KL and SAL were
involved in analysis of data, and reviewing and editing of the
manuscript. SKK and JK were involved in acquisition and analysis of
data. IK and SL confirmed the authenticity of all the raw data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Animal experimental procedures were reviewed and
approved by the CHA University Animal Care and Use Committee
(approval no. 210152).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, Van De Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takahashi RU, Miyazaki H and Ochiya T: The
roles of microRNAs in breast cancer. Cancers (Basel). 7:598–616.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W,
Yu M, Lin J and Cui Q: MicroRNAs involved in carcinogenesis,
prognosis, therapeutic resistance and applications in human
triple-negative breast cancer. Cells. 8:14922019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Loh HY, Norman BP, Lai KS, Rahman NMANA,
Alitheen NBM and Osman MA: The regulatory role of microRNAs in
breast cancer. Int J Mol Sci. 20:49402019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yoshiko Y and Aubin JE: Stanniocalcin 1 as
a pleiotropic factor in mammals. Peptides. 25:1663–1669. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin F, Li X and Wang X, Sun H, Wang Z and
Wang X: Stanniocalcin 1 promotes metastasis, lipid metabolism and
cisplatin chemoresistance via the FOXC2/ITGB6 signaling axis in
ovarian cancer. J Exp Clin Cancer Res. 41:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bai S, Zhao Y, Chen W, Peng W, Wang Y,
Xiong S, Arun A, Li Y, Yang Y, Chen S, et al: The stromal-tumor
amplifying STC1-Notch1 feedforward signal promotes the stemness of
hepatocellular carcinoma. J Transl Med. 21:2362023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang AC, Doherty J, Huschtscha LI,
Redvers R, Restall C, Reddel RR and Anderson RL: STC1 expression is
associated with tumor growth and metastasis in breast cancer. Clin
Exp Metastasis. 32:15–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu A, Li Y, Lu S, Cai C, Zou F and Meng
X: Stanniocalcin 1 promotes lung metastasis of breast cancer by
enhancing EGFR-ERK-S100A4 signaling. Cell Death Dis. 14:3952023.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chandran UR, Medvedeva OP, Barmada MM,
Blood PD, Chakka A, Luthra S, Ferreira A, Wong KF, Lee AV, Zhang Z,
et al: TCGA expedition: A data acquisition and management system
for TCGA data. PLoS One. 11:e01653952016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lasham A, Fitzgerald SJ, Knowlton N, Robb
T, Tsai P, Black MA, Williams L, Mehta SY, Harris G, Shelling AN,
et al: A predictor of early disease recurrence in patients with
breast cancer using a cell-free RNA and protein liquid biopsy. Clin
Breast Cancer. 20:108–116. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang H, Zhang W, Ding Z, Xu T, Zhang X and
Xu K: Comprehensive exploration of the expression and prognostic
value of AQPs in clear cell renal cell carcinoma. Medicine
(Baltimore). 101:e293442022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yao Y, Chu Y, Xu B, Hu Q and Song Q: Risk
factors for distant metastasis of patients with primary
triple-negative breast cancer. Biosci Rep. 39:BSR201902882019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
De Laurentiis M, Cianniello D, Caputo R,
Stanzione B, Arpino G, Cinieri S, Lorusso V and De Placido S:
Treatment of triple negative breast cancer (TNBC): Current options
and future perspectives. Cancer Treat Rev. 36 (Suppl 3):S80–S86.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu M, Wang G, Tian W, Deng Y and Xu Y:
MiRNA-based therapeutics for lung cancer. Curr Pharm Des.
23:5989–5996. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mishra S, Yadav T and Rani V: Exploring
miRNA based approaches in cancer diagnostics and therapeutics. Crit
Rev Oncol Hematol. 98:12–23. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang AC, Jeffrey KJ, Tokutake Y,
Shimamoto A, Neumann AA, Dunham MA, Cha J, Sugawara M, Furuichi Y
and Reddel RR: Human stanniocalcin (STC): Genomic structure,
chromosomal localization, and the presence of CAG trinucleotide
repeats. Genomics. 47:393–398. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Madsen KL, Tavernini MM, Yachimec C,
Mendrick DL, Alfonso PJ, Buergin M, Olsen HS, Antonaccio MJ,
Thomson AB and Fedorak RN: Stanniocalcin: A novel protein
regulating calcium and phosphate transport across mammalian
intestine. Am J Physiol. 274:G96–G102. 1998.PubMed/NCBI
|
|
24
|
Chang AC, Jellinek DA and Reddel RR:
Mammalian stanniocalcins and cancer. Endocr Relat Cancer.
10:359–373. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deol HK, Varghese R, Wagner GF and
Dimattia GE: Dynamic regulation of mouse ovarian stanniocalcin
expression during gestation and lactation. Endocrinology.
141:3412–3421. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zlot C, Ingle G, Hongo J, Yang S, Sheng Z,
Schwall R, Paoni N, Wang F, Peale FV Jr and Gerritsen ME:
Stanniocalcin 1 is an autocrine modulator of endothelial angiogenic
responses to hepatocyte growth factor. J Biol Chem.
278:47654–47659. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Block GJ, Ohkouchi S, Fung F, Frenkel J,
Gregory C, Pochampally R, DiMattia G, Sullivan DE and Prockop DJ:
Multipotent stromal cells are activated to reduce apoptosis in part
by upregulation and secretion of stanniocalcin-1. Stem Cells.
27:670–681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan X, Jiang B, Liu J, Ding J, Li Y, Sun
R, Peng L, Qin C, Fang S and Li G: STC1 promotes cell apoptosis via
NF-κB phospho-P65 Ser536 in cervical cancer cells. Oncotarget.
8:46249–46261. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peña C, Céspedes MV, Lindh MB, Kiflemariam
S, Mezheyeuski A, Edqvist PH, Hägglöf C, Birgisson H, Bojmar L,
Jirström K, et al: STC1 expression by cancer-associated fibroblasts
drives metastasis of colorectal cancer. Cancer Res. 73:1287–1297.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leung CC and Wong CK: Effects of STC1
overexpression on tumorigenicity and metabolism of hepatocellular
carcinoma. Oncotarget. 9:6852–6861. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen F, Zhang Z and Pu F: Role of
stanniocalcin-1 in breast cancer. Oncol Lett. 18:3946–3953.
2019.PubMed/NCBI
|
|
32
|
Murai R, Tanaka M, Takahashi Y,
Kuribayashi K, Kobayashi D and Watanabe N: Stanniocalcin-1 promotes
metastasis in a human breast cancer cell line through activation of
PI3K. Clin Exp Metastasis. 31:787–794. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
McCudden CR, Majewski A, Chakrabarti S and
Wagner GF: Co-localization of stanniocalcin-1 ligand and receptor
in human breast carcinomas. Mol Cell Endocrinol. 213:167–172. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Joensuu K, Heikkilä P and Andersson LC:
Tumor dormancy: Elevated expression of stanniocalcins in late
relapsing breast cancer. Cancer Lett. 265:76–83. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Avalle L, Raggi L, Monteleone E, Savino A,
Viavattene D, Statello L, Camperi A, Stabile SA, Salemme V, De
Marzo N, et al: STAT3 induces breast cancer growth via ANGPTL4,
MMP13 and STC1 secretion by cancer associated fibroblasts.
Oncogene. 41:1456–1467. 2022. View Article : Google Scholar : PubMed/NCBI
|