Introduction
Osteosarcoma is the most common primary malignant
bone tumor in adolescents, with a mean presenting age of 16 years
and a male predominance (1–3). Pathologically, osteosarcomas are
malignant mesenchymal tumors characterized by pleomorphic
spindle-shaped cells capable of producing bone-like stroma
(4–6). In addition, osteosarcomas present a
high degree of malignancy, rapid growth and metastasis
susceptibility and are difficult to treat, resulting in a
relatively high rate of disability and mortality (7,8).
Clinical treatments for osteosarcoma include surgery, adjuvant
chemotherapy, targeted therapies and immunotherapy (9,10).
However, the average 5-year survival rates for patients with
primary osteosarcoma and distant metastases are <65 and 25%,
respectively (11,12). Methotrexate, doxorubicin and
cisplatin have become the standard regimens for the treatment of
osteosarcoma in clinical practice. However, their clinical
application is limited due to their toxicity and side effects
(13). On the other hand, patients
with metastases, recurrences and unresectable tumors often become
resistant to current standard treatment regimens (14,15).
Therefore, novel therapeutic agents for advanced osteosarcoma with
less toxicity remain to be elucidated. Natural effective components
of plants are potentially useful against tumors due to their
multi-target and multi-pathway synergistic modulatory effects that
can lead to multiple therapeutic effects at different stages of
tumorigenesis, development, metastasis and immune regulation
(16). Asiatic acid
(C30H48O5) is a pentacyclic
triterpenoid from Centella asiatica with anti-inflammatory,
neuroprotective, anti-diabetic, antitumor and antibacterial
characteristics (17–19). The results of a phase I clinical
trial of asiatic acid capsules (ECa 233) in healthy volunteers
indicated a lack of adverse reactions, suggesting that asiatic acid
is medicinally safe (20).
Therefore, asiatic acid has the characteristics of fewer toxic side
effects and antitumor effects, which can provide a reference value
for the treatment of osteosarcoma. However, the potential targets
and mechanisms of the treatment of osteosarcoma with asiatic acid
remain to be elucidated.
Cell cycle regulation mechanism proteins include
cyclin, CDK and CDK inhibitors (21). Cancer therapy often targets the
inhibition of cyclins and CDKs (22). During apoptosis, cells are
stimulated by specific intracellular or extracellular signals to
undergo programmed cell death (23)
and the process results in chromatin pyknosis, DNA fragmentation,
cell shrinkage and apoptotic body formation (24). Classical apoptosis mechanisms are
triggered by intrinsic or extrinsic signaling pathways (25). The extrinsic apoptotic pathway
starts with the binding of specific death receptors (such as Fas,
tumor necrosis factor receptor 1 and death receptor 5) to their
corresponding ligands, and recruitment of the adaptor proteins Fas
associated death domain and caspase-8 to form a death-inducing
signaling complex that initiates the subsequent caspase cascade to
promote cell death (26). In the
intrinsic apoptotic pathway, a Bcl-2 family protein imbalance leads
to a decreased mitochondrial membrane potential (MMP) and the
release of apoptosis-related proteins, resulting in activation of
the caspase cascade and apoptosis induction (27). Autophagy is a type II cell death
mechanism (28). This dynamic
process is tightly regulated to allow cells to maintain their
cellular nutrition and energy balance by phagocytosing and breaking
down damaged or senescent proteins, organelles and harmful
components (29). Autophagy
mechanisms are frequently linked to apoptosis, inflammatory
responses and cancer chemoresistance (30–32).
The role of autophagy in tumor growth is two-sided. On the one
hand, tumor cells can recover damaged or excess cytoplasmic
contents in a lysosome-dependent manner through the autophagy
pathway to overcome an undernourished environment and promote tumor
growth. On the other hand, the autophagy regulation can induce
apoptosis (33,34). Thus, cancer treatments can induce
apoptosis by inhibiting autophagy in different tumor cell types and
environments (35,36). Apoptosis-related proteins [Bcl-2,
caspase-3 (Caspase-3) and P53] and diverse signaling pathways are
activated during autophagy (37,38).
Network pharmacology can be used to predict
potential targets and pathways by constructing complex networks
among a drug, its targets and a disease, to improve the success
rate of novel drug trials (39).
Molecular docking is a theoretical simulation method that can be
used to evaluate ligand-receptor interactions and find the best
binding models for ligands (40).
The present study used network pharmacology to predict the targets
and signaling pathways associated with asiatic acid in
osteosarcoma. Finally, the predicted targets and pathways were
assessed via molecular docking and in vitro experiments to
identify novel treatment strategies for osteosarcoma.
Materials and methods
Analysis of the disease-target-drug
network
The molecular structure of asiatic acid was obtained
from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) and entered into
the PharmMapper (http://www.lilab-ecust.cn/pharmmapper/), Swiss Target
Prediction (http://swisstargetprediction.ch/) and Uniprot
(https://www.uniprot.org/) databases to identify
drug targets with scores >0 after eliminating repeated targets.
In addition, the Online Mendelian Inheritance in Man (OMIM;
http://omim.org/), Therapeutic Target Database (TTD;
http://db.idrblab.net/ttd/) and
Genecards (https://www.genecards.org/) databases
were searched to identify disease targets based on the keyword
‘osteosarcoma’. Duplicated disease targets were removed and Venn
diagrams of common drug-disease targets were drawn using the Venny
software mapping tool (https://bioinfogp.cnb.csic.es/tools/venny/). Finally,
Cytoscape software (3.8.2 version) was used to draw a network
diagram of ‘disease-target-drug’ interactions.
Protein-protein interaction (PPI)
network construction
The aforementioned common targets were imported into
the Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING) database (https://string-db.org/) and Cytoscape software
(https://cytoscape.org/; version 3.8.2) was used
to draw the resulting PPI network maps. Network analyzer was used
to perform a topological analysis of the PPI networks. The core
targets selected according to a degree value were mapped using R
software (http://www.R-project.org).
Gene ontology (GO) and Kyoto
encyclopedia of genes and genomes (KEGG) enrichment analyses
GO and KEGG functional enrichment analyses of
critical target genes were performed using the Bioconductor
bioinformatics package (https://www.bioconductor.org/; version 3.17) of R
software (http://www.R-project.org). GO and
KEGG enrichment analysis results were visualized in bar and bubble
plots. P-value <0.05 was employed to identify statistically
significant GO terms and KEGG pathways. The significance of GO and
KEGG enrichment was represented by employing the fold change
threshold=-log10 (P-value) for a more comprehensive
depiction of the figure.
Molecular docking
The asiatic acid structure obtained from the PubChem
database was imported into Chem3D software and the optimized
structures were further imported into Schrodinger software
(https://www.schrodinger.com/; version
2019.1) for hydrogenation and energy minimization. The protein
structures of EGFR (1M17), SRC proto-oncogene, non-receptor
tyrosine kinase (SRC; 1YOL), Caspase-3 (2CNK), heat shock protein
90α family class A member 1 (HSP90AA1; 4BQG), Estrogen Receptor 1
(ESR1; 7UJF) and Interleukin 6 (IL-6; 4O9H) were obtained from the
RCSB database (https://www.rcsb.org/), and entered
into Schrodinger Maestro's Protein Preparation Wizard for
hydrogenation, peptide repair, energy minimization and structure
optimization. The molecular docking was then optimized in the Glide
platform of the software by determining sites on the basis of
protein structure and ligands in 15 Å × 15 Å × 15 Å boxes. Finally,
molecular docking and screening were conducted using the Standard
Precision Glide Docking method.
Cell culture
MG63 human osteosarcoma cells (cat. no. TCHu124),
HOS human osteosarcoma cells (cat. no. TCHu167) and MC3T3-E1
mouse-derived normal osteoblast cells (cat. no. GNM15) were
purchased from The Cell Bank of Type Culture Collection of The
Chinese Academy of Sciences. 143B human osteosarcoma cells (cat.
no. CRL-8303) were obtained from American Type Culture Collection.
Human osteosarcoma cells were cultured in Minimum Essential Medium
(MEM; Gibco; Thermo Fisher Scientific, Inc.) containing 1%
penicillin-streptomycin (Beijing Solarbio Science & Technology
Co., Ltd.) and 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in a
sterile incubator containing 5% CO2 at 37°C. MC3T3-E1
cells were cultured in α-MEM (Gibco; Thermo Fisher Scientific,
Inc.) containing 10% FBS and 1% penicillin-streptomycin under the
same controlled culture conditions. The osteosarcoma and MC3T3-E1
cells were passaged when reaching 80–90% confluency in culture
flasks.
Antibodies and reagents
Asiatic acid (high-performance liquid chromatography
≥98%; Shanghai Yuanye Biotechnology Co., Ltd.) was dissolved in
DMSO. An appropriate concentration of DMSO was used for the
experimental controls. Antibodies against Bcl-2 (cat. no. 4223),
Bax (cat. no. 2772), phosphorylated (p-)ERK1/2 (cat. no. 4370),
ERK1/2 (cat. no. 4695), p38 (cat. no. 8690), p-p38 (cat. no. 4511),
JNK (cat. no. 9252), p-JNK (cat. no. 4668), p-PI3K (cat. no.
17366), CDK2 (cat. no. 18048) and cyclin A2 (cat. no. 67955) and
the goat anti-rabbit IgG (cat. no. 7074) and anti-mouse IgG (cat.
no. 7076) secondary antibodies were purchased from Cell Signaling
Technology, Inc. The voltage dependent anion channel 1 (VDAC1; cat.
no. ab14734), p-AKT (cat. no. ab192623), AKT (cat. no. ab179463),
PI3K (cat. no. ab191606), LC3 (cat. no. ab48394) and p62 (cat. no.
ab56416) antibodies were obtained from Abcam. The GAPDH antibody
(cat. no. 21612) was purchased from Signalway Antibody LLC.
Cell Counting Kit 8 (CCK8) cell
viability assay
Growing osteosarcoma and MC3T3-E1 cells were
trypsinized, centrifuged (180 × g; 25°C; 3 min) and resuspended in
complete MEM or α-MEM to prepare cell suspensions. Cells
(~4.0×103) were seeded into 96-well plates and incubated
at 37°C for 24 h. Subsequently, the osteosarcoma cells were treated
with asiatic acid (0, 10, 20 and 40 µM) at 37°C for 24 and 48 h.
The culture medium was replaced with 10 µl CCK8 (Beyotime Institute
of Biotechnology) in MEM and cells were incubated at 37°C for 2–4
h. OD values were determined using a microplate reader (MK3; Thermo
Fisher Scientific, Inc.) at 450 nm.
Cell cycle analysis
The osteosarcoma cells were treated with asiatic
acid (0, 20 and 40 µM) at 37°C for 24 h, then collected by
centrifugation (180 × g; 4°C; min) and fixed with 70% chilled
ethanol at 4°C for 12 h. The fixed cells were stained with PI dye
(Beyotime Institute of Biotechnology) and incubated at 37°C for 30
min. A flow cytometer (FACSCelesta; BD Biosciences) and Modfit LT
software (5.0 version; Verity Software House, Inc.) were used to
analyze the cell cycle phases.
Apoptosis analysis
The osteosarcoma cells (~15.0×104) were
seeded into six-well plates and incubated with asiatic acid (0, 10,
20 and 40 µM) at 37°C for 24 h. The adherent cells from each well
were collected, and suspended and placed in a flow tube. To stain
the cells, the cells were resuspended in 5 µl annexin V-FITC and 10
µl PI (Beyotime Institute of Biotechnology) and incubated at room
temperature for 30 min. Finally, the stained cells were analyzed by
flow cytometry (FACSCelesta; BD Biosciences) and BD FACSDiva
Software (version 6.1.3; BD Biosciences). The apoptotic rate was
calculated by the percentage of early + late apoptotic cells.
Measurement of MMP
The osteosarcoma cells were treated with asiatic
acid for 24 h and then resuspended in 0.5 ml cell culture medium.
JC-1 (0.5 ml; Beyotime Institute of Biotechnology) staining working
solution was added to the culture medium and the cultures were
placed in a cell incubator at 37°C for 20 min. Subsequently, the
cells were washed twice with staining buffer and analyzed using a
flow cytometer (FACSCelesta; BD Biosciences) and BD FACSDiva
Software (version 6.1.3; BD Biosciences).
Measurement of intracellular reactive
oxygen species (ROS) content
The 143B and HOS human osteosarcoma cell lines were
treated with asiatic acid (0 and 40 µM) at 37°C for 24 h.
Dichlorodihydrofluorescein diacetate (DCFH-DA; Beyotime Institute
of Biotechnology) was then diluted with serum-free MEM to a final
working solution concentration of 10 µM, and the cells were
resuspended in the diluted DCFH-DA and incubated at 37°C for 20
min. After washing the cells three times with serum-free MEM,
intracellular ROS levels were measured using a flow cytometer
(FACSCelesta; BD Biosciences) and BD FACSDiva Software (version
6.1.3; BD Biosciences).
Transmission electron microscopy
The preparation and observation of electron
microscope specimens was completed in the following seven steps. In
the first step, the collected cells were fixed with 3%
glutaraldehyde (SPI-CHEM Inc.) for 4 h at 4°C, and then fixed with
0.1 M sodium dimethylarsenate buffer at 4°C for three times, with a
2 h interval between changes. In the second step, the samples were
subsequently immersed with 1% osmium acid (SPI-CHEM Inc.) for 2 h
at 4°C, then rinsed with 0.1 M sodium dimethylarsenate buffer
twice, each time at 4°C for 15 min. Step three was staining with
saturated uranyl acetate dye (SPI-CHEM Inc.) for 2 h at room
temperature. In the fourth step, the stained samples were soaked in
50% alcohol at 4°C for 10 min, 70% alcohol at 4°C for 10 min, 80%
alcohol at room temperature for 10 min, 90% alcohol at room
temperature for 10 min, anhydrous ethanol soaks twice at room
temperature for 10 min, times acetone permeations twice at room
temperature for 15 min, a complete encapsulation solution
(Eponate12 epoxy resin; Ted Pella, Inc.) and acetone in a 1:1 ratio
at room temperature for 3 h, then changing the above ratio to a 1:2
penetration for 3 h, and then overnight penetration of the complete
embedding solution at room temperature. In the fifth step, the
samples soaked with the complete embedding solution were placed in
a 40°C incubator for 12 h, and then transferred to an embedding
plate still soaked with the complete embedding solution and placed
in a 60°C incubator for 24 h. In the sixth step, the samples were
sectioned by using an ultrathin sectioning machine (UC7; Leica
Microsystems GmbH), with a section thickness of 90 nm. Finally, the
samples were observed by a JEM-1400 transmission electron
microscope (JEOL Ltd.) with an operating voltage of 80 kV, and
image acquisition was performed with DigitalMicrograph Software
version 832 (Gatan, Inc.; Thermo Fisher Scientific, Inc.).
Western blot analysis
Osteosarcoma cells were trypsinized and collected by
centrifugation (180 × g; 4°C; 3 min) before mixing with appropriate
amounts of protein lysate (PMSF; RIPA, 1:100; Beyotime Institute of
Biotechnology). The cells were lysed on ice, sonicated and
centrifuged at high speed (13,800 × g; 4°C; 15 min) to collect the
supernatant protein. Next, the protein concentration of each sample
was measured using a BCA protein assay kit (Beyotime Institute of
Biotechnology) and each protein sample was adjusted to the same
concentration. Subsequently, 5X SDS-PAGE protein loading buffer was
added to the protein solutions and these were boiled for 10 min.
Proteins (20 µg/lane) were separated by SDS-PAGE on 10 and 12%
separation gels and then transferred (250 mA; 2.5 h) to PVDF
membranes (pore size, 0.2 µm; Merck KGaA). Fresh 5% skimmed milk
was used to block the membranes for 60 min. Subsequently, the
membranes were cut and incubated with primary antibodies (dilution,
1:1,000) at 4°C for 12 h and with secondary antibodies (dilution,
1:3,000) at room temperature for 2 h. The ECL reagents (Zeta-Life
Inc.) and Fluorescent Imaging System (Tanon 5200; Tanon Science and
Technology Co., Ltd.) were used to visualize the protein bands. In
addition, grey value intensities were calculated using ImageJ
software (v1.8.0; National Institutes of Health).
Statistical analysis
All data are presented as the mean ± SD of at least
three independent experiments, and all data were analyzed and
graphs were plotted using the Figdraw Platform (https://www.figdraw.com/), SPSS Statistics 25.0 (IBM
Corp.) and GraphPad Prism 8.0 (GraphPad Software; Dotmatics)
software. Unpaired Student's t-tests were used for comparisons
between 2 groups. Statistical differences among ≥3 groups were
determined using one-way ANOVA followed by Tukey's post hoc tests.
P<0.05 was considered to indicate a statistically significant
difference.
Results
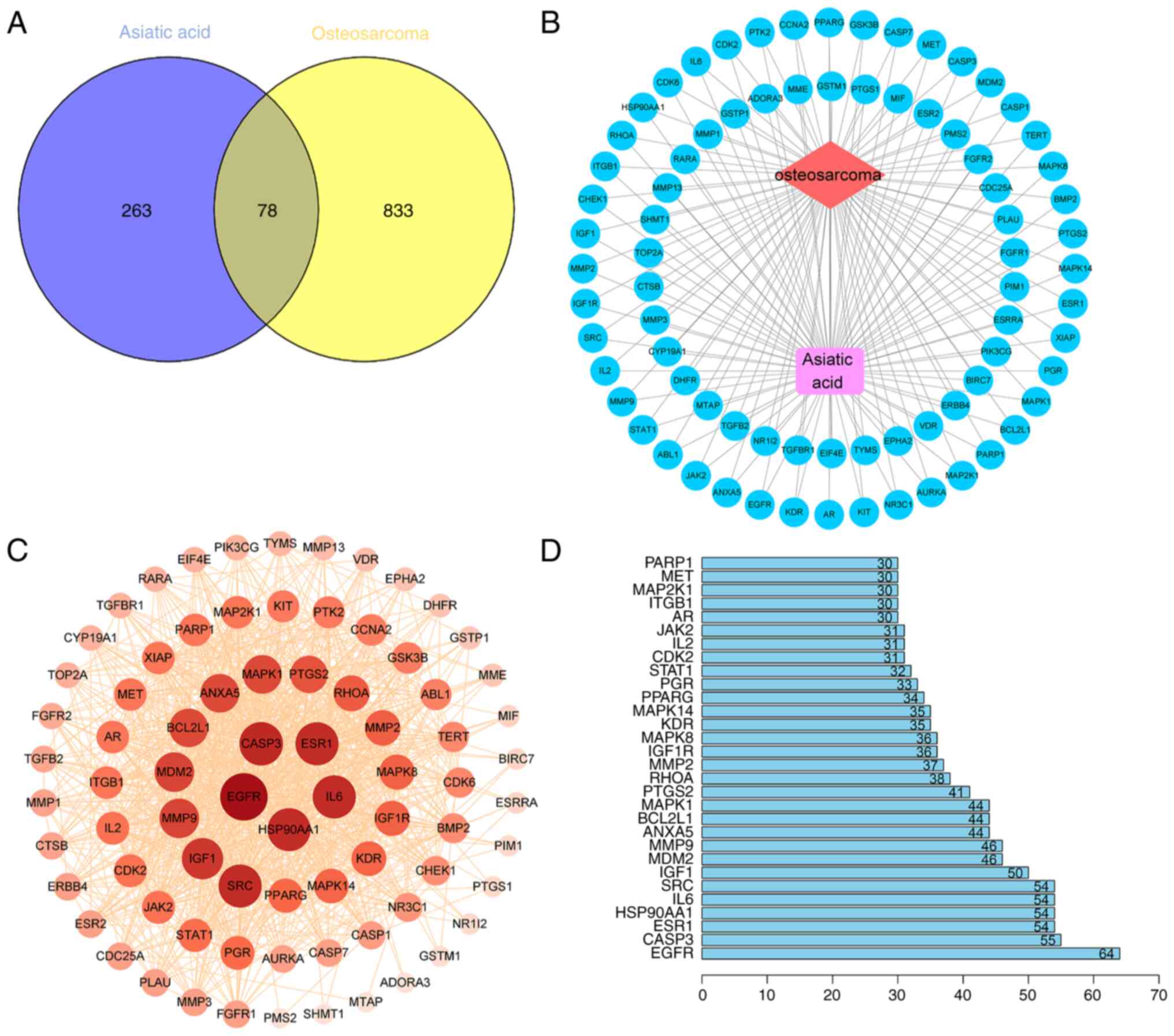
Potential targets of asiatic acid in
osteosarcoma
The asiatic acid structure was obtained from the
PubChem database, 341 drug targets after de-duplication were
obtained from the PharmMapper, Swiss Target Prediction and Uniprot
databases (Table SI), and 911
disease targets were obtained from the OMIM, TTD and Genecards
databases (Table SII). A Venn
diagram (Fig. 1A) identified 78
common targets, which were entered into Cytoscape to draw a network
diagram of ‘osteosarcoma-target-asiatic acid’ interactions
(Fig. 1B). The common targets of
asiatic acid were entered into the STRING database and a PPI
network graph with 78 nodes, 969 edges and 4 concentric circles was
obtained (Fig. 1C). Node size,
color and depth represent the degree values. The PPI network data
were then subjected to topological analysis and 30 core targets
were identified based on mean degree values. Among the core target
genes, EGFR (64 edges), Caspase-3 (55 edges), ESR1 (54 edges),
HSP90AA1 (54 edges), IL-6 (54 edges) and SRC (54 edges) had a high
node degree, indicating their close association with the effect of
asiatic acid against osteosarcoma (Fig.
1D).
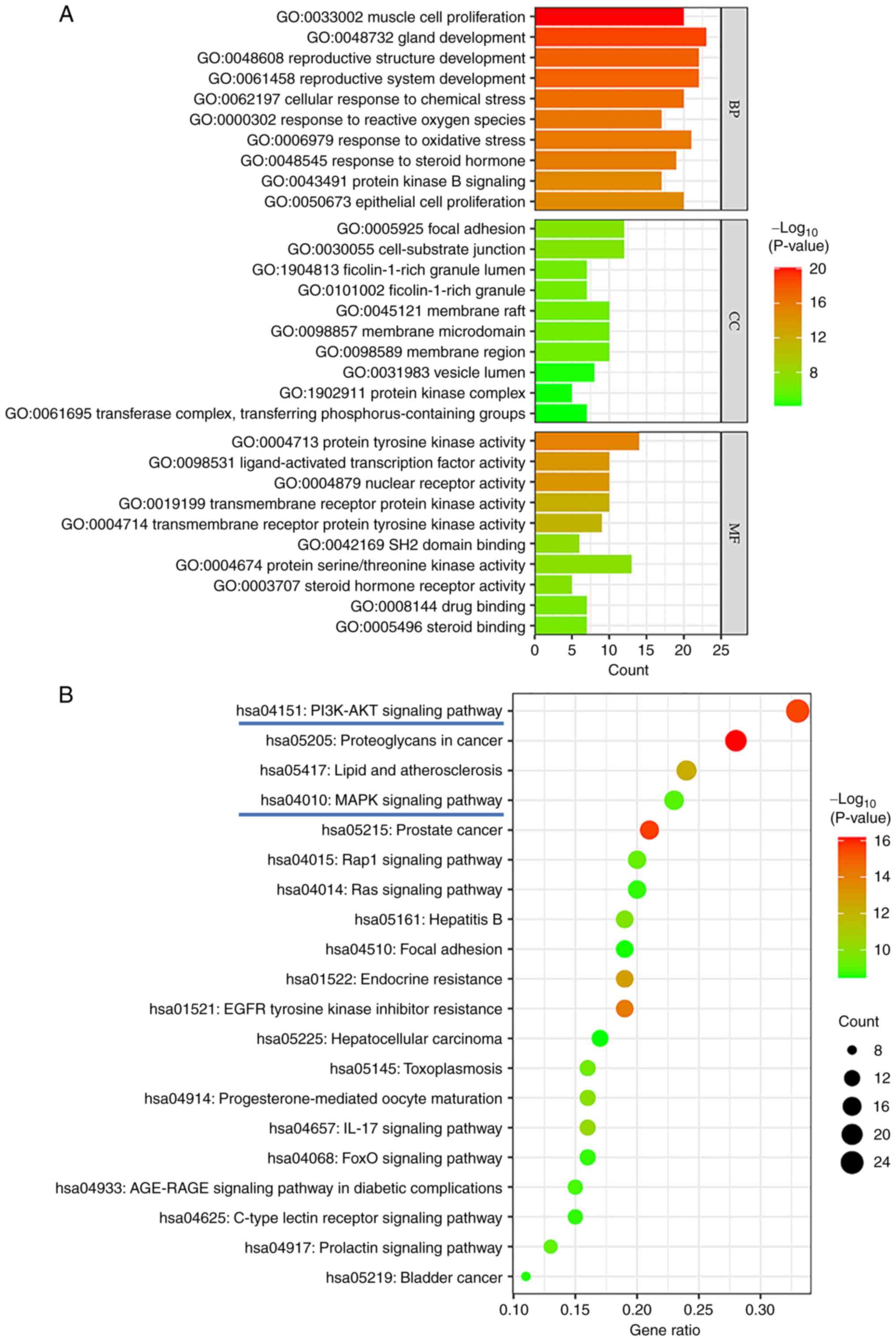
GO and KEGG analysis of asiatic acid
treatment-associated genes in osteosarcoma
Common targets were analyzed via GO enrichment
analysis after running the R programming language to select 1,581
biological process (BP) pathways, 27 items associated with cellular
component (CC) expression processes and 110 items associated with
molecular function (MF) processes. Fig.
2A shows the top 10 BP, CC and MF items. KEGG analysis of the
common targets revealed 134 related pathways, enriched in
cancer-related signaling pathways such as ‘Proteoglycans in cancer’
(hsa05205), ‘PI3K-AKT signaling pathway’ (hsa04151), ‘Rap1
signaling pathway’ (hsa04015) ‘MAPK signaling pathway’ (hsa04010)
and ‘Ras signaling pathway’ (hsa04014). Fig. 2B shows the top 20 results from a
KEGG enrichment bubble map. The present results suggested that the
PI3K-AKT and MAPK signaling pathways may be associated with the
effects of asiatic acid against osteosarcoma.
Molecular docking analysis of the core
targets
The PPI network analysis of asiatic acid and
osteosarcoma revealed target genes with a significant degree value
of association with anti-osteosarcoma effects. Therefore, the top
six highest-ranking target genes were selected for molecular
docking. Binding energy and hydrogen bond interactions were
assessed to evaluate the affinity and binding capacity between
asiatic acid and target proteins. Molecular docking visualization
showed hydrogen bonds between asiatic acid and amino acids in the
active sites of the target proteins. asiatic acid binds the
following proteins: EGFR (binding energy, −7.01 kcal/mol) forming
hydrogen bonds with LYS-721, ASP-776, CYS-773 and ASP-831 amino
acids in its active site (Fig. 3A);
Caspase-3 (binding energy, −6.51 kcal/mol) forming hydrogen bonds
with TRP-206, GLY-122 and THR-62 amino acids in its active site
(Fig. 3B); ESR1 (binding energy,
−6.85 kcal/mol) forming hydrogen bonds with VAL-533, THR-347 and
ASP-351 amino acids in its active site (Fig. 3C); HSP90AA1 (binding energy, −5.83
kcal/mol) forming hydrogen bonds with LYS-581, ASN-51, GLY-135 and
LEU-107 amino acids in its active site (Fig. 3D); IL-6 (binding energy, −6.69
kcal/mol) forming hydrogen bonds with LYS-86, TYR-97 and ASN-63
amino acids in its active site (Fig.
3E); and SRC (binding energy, −7.02 kcal/mol) forming hydrogen
bonds with GLY-346, SER-347 and ASN-393 amino acids in its active
site (Fig. 3F). In summary,
molecular docking revealed that asiatic acid exhibited strong
affinity and binding potential with the target proteins EGFR,
Caspase-3, ESR1, HSP90AA1, IL-6 and SRC.
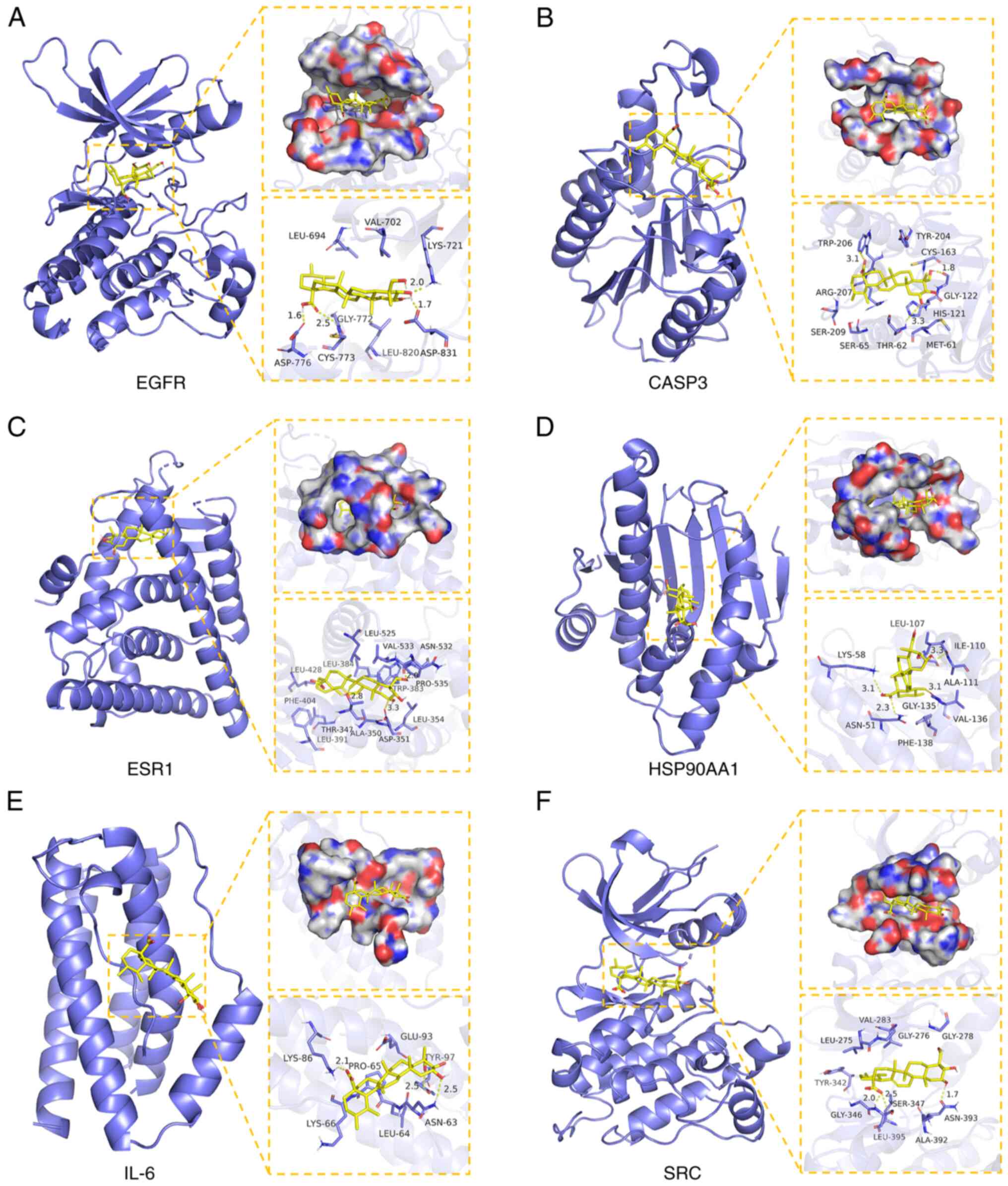 | Figure 3.Molecular Docking Analysis of asiatic
acid with its core target. (A) Asiatic acid binds four amino acid
residues (LYS-721, ASP-776, CYS-773, and ASP-831) in EGFR protein
via hydrogen bonds. (B) Asiatic acid binds three amino acid
residues (TRP-206, GLY-122, and THR-62) in Caspase-3 protein via
hydrogen bonds. (C) Asiatic acid binds to three amino acid residues
(VAL-533, THR-341, and ASP-351) in the ESR1 protein via hydrogen
bonds. (D) Asiatic acid binds four amino acid residues (LYS-581,
ASN-51, GLY-135, and LEU-107) in the HSP90AA1 protein via hydrogen
bonds. (E) Asiatic acid binds three amino acid residues (LYS-86,
TYR-97, and ASN-63) in the IL-6 protein via hydrogen bonds. (F)
Asiatic acid binds three amino acid residues (GLY-346, SER-347, and
ASN-393) in the SRC protein via hydrogen bonds. Yellow dashed lines
represent hydrogen bonds. |
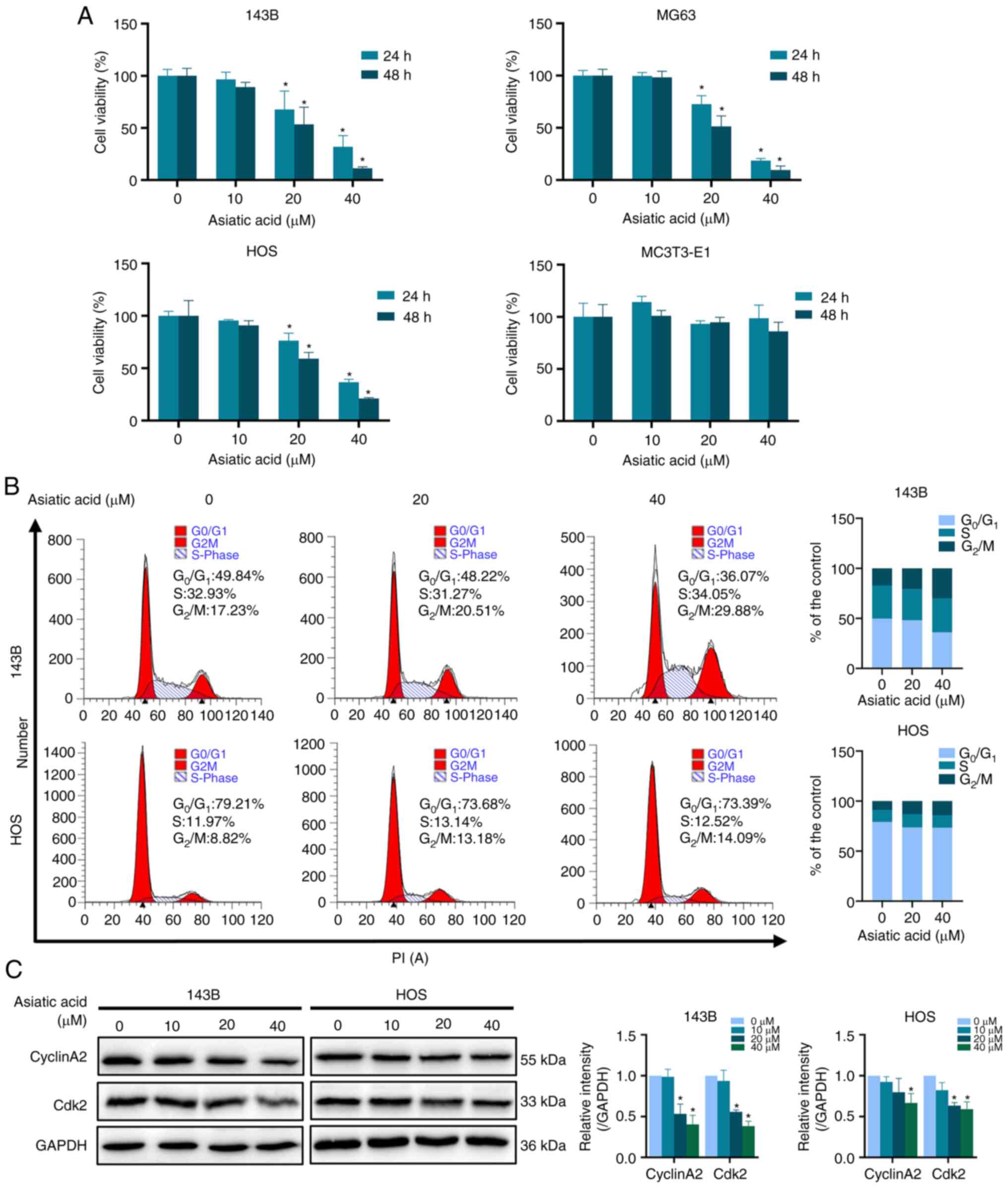
Asiatic acid inhibits proliferation
and induces cell cycle arrest of osteosarcoma cells
Asiatic acid inhibited the proliferation of 143B,
MG63 and HOS human osteosarcoma cell lines in a dose- and
time-dependent manner. No cytotoxicity was observed in MC3T3-E1
normal osteoblast cells treated with asiatic acid under the same
conditions (Fig. 4A). Cell cycle
analysis revealed that the proportion of cells in the
G0/G1 phase was decreased and that in the
G2/M phase was increased (Fig. 4B). Western blot analysis further
demonstrated that asiatic acid suppressed the expression of cyclin
A2 and CDK2 in osteosarcoma cells (Fig.
4C).
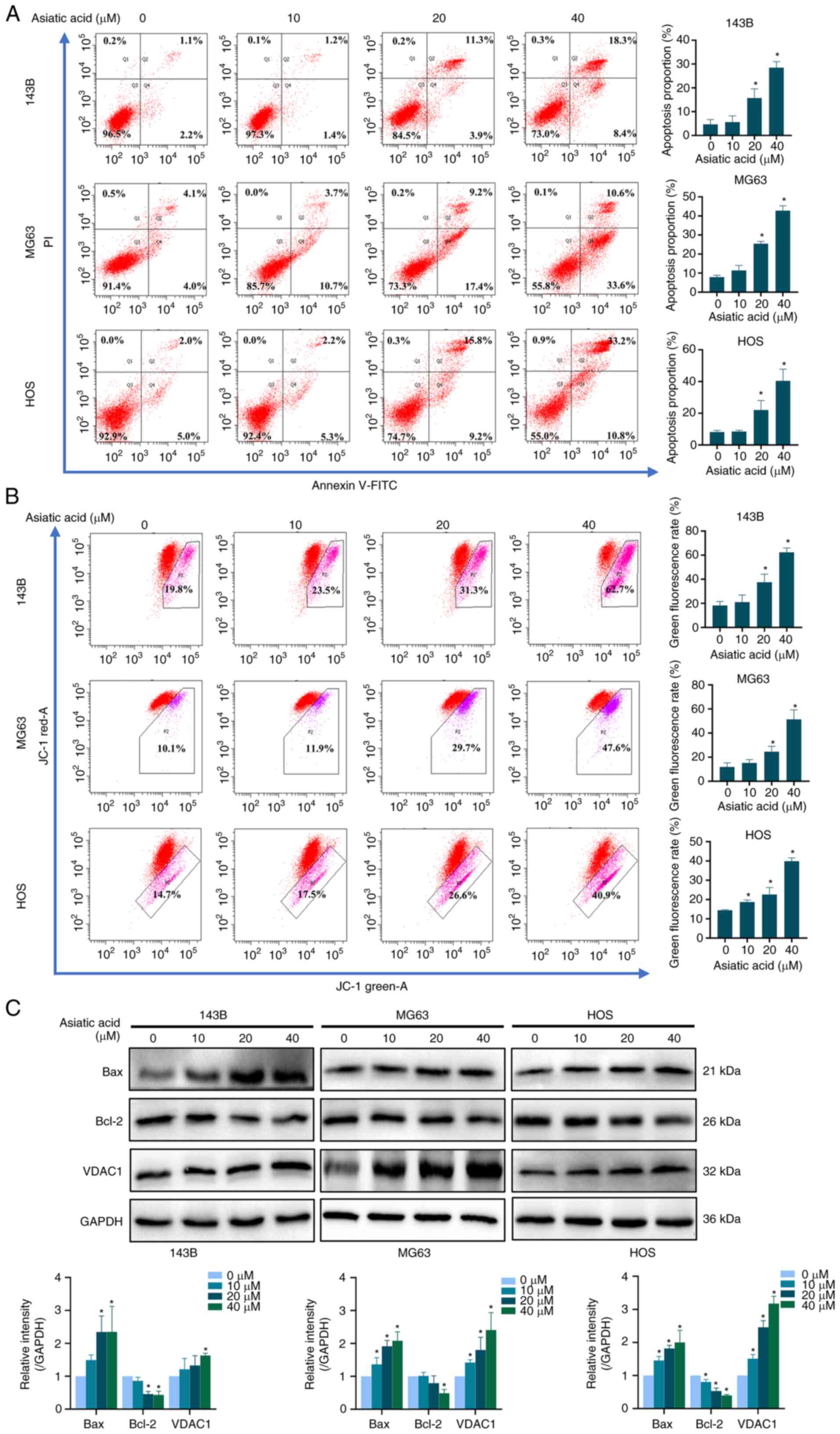
Asiatic acid induces apoptosis in
osteosarcoma cells
To investigate whether asiatic acid induced
apoptosis in 143B, MG63 and HOS osteosarcoma cells, annexin
V-FITC/PI double staining reagent was used to analyze the
proportion of apoptotic cells. The present results demonstrated
that the proportion of apoptotic cells increased with increasing
asiatic acid concentrations (Fig.
5A). MMP level reductions are an essential feature of early
apoptosis stages (41). Therefore,
the MMP was examined and an increase in green fluorescence was
observed in JC-1-labeled mitochondrial membranes, indicating early
apoptosis with a reduced MMP (Fig.
5B). In addition, changes in apoptosis-related proteins
validated these findings. The present results indicated that
asiatic acid increased the expression levels of Bax and VDAC1, and
decreased the expression levels of Bcl-2 (Fig. 5C). Thus, asiatic acid promoted
mitochondrial dysfunction and induced apoptosis in osteosarcoma
cells.
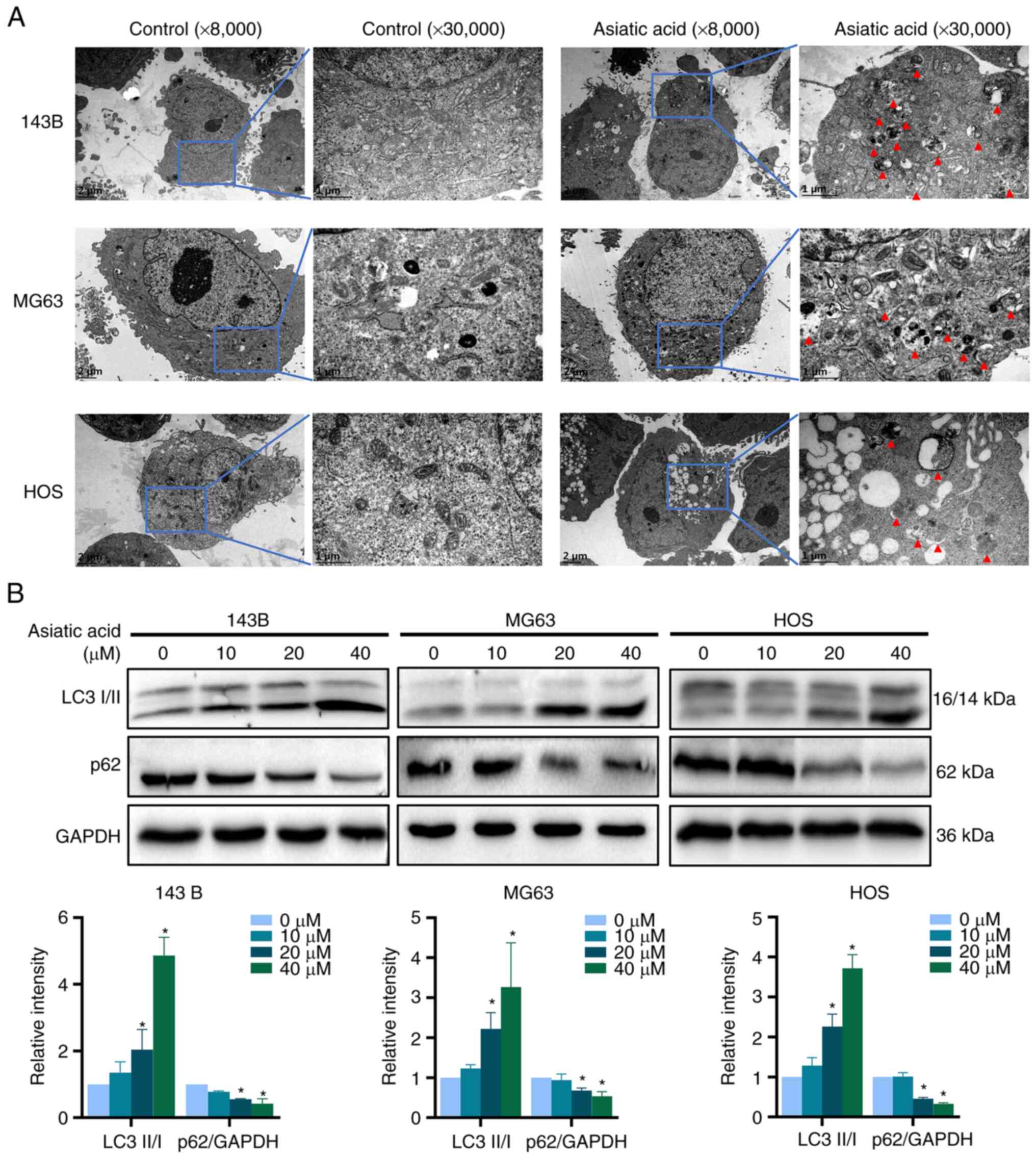
Asiatic acid triggers autophagy in
osteosarcoma cells
Autophagy is closely related to apoptosis and both
processes are involved in tumor cell death mechanisms (42,43).
Therefore, the present study next investigated whether asiatic acid
triggers autophagy in 143B, MG63 and HOS osteosarcoma cells.
Transmission electron microscopy is the gold standard for examining
autophagy by confirming the presence of autophagosomes at the
subcellular level. Osteosarcoma cells were treated with 40 µM
asiatic acid. The transmission electron microscopy images revealed
marked autophagosome increases in all the treated cells (Fig. 6A). Furthermore, the present study
assessed the effects of asiatic acid on the levels of
autophagy-related proteins LC3 and p62, which are marker proteins,
and the western blot analysis results demonstrated that asiatic
acid treatment markedly increased the LC3-II/I ratio and decreased
the p62 levels, indicating that asiatic acid could trigger
autophagy in osteosarcoma cells (Fig.
6B).
Asiatic acid inhibits the PI3K/AKT
signaling pathway and activates the ROS/MAPK signaling pathway in
osteosarcoma cells
The KEGG enrichment analysis results suggested that
the PI3K/AKT and the MAPK signaling pathways were associated with
the anti-osteosarcoma effects of asiatic acid. Thus, the present
study examined the expression levels of PI3K/AKT signaling
pathway-related proteins. Fig. 7A
shows that asiatic acid treatment markedly decreased the levels of
p-PI3K/PI3K and p-AKT/AKT. ROS analysis using DCFH-DA staining
(Fig. 7B) revealed that asiatic
acid treatment markedly increased intracellular ROS content.
Western blot analysis results further demonstrated that asiatic
acid upregulated the protein levels of p-ERK1/2/ERK, p-p38/p38 and
p-JNK/JNK (Fig. 7C). Consequently,
asiatic acid may inhibit the PI3K/AKT signaling pathway and
activate the ROS/MAPK signaling pathway to regulate osteosarcoma
cell death mechanisms.
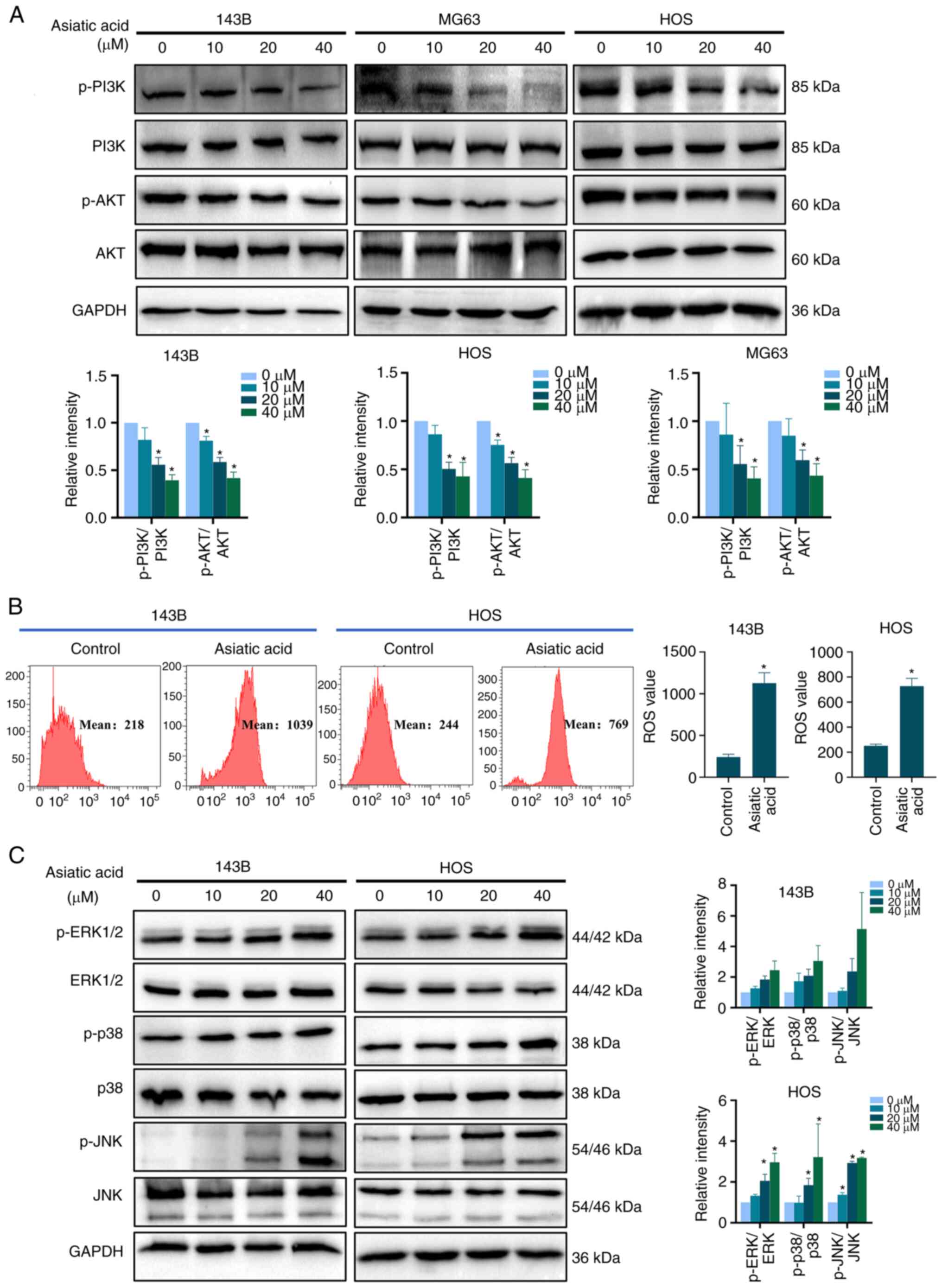 | Figure 7.Asiatic acid inhibits PI3K/AKT and
activates ROS/MAPK pathways in osteosarcoma cells. Osteosarcoma
cells were treated with asiatic acid for 24 h. (A) Protein levels
of p-PI3K, PI3K, p-AKT, and AKT as measured by western blot
analyses. (B) Intracellular ROS content by flow cytometry. (C)
Protein levels of p-ERK1/2, ERK1/2, p-p38, p38, p-JNK, and JNK by
western blot analyses. *P<0.05 vs. control group. ROS, reactive
oxygen species; p-, phosphorylated. |
Discussion
Doxorubicin, cisplatin and methotrexate are common
chemotherapy agents used against osteosarcoma (44); however, their cytotoxic effects and
drug resistance limit their use in the clinic (45,46).
Therefore, uncovering the pathogenesis of osteosarcoma and
developing safe and effective therapeutic agents is important. In
the present study, the potential targets and pathways of asiatic
acid for osteosarcoma treatment were predicted using network
pharmacology and molecular docking, and validated by in
vitro experiments in osteosarcoma cells. The results indicated
78 common targets of asiatic acid and osteosarcoma. PPI network
analysis identified EGFR, Caspase-3, ESR1, HSP90AA1, IL-6 and SRC
as potential targets of asiatic acid in osteosarcoma. KEGG pathway
analysis suggested a mechanism for asiatic acid against
osteosarcoma associated with cancer-related pathways such as
‘Proteoglycans in cancer’ (hsa05205), ‘PI3K-AKT signaling pathway’
(hsa04151), ‘Rap1 signaling pathway’ (hsa04015) ‘MAPK signaling
pathway’ (hsa04010) and ‘Ras signaling pathway’ (hsa04014).
Numerous types of cancer develop after genetic
aberrations in EGFR facilitate growth, motility and metastasis of
tumor cells (47,48). Abnormal expression and mutations of
EGFR are associated with the development of osteosarcoma (49,50).
Upregulation of EGFR expression is prevalent in osteosarcoma
samples and EGFR expression is associated with chemotherapy-induced
stress survival of tumor cells (51,52).
Caspase-3 is the main terminal cleaving enzyme of apoptosis and is
an essential target for cancer therapy (53). ESR1 is positive in most osteosarcoma
specimens, its level is associated with the tumor volume and
inhibition of the ESR1 enhances chemotherapeutic effects in P53(+)
osteosarcomas (54). HSP90AA1, a
heat shock protein, promotes cancer metastases and drug resistance
in multiple tumors (55–57). In addition, as an essential
autophagy regulator, it participates in the drug resistance of
osteosarcoma cells (58). Abnormal
IL-6 expression is associated with tumor angiogenesis, invasion,
metastases, diagnoses and prognoses (59,60).
IL-6 can improve invasion by promoting the expression of
intercellular adhesion molecule-1 in osteosarcoma cells (61). It also promotes osteosarcoma
stemness and carcinogenicity by upregulating the STAT3 signaling
pathway (62). SRC is a member of
the non-receptor protein tyrosine kinase family. Its abnormal
expression can cause the development of certain tumors (63), and the protein has been associated
with malignant features of osteosarcomas (64,65).
The activated SRC kinase activates the MAPK and PI3K/AKT signaling
pathways by phosphorylating target tyrosine residues (65,66).
Based on PPI network analysis, EGFR, Caspase-3, ESR1, HSP90AA1,
IL-6 and SRC proteins were selected for molecular docking. The
present findings demonstrated that asiatic acid had a strong
affinity for these targets, indicating its anti-osteosarcoma effect
through these targets and related pathways.
Cyclins and CDKs determine cell cycle progression
(67). Numerous anticancer drugs
arrest cancer cell cycles (68).
CDK2/cyclin E and CDK2/cyclin A promote the initiation and
progression of DNA replication through the S phase (69), while CDK1/cyclin A and CDK1/cyclin B
complexes activate the expression of genes essential for the
mitotic process (70) during the
G2/M phase. Through in vitro validation, the
present results indicated that asiatic acid inhibited
proliferation, induced G2/M arrest and suppressed cyclin
A2 and CDK2 levels in osteosarcoma cells. The utilization of
MC3T3-E1 mouse-derived normal osteoblast cells as controls in safe
concentration trials for osteosarcoma has been extensively
documented (71–74). Therefore, the present study
purposively opted for MC3T3-E1 normal osteoblast cells as a
reference acid on normal cells. However, it would be preferable to
select human normal osteoblast cells, such as the hFOB 1.19 cell
line. Unfortunately, the lack of toxicity assessment of asiatic
acid on normal human osteoblast cells is a limitation in the
present study. In addition, it was observed that the viability of
MC3T3-E1 normal osteoblasts remained unaffected under the same
conditions, thereby demonstrating the anti-osteosarcoma effects of
asiatic acid within a safe concentration range. Thus, asiatic acid
inhibited osteosarcoma cell proliferation through G2/M
cell cycle arrest.
Elevations in the Bax/Bcl-2 ratio reduce the MMP in
the early apoptosis stages (75),
triggering the release of cytochrome C, which further activates the
caspase family and induces cell death (76,77).
The channel protein VDAC1 on the outer mitochondrial membrane
controls the entry and exit of substances and energy into and out
of mitochondria (78), including
the entry of cytochrome C into the cytoplasm. VDAC1 is also a
crucial component of apoptosis (79,80)
regulating related proteins to induce apoptosis (81). The present flow cytometry
experiments revealed that asiatic acid treatment reduced the MMP
and induced apoptosis in osteosarcoma cells, as demonstrated by
increased Bax and VDAC1 levels and decreased Bcl-2 levels,
indicating mitochondria-dependent apoptosis. Autophagy includes
four stages: Initiation, autophagosome formation, binding of
autophagosomes to lysosomes and autophagosome degradation (82,83).
During autophagy, cytoplasmic LC3 becomes hydrolyzed to LC3-I,
which is conjugated with phosphatidylethanolamine to form LC3-II,
which is found in autophagosome membranes (84,85).
During this dynamic process, p62 is selectively encapsulated into
autophagosomes and later degraded by autolysosomes (86). The present results revealed an
increase in the LC3-II/LC3-I ratio and a decrease in p62 protein
expression in osteosarcoma cells treated with asiatic acid,
indicating that autophagy had been triggered. In addition, the high
autophagosome abundance observed in asiatic acid-treated cells
under the electron microscope further confirmed that asiatic acid
induced autophagy in osteosarcoma cells.
KEGG enrichment analysis of network pharmacology
revealed that the PI3K/AKT and MAPK signaling pathways are
essential for asiatic acid to combat osteosarcomas. The PI3K/AKT
signaling pathway is fixed and activated in various tumors and
promotes tumor development (87).
It mediates the progression of osteosarcoma cell proliferation,
metastasis, apoptosis and autophagy (88,89).
Furthermore, studies have demonstrated that inhibiting the activity
of this pathway and its related upstream and downstream molecules
via small molecule inhibitors is vital for treating osteosarcomas
(90,91). The ERK1/2, JNK and p38 proteins of
the MAPK cascade pathway are sensitive to intracellular oxidative
stress (92–94). ROS within osteosarcoma cells
activate the MAPK signaling pathway, which induces apoptosis and
autophagy (95,96). Notably, asiatic acid has antioxidant
properties in osteoporotic mice, RAW264.7 cells, H9c2 rat
cardiomyocytes and HepG2 human hepatoma cells, mainly involving
peroxisome proliferator-activated receptor γ expression and the
AKT/GSK-3β/hypoxia-inducible factor-1α, sirtuin 1/FOXO1 and nuclear
factor erythroid 2-related factor 2/heme oxygenase 1 (Nrf2/HO-1)
pathways (97–100). However, asiatic acid also promotes
ROS generation in MCF7 human breast cancer cells, A549 and H1299
human lung cancer cells, and SK-MEL-2 human melanoma cells
(101–103). Antioxidant and pro-oxidant effects
in different cellular biological environments, probably due to
different targets and mechanisms of asiatic acid. The present
findings suggested that asiatic acid decreased the phosphorylation
of PI3K and AKT, increased intracellular ROS levels, and resulted
in markedly higher p-ERK1/2/ERK1/2, p-p38/p38 and p-JNK/JNK levels
in osteosarcoma cells. Regrettably, the absence of evaluation for
transfection of target gene overexpression, mutation and inhibition
experiments and in vivo experiments are limitations of the
present study. Based on the present results, it was hypothesized
that asiatic acid may induce apoptosis and autophagy in
osteosarcoma cells by inhibiting PI3K/AKT and activating ROS/MAPK
pathways.
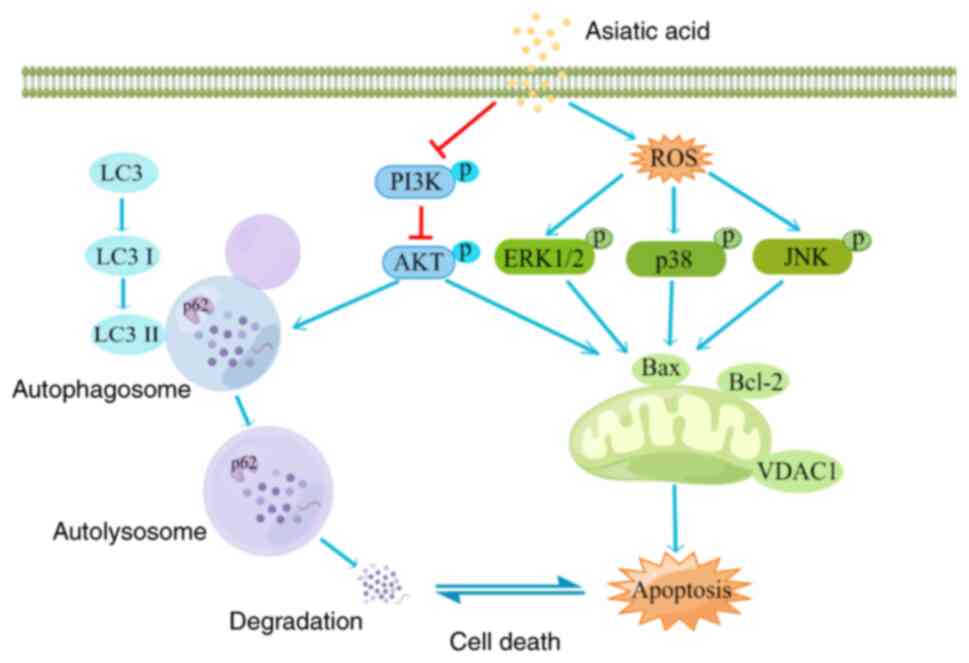
In conclusion, the present study explored the
multi-target and multi-signaling pathways of asiatic acid against
osteosarcoma using network pharmacology and molecular docking
techniques. Subsequently, the present in vitro experiment
results demonstrated that asiatic acid mediated apoptosis and
autophagy in osteosarcoma cells by inhibiting the PI3K/AKT
signaling pathways and activating the ROS/MAPK signaling pathways.
The excellent anti-osteosarcoma efficacy and associated mechanisms
of asiatic acid (Fig. 8) make it a
promising novel anti-osteosarcoma agent.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The Priming Fund for Scientific Research of High-level Talents
in Guangdong Medical University (grant no. 1037Z20220030), The
Young Innovative Talents Project of Guangdong Higher Education
Institution (grant no. 2021KQNCX023) and the Department of Science
and Technology of Guangdong Medical University (grant no.
2001/2XK17006) and the Zhanjiang Science and Technology Bureau
(grant no. 200513174547221) supported the present study.
Availability of data and materials
The datasets used and/or analyzed during this study
are available from the corresponding author upon reasonable
request.
Authors' contributions
BW, LS, and HP conducted and designed the research.
HP, HW, and ZZ performed the experiments, analyzed data and wrote
the paper. HP, MX and TW provided technical support and purchased
all the reagents and chemicals needed. ZW, HP, HW, and TW designed
the figures. HP and HW contributed equally to this work and should
be considered co-first authors. HP and HW confirm the authenticity
of all the raw data. All authors contributed to the article and all
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The PubChem, PharmMapper, Swiss Target Prediction,
Uniprot, OMIM, TTD, Genecards, and STRING databases are publicly
available and allows researchers to download and analyze. Thus, our
Ethics Committee of Affiliated Hospital of Guangdong Medical
University waived ethical approval for open public databases that
were used in this work.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Karadurmus N, Sahin U, Bahadir Basgoz B
and Demirer T: Is there a role of high dose chemotherapy and
autologous stem cell transplantation in the treatment of Ewing's
sarcoma and osteosarcomas? J BUON. 23:1235–1241. 2018.PubMed/NCBI
|
|
2
|
Zhu T, Han J, Yang L, Cai Z, Sun W, Hua Y
and Xu J: Immune microenvironment in osteosarcoma: Components,
therapeutic strategies and clinical applications. Front Immunol.
13:9075502022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng C, Tang F, Min L, Hornicek F, Duan Z
and Tu C: PTEN in osteosarcoma: Recent advances and the therapeutic
potential. Biochim Biophys Acta Rev Cancer. 1874:1884052020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cortini M, Avnet S and Baldini N:
Mesenchymal stroma: Role in osteosarcoma progression. Cancer Lett.
405:90–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mutsaers A and Walkley C: Cells of origin
in osteosarcoma: Mesenchymal stem cells or osteoblast committed
cells? Bone. 62:56–63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cascini C and Chiodoni C: The immune
landscape of osteosarcoma: Implications for prognosis and treatment
response. Cells. 10:16682021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ghafouri-Fard S, Shirvani-Farsani Z,
Hussen B and Taheri M: The critical roles of lncRNAs in the
development of osteosarcoma. Biomed Pharmacother. 135:1112172021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Osborne T and Khanna C: A review of the
association between osteosarcoma metastasis and protein
translation. J Comp Pathol. 146:132–142. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen C, Xie L, Ren T, Huang Y, Xu J and
Guo W: Immunotherapy for osteosarcoma: Fundamental mechanism,
rationale, and recent breakthroughs. Cancer Lett. 500:1–10. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prudowsky Z and Yustein J: Recent insights
into therapy resistance in osteosarcoma. Cancers (Basel).
13:832020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Berner K, Johannesen TB, Berner A,
Haugland HK, Bjerkehagen B, Bøhler PJ and Bruland ØS: Time-trends
on incidence and survival in a nationwide and unselected cohort of
patients with skeletal osteosarcoma. Acta Oncol. 54:25–33. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Grinberg S, Posta A, Weber K and Wilson R:
Limb salvage and reconstruction options in osteosarcoma. Adv Exp
Med Biol. 1257:13–29. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bielack S, Jürgens H, Jundt G, Kevric M,
Kühne T, Reichardt P, Zoubek A, Werner M, Winkelmann W and Kotz R:
Osteosarcoma: The COSS experience. Cancer Treat Res. 152:289–308.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bielack S, Kempf-Bielack B, Delling G,
Exner GU, Flege S, Helmke K, Kotz R, Salzer-Kuntschik M, Werner M,
Winkelmann W, et al: Prognostic factors in high-grade osteosarcoma
of the extremities or trunk: An analysis of 1,702 patients treated
on neoadjuvant cooperative osteosarcoma study group protocols. J
Clin Oncol. 20:776–790. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kager L, Zoubek A, Kastner U,
Kempf-Bielack B, Potratz J, Kotz R, Exner GU, Franzius C, Lang S,
Maas R, et al: Skip metastases in osteosarcoma: Experience of the
cooperative osteosarcoma study group. J Clin Oncol. 24:1535–1541.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Lou Y, Wang J, Yu C and Shen W:
Research status and molecular mechanism of the traditional chinese
medicine and antitumor therapy combined strategy based on tumor
microenvironment. Front Immunol. 11:6097052020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mioc M, Milan A, Malița D, Mioc A, Prodea
A, Racoviceanu R, Ghiulai R, Cristea A, Căruntu F and Șoica C:
Recent advances regarding the molecular mechanisms of triterpenic
acids: A review (Part I). Int J Mol Sci. 23:77402022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hong G, Zhou L, Han X, Sun P, Chen Z, He
W, Tickner J, Chen L, Shi X and Xu J: Asiatic acid inhibits
OVX-induced osteoporosis and osteoclastogenesis regulating
RANKL-mediated NF-κb and Nfatc1 signaling pathways. Front
Pharmacol. 11:3312020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sycz Z, Tichaczek-Goska D and Wojnicz D:
Anti-planktonic and Anti-biofilm properties of pentacyclic
triterpenes-asiatic acid and ursolic acid as promising
antibacterial future pharmaceuticals. Biomolecules. 12:982022.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Songvut P, Chariyavilaskul P, Tantisira M
and Khemawoot P: Safety and pharmacokinetics of standardized
extract of centella asiatica (ECa 233) Capsules in healthy thai
volunteers: A phase 1 clinical study. Planta Med. 85:483–490. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Palmer N and Kaldis P: Less-well known
functions of cyclin/CDK complexes. Semin Cell Dev Biol. 107:54–62.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jirawatnotai S, Dalton S and
Wattanapanitch M: Role of cyclins and cyclin-dependent kinases in
pluripotent stem cells and their potential as a therapeutic target.
Semin Cell Dev Biol. 107:63–71. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu W, Jin W, Zhu S, Chen Y and Liu B:
Targeting regulated cell death (RCD) with small-molecule compounds
in cancer therapy: A revisited review of apoptosis,
autophagy-dependent cell death and necroptosis. Drug Discov Today.
27:612–625. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morana O, Wood W and Gregory C: The
apoptosis paradox in cancer. Int J Mol Sci. 23:13282022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carneiro B and El-Deiry WS: Targeting
apoptosis in cancer therapy. Nat Rev Clin Oncol. 17:395–417. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fulda S: Targeting extrinsic apoptosis in
cancer: Challenges and opportunities. Semin Cell Dev Biol.
39:20–25. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tan Y, Zhang X, Zhang S, Zhu T, Garg M,
Lobie PE and Pandey V: Mitochondria: The metabolic switch of
cellular oncogenic transformation. Biochim Biophys Acta Rev Cancer.
1876:1885342021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Noguchi M, Hirata N, Tanaka T, Suizu F,
Nakajima H and Chiorini J: Autophagy as a modulator of cell death
machinery. Cell Death Dis. 11:5172020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gerada C and Ryan K: Autophagy, the innate
immune response and cancer. Mol Oncol. 14:1913–1929. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Braicu C, Zanoaga O, Zimta AA, Tigu AB,
Kilpatrick KL, Bishayee A, Nabavi SM and Berindan-Neagoe I: Natural
compounds modulate the crosstalk between apoptosis- and
autophagy-regulated signaling pathways: Controlling the
uncontrolled expansion of tumor cells. Semin Cancer Biol.
80:218–236. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
du Plessis M, Davis T, Loos B, Pretorius
E, de Villiers W and Engelbrecht A: Molecular regulation of
autophagy in a pro-inflammatory tumour microenvironment: New
insight into the role of serum amyloid A. Cytokine Growth Factor
Rev. 59:71–83. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jing Y, Liang W, Liu J, Zhang L, Wei J,
Yang J, Zhang Y and Huang Z: Autophagy-mediating microRNAs in
cancer chemoresistance. Cell Biol Toxicol. 36:517–536. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miller D and Thorburn A: Autophagy and
organelle homeostasis in cancer. Dev Cell. 56:906–918. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ning B, Liu Y, Huang T and Wei Y:
Autophagy and its role in osteosarcoma. Cancer Med. 12:5676–5687.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Das C, Banerjee I and Mandal M:
Pro-survival autophagy: An emerging candidate of tumor progression
through maintaining hallmarks of cancer. Semin Cancer Biol.
66:59–74. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Long M and McWilliams T: Monitoring
autophagy in cancer: From bench to bedside. Semin Cancer Biol.
66:12–21. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gupta R, Ambasta R and Pravir K: Autophagy
and apoptosis cascade: Which is more prominent in neuronal death?
Cell Mol Life Sci. 78:8001–8047. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu K, Ren T, Huang Y, Sun K, Bao X, Wang
S, Zheng B and Guo W: Apatinib promotes autophagy and apoptosis
through VEGFR2/STAT3/BCL-2 signaling in osteosarcoma. Cell Death
Dis. 8:e30152017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hao DC and Xiao P: Network pharmacology: A
Rosetta stone for traditional Chinese medicine. Drug Dev Res.
75:299–312. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Stanzione F, Giangreco I and Cole J: Use
of molecular docking computational tools in drug discovery. Prog
Med Chem. 60:273–343. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bock FJ and Tait SWG: Mitochondria as
multifaceted regulators of cell death. Nat Rev Mol Cell Biol.
21:85–100. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
42. Mariño G, Niso-Santano M, Baehrecke E
and Kroemer G: Self-consumption: The interplay of autophagy and
apoptosis. Nat Rev Mol Cell Biol. 15:81–94. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol
Cancer. 14:482015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ritter J and Bielack S: Osteosarcoma. Ann
Oncol. 21 (Suppl 7):vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
A sleeping beauty screen highlights cancer
drivers in osteosarcoma. Cancer Discov. 5:6902015. View Article : Google Scholar
|
|
46
|
Spalato M and Italiano A: The safety of
current pharmacotherapeutic strategies for osteosarcoma. Expert
Opin Drug Safety. 20:427–438. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Sevelda F, Mayr L, Kubista B, Lötsch D,
van Schoonhoven S, Windhager R, Pirker C, Micksche M and Berger W:
EGFR is not a major driver for osteosarcoma cell growth in vitro
but contributes to starvation and chemotherapy resistance. J Exp
Clin Cancer Res. 34:1342015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kato S, Lippman S, Flaherty K and Kurzrock
R: The conundrum of genetic ‘Drivers’ in benign conditions. J Natl
Cancer Inst. 108:djw0362016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang W, Zhao HF, Yao TF and Gong H:
Advanced development of ErbB family-targeted therapies in
osteosarcoma treatment. Invest New Drugs. 37:175–183. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wan Z, Huang S, Mo F, Yao Y, Liu G, Han Z,
Chen M and Zhiyun L: CSN5 controls the growth of osteosarcoma via
modulating the EGFR/PI3K/Akt axis. Exp Cell Res. 384:1116462019.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kersting C, Gebert C, Agelopoulos K,
Schmidt H, van Diest PJ, Juergens H, Winkelmann W, Kevric M,
Gosheger G, Brandt B, et al: Epidermal growth factor receptor
expression in high-grade osteosarcomas is associated with a good
clinical outcome. Clin Cancer Res. 13:2998–3005. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang SL, Zhong GX, Wang XW, Yu FQ, Weng
DF, Wang XX and Lin JH: Prognostic significance of the expression
of HER family members in primary osteosarcoma. Oncol Lett.
16:2185–2194. 2018.PubMed/NCBI
|
|
53
|
Yadav P, Yadav R, Jain S and Vaidya A:
Caspase-3: A primary target for natural and synthetic compounds for
cancer therapy. Chem Biol Drug Des. 98:144–165. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang J, Chen C, Chen C, Wu P and Chen WM:
Suppression of estrogen receptor alpha inhibits cell proliferation,
differentiation and enhances the chemosensitivity of P53-positive
U2OS osteosarcoma cell. Int J Mol Sci. 22:112382021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Taipale M, Jarosz D and Lindquist S: HSP90
at the hub of protein homeostasis: Emerging mechanistic insights.
Nat Rev Mol Cell Biol. 11:515–528. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang M, Peng Y, Yang Z, Zhang H, Xu C,
Liu L, Zhao Q, Wu J, Wang H and Liu J: DAB2IP down-regulates
HSP90AA1 to inhibit the malignant biological behaviors of
colorectal cancer. BMC Cancer. 22:5612022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chu S, Liu Y, Zhang L, Liu B and Li L, Shi
JZ and Li L: Regulation of survival and chemoresistance by HSP90AA1
in ovarian cancer SKOV3 cells. Mol Biol Rep. 40:1–6. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Xiao X, Wang W, Li Y, Yang D, Li X, Shen
C, Liu Y, Ke X, Guo S and Guo Z: HSP90AA1-mediated autophagy
promotes drug resistance in osteosarcoma. J Exp Clin Cancer Res.
37:2012018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Szulc-Kielbik I, Kielbik M, Nowak M and
Klink M: The implication of IL-6 in the invasiveness and
chemoresistance of ovarian cancer cells. Systematic review of its
potential role as a biomarker in ovarian cancer patients. Biochim
Biophys Acta Rev Cancer. 1876:1886392021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tzeng HE, Tsai CH, Chang ZL, Su CM, Wang
SW, Hwang WL and Tang CH: Interleukin-6 induces vascular
endothelial growth factor expression and promotes angiogenesis
through apoptosis signal-regulating kinase 1 in human osteosarcoma.
Biochem Pharmacol. 85:531–540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Itoh H, Kadomatsu T, Tanoue H, Yugami M,
Miyata K, Endo M, Morinaga J, Kobayashi E, Miyamoto T, Kurahashi R,
et al: TET2-dependent IL-6 induction mediated by the tumor
microenvironment promotes tumor metastasis in osteosarcoma.
Oncogene. 37:2903–2920. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang C, Ma K and Li WY: IL-6 promotes
cancer stemness and oncogenicity in U2OS and MG-63 Osteosarcoma
Cells by Upregulating the OPN-STAT3 pathway. J Cancer.
10:6511–6525. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Parkin A, Man J, Timpson P and Pajic M:
Targeting the complexity of Src signalling in the tumour
microenvironment of pancreatic cancer: From mechanism to therapy.
FEBS J. 286:3510–3539. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang Z, Xie J, Fang J, Lv M, Yang M, Deng
Z, Xie Y and Cai L: Nigericin exerts anticancer effects through
inhibition of the SRC/STAT3/BCL-2 in osteosarcoma. Biochem
Pharmacol. 198:1149382022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Urciuoli E, Coletta I, Rizzuto E, De Vito
R, Petrini S, D'Oria V, Pezzullo M, Milano GM, Cozza R, Locatelli F
and Peruzzi B: Src nuclear localization and its prognostic
relevance in human osteosarcoma. J Cell Physiol. 233:1658–1670.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhao G, Gao Z, Zhang Q, Tang XF, Lv YF,
Zhang ZS, Zhang Y, Tan QL, Peng DB, Jiang DM and Guo QN: TSSC3
promotes autophagy via inactivating the Src-mediated PI3K/Akt/mTOR
pathway to suppress tumorigenesis and metastasis in osteosarcoma,
and predicts a favorable prognosis. J Exp Clin Cancer Res.
37:1882018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Klein MJ: Cyclin-dependent kinase
inhibition: An opportunity to target protein-protein interactions.
Adv Protein Chem Struct Biol. 121:115–141. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zou T and Lin Z: The involvement of
ubiquitination machinery in cell cycle regulation and cancer
progression. Int J Mol Sci. 22:57542021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Han C, Wang Z, Chen S, Li L, Xu Y, Kang W,
Wei C, Ma H, Wang M and Jin X: Berbamine suppresses the progression
of bladder cancer by modulating the ROS/NF-κ B axis. Oxid Med Cell
Longev. 2021:88517632021. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Fischer M and Müller GJ: Cell cycle
transcription control: DREAM/MuvB and RB-E2F complexes. Crit Rev
Biochem Mol Biol. 52:638–662. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang C, Huang C, Yang P, Li C and Li M:
Eldecalcitol induces apoptosis and autophagy in human osteosarcoma
MG-63 cells by accumulating ROS to suppress the PI3K/Akt/mTOR
signaling pathway. Cell Signal. 78:1098412021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wirries A, Jabari S, Jansen EP, Roth S,
Figueroa-Juárez E, Wissniowski TT, Neureiter D, Klieser E, Lechler
P, Ruchholtz S, et al: Panobinostat mediated cell death: A novel
therapeutic approach for osteosarcoma. Oncotarget. 9:32997–33010.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Mickymaray S, Alfaiz FA, Paramasivam A,
Veeraraghavan VP, Periadurai ND, Surapaneni KM and Niu G:
Rhaponticin suppresses osteosarcoma through the inhibition of
PI3K-Akt-mTOR pathway. Saudi J Biol Sci. 28:3641–3649. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tung FI, Chen LC, Wang YC, Chen MH, Shueng
PW and Liu TY: Using a hybrid radioenhancer to discover tumor
cell-targeted treatment for osteosarcoma: An in vitro study. Curr
Med Chem. 28:3877–3889. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Burke PJ: Mitochondria, bioenergetics and
apoptosis in cancer. Trends Cancer. 3:857–870. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Praharaj PP, Naik PP, Panigrahi DP, Bhol
CS, Mahapatra KK, Patra S, Sethi G and Bhutia SK: Intricate role of
mitochondrial lipid in mitophagy and mitochondrial apoptosis: Its
implication in cancer therapeutics. Cell Mol Life Sci.
76:1641–1652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Gibson CJ and Davids MS: BCL-2 antagonism
to target the intrinsic mitochondrial pathway of apoptosis. Clin
Cancer Res. 21:5021–5029. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Shoshan-Barmatz V, Shteinfer-Kuzmine A and
Verma A: VDAC1 at the intersection of cell metabolism, apoptosis,
and diseases. Biomolecules. 10:14852020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shoshan-Barmatz V, Krelin Y and Chen Q:
VDAC1 as a player in mitochondria-mediated apoptosis and target for
modulating apoptosis. Curr Med Chem. 24:4435–4446. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Shoshan-Barmatz V, De S and Meir A: The
mitochondrial voltage-dependent anion channel 1, Ca2+ transport,
apoptosis, and their regulation. Front Oncol. 7:602017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shoshan-Barmatz V, Krelin Y,
Shteinfer-Kuzmine A and Arif T: Voltage-dependent anion channel 1
as an emerging drug target for novel anti-cancer therapeutics.
Front Oncol. 7:1542017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Xia H, Green D and Zou W: Autophagy in
tumour immunity and therapy. Nat Rev Cancer. 21:281–297. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Levy J, Towers C and Thorburn A: Targeting
autophagy in cancer. Nat Rev Cancer. 17:528–542. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jacquet M, Guittaut M, Fraichard A and
Despouy G: The functions of Atg8-family proteins in autophagy and
cancer: Linked or unrelated? Autophagy. 17:599–611. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Heckmann BL and Green DR: LC3-associated
phagocytosis at a glance. J Cell Sci. 132:2019. View Article : Google Scholar
|
|
86
|
Moscat J, Karin M and Diaz-Meco MT: p62 in
cancer: Signaling adaptor beyond autophagy. Cell. 167:606–609.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
He Y, Sun MM, Zhang GG, Yang J, Chen KS,
Xu WW and Li B: Targeting PI3K/Akt signal transduction for cancer
therapy. Signal Transduct Target Ther. 6:4252021. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhang J, Yu X, Yan Y, Wang C and Wang WJ:
PI3K/Akt signaling in osteosarcoma. Clin Chim Acta. 444:182–192.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Liu M, Liu F, Li YJ, Yin JN, Gao YL, Wang
XY, Yang C, Liu JG and Li HJ: Ginsenoside Rg5 inhibits human
osteosarcoma cell proliferation and induces cell apoptosis through
PI3K/Akt/mTORC1-Related LC3 autophagy pathway. Oxid Med Cell
Longev. 2021:50403262021.PubMed/NCBI
|
|
90
|
Angulo P, Kaushik G, Subramaniam D,
Dandawate P, Neville K, Chastain K and Anant S: Natural compounds
targeting major cell signaling pathways: A novel paradigm for
osteosarcoma therapy. J Hematol Oncol. 10:102017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Khezri MR, Jafari R, Yousefi K and
Zolbanin NM: The PI3K/AKT signaling pathway in cancer: Molecular
mechanisms and possible therapeutic interventions. Exp Mol Pathol.
127:1047872022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Rezatabar S, Karimian A, Rameshknia V,
Parsian H, Majidinia M, Kopi TA, Bishayee A, Sadeghinia A, Yousefi
M, Monirialamdari M and Yousefi B: RAS/MAPK signaling functions in
oxidative stress, DNA damage response and cancer progression. J
Cell Physiol. 234:14951–14965. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Wagner E and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Ding S, Pang ZY, Chen XM, Li Z, Liu XX,
Zhai QL, Huang JM and Ruan ZY: Urolithin a attenuates IL-1β-induced
inflammatory responses and cartilage degradation via inhibiting the
MAPK/NF-κB signaling pathways in rat articular chondrocytes. J
Inflamm (Lond). 17:132020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lv H, Zhen C, Liu J and Shang PJ:
β-Phenethyl isothiocyanate induces cell death in human osteosarcoma
through altering iron metabolism, disturbing the redox balance, and
activating the MAPK signaling pathway. Oxid Med Cell Longev.
2020:50219832020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Zhu J, Yu W, Liu B, Wang Y, Shao J, Wang
J, Xia K, Liang C, Fang W, Zhou C and Tao H: Escin induces
caspase-dependent apoptosis and autophagy through the ROS/p38 MAPK
signalling pathway in human osteosarcoma cells in vitro and in
vivo. Cell Death Dis. 8:e31132017. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen X, Han D, Liu T, Huang C, Hu Z, Tan X
and Wu S: Asiatic acid improves high-fat-diet-induced osteoporosis
in mice via regulating SIRT1/FOXO1 signaling and inhibiting
oxidative stress. Histol Histopathol. 37:769–777. 2022.PubMed/NCBI
|
|
98
|
Huang X, Zuo L, Lv Y, Chen C, Yang Y, Xin
H, Li Y and Qian Y: Asiatic acid attenuates myocardial
ischemia/reperfusion injury via Akt/GSK-3β/HIF-1α signaling in rat
H9c2 cardiomyocytes. Molecules. 21:12482016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Qi Z, Ci X, Huang J, Liu Q, Yu Q, Zhou J
and Deng X: Asiatic acid enhances Nrf2 signaling to protect HepG2
cells from oxidative damage through Akt and ERK activation. Biomed
Pharmacother. 88:252–259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xu Y, Yao J, Zou C, Zhang H, Zhang S, Liu
J, Ma G, Jiang P and Zhang W: Asiatic acid protects against hepatic
ischemia/reperfusion injury by inactivation of Kupffer cells via
PPARγ/NLRP3 inflammasome signaling pathway. Oncotarget.
8:86339–86355. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Dutta S, Chakraborty P, Basak S, Ghosh S,
Ghosh N, Chatterjee S, Dewanjee S and Sil PC: Synthesis,
characterization, and evaluation of in vitro cytotoxicity and in
vivo antitumor activity of asiatic acid-loaded poly
lactic-co-glycolic acid nanoparticles: A strategy of treating
breast cancer. Life Sci. 307:1208762022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Wu T, Geng J, Guo W, Gao J and Zhu XJ:
Asiatic acid inhibits lung cancer cell growth in vitro and in vivo
by destroying mitochondria. Acta Pharm Sin B. 7:65–72. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Park B, Bosire K, Lee E, Lee Y and Kim JJ:
Asiatic acid induces apoptosis in SK-MEL-2 human melanoma cells.
Cancer Lett. 218:81–90. 2005. View Article : Google Scholar : PubMed/NCBI
|