Introduction
Lung cancer is considered being one of the most
threatening malignant tumors in the health and life of individuals,
with the highest fatality rate in the world (1). It can be histologically divided into
two categories: Small cell lung cancer (SCLC) and non-small cell
lung cancer (NSCLC), of which NSCLC accounts for ~85% (2). NSCLC is characterized by lack of
symptoms when tumors are small in the early stages, so most
patients have locally advanced or metastatic lesions at diagnosis
(3). Therefore, it is of great
importance to understand the mechanism of NSCLC development to
guide the development and use of lung cancer-targeted drugs,
including the combination of clinical drugs.
In the past decade, scholars have generally
recognized the importance of tumor-specific metabolic patterns in
the pathogenesis of tumors. Among them, the Warburg effect
indicates that tumor cells tend to obtain energy through aerobic
glycolysis along with lactate production, even in the presence of
oxygen (4). In addition, hypoxia is
an important feature of solid tumors and induces cells to undergo
glycolysis to produce energy (5).
Therefore, efficient glycolysis is the main function of tumor
cells. The occurrence of this phenomenon depends on the regulation
of intracellular oncogene and tumor suppressor gene signaling
pathways on cellular metabolic pathways. The central regulator in
the above regulatory process is the hypoxia-inducible factor (HIF)
(6).
HIF is a type of transcription factor that is
rapidly produced and accumulated in the cell environment under
hypoxic conditions. It was initially discovered that it can
significantly increase the transcription of erythropoietin by
interacting with its enhancer (7).
HIF is a heterodimer consisting of one α-subunit and one β-subunit.
It is currently known that there are three types of both a-subunit
and β-subunit which are HIF-1α, HIF-2α, HIF-3α, HIF-1β, HIF-2β and
HIF-3β (8). Among them, HIF-1α is
the most oxygen-sensitive active subunit, which is the main part of
HIF-1 to fulfill its function, and it is the earliest discovered
and the more thoroughly studied subunit at present (9). Under physiological conditions when
oxygen supply is sufficient, HIF-1α is easily degraded by the
proteasome through the complex formed by oxygen-dependent
hydroxylation and E3 ubiquitin ligase, so its half-life is only
5–10 min (10). As a result,
signaling pathways downstream of HIF are not activated and
transcription of the more than 40 genes it regulates, including
erythropoietin, glucose transporter proteins, glycolytic enzymes,
vascular endothelial growth factor and other genes and protein
products that increase oxygen delivery or promote hypoxic
metabolism, is at a low level (11).
Generally, regulation of HIF-1α expression is
complex and can be categorized into oxygen-dependent and
oxygen-independent components. The oxygen-independent mechanisms
include the transcription regulation of HIF-1α expression through
the action of transcription factors such as nuclear factor-κB
(NF-κB), specificity protein 1 (SP1) and signal transducer and
activator of transcription 3 (STAT3) (12). The actions of these transcription
factors are in turn regulated by reactive oxygen species (ROS),
cytokines and/or lipopolysaccharide (LPS)-dependent
signaling-activated protein kinase C (PKC), NF-κB kinase inhibitor
(IKK), and/or phosphatidylinositol 3-kinase (PI3K) pathways
(13). HIF-1α protein translation
can be regulated by microRNAs (miRNAs), long non-coding RNAs
(lncRNAs), and/or angiotensin II-mediated signaling involving PI3K.
The oxygen-dependent mechanism affecting the half-life of HIF
protein in the normal state is mainly due to the von Hippel-Lindau
protein (pVHL) degradation pathway. In this degradation pathway,
pVHL catalyzes the ubiquitination of the hydroxylated HIF-1α
(14). While hydroxylation of the
HIF-1α is catalyzed by two currently known enzymes, prolyl
hydroxylase domain-containing proteins (PHDs) and asparagine
hydroxylase factor inhibiting HIF (FIH) (15,16),
their activity is reduced by a decrease in oxygen tension and thus
the stability of the HIF-1α is affected by oxygen tension.
From the clinical data, HIF-1α is highly expressed
in NSCLC and is strongly associated with poor clinical prognosis
(17–19). Therefore, studying the role of
HIF-1α in NSCLC is of great significance for NSCLC treatment. The
current review focused on HIF-1α mediated glycolysis in NSCLC,
described the intracellular regulatory mechanisms and summarized
how they affect the development and treatment of tumors.
Regulation of HIF-1α expression in
NSCLC
A number of clinical investigations have shown that
the positive rate of HIF-1α expression in NSCLC is significantly
higher (up to 15–20 times) than that in normal tissues (20–22).
This phenomenon is closely related to the molecular signaling and
cells in the tumor microenvironment described in the following
sections.
Molecular signaling regulating HIF-1α
level
Several molecules that are differentially expressed
in NSCLC compared with normal cells can alter the HIF-1α expression
in NSCLC by affecting the transcription, translation or protein
degradation of HIF-1α (Fig. 1). In
the HIF-1α protein degradation signaling pathway, it has been found
that in primary NSCLC, the content of intracellular HIF-1α and the
expression of its downstream genes are positively correlated with
the expression of PHD protein family (23). Aldolase A is also highly expressed
in NSCLC and is positively correlated with the expression of HIF-1α
in the nucleus. It can cause HIF-1α accumulation by promoting the
release of lactic acid, which causes the decrease in PHD activity
(24). ROS can also inhibit PHD to
stabilize HIF-1α (25) and the
depletion of intracellular succinate dehydrogenase 5 in high
glucose environment leads to the accumulation of ROS and the
increase of HIF-1α protein level (26) (Fig.
1 left; bottom orange section).
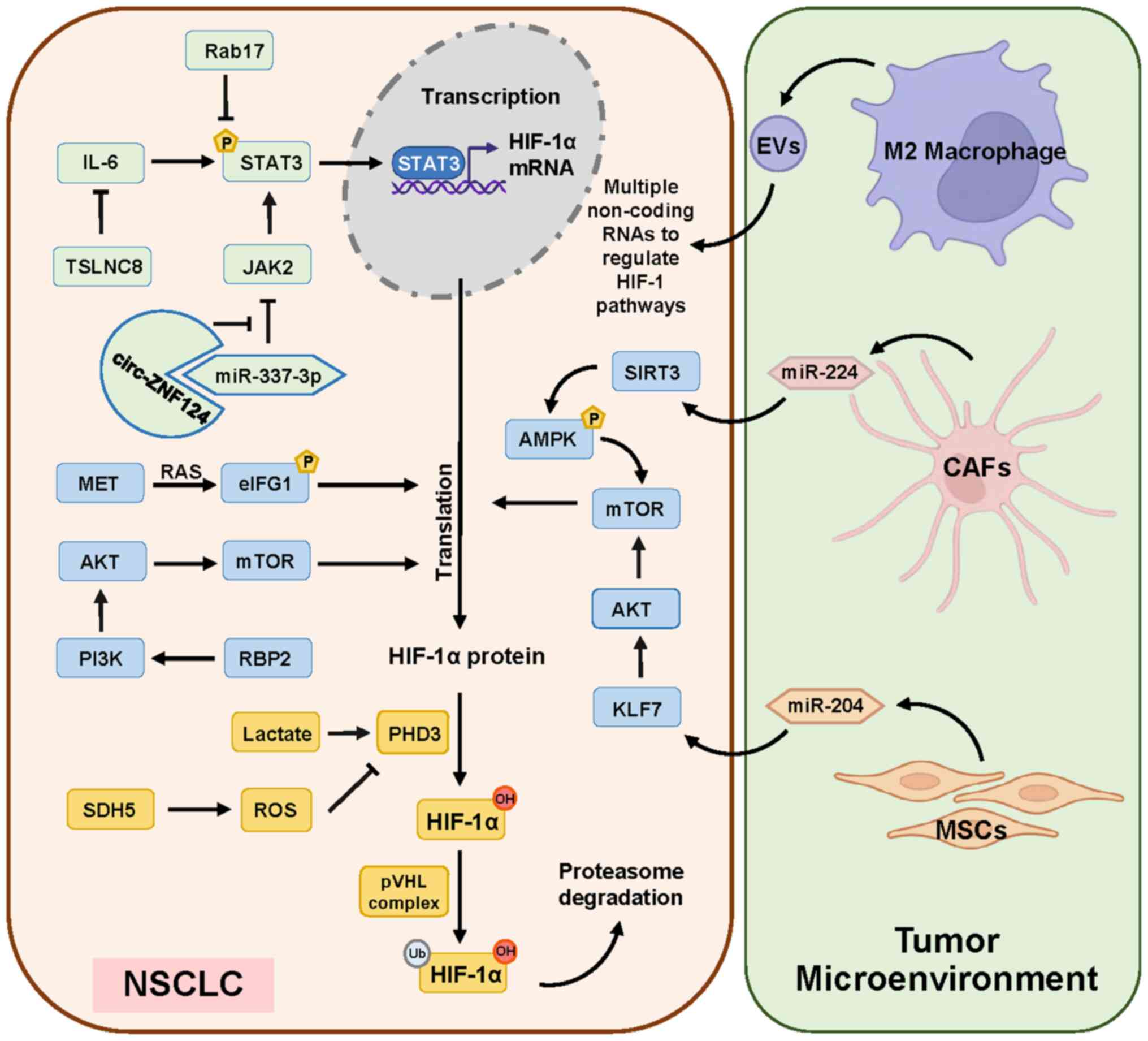 | Figure 1.Molecular signaling and cells in
tumor microenvironment regulating HIF-1α level in NSCLC. Currently
demonstrated data show that STAT3 plays a major role in the
transcriptional regulation of the HIF-1α expression (green). The
pathways that promote the translation regulation of HIF-1α are
mainly the PI3K/AKT and the mTOR pathways (blue). The enzymes PHD3
and pVHL influence the promotion of ubiquitinated degradation of
HIF-1α (orange). Several types of cells in the tumor cell
microenvironment are also involved in the regulation of HIF-1α
expression inside NSCLC (right panel). HIF-1α, hypoxia-inducible
factor 1α; NSCLC, non-small cell lung cancer; PHD3, prolyl
hydroxylate 3; pVHL, von Hippel-Lindau protein; miR, microRNA;
circ, circularRNA; ROS, reactive oxygen species; RBP2,
retinoblastoma binding protein 2; SDH5, succinate dehydrogenase 5;
KLF7, Krüppel-like factor 7; EVs, extracellular vesicles; CAFs,
cancer-associated fibroblasts; MSCs, mesenchymal stem cells. |
The main factor on the transcriptional regulation of
HIF-1α is STAT3 signaling pathway (Fig.
1 left; top green section) in NSCLC. The JAK2/STAT3 pathway is
inhibited by miR-337-3p, which is downregulated in NSCLC. At the
same time, circular (circ)RNA zinc finger protein 124 is highly
expressed and binds with miR-337-3p to further weaken its
inhibitory effect on JAK2/STAT3 pathway (27). In addition, Ras-related protein
Rab-17 is downregulated in NSCLS, which could reduce the inhibition
of STAT3 phosphorylation (28). The
non-coding (nc)RNA TSLNC8 is also downregulated, which can inhibit
the IL-6/STAT3 signaling pathway. Taken together, all these three
signaling pathways can promote the transcription expression of
HIF-1α via STAT3 pathway in NSCLC cells (29).
The translational regulation of HIF-1α is promoted
by eukaryotic initiation factor 4G1(eIF4G1) and mTOR signaling
pathways (Fig. 1 left; middle blue
section). Eukaryotic initiation factor complex F4 (eIF4F) includes
eIF4G1 and eIF4E. MET significantly increases the phosphorylation
level on Ser-1232 of eIF4G1 via MAPK, which leads to the
translational expression of HIF-1α (30). Activation of PI3K/AKT/mTOR pathway
also promotes HIF-1α translation. Retinoblastoma binding protein 2
(RBP2) is highly expressed in NSCLC and is associated with poor
prognosis. By stimulating this pathway, RBP2 can upregulate the
expression of HIF-1α to promote the growth of tumor blood vessels
(31).
Cells in tumor microenvironment
affecting HIF-1α level
Several types of cells in the tumor cell
microenvironment are also involved in the regulation of HIF-1α
expression inside NSCLC (Fig. 1
right panel). Cancer-associated fibroblasts (CAFs) are important
stromal cell components in the solid tumor microenvironment and
significantly accelerate the proliferation, invasion and
epithelial-mesenchymal transition of NSCLC cells (32). miR-224 is significantly upregulated
in both CAFs and CAFs co-cultured NSCLC cells and Sirtuins 3
(SIRT3)/AMPK axis is inhibited by miR-224-targeted SIRT3
untranslated region in NSCLC, thereby activating mTOR and
increasing HIF-1α expression. In turn, high levels of HIF-1α can
promote the high expression of miR-224, forming a positive feedback
loop (33). Mesenchymal stem cells
(MSCs)-derived exosomal miR-204 acts on Krüppel-like factor 7
(KLF7) in NSCLC to downregulate the KLF7/AKT/mTOR/HIF-1α axis to
play an anticancer role (34). M2
macrophage-derived extracellular vesicles regulate the Hippo/HIF-1
axis to enhance the cell viability and migration ability of NSCLC
under hypoxic conditions (35).
Regulatory mechanism of HIF-1α/glycolysis
axis in NSCLC
As HIF-1α is a transcription factor, its activation
can promote the transcription of multiple genes, including energy
metabolism, angiogenesis and apoptosis, among which
glycolysis-related genes are the most important in NSCLC. The
currently known ways in which HIF-1α promotes glycolysis can be
simply classified into three aspects: Promoting glucose uptake by
cells, inhibiting the tricarboxylic acid (TCA) cycle and regulating
the activity of glycolysis-related enzymes. The following are the
signaling pathways regulated by HIF-1α found in the field of
glycolytic metabolism in recent years.
ncRNAs alterations as initiating
factors of HIF-1α/glycolysis axis
ncRNAs play an important regulatory role in
HIF-1α-mediated glucose metabolism. Among them, miRNA, circRNA and
lncRNA are common molecules involved in the regulation (Fig. 2). miR-182 promotes the mRNA
expression of glycolysis-related enzymes alpha-enolase, glucose
transporter 1 (GLUT1), hexokinase (HK) 1, HK2, lactate
dehydrogenase (LDHA) and pyruvate dehydrogenase kinase-1 (PDK1) by
upregulating HIF-1α (36). In
addition, miRNA-31-5p overexpressed in NSCLC targets HIF-1α
inhibitors to increase the activity of HIF-1α and then increase the
expression of GLUT1, GAPDH, and LDHA to promote glycolysis
(37). Meanwhile, miRNA-199a is
downregulated in NSCLC, which reduces its target inhibitory effect
on HIF-1α, causing the increased expression of HIF-1α. This further
increases the expression of PDK1 to inhibit the TCA cycle (38).
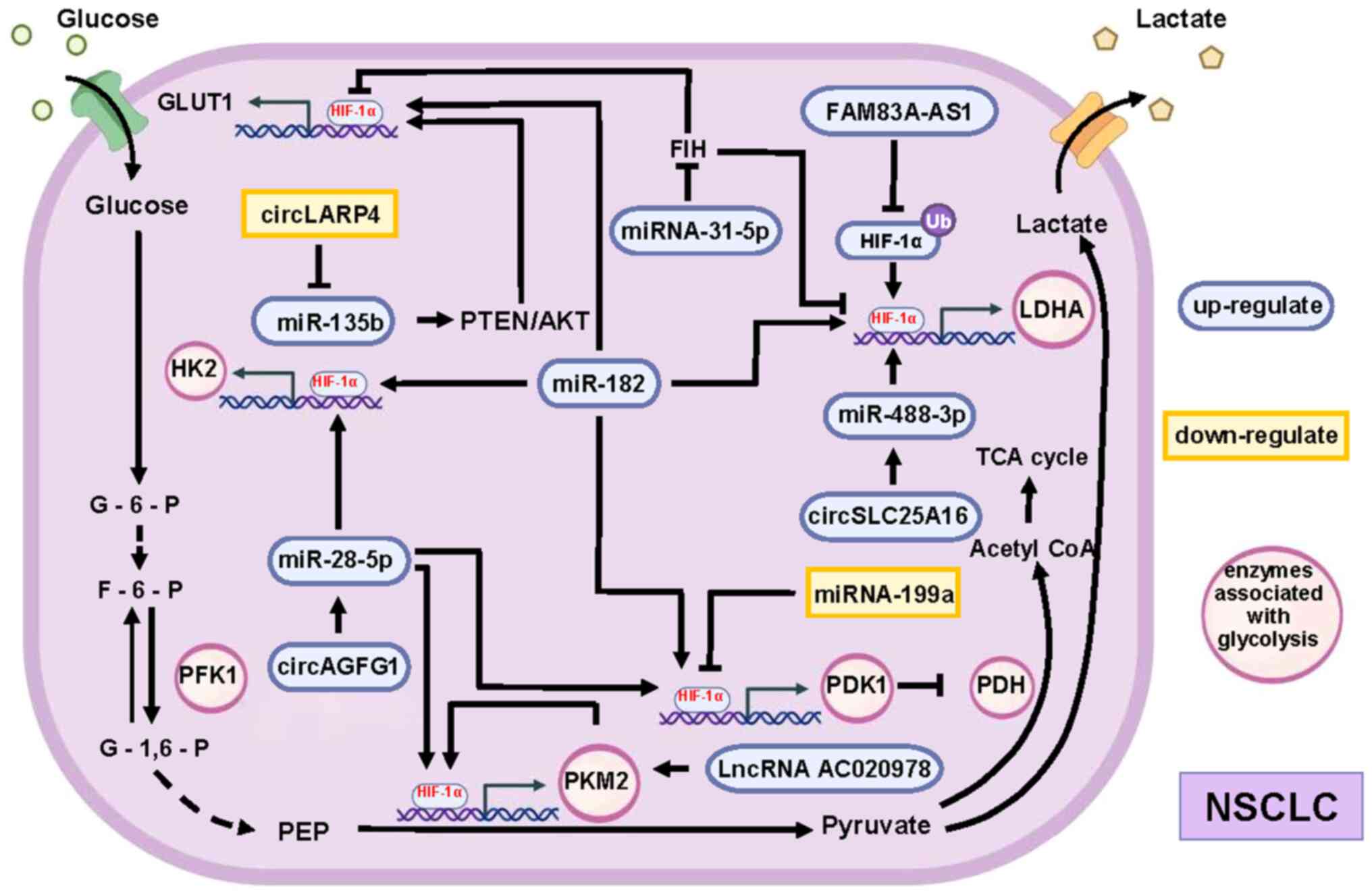 | Figure 2.Non-coding RNAs alterations as
initiating factors of HIF-1α/glycolysis axis. Various non-coding
RNAs can affect the expression of glycolysis-related enzymes by
altering the expression of HIF-1α, and these different pathways
work together to promote glycolysis in NSCLC. Compared with normal
cells, the expression of these non-coding RNAs is altered in NSCLC,
with some upregulated (purple) and others downregulated (orange).
The roles of different non-coding RNA expression changes in
promoting the expression of all key enzymes of HIF-1α-mediated
glycolysis are synergistic and complementary. For example, the
increase in HK2 expression is facilitated by the increase in
miR-182 and miR-28-5p content. At the same time, the increased
levels of miR-182 and miR-28-5p also synergizes with the decreased
levels of miRNA-199a to promote PDK1 expression. HIF-1α,
hypoxia-inducible factor 1α; NSCLC, non-small cell lung cancer; HK,
hexokinase; miR, microRNA; PDK1, pyruvate dehydrogenase kinase-1;
circ, circularRNA; PFK1, phosphofructokinase; lnc, long non-coding;
LDHA, lactate dehydrogenase; PKM2, pyruvate kinase 2; TCA,
tricarboxylic acid; acetyl CoA, acetyl coenzyme A; PDH, pyruvate
dehydrogenase. |
In addition to the regulatory effect of changes in
the expression of miRNAs themselves, some circRNAs can indirectly
regulate the HIF-1α/glycolysis axis by regulating the expression of
miRNAs. Among the upregulated circRNAs, circSLC25A16 and circAGFG1
promote glycolysis via miR-488-3p/HIF-1α/LDHA (39) and miR-28-5p/HIF-1α/GLUT1,
phosphoglycerate kinase 1 and Pyruvate kinase M2 (PKM2) (40), respectively. Exosomal circSHKBP1
promotes the expression of PKM2 by inhibiting miR-1294 (41). PKM2 can not only accelerate
glycolysis as a key enzyme but also promote the expression of
HIF-1α-dependent glycolytic enzymes by activating HIF-1α (42). In addition, the expression of
circLARP4 reduces the activity of HK2 and reduces the amount of
glucose uptake and lactate excretion by cells. Downregulation of
circLARP4 affects the miR-135b/PTEN/AKT/HIF-1α axis to promote
glycolysis in NSCLC (43).
Furthermore, lncRNA-AC020978 is upregulated during
hypoxia, increasing the stability of PKM2 by directly interacting
with PKM2 to participate in the regulation of PKM2-enhanced HIF-1α
transcription activity (44).
lncRNA FAM83A-AS1 promotes glycolysis by inhibiting the
ubiquitination of HIF-1α, which accumulates in cells and then
promotes the expression of HK2 and LDHA (45).
Protein molecules alterations as
initiating factors of HIF-1α/glycolysis axis
Several protein molecules are involved in specific
HIF-1α-dependent regulation of glycolysis in NSCLC (Fig. 3). Part of the mechanism is centered
on the altered expression of HK2 as the core of regulation.
Echinoderm microtubule-associated protein-like 4-anaplastic
lymphoma kinase (EML4-ALK) is a fusion protein found in 3–7% of
NSCLC. EML4-ALK induces hypoxia-independent but glucose-dependent
accumulation of HIF-1α via both transcriptional activation of
HIF-1α mRNA and the PI3K-AKT pathway to enhance HIF-1α synthesis.
In addition, the high expression of EML4-ALK promotes the binding
of HIF-1α to HK2 promoter, which increases the expression of HK2 to
promote glycolysis (46). Tumor
necrosis factor receptor-associated factor 6 (TRAF6) is achieved by
direct activation of AKT, which also causes the upregulation of
HIF-1α/HK2 and promotes glycolysis (47).
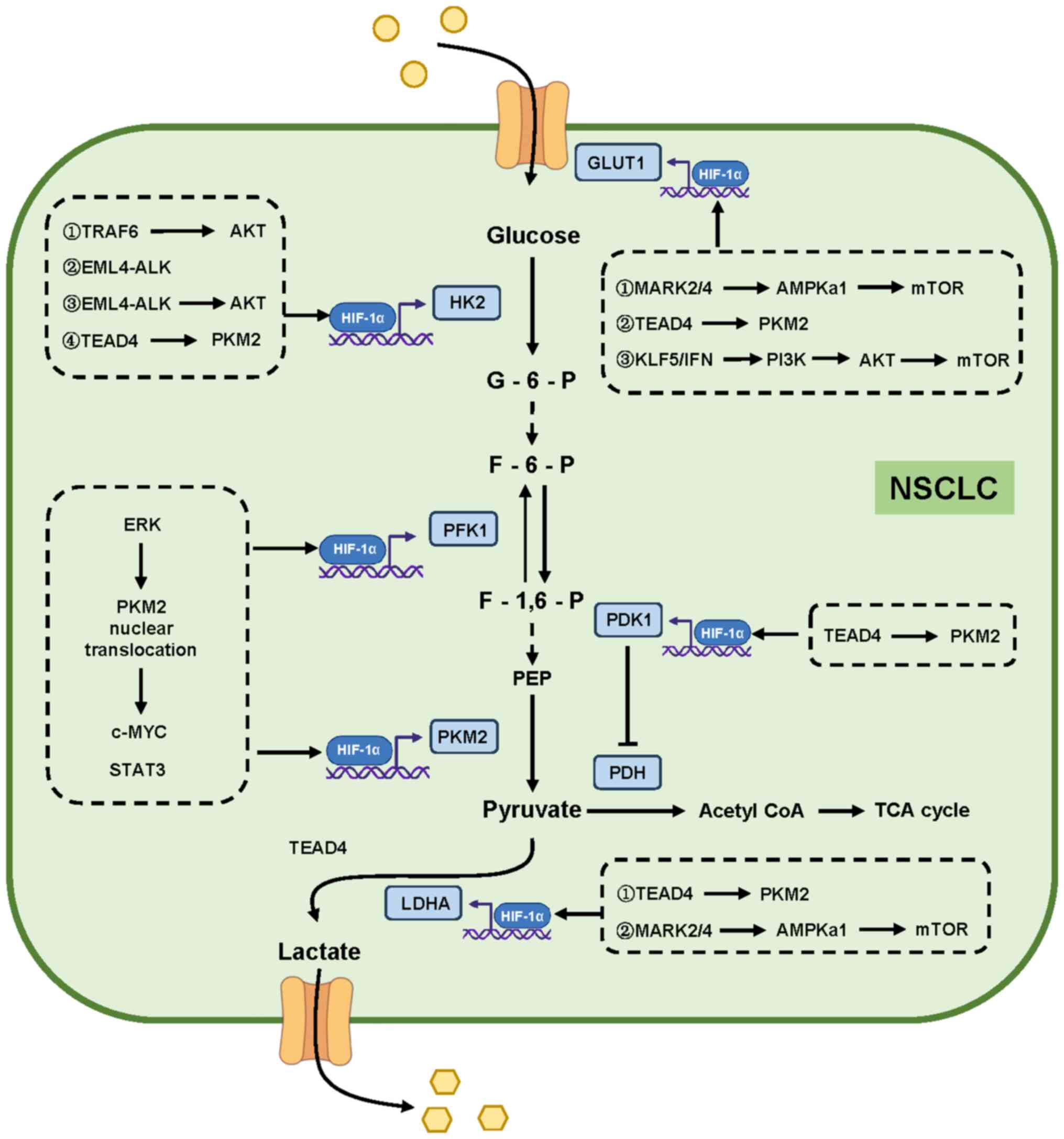 | Figure 3.Protein molecules alterations as
initiating factors of HIF-1α/glycolysis axis. Activation of
different signaling pathways can promote the expression of
glycolysis-related enzymes (HK2, PFK1, PKM2, LDHA, GLUT1, and PDK1)
by increasing the expression of HIF-1α, and these different
pathways work together to promote glycolysis in NSCLC. The
activation of different signaling pathways plays a synergistic and
complementary role in promoting the expression of all key enzymes
of HIF-1α-mediated glycolysis. HIF-1α, hypoxia-inducible factor 1α;
NSCLC, non-small cell lung cancer; HK2, hexokinase 2; PFK1,
phosphofructokinase; PKM2, pyruvate kinase 2; LDHA, lactate
dehydrogenase; GLUT1, glucose transporter 1; PDK1, pyruvate
dehydrogenase kinase-1. |
Another key enzyme is PKM2. ERK in the MAPK
signaling pathway promotes the endonuclear translocation of
low-activity PKM2 and activates the Wnt pathway to promote the
induced expression of HIF-1α by c-MYC and STAT3, thereby increasing
the expression of multiple glycolytic related enzymes
[phosphofructokinase (PFK1), PKM2, pyruvate dehydrogenase kinase 1
(PDK1) and LDHA] (48). Moreover,
TEA domain 4 (TEAD4) can act as a transcription factor to promote
the transcription of PKM2, which accelerates glycolysis and
increases the activity of HIF-1α to increase the expression of
GLUT1 and HK2 (49).
Finally, there are mechanisms associated with mTOR,
which is the classical molecule that regulates HIF-1α translation.
Under the condition of hypoxia in NSCLC, the expression of KLF
transcription factor 5 (KLF5) increases to activate the
PI3K/AKT/mTOR/HIF-1α pathway, increasing the glucose consumption
and lactate production of tumor cells (50). Inflammatory interferon, such as
IL-6, promotes the expression of glycolytic related genes by
stimulating PI3k/AKT/mTOR to activate HIF-1α (51). At the same time, phosphorylated
AMPKa1 can also stimulate mTOR/HIF-1α to increase the expression of
GLUT1 and LDHA to promote glycolysis. Microtubule affinity
regulating kinase 2/4 is highly expressed in advanced NSCLC to
maintain the phosphorylation level of AMPKa1 and promote tumor
cells to glycolysis (52).
The aforementioned studies show that most molecules
typically act on HIF-1α ubiquitination and PI3K/AKT/mTOR signaling
pathways that alter intracellular HIF-1α content and HIF-1α
activity. At the same time, more ncRNAs and protein molecules
upstream of HIF-1α in NSCLC tend to regulate HK2, LDHA and PKM2 to
alter glycolytic processes. It is evident that blocking HIF-1α for
the decreasing of HK2, PKM2 and LDHA transcription has the
potential approach to achieve improved therapeutic effects.
Progress in targeting the HIF-1α/glycolysis
axis for NSCLC treatment
Recent discoveries have elucidated mechanisms by
which stable HIF-1α expression in hypoxic environments contributes
to drug resistance and treatment ineffectiveness in NSCLC. Novel
therapeutic strategies have emerged, focusing on targeting the
HIF-1α-mediated hypoxic metabolic characteristics (Table I).
 | Table I.Mechanisms of chemotherapy resistance
and radiotherapy tolerance associated with HIF-1a/glycolysis. |
Table I.
Mechanisms of chemotherapy resistance
and radiotherapy tolerance associated with HIF-1a/glycolysis.
| Author(s),
year | Treatment mode | Result or
achievement | Mechanism | (Refs.) |
|---|
| Gong et al,
2018 | Chemotherapy | Resistance to
cisplatin | KLF5 →
PI3K/AKT/mTOR→ HIF-1a/glycolysis → resistance | (50) |
| Sun et al,
2021 |
|
| miRNA-21 →
PI3K/AKT/mTOR pathway → HIF-1a/glycolysis → resistance | (54) |
| Xu et al,
2020 |
|
| circAKT3 →
miR-516b-5p/STAT3 → resistance (inhibition of HIF-1a/glycolysis can
attenuate this effect) | (55) |
| Dong et al,
2020 |
| Resistance to
etoposide | Etoposide→ ROS →
HIF-1a/glycolysis → Lactic acid → TGF-β1/Snail and TAZ/AP-1 pathway
→ MRP1/ABCC1 proteins → resistance | (56) |
| Sun et al,
2017 |
| Resistance to
paclitaxel | Glucose uptake in
paclitaxel resistant tumor cells is more dependent on the promotion
of GLUT1 expression by PDK2 activated c-Myc and HIF-1a | (57) |
| Grosso et
al, 2013 | Radiotherapy | Low sensitivity to
radiotherapy | miR-210 →
HIF-1a/glycolysis → low sensitivity | (59) |
| Zhang et al,
2019 |
| Enhance
sensitivity | Cyclocarya paliurus
polysaccharide inhibits mTOR/AKT/PI3K pathway →HIF-1a↓ → apoptosis
of hypoxic cells → sensitivity enhanced | (60) |
| You et al,
2022 | TKI | Resistance to
erlotinib | SIRT6 → HIF-1a/HK2
→ resistance | (62) |
| Zhang et al,
2021 | New methods to
treat tumor through metabolic pathway (Reduce the energy source of
tumor cells) | Hyperbaric oxygen
therapy | Inhibit HIF-1a/PFKP
axis | (67) |
| Kim et al,
2015 |
| PA-12 | PA-12 → Inhibit
nuclear translocation of PKM2 → HIF-1a/glycolysis→ | (68) |
| Zhao et al,
2014 |
| YC-1 | YC-1 → HIF-1↓→ The
activity of LDHA and pyruvate dehydrogenase was downregulated →
glycolysis→ | (69) |
| Zhou et al,
2017. |
| Albendazole | Albendazole →
HIF-1a↓ → glycolysis→ | (70) |
| Yang et al,
2021 |
|
Deoxypodophyllotoxin |
Deoxypodophyllotoxin→ parkin mediated
protein degradation→ HIF-1a↓ → GLUT1/HK2/LDHA→ | (71) |
| Liu et al,
2021 |
| Huaier | Huaier→ inhibit
PI3K/AKT/HIF-1a signaling pathway → glycolysis/glucose uptake↓ | (72) |
| Huang et al,
2022 |
| Nanoparticle | Deliver EGFR-TKI
and YAP-siRNA → HIF-1a↓ → Glycolysis↓ | (75) |
| Alkhathami et
al, 2023 |
|
| Deliver circRNA→
HIF-1a↓→Glycolysis↓ | (76) |
| Kopecka et
al, 2015 |
|
| Deliver Zoledronic
acid→ inhibit Ras/erk1/2→ activation of HIF-1a ↓→glycolysis↓ | (53) |
Therapeutic desensitization
Chemotherapy is one of the major ways to treat
tumors, but inherent or acquired chemo-resistance limits its
clinical application. Platinum-based drugs, central to NSCLC
chemotherapy, face resistance mechanisms that are multifaceted,
including factors such as platinum transporters, detoxification
systems and DNA repair processes (53). These resistance mechanisms are
intricately linked to glucose metabolism regulation. It was found
that knocking down KLF5 in NSCLC cells alleviated resistance to
cisplatin (50). This process is
closely linked to the promotion of HIF-1α-dependent glycolysis
following deregulation of the KLF5/PI3K/AKT/mTOR pathway activation
(50). miR-21 activates the
PI3K/AKT/mTOR/HIF-1α pathway, leading to the upregulation of PKM2
and LDHA2 expression. These enzymes are glycolysis-related and are
notably abundant in cisplatin-resistant NSCLC cells. Their
heightened presence enhances glucose metabolism in tumor cells,
contributing to their resistance against cisplatin. Consequently,
the elevated expression of miR-21 emerges as a pivotal factor in
the development of cisplatin resistance in NSCLC (54). circAKT3 is also a non-coding RNA
that is highly expressed in cisplatin-resistant NSCLC. It
diminishes the sensitivity of tumor tissue to cisplatin through the
regulation of miR-516b-5p/STAT3. However, this effect can be
alleviated by suppressing HIF-1α-mediated glycolysis (55). The utilization of etoposide in NSCLC
treatment induces elevated intracellular ROS production. This
effect activates HIF-1α-mediated metabolic reprogramming, resulting
in an upsurge in glycolysis and subsequently leading to increased
lactate production. Lactate can regulate the TGF-β1/Snail and
TAZ/AP-1 pathways to activate the expression of MRP1/ABCC1 proteins
that increase etoposide resistance (56). Scholars have also found that the
resistance of NSCLC to paclitaxel is related to the glycolysis
promoted by HIF-1α. Evidence suggests that in paclitaxel-resistant
tumor cells, glucose uptake relies more on the promotion of GLUT1
expression by pyruvate dehydrogenase kinase 2-activated c-Myc and
HIF-1α (57). The increase of
HIF-1α-induced glycolysis always promotes the occurrence of drug
resistance, which is in line with the current general hypothesis
that inhibiting glycolysis can improve chemotherapy (58). However, studies on the relationship
between HIF-1α-induced glycolysis and NSCLC resistance to
chemotherapeutic agents are fragmented. Further exploration is
needed to systematically refine the specific mechanisms
involved.
Reduced sensitivity has also been seen in NSCLC
radiotherapy. Alternative methods exist to reduce tumor
responsiveness to radiation. Particularly, the regulation of the
HIF-1α/glycolysis axis has been identified as a promising approach.
Researchers have indicated that the stable expression of HIF-1 in
NSCLC, achieved through the high expression of miR-210, further
prompts cells to manifest mitochondrial defects and glycolytic
phenotypes. The existence of this metabolic state is crucial for
miR-210 to repair gene double-strand breaks in radiation-resistant
NSCLC (59). Cyclocarya paliurus
polysaccharide is mainly used to regulate blood glucose and has
potential radiosensitization effects when combined with
radiotherapy. The specific mechanism is that cyclocarya paliurus
polysaccharide inhibits mTOR/AKT/PI3K pathway and reduces HIF-1α
expression, which leads to apoptosis of hypoxic NSCLC cells
(60). In conclusion, blocking
HIF-1α-mediated Warburg effect can effectively enhance the
sensitivity of hypoxic tumor tissue to radiotherapy.
EGFR mutants often appear in NSCLC, making EGFR
tyrosine kinase inhibitor (TKI)-targeted therapy a prevalent
treatment option, though it often encounters resistance issues. In
normal cells, SIRT6 acts as a histone H3K9 deacetylase to control
the expression of multiple glycolytic genes (61). However, in erlotinib-resistant NSCLC
cells, elevated SIRT6 levels enhance glycolysis via the HIF-1α/HK2
axis, reducing the sensitivity of cells to erlotinib. HIF-1α
blocker PX478-2HCL blocks this process to alleviate NSCLC
resistance to erlotinib (62).
Currently, few studies have explored the relationship between the
mechanism of EGFR-TKI resistance and the HIF-1α/glycolytic axis.
This scarcity might stem from variations in the understanding of
tumor metabolic characteristics and their correlation with EGFR-TKI
resistance across different academic findings. Some consider that
glycolysis promotes EGFR-TKI resistance, while others consider that
the metabolic characteristics of drug-resistant tissues tend to be
oxidative phosphorylation (63–66).
New treatment options
In recent years, several new NSCLC therapies
targeting HIF-1α/glycolysis axis have been discovered (Table I). In hypoxic NSCLC cells,
hyperbaric oxygen therapy disrupts the HIF-1α/phosphofructokinase,
platelet axis, thus impeding the Warburg effect. This reduction in
glycolytic capacity due to the high oxygen concentration
effectively inhibits excessive tumor cell proliferation (67). PA-12, an activator of PKM2, can
inhibit the nuclear translocation of PKM2 to suppress the
expression of HIF-1. Therefore, the glycolysis promoted by HIF-1 is
blocked, which affects the proliferation of tumor cells with
limited energy supply during hypoxia (68). The HIF-1 inhibitor YC-1 targets the
transient activation of LDHA and phosphorylation of the E1a subunit
of pyruvate dehydrogenase, effectively inhibiting the metabolic
shift from oxidative phosphorylation to glycolysis in tumors. This
process impedes the metastatic potential of lung cancer (69). Albendazole is primarily employed as
an anthelmintic and insect repellent (70). However, recent research has revealed
its additional potential. Albendazole has been shown to
downregulate the expression of HIF-1α and decrease the levels of
HK, PK and LDH in tumor cells under hypoxic conditions. This
mechanism inhibits glycolysis in NSCLC, ultimately impeding their
proliferation (70).
Deoxypodophyllotoxin hinders NSCLC progression through a two-fold
mechanism. Initially, it disrupts angiogenesis in the tumor
vicinity. Second, it facilitates parkin-mediated protein
degradation, resulting in decreased expression of HIF-1α and
subsequently reducing the levels of HIF-1α target genes such as
GLUT1, HK2, and LDHA (71). The
Chinese medicine Huaier is currently an adjunct drug in the
treatment of a number of cancers. It can inactivate PI3K/AKT/HIF-1α
pathway, downregulate glycolysis and glucose transport in NSCLC and
exert anti-tumor effects (72). For
squamous NSCLC, Fibroblast Growth Factor Receptor 1 (FGFR1) is
often overexpressed. Targeting FGFR1 may prevent cancer cell growth
by inhibiting glucose metabolism, as FGFR1 activates the AKT/mTOR
pathway for HIF-1α accumulation and thus increases glucose uptake
and glycolysis (73).
In addition, nanoparticles are gaining traction in
tumor medicine, with ongoing clinical translation efforts (74). A newly invented nanodrug can
simultaneously target the delivery of EGFR-TKI and yes associated
protein (YAP)-siRNA combination drugs, addressing the role of YAP
in epidermal growth factor receptor-TKI resistance in NSCLC. This
nanodrug diminishes the glycolysis function by downregulating
HIF-1α, enhancing tumor cell sensitivity to photodynamic
therapy-induced apoptosis (75).
Nanoparticles can be utilized to deliver circRNAs for the treatment
of NSCLC. These circRNAs, either directly or through the inhibition
of glycolysis via HIF-1α, hold promise as a therapeutic approach
for NSCLC (76). Zoledronic
acid-loaded nanoparticles suppress isoprenoid synthesis and HIF-1α
activation through the Ras/erk1/2 pathway, decreasing glucose
transport and glycolytic enzyme transcriptional activity. This
method shows promise in combating multidrug-resistant tumors
(53).
Conclusions
HIF is an important metabolic regulatory molecule in
tumors and the dysregulation of HIF expression is necessary for
tumor survival and growth (77).
The expression of HIF-1α mainly affects the balance between
glycolysis and oxidized phosphoric acid in higher metazoan tumor
cells. However, there is no universal mechanism for reprogramming
glucose metabolism in all cancers (58), so the present review focused on
NSCLC. The present review summarized the regulatory mechanism of
HIF-1α/glycolysis in order to discover the regulatory
characteristics of glucose metabolism in NSCLC and paved the way
for finding the mechanism that is common in different cancers. It
characterized the regulatory mechanisms of HIF-1α in NSCLC
according to different stages affecting HIF-1α expression,
reflecting the unique regulatory pattern of HIF-1α in NSCLC. Part
of the mechanism can be reflected in the mechanism of the
regulation of HIF-1α-mediated glycolysis. For example, the
inflammatory factor IL-6 can increase the expression of HIF-1α
through STAT3 and promote glycolysis (29). The section ‘Regulatory mechanism of
HIF-1α/glycolysis axis in NSCLC’ summarized the types of regulatory
molecules and classical pathways as clues. It was found that ncRNA
and PI3K/AKT/mTOR signaling pathways play an important role in
regulating the HIF-1α/glycolysis axis, and most of them regulate
the glycolysis process by affecting the downstream HK2 content. In
addition, it was found that the accumulation of HIF-1α and the
overexpression of HIF-1α promote glycolysis to form a cycle
promoting a mutually stable relationship. Taking the regulatory
mechanism in NSCLC as an example, HIF-1α can promote the expression
of PKM2 to accelerate glycolysis. At the same time, low PKM2
activity leads to decreased glycolysis but can induce HIF-1α
expression to solve this conundrum.
The mechanism of glucose metabolism reprogramming is
gradually being analyzed, and it is generally considered that the
regulation of glucose metabolism may be one of the important
development directions for cancer treatment in the future (78). A variety of NSCLC treatment
modalities and resistance mechanisms related to the
HIF-1α/glycolysis axis were summarized in part three of the present
review. The central idea behind these approaches is to alter the
specific metabolic pattern of tumors by blocking HIF-1α expression
and activation or disabling glycolytic rate-limiting enzyme to aid
classical NSCLC therapy. However, in the case of tumor development
led by HIF-1α/glycolysis, a more ideal therapeutic target is that
of selective intervention of HIF-1 pathway (79). Selectivity can be manifested in the
development of inhibitors that target molecules that regulate
HIF-1α to initiate the transcription process of glycolytic enzymes,
rather than HIF-1α or glycolytic rate-limiting enzymes or upstream
molecules that regulate HIF-1α themselves. This allows for more
precise regulation of HIF-1α-mediated changes in tumor metabolism
without affecting other roles of HIF-1α and other molecules in
complex signaling pathway networks. There are few relevant studies
on HIF-1α initiating the transcription of glycolysis-related genes
in NSCLC (46). If more targets
that selectively inhibit this axis can be found in the future, it
will promote the research of methods to treat NSCLC by regulating
metabolism.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Training Program
of Undergraduate Innovation and Entrepreneurship (grant no. 2022G19
to YS); Special Funds for the Cultivation of Guangdong College
Students Scientific and Technological Innovation (grant no.
Pdjh2022c0098 to XL); Shenzhen Municipal Science and Technology
Innovation Commission Foundation (grant no. JCYJ20210324104800001
and JCYJ20220530114415036 to GC).
Availability of data and materials
Not applicable.
Authors' contributions
YS and GC conceived and designed the article. YS and
XL surveyed the literature and wrote the manuscript. JW, ZZ and SC
surveyed the literature and provided suggestions. All authors read
and approved the final version of the manuscript. Data
authentication is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
General Office of National Health
Commission of the People's Republic of China, . Primary lung cancer
diagnosis and treatment guidelines (2022 edition). Med J Peking
Union Med Coll Hosp. 13:549–570. 2022.
|
|
3
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zahra K, Dey T, Ashis H, Mishra SP and
Pandey U: Pyruvate Kinase M2 and Cancer: The Role of PKM2 in
promoting tumorigenesis. Front Oncol. 10:1592020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Emami Nejad A, Najafgholian S, Rostami A,
Sistani A, Shojaeifar S, Esparvarinha M, Nedaeinia R, Haghjooy
Javanmard S, Taherian M, Ahmadlou M, et al: The role of hypoxia in
the tumor microenvironment and development of cancer stem cell: A
novel approach to developing treatment. Cancer Cell Int. 21:622021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Courtnay R, Ngo DC, Malik N, Ververis K,
Tortorella SM and Karagiannis TC: Cancer metabolism and the Warburg
effect: The role of HIF-1 and PI3K. Mol Biol Rep. 42:841–851. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang GL and Semenza GL: General
involvement of hypoxia-inducible factor 1 in transcriptional
response to hypoxia. Proc Natl Acad Sci USA. 90:4304–4308. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McGettrick AF and O'Neill LAJ: The role of
HIF in immunity and inflammation. Cell Metab. 32:524–536. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Semenza GL, Agani F, Booth G, Forsythe J,
Iyer N, Jiang BH, Leung S, Roe R, Wiener C and Yu A: Structural and
functional analysis of hypoxia-inducible factor 1. Kidney Int.
51:553–555. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kierans SJ and Taylor CT: Regulation of
glycolysis by the hypoxia-inducible factor (HIF): Implications for
cellular physiology. J Physiol. 599:23–37. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Albadari N, Deng S and Li W: The
transcriptional factors HIF-1 and HIF-2 and their novel inhibitors
in cancer therapy. Expert Opin Drug Discov. 14:667–682. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Palazón A, Aragonés J, Morales-Kastresana
A, de Landázuri MO and Melero I: Molecular pathways: Hypoxia
response in immune cells fighting or promoting cancer. Clin Cancer
Res. 18:1207–1213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Taylor CT and Scholz CC: The effect of HIF
on metabolism and immunity. Nat Rev Nephrol. 18:573–587. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kaelin WG: The von Hippel-Lindau tumor
suppressor protein: Roles in cancer and oxygen sensing. Cold Spring
Harb Symp Quant Biol. 70:159–166. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hampton-Smith RJ, Davenport BA, Nagarajan
Y and Peet DJ: The conservation and functionality of the
oxygen-sensing enzyme Factor Inhibiting HIF (FIH) in
non-vertebrates. PLoS One. 14:e02161342019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nguyen TL and Durán RV: Prolyl hydroxylase
domain enzymes and their role in cell signaling and cancer
metabolism. Int J Biochem Cell Biol. 80:71–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Q, Hu DF, Rui Y, Jiang AB, Liu ZL and
Huang LN: Prognosis value of HIF-1α expression in patients with
non-small cell lung cancer. Gene. 541:69–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin X, Xia J, Sun Y and Zhang Z: CHCHD2 is
a potential prognostic factor for NSCLC and is associated with
HIF-1α expression. BMC Pulm Med. 20:402020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu R, Du W, Ding Z, Wang Y, Li Y, Zhu J,
Zeng Y, Zheng Y, Liu Z and Huang JA: HIF-1α promoted vasculogenic
mimicry formation in lung adenocarcinoma through NRP1 upregulation
in the hypoxic tumor microenvironment. Cell Death Dis. 12:3942021.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y and Zhang Y, Li X, Huang Y, Chen W,
He L and Zhang Y: Status of hypoxia-inducible factor-1α expression
in non-small cell lung cancer. Pharmazie. 76:404–411.
2021.PubMed/NCBI
|
|
21
|
Qi Q, Wang C, Yang J, Tian Y and Feng C:
Correlation between HIF-1α, PD-L1 and lymphatic metastasis in
non-small cell lung cancer. Chin J Lung Dis (Electronic Edition).
13:242–246. 2020.
|
|
22
|
Yang SL, Ren QG, Wen L and Hu JL:
Clinicopathological and prognostic significance of
hypoxia-inducible factor-1 alpha in lung cancer: A systematic
review with meta-analysis. J Huazhong Univ Sci Technol Med Sci.
36:321–327. 2016. View Article : Google Scholar
|
|
23
|
Koren A, Rijavec M, Krumpestar T, Kern I,
Sadikov A, Čufer T and Korošec P: Gene expression levels of the
prolyl hydroxylase domain proteins PHD1 and PHD2 but not PHD3 are
decreased in primary tumours and correlate with poor prognosis of
patients with surgically resected Non-Small-Cell lung cancer.
Cancers (Basel). 13:23092021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang YC, Chan YC, Chang WM, Lin YF, Yang
CJ, Su CY, Huang MS, Wu ATH and Hsiao M: Feedback regulation of
ALDOA activates the HIF-1α/MMP9 axis to promote lung cancer
progression. Cancer Lett. 403:28–36. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kietzmann T and Görlach A: Reactive oxygen
species in the control of hypoxia-inducible factor-mediated gene
expression. Semin Cell Dev Biol. 16:474–486. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Tuo Z, Zong Y and Liu J: Succinate
dehydrogenase 5 regulates lung cancer metastasis by reprogramming
glucose metabolism. J Thorac Dis. 13:6427–6438. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Q, Huang Q, Cheng S, Wu S, Sang H and
Hou J: Circ_ZNF124 promotes non-small cell lung cancer progression
by abolishing miR-337-3p mediated downregulation of JAK2/STAT3
signaling pathway. Cancer Cell Int. 19:2912019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang M, Wang W, Ding J, Wang J and Zhang
J: Downregulation of Rab17 promotes cell proliferation and invasion
in non-small cell lung cancer through STAT3/HIF-1α/VEGF signaling.
Thorac Cancer. 11:379–388. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fan H, Li J, Wang J and Hu Z: Long
Non-Coding RNAs (lncRNAs) Tumor-Suppressive Role of lncRNA on
Chromosome 8p12 (TSLNC8) inhibits tumor metastasis and promotes
apoptosis by regulating interleukin 6 (IL-6)/Signal transducer and
activator of transcription 3 (STAT3)/Hypoxia-Inducible factor
1-alpha (HIF-1α) signaling pathway in Non-Small cell lung cancer.
Med Sci Monit. 25:7624–7633. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Glück AA, Orlando E, Leiser D, Poliaková
M, Nisa L, Quintin A, Gavini J, Stroka DM, Berezowska S, Bubendorf
L, et al: Identification of a MET-eIF4G1 translational regulation
axis that controls HIF-1α levels under hypoxia. Oncogene.
37:4181–4196. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qi L, Zhu F, Li SH, Si LB, Hu LK and Tian
H: Retinoblastoma binding protein 2 (RBP2) promotes
HIF-1α-VEGF-induced angiogenesis of non-small cell lung cancer via
the Akt pathway. PLoS One. 9:e1060322014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang D, Liu J, Qian H and Zhuang Q:
Cancer-associated fibroblasts: From basic science to anticancer
therapy. Exp Mol Med. 55:1322–1332. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang J, Han L, Yu J, Li H and Li Q:
miR-224 aggravates cancer-associated fibroblast-induced progression
of non-small cell lung cancer by modulating a positive loop of the
SIRT3/AMPK/mTOR/HIF-1α axis. Aging (Albany NY). 13:10431–10449.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu XN, Zhang CB, Lin H, Tang XY, Zhou R,
Wen HL and Li J: microRNA-204 shuttled by mesenchymal stem
cell-derived exosomes inhibits the migration and invasion of
non-small-cell lung cancer cells via the KLF7/AKT/HIF-1α axis.
Neoplasma. 68:719–731. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chu X, Wang Z, Wang W, Liu W, Cao Y and
Feng L: Roles of hypoxic environment and M2 macrophage-derived
extracellular vesicles on the progression of non-small cell lung
cancer. BMC Pulm Med. 23:2392023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang M, Wang W, Wang J and Zhang J:
MiR-182 promotes glucose metabolism by upregulating
hypoxia-inducible factor 1α in NSCLC cells. Biochem Biophys Res
Commun. 504:400–405. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhu B, Cao X, Zhang W, Pan G, Yi Q, Zhong
W and Yan D: MicroRNA-31-5p enhances the Warburg effect via
targeting FIH. FASEB J. 33:545–556. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ding G, Huang G, Liu HD, Liang HX, Ni YF,
Ding ZH, Ni GY and Hua HW: MiR-199a suppresses the hypoxia-induced
proliferation of non-small cell lung cancer cells through targeting
HIF1α. Mol Cell Biochem. 384:173–180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shangguan H, Feng H, Lv D, Wang J, Tian T
and Wang X: Circular RNA circSLC25A16 contributes to the glycolysis
of non-small-cell lung cancer through epigenetic modification. Cell
Death Dis. 11:4372020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma X, Wang C, Chen J, Wei D, Yu F and Sun
J: circAGFG1 sponges miR-28-5p to promote non-small-cell lung
cancer progression through modulating HIF-1α level. Open Med
(Wars). 16:703–717. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen W, Tang D, Lin J, Huang X, Lin S,
Shen G and Dai Y: Exosomal circSHKBP1 participates in non-small
cell lung cancer progression through PKM2-mediated glycolysis. Mol
Ther Oncolytics. 24:470–485. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Luo W, Hu H, Chang R, Zhong J, Knabel M,
O'Meally R, Cole RN, Pandey A and Semenza GL: Pyruvate kinase M2 is
a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell.
145:732–744. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lu H, Guo Q, Mao G, Zhu J and Li F:
CircLARP4 suppresses cell proliferation, invasion and glycolysis
and promotes apoptosis in non-small cell lung cancer by targeting
miR-135b. Onco Targets Ther. 13:3717–3728. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hua Q, Mi B, Xu F, Wen J, Zhao L, Liu J
and Huang G: Hypoxia-induced lncRNA-AC020978 promotes proliferation
and glycolytic metabolism of non-small cell lung cancer by
regulating PKM2/HIF-1α axis. Theranostics. 10:4762–4778. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen Z, Hu Z, Sui Q, Huang Y, Zhao M, Li
M, Liang J, Lu T, Zhan C, Lin Z, et al: LncRNA FAM83A-AS1
facilitates tumor proliferation and the migration via the
HIF-1α/glycolysis axis in lung adenocarcinoma. Int J Biol Sci.
18:522–535. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Ma Y, Yu C, Mohamed EM, Shao H, Wang L,
Sundaresan G, Zweit J, Idowu M and Fang X: A causal link from ALK
to hexokinase II overexpression and hyperactive glycolysis in
EML4-ALK-positive lung cancer. Oncogene. 35:6132–6142. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Feng L, Feng S, Nie Z, Deng Y, Xuan Y,
Chen X, Lu Y, Liang L and Chen Y: TRAF6 promoted tumor glycolysis
in non-small-cell lung cancer by activating the Akt-HIFα Pathway.
Biomed Res Int. 2021:34312452021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Icard P, Simula L, Fournel L, Leroy K,
Lupo A, Damotte D, Charpentier MC, Durdux C, Loi M, Schussler O, et
al: The strategic roles of four enzymes in the interconnection
between metabolism and oncogene activation in non-small cell lung
cancer: Therapeutic implications. Drug Resist Updat. 63:1008522022.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hu Y, Mu H and Deng Z: The transcription
factor TEAD4 enhances lung adenocarcinoma progression through
enhancing PKM2 mediated glycolysis. Cell Biol Int. 45:2063–2073.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gong T, Cui L, Wang H, Wang H and Han N:
Knockdown of KLF5 suppresses hypoxia-induced resistance to
cisplatin in NSCLC cells by regulating HIF-1α-dependent glycolysis
through inactivation of the PI3K/Akt/mTOR pathway. J Transl Med.
16:1642018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yeh YH, Hsiao HF, Yeh YC, Chen TW and Li
TK: Inflammatory interferon activates HIF-1α-mediated
epithelial-to-mesenchymal transition via PI3K/AKT/mTOR pathway. J
Exp Clin Cancer Res. 37:702018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Natarajan SR, Ponnusamy L and Manoharan R:
MARK2/4 promotes Warburg effect and cell growth in non-small cell
lung carcinoma through the AMPKα1/mTOR/HIF-1α signaling pathway.
Biochim Biophys Acta Mol Cell Res. 1869:1192422022. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kopecka J, Porto S, Lusa S, Gazzano E,
Salzano G, Giordano A, Desiderio V, Ghigo D, Caraglia M, De Rosa G
and Riganti C: Self-assembling nanoparticles encapsulating
zoledronic acid revert multidrug resistance in cancer cells.
Oncotarget. 6:31461–31478. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sun Y, Liu W, Zhao Q, Zhang R, Wang J, Pan
P, Shang H, Liu C and Wang C: Downregulating the expression of
miRNA-21 inhibits the glucose metabolism of A549/DDP cells and
promotes cell death through the PI3K/AKT/mTOR/HIF-1α pathway. Front
Oncol. 11:6535962021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xu Y, Jiang T, Wu C and Zhang Y: CircAKT3
inhibits glycolysis balance in lung cancer cells by regulating
miR-516b-5p/STAT3 to inhibit cisplatin sensitivity. Biotechnol
Lett. 42:1123–1135. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Dong Q, Zhou C, Ren H, Zhang Z, Cheng F,
Xiong Z, Chen C, Yang J, Gao J, Zhang Y, et al: Lactate-induced
MRP1 expression contributes to metabolism-based etoposide
resistance in non-small cell lung cancer cells. Cell Commun Signal.
18:1672020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sun H, Zhu A, Zhou X and Wang F:
Suppression of pyruvate dehydrogenase kinase-2 re-sensitizes
paclitaxel-resistant human lung cancer cells to paclitaxel.
Oncotarget. 8:52642–52650. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Chelakkot C, Chelakkot VS, Shin Y and Song
K: Modulating glycolysis to improve cancer therapy. Int J Mol Sci.
24:26062023. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Grosso S, Doyen J, Parks SK, Bertero T,
Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noël A, Pouysségur
J, et al: MiR-210 promotes a hypoxic phenotype and increases
radioresistance in human lung cancer cell lines. Cell Death Dis.
4:e5442013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang F, Fan B and Mao L: Radiosensitizing
effects of Cyclocarya paliurus polysaccharide on hypoxic A549 and
H520 human non-small cell lung carcinoma cells. Int J Mol Med.
44:1233–1242. 2019.PubMed/NCBI
|
|
61
|
Zhong L, D'Urso A, Toiber D, Sebastian C,
Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD,
Nir T, et al: The histone deacetylase Sirt6 regulates glucose
homeostasis via Hif1alpha. Cell. 140:280–293. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
You Q, Wang J, Yu Y, Li F, Meng L, Chen M,
Yang Q, Xu Z, Sun J, Zhuo W and Chen Z: The histone deacetylase
SIRT6 promotes glycolysis through the HIF-1α/HK2 signaling axis and
induces erlotinib resistance in non-small cell lung cancer.
Apoptosis. 27:883–898. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang J, Liu X, Huang Y, Li P, Yang M, Zeng
S, Chen D, Wang Q, Liu H, Luo K and Deng J: Targeting nicotinamide
N-methyltransferase overcomes resistance to EGFR-TKI in non-small
cell lung cancer cells. Cell Death Discov. 8:1702022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kim S, Im JH, Kim WK, Choi YJ, Lee JY, Kim
SK, Kim SJ, Kwon SW and Kang KW: Enhanced sensitivity of nonsmall
cell lung cancer with acquired resistance to epidermal growth
factor receptor-tyrosine kinase inhibitors to phenformin: The roles
of a metabolic shift to oxidative phosphorylation and redox
balance. Oxid Med Cell Longev. 2021:54283642021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dyrstad SE, Lotsberg ML, Tan TZ, Pettersen
IKN, Hjellbrekke S, Tusubira D, Engelsen AST, Daubon T, Mourier A,
Thiery JP, et al: Blocking aerobic glycolysis by targeting pyruvate
dehydrogenase kinase in combination with EGFR TKI and ionizing
radiation increases therapeutic effect in non-small cell lung
cancer cells. Cancers (Basel). 13:9412021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cheng FJ, Chen CH, Tsai WC, Wang BW, Yu
MC, Hsia TC, Wei YL, Hsiao YC, Hu DW, Ho CY, et al: Cigarette
smoke-induced LKB1/AMPK pathway deficiency reduces EGFR TKI
sensitivity in NSCLC. Oncogene. 40:1162–1175. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang L, Ke J, Min S, Wu N, Liu F, Qu Z,
Li W, Wang H, Qian Z and V Wang X: Hyperbaric oxygen therapy
represses the warburg effect and epithelial-mesenchymal transition
in hypoxic NSCLC cells via the HIF-1α/PFKP Axis. Front Oncol.
11:6917622021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kim DJ, Park YS, Kim ND, Min SH, You YM,
Jung Y, Koo H, Noh H, Kim JA, Park KC and Yeom YI: A novel pyruvate
kinase M2 activator compound that suppresses lung cancer cell
viability under hypoxia. Mol Cells. 38:373–379. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhao T, Zhu Y, Morinibu A, Kobayashi M,
Shinomiya K, Itasaka S, Yoshimura M, Guo G, Hiraoka M and Harada H:
HIF-1-mediated metabolic reprogramming reduces ROS levels and
facilitates the metastatic colonization of cancers in lungs. Sci
Rep. 4:37932014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhou F, Du J and Wang J: Albendazole
inhibits HIF-1α-dependent glycolysis and VEGF expression in
non-small cell lung cancer cells. Mol Cell Biochem. 428:171–178.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yang Y, Liu L, Sun J, Wang S, Yang Z, Li
H, Huang N and Zhao W: Deoxypodophyllotoxin inhibits non-small cell
lung cancer cell growth by reducing HIF-1α-Mediated glycolysis.
Front Oncol. 11:6295432021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Liu X, Liu L, Chen K, Sun L, Li W and
Zhang S: Huaier shows anti-cancer activities by inhibition of cell
growth, migration and energy metabolism in lung cancer through
PI3K/AKT/HIF-1α pathway. J Cell Mol Med. 25:2228–2237. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Fumarola C, Cretella D, La Monica S,
Bonelli MA, Alfieri R, Caffarra C, Quaini F, Madeddu D, Falco A,
Cavazzoni A, et al: Enhancement of the anti-tumor activity of FGFR1
inhibition in squamous cell lung cancer by targeting downstream
signaling involved in glucose metabolism. Oncotarget.
8:91841–91859. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sun L, Liu H, Ye Y, Lei Y, Islam R, Tan S,
Tong R, Miao YB and Cai L: Smart nanoparticles for cancer therapy.
Signal Transduct Target Ther. 8:4182023. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Huang J, Zhuang C, Chen J, Chen X, Li X,
Zhang T, Wang B, Feng Q, Zheng X, Gong M, et al: Targeted
Drug/Gene/Photodynamic therapy via a stimuli-responsive
dendritic-polymer-based nanococktail for treatment of
EGFR-TKI-resistant Non-small-cell lung cancer. Adv Mater.
34:e22015162022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Alkhathami AG, Sahib AS, Al Fayi MS,
Fadhil AA, Jawad MA, Shafik SA, Sultan SJ, Almulla AF and Shen M:
Glycolysis in human cancers: Emphasis circRNA/glycolysis axis and
nanoparticles in glycolysis regulation in cancer therapy. Environ
Res. 234:1160072023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Martínez-Reyes I and Chandel NS: Cancer
metabolism: Looking forward. Nat Rev Cancer. 21:669–680. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Xiao Y, Yu TJ, Xu Y, Ding R, Wang YP,
Jiang YZ and Shao ZM: Emerging therapies in cancer metabolism. Cell
Metab. 35:1283–1303. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zheng F, Chen J, Zhang X, Wang Z, Chen J,
Lin X, Huang H, Fu W, Liang J, Wu W, et al: The HIF-1α antisense
long non-coding RNA drives a positive feedback loop of HIF-1α
mediated transactivation and glycolysis. Nat Commun. 12:13412021.
View Article : Google Scholar : PubMed/NCBI
|

















