Introduction
Glomerular hypertrophy is a characteristic
pathological change of diabetic nephropathy (DN), which
predominantly appears as hypertrophy of renal cells and the
progressive accumulation of external extracellular matrix. Cellular
hypertrophy appears as an increase in cell size, rather than cell
number, and its occurrence is closely associated with regulation of
the cell cycle (1). In diabetes,
cyclin-dependent kinase inhibitors, represented by the p21 protein,
are overexpressed, thus the DNA replication is affected and cell
proliferation is inhibited (2),
which further leads to the occurrence of hypertrophy.
12-lipoxygenase (12-LO) is a type of oxygenase,
which is involved in in vivo oxidation reactions of
polyunsaturated fatty acids, including arachidonic acid and
linoleic acid, and its metabolites may be involved in the
occurrence and development of diabetes through oxidative stress and
inflammatory responses (3). A
previous study confirmed that 12-LO was involved in the
pathological changes of diabetic kidney hypertrophy, and was
identified as an important factor in DN progression and glomerular
fibrosis. Therefore, controlling the expression of 12-LO may assist
in reducing the development of renal cell hypertrophy and delay the
progression of glomerular sclerosis (4,5). Our
preliminary investigations confirmed that the metabolite of 12-LO,
12 (S)-hydroxyeicosatetraenoic acid [12 (S)-HETE], can induce an
increase in the protein expression levels of p21 and p27 in
mesangial cells, and upregulate the gene expression of p21 through
transcription (6). However, the
exact regulatory mechanisms remain to be elucidated.
The caudal amino acid residues of histones can
affect the density of the DNA double helix through acetylation,
methylation, phosphorylation and other modifications, and are thus
involved in the transcriptional regulation of genes, known as
epigenetic regulation. Previous investigations have confirmed that
high blood sugar levels are involved in the occurrence of DN and
other vascular complications by affecting the apparent
transcriptional regulation (7,8). In
addition, the angiotensin II receptor antagonist, losartan, can
affect the chemical modification of histones, indicating the
effects of reducing proteinuria and inhibiting glomerular
sclerosis, and suggesting that the chemical modification of
histones may be a novel mechanism and target in the intervention of
vascular complications in diabetes (9,10).
Our preliminary results confirmed that the acetylation of histones
involved the regulation by transforming growth factor (TGF-β1) on
the expression of the pro-fibrotic gene, PAI-1 (11,12).
As the downstream regulatory factor of the TGF-β1 pathway, 12-LO
and the TGF-β1 pathway may mutually activate each other, thus
jointly mediating the high expression levels of PAI-1, collagen and
fibronectin under high glucose conditions (6). However, whether 12 (S)-HETE is
involved in the gene expression of p21 through epigenetic
regulation (chemical modification of histones) has not been
reported. For the first time, to the best of our knowledge, the
present study used chromatin co-immunoprecipitation technology to
examine the effects and mechanisms of 12-LO, and its metabolites,
on the chemical modification (acetylation and methylation) of
histones in the p21 gene. Thus, the present study demonstrated the
epigenetic modification mechanism associated with the
overexpression of cyclin-dependent kinase inhibitor p21 under
diabetic conditions, and provides a novel-theoretical basis and
intervention targets to delay diabetic glomeruli hypertrophy.
Materials and methods
Cell culture
The primary cultured rat MCs were isolated from
Sprague-Dawley rats and cultured as previously described (13) and were seeded into culture dishes
with RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and cultured at 37°C in 5% CO2. When
the cells fused and covered 80% of the bottom of the petri dish,
the serum-free medium was replaced for synchronic culture for
another 24 h for the subsequent experiments. The present study was
approved by the ethics committee of the First Affiliated Hospital
of Jilin University (Changchun, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
An RNA STAT60 kit (AMS Biotechnology Europe Ltd.,
Abingdon, UK) was used to extract the total mRNA from the MCs, and
a TaqMan PreAmp Master mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) was then used to reverse transcribe 1.5 µg
mRNA into cDNA, the final product was diluted 10-fold and used as
the template for PCR amplification. An ABI-7500 real-time
quantitative PCR instrument (Thermo Fisher Scientific, Inc.) was
used, together with the SYBR Green PCR Master Mix kit (Thermo
Fisher Scientific, Inc.). The reaction volume, of 20 µl,
comprised 10 µl SYBR Green mix, 1 µl of the positive
and negative primer strands (50 pmol), respectively, 5 µl
cDNA template and 3 µl DEPC water. The thermocycling
conditions were as follows: 50°C for 2 min and 95°C for 10 min; 40
cycles of 95°C for 15 sec, 60°C for 1 min; followed by melt curve
analysis at 95°C for 15 sec, 60°C for 1 min, 95°C for 30 sec and
60°C for 1 min. Primers are presented in Table I. β-actin was selected as the
internal control, and the 2−ΔΔCq method was used for the
analysis as described previously (13).
 | Table IPrimer sequences. |
Table I
Primer sequences.
| Primer | Forward (5′-3′) | Reverse (5′-3′) |
|---|
| ChIP assay | | |
| p21 P1 (R) |
AGTGGATTCAAACATATGAGCCACT |
CCCTCCATCCCCCAAGGCCCTG |
| p21 P2 (R) |
GTTCAGCCCTGGAACCGAAG |
GTACCAAACACCCTTCACCTGGTAC |
| p21 T1 (R) |
GGCGCTCAGCCATTCAGTAT |
CGTGACCAGGGATAGGCTGTCACG |
| p21 T2 (R) |
CGGCCAGTGAGCAGTTGAGCcG |
CCCTCCAGTGGCGTCTCAGT |
| cDNA | | |
| p21 (R) |
GTGGCCTTGTCGCTGTCTTG |
CGATTCTTGCAGAAGACCAATCG |
| β-actin (R) |
CCCTGTATGCCTCTGGTCGT |
CGGACGCAGCTCAGTAACAGTCCG |
Western blot analysis
Cells were lysed in 1.5X sodium dodecyl sulfate
(SDS) buffer and protein concentration was estimated using the
Lowry method. Equal quantities of the samples (50 µg) were
separated by electrophoresis on a 10% SDS gel and transferred onto
a nylon membrane. Following treatment with closure fluid, rabbit
antibodies against p21 (1:500; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA; cat. no. sc-756) lysine-specific demethylase
(LSD1; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA;
cat. no. 2139) and mouse antibodies against p300 (1:500; EMD
Millipore, Billerica, MA, USA; cat. no. 05-257) were added for
overnight incubation at 4°C, and the membrane was rinsed.
Horseradish peroxidase-labeled goat anti-rabbit (1:1,000; cat. no.
ER48616) and goat anti-mouse (1:1,000; cat. no. ER48627) secondary
antibodies (Sigma-Aldrich, St. Louis, MO, USA) were then added for
hybridization at room temperature for 1 h, following which the
membrane was washed and ECL reagent was added for staining and
developing images as described previously (6). Images of the blots were analyzed
using Quantity One software (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The nucleoprotein extraction method was performed,
according to the protocol of the Nucleoprotein Extraction kit
(Thermo Fisher Scientific, Inc.).
Chromatin immunoprecipitation (ChIP
assay)
Following stimulation with 12 (S)-HETE, the MCs were
fixed in 1% formalin, sonicated and pretreated with protein
A/G-immunomagnetic beads (Invitrogen; Thermo Fisher Scientific,
Inc.), followed by the addition of specific histone-3-lysine
(H3K)9Ac (1:50; EMD Millipore; cat. no. 06-599), H3K4Me1 (1:50;
Abcam, Cambridge, MA, USA; cat. no. ab8895), H3K9Me3 (1:50; Abcam;
cat. no. ab8898), p300 (1:50) and LSD1 (1:50) antibodies or the
internal reference antibody (mouse and rabbit IgG, cat. nos. 12-371
and 12-370, respectively), obtained from EMD Millipore for
overnight incubation (EZ ChIP kit; EMD Millipore). The precipitated
and separated DNA fragments were then used as the template for qPCR
amplification using the ABI-7500 real-time quantitative PCR
instrument. The 2−ΔΔCq method was used to analyze the
results. The primer sequences and the amplification regions are
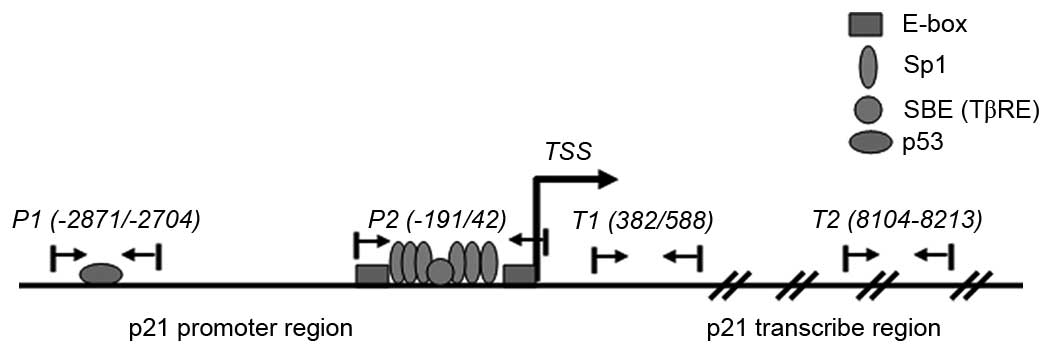
shown in Table I and Fig. 1.
Construction and amplification of
plasmids
According to the nucleotide sequence of the rat p21
gene promoter region (GenBank), Invitrogen primer design software
(Thermo Fisher Scientific, Inc.) was used to amplify the nucleotide
sequence of the p21 genome between DNA −4,542 and +113; the
amplified DNA fragment was then inserted using the enzyme-linked
method, into a pGL3 vector (Promega Corporation, Madison, WI, USA),
which contained the luciferase reporter gene, to synthesize
pGL3-p21 according to our previous study (11). The recombinant was then transformed
into competent Escherichia coli (donated by Professor Wang
Fang, Department of Microbiology, Bethune Medical School of Jilin
University, Changchun, China), which were extracted using
resistance screening for identification and future use. The p300
expression plasmid was provided by Dr B. Forman (Beckman Research
Center, Duarte, CA, USA).
Cell transfection and analysis of
luciferase activity
FuGene 6 (Roche, Basel, Switzerland) was used as the
transfection reagent for the transfection procedures. The cells in
the control group were transfected with a plasmid containing the
green fluorescent protein sequence, according to the protocol of
the Nucleofecter™ kit (Lonza Group, Basel, Switzerland). A
luciferase activity detection kit (Promega Corporation) and
TD-20/20 luciferase activity analyzer (Promega Corporation) were
used to analyze the luciferase activities of pGL-3 and pRL-TK,
according to the kit protocol.
Statistical analysis
Data were expressed as the mean ± standard error of
the mean from multiple experiments. GraphPad Prism 5.0 software
(GraphPad Software, Inc., La Jolla, CA, USA) was used for data
analysis. Intergroup comparison was performed using paired
Student's t-test; multigroup comparisons were performed using
analysis of variance with Dunnett's post-hoc tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression and transcriptional activity
of the p21 gene
Following synchronic culture in serum-free medium
for 24 h, 12 (S)-HETE (10−6, 10−7 and
10−8) was added to the MCs for 2, 4 and 8 h, the cells
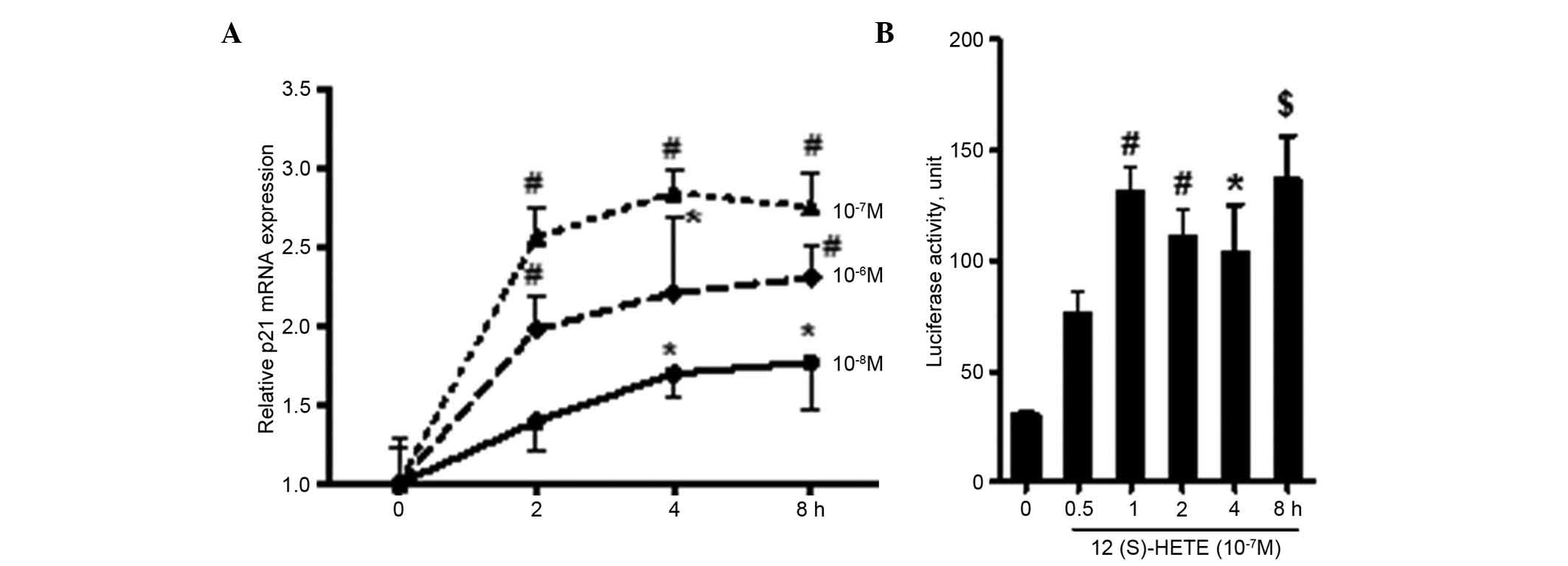
were collected to extract mRNA for RT-qPCR analysis. The results
(Fig. 2A) showed that 12 (S)-HETE
promoted the intracellular mRNA expression of p21, and the effects
stimulated by the 10−7 M concentration were the most
marked (P<0.01). Within the stimulation period (2–8 h), the mRNA
expression of p21 was increased by 2.5–2.8, compared with the
control group. In order to determine whether the regulation was
through the transcriptional pathway, when the MCs grew and covered
70–80% of the bottom of the petri dish, the pGL3-p21 plasmid was
transfected into the cells using Fugene 6. After 6 h, the
serum-free medium was replaced prior to incubation for another 24
h, following which 12 (S)-HETE (10−7 M) was added for
0.5–8 h stimulation. The cells were then collected for the
detection of luciferase activity. As shown in Fig. 2B, following stimulation with 12
(S)-HETE for 0.5 h, the transcriptional activity of p21 began to
increase, which continued until 8 h (P<0.001). This further
confirmed that 12 (S)-HETE promoted the gene expression of p21 via
the transcriptional pathway.
Modification of the p21 gene promoter and
transcription regions of H3K9Ac, H3K4Me1 and H3K9Me3
In order to confirm whether the chemical
modification of histones, via acetylation and methylation, was
involved in the transcriptional regulation of 12 (S)-HETE towards
the p21 gene, a ChIP assay was performed to observe the changes in
the levels of chemical modification of H3K in the p21 gene promoter
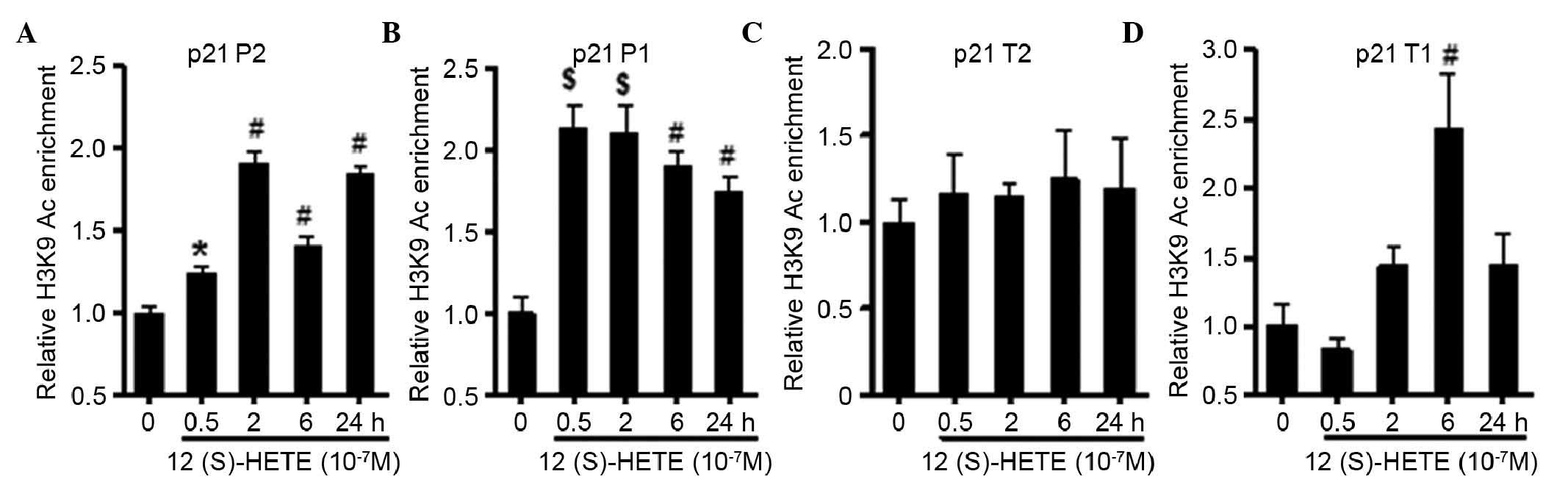
(P) and transcription (T) regions. As shown in Fig. 3, the results showed that 12
(S)-HETE significantly increased the level of H3K9Ac in P1 and P2
of the p21 gene (P<0.001). The level of H3K9Ac was also
significantly increased in T1 (P<0.01), whereas the level of
H3K9Ac in T2 (8,104–8,213 bp), far from the transcription start
site (TSS), did not alter significantly. Compared with acetylation,
the mechanisms underlying the effects of methylated histones in the
regulation of gene expression were relatively complex. It was
confirmed that methylation at the H3K4 site was involved in the
transcriptional activation, whereas the methylation at H3K9
suppressed transcriptional activation. As shown in Fig. 4A, 12 (S)-HETE increased the
modification of H3K4Me1 in P2 and T1, close to TSS, and in P1,
however, no significant difference was found in T2. It was found
that the stimulation of 12 (S)-HETE in P1 and P2 significantly
reduced the modification of H3K9Me3 (P<0.01; Fig. 4B), whereas no significant
difference was found in the transcription region. These results
suggested that 12 (S)-HETE increased the modification of H3K9Ac and
H3K4Me1 in p21, and reduced the modification of H3K9Me3 in the
promoter region, thus promoting the transcriptional expression of
p21.
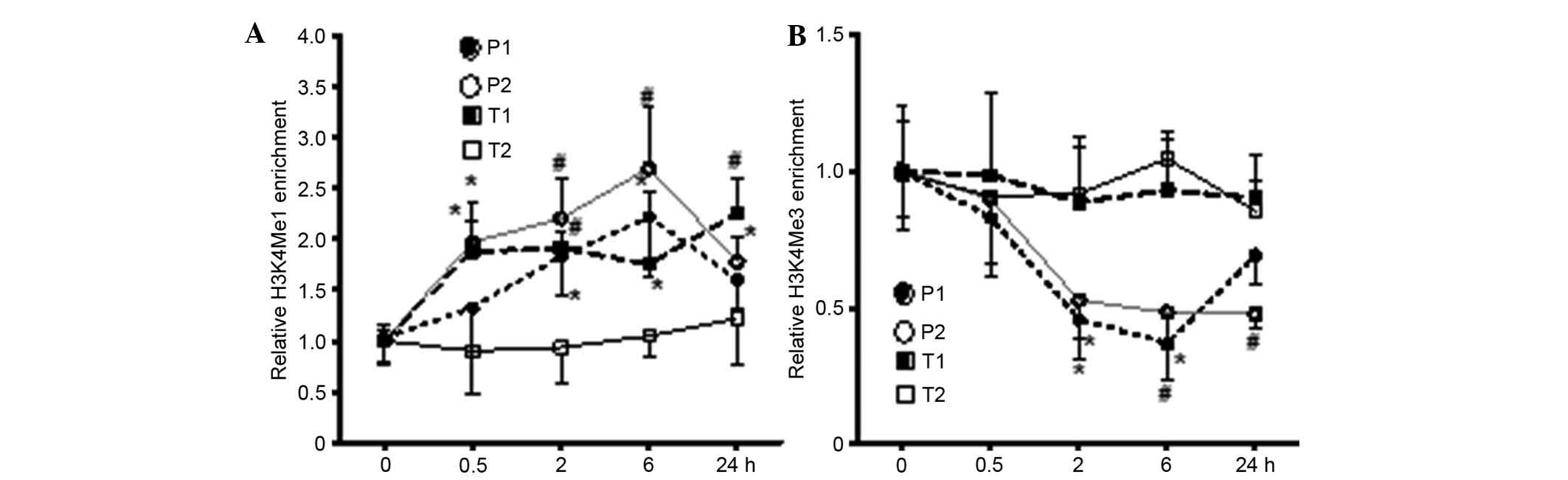 | Figure 412 (S)-HETE induces H3K4Me1 and
H3K9Me3 epigenetic modifications in the p21 promoter and
transcription regions. Quiescent rat MCs were treated with 12
(S)-HETE (10-7 M) for the indicated time course. (A) H3K4Me1 and
(B) H3K9Me3 enrichment at indicated promoter and transcription
regions were detected using chromatin immunoprecipitation assays.
Data from quantitative-polymerase chain reaction analysis were
analyzed using the 2−ΔΔCq method, and results normalized
to input DNA were expressed as the fold change over the respective
untreated control cells. Values are presented as the mean ±
standard error of the mean (n=3). *P<0.05,
#P<0.01 vs. corresponding untreated control. MCs,
mesangial cells; 12 (S)-HETE, 12 (S)-hydroxyeicosatetraenoic acid;
H3K, histone-3-lysine; P1, protomer 1, P2, promoter 2; T1,
transcription region 1; T2, transcription region 2. |
Effect of the level of histoneacetyl
transferase (HAT; p300) on the transcriptional regulation of 12
(S)-HETE on the expression of p21
As shown in Fig.
5A, the results showed that, following transfection with the
p300 expression vector, the intracellular protein level of p300 was
significantly increased, and the protein level of p21 was also
increased. In the group of cells expressing a high level of P300, 2
h stimulation with 12 (S)-HETE (10−7 M) significantly
increased the transcriptional activity of the p21 promoter region
(Fig. 5B) and its mRNA level
(Fig. 5C), compared with the
control group and 12 (S)-HETE stimulation only group, suggesting
that increasing the intracellular level of HAT (p300) significantly
promotes the 12 (S)-HETE-induced transcriptional expression of p21.
The results of the western blot analysis (Fig. 5D) further confirmed that the
increased expression of HAT (p300) increased the intracellular
protein expression of p21 and the promoting effect of 12 (S)-HETE
on the expression of p21.
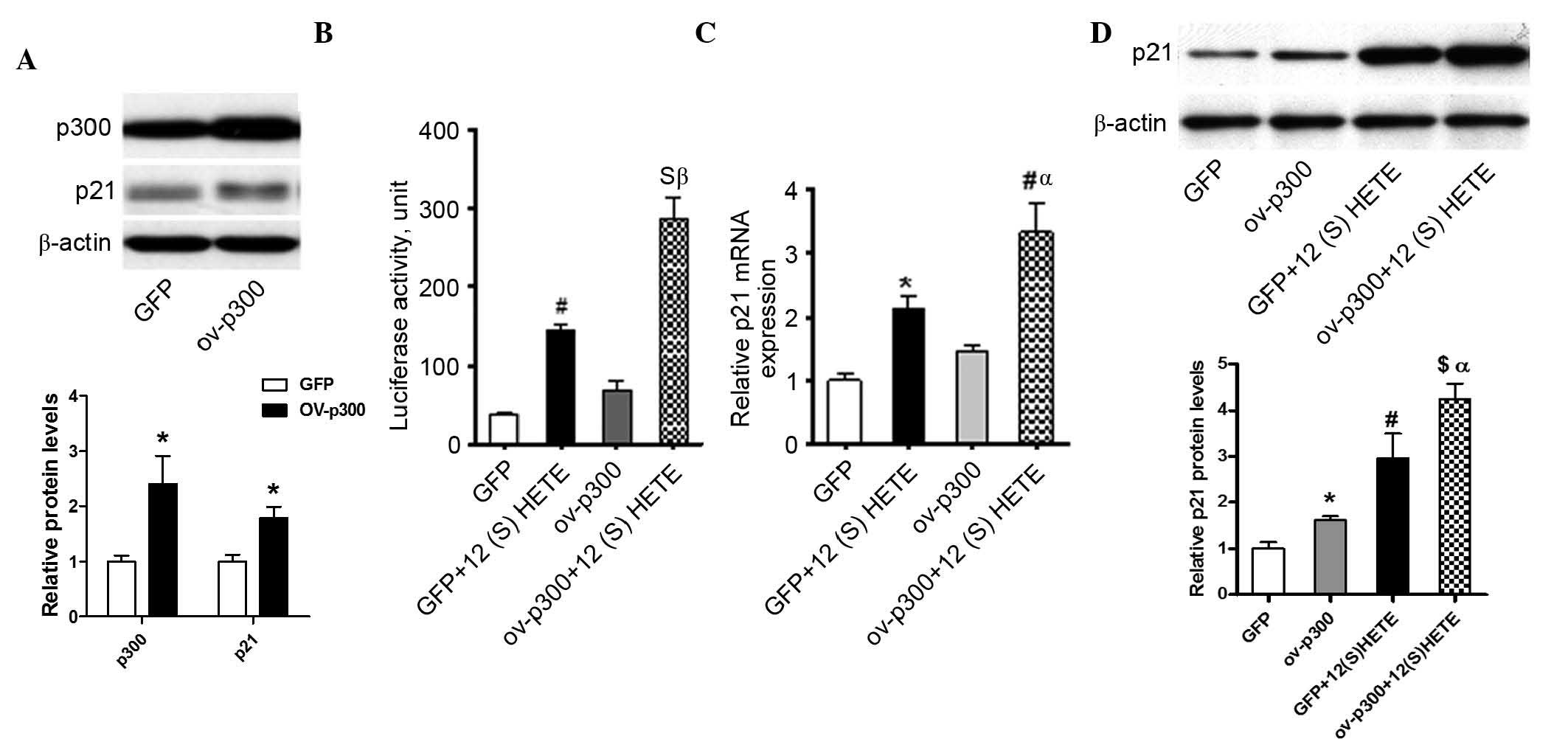 | Figure 5Overexpression of p300 enhances basal
and 12 (S)-HETE-associated gene expression and transcriptional
activity of p21 (A) Rat MCs were transiently transfected with
either a p300 expression vector or a control enhanced GFP
expression vector, followed by western blot analysis. Bar graph
demonstrates quantification of immunoblotting, a significant
increase in p300 and p21 protein expression levels in
p300-overexpressing cells relative to GFP was observed. (B) Rat MCs
were co-transfected with the p300 expression vector or GFP
expression vector, combined with a p21 promoter-luciferase
construct, and then treated without or with 12 (S)-HETE
(10−7 M) for 2 h. The luciferase activity in the cell
lysates were determined. Rat MCs were co-transfected with the p300
expression vector or a GFP expression vector, and then treated with
12 (S)-HETE (10−7 M) for 2 h. Total RNA and proteins
were extracted for (C) quantitative polymerase chain reaction and
(D) western blot analyses to determine the changes in the
expression of p21. Bar graph represent quantification of
immunoblotting. Values are presented as the mean ± standard error
of the mean (n=3). *P<0.05, #P<0.01 and
$P<0.001, vs. GFP; αP<0.05 and
βP<0.01, vs. GFP+12 (S)-HETE. MCs, mesangial cells;
12 (S)-HETE, 12 (S)-hydroxyeicosatetraenoic acid; GFP, green
fluorescence protein. |
Level of LSD1 in the nuclei and its
binding in the promoter region of the p21 gene
To further investigate the mechanism of 12
(S)-HETE-induced chemical modification of histones in the p21 gene,
the present study examined the binding of HAT (p300) and
demethylase LSD1 in the p21 promoter region following stimulation
with 12 (S)-HETE (10−7 M). The ChIP assay results
(Fig. 6A) showed that 12 (S)-HETE
promoted the binding of p300 in P2 (P<0.01), whereas no
significant changes were observed in the P1 or transcription
regions, suggesting that the modification of H3K9Ac in P2 was
associated with the increased binding of p300 in this area. As
shown in Fig. 6B, 12 (S)-HETE
significantly inhibited the binding of LSD1 in P1 and P2
(P<0.01), whereas no significant difference in the transcription
region was observed, compared with the control group, suggesting
that 12 (S)-HETE inhibited the binding of LSD1 in the promoter
region, thereby inducing the modification of H3K4Me1 in P2. Further
investigation (Fig. 6C) showed
that 12 (S)-HETE (10−7 M) stimulated the reduction in
the protein level of LSD1 in the nuclei, suggesting that 12
(S)-HETE induced the modification of H3K4Me1 in p21 through
inhibiting the expression of LSD1 in the nuclei, thus contributing
to the expression of p21.
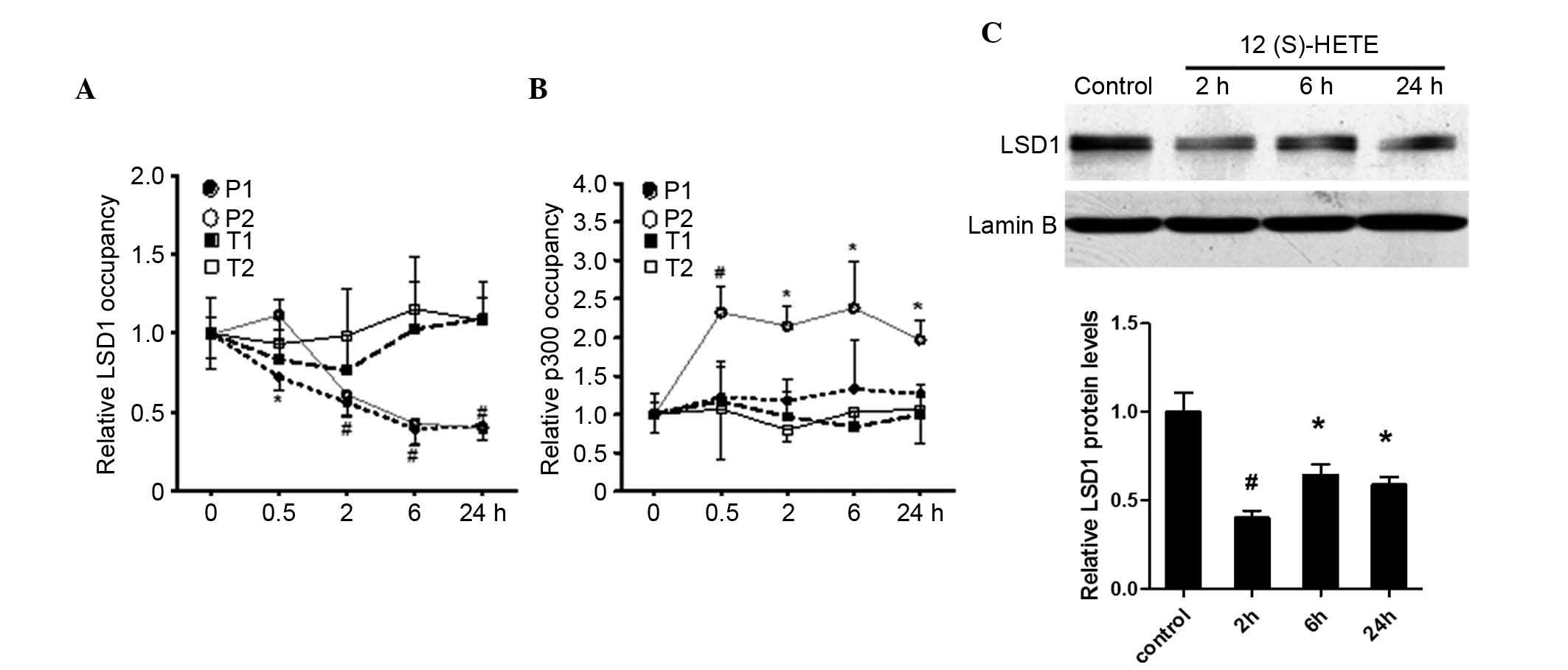 | Figure 6Effect of 12 (S)-HETE on the levels of
p300 and LSD1 in the p21 promoter and transcription regions.
Quiescent rat MCs were treated with 12 (S)-HETE (10-7 M) for the
indicated time course. The relative occupancy of (A) p300 and (B)
LSD1 at indicated promoter and transcription regions of p21 were
detected using ChIP assays. Quantitative polymerase chain reaction
data were analyzed using the 2−ΔΔCq method and results,
normalized to input DNA, are expressed as the fold change over the
respective untreated control cells. Values are presented as the
means ± standard error of the mean (n=3). (C) Quiescent rat MCs
were treated with 12 (S)-HETE (10-7 M) for the indicated time
course, followed by western blot analysis of nuclear protein
lysates from the untreated control and 12 (S)-HETE-treated MCs
using LSD1 and Lamin B antibodies. Bar graph quantification showing
a significant decrease in LSD1 protein levels by 12(S)-HETE
treatment. *P<0.05, #P<0.01 vs. control
cells. MCs, mesangial cells; H3K, histone-3-lysine; 12 (S)-HETE, 12
(S)-hydroxyeicosatetraenoic acid; LSD1, lysine-specific
demethylase; ChIP,. chromatin immunoprecipitation; P1, protomer
region 1, P2, promoter region 2; T1, transcption region 1; T2,
transcription region 2. |
Discussion
DM is a chronic metabolic disease, which is affected
by multiple and complex factors, and the occurrence of which can be
caused by alterations in genetic and environmental factors. Studies
have confirmed that, prior to the diagnosis of DM, patients present
with abnormal glucose tolerance and the body has already formed
metabolic memory, presenting potential hidden risks for the
occurrence of DM and its complications (14–16),
however, it also indicates that abnormal epigenetic modifications
may be involved in the pathogenesis of DM and its complications
(17,18).
The acetylation of histones occurs predominantly
under the coordination of HATs and histone deacetylases (HDACs).
HATs can catalyze the acetylation of histones, resulting in a loose
chromatin structure and the promotion of gene transcription,
whereas HDACs induce the deacetylation of histones, resulting in
the condensation of chromatin and inhibition of gene transcription.
Compared with the acetylation of histones, the methylation of
histones is more stable and lasting, according to the methylation
patterns, sites and modified amino acid residues. The methylation
of histones can lead to transcriptional activation or inhibition,
for example H3K9Me can inhibit transcription, whereas H3K4Me can
lead to transcriptional activation (19). Different types of chemical
modification at the same site of histones can affect each other,
thus co-acting on the regulation of gene expression. H3K9Me3 is
important in gene silencing, which can inhibit transcriptional
activity by increasing the binding of HDAC on the DNA chain
(20).
TGF-β1 is an important pathogenic factor in the
pathogenesis of DN, which can induce the modifications of H3K9Ac
and H3K4Me, thus affecting the transcription level of plasminogen
activator inhibitor-1 and other genes, and inducing the occurrence
of renal fibrosis (11,21). Our previous study found that 12
(S)-HETE increases the protein expression levels of cyclin
inhibitor p21 and p27, and is involved in the occurrence and
development of DN (6). In the
present study, the ChIP assay revealed that 12 (S)-HETE induced the
modification of H3K9Ac in the p21 gene promoter region and the
TSS-adjacent transcription region. 12 (S)-HETE also stimulated and
increased the modification of H3K4Me1 in TSS-adjacent P2 and T1,
and reduced the level of H3K9Me3 modification in the promoter
region.
In investigating the mechanism of the chemical
modification of histones in regulating the gene expression of p21,
the present study found that the increased expression of HAT (p300)
significantly increased the promoting effect of 12 (S)-HETE on the
gene expression of p21, and this further confirmed the role of
acetylation in the transcriptional regulation of the p21 gene. The
ChIP assay showed that 12 (S)-HETE induced the binding of p300 in
P2, suggesting that the 12 (S)-HETE P2-induced modification of
H3K9Ac was associated with the binding of the p300 protein in p21.
The acetylation in P1 may be due to fact that this region contains
the binding sites of p53, and the p53 protein can induce
modifications in the binding region through acetylation or binding
with other HATs (22–24).
The methylation of histone H3K4 is usually
associated with active gene expression. H3K4 can be monomethylated,
dimethylated or trimethylated on lysine residues by specific
histone H3K4 methyltransferases, including SET7/9 or mixed-lineage
leukemia (20). The previous
identification of LSD1, which specifically removes H3K4me1 and
H3K4me2, demonstrated the dynamic nature of H3K methylation
(25,26). In the present study, it was found
that 12 (S)-HETE reduced the protein level of LSD1 within nuclei
and inhibited its binding in the promoter region of the p21 gene,
suggesting that it may be the primary cause of the H3K4Me1
modification in this region. Previous studies have shown that the
H3K4Me modifications of histones can competitively inhibit the
binding of HDAC and DNA, thus promoting H3K9Ac and suppressing the
occurrence of H3K9Me (27,28). The present study showed that, in
addition to 12 (S)-HETE-induced H3K4Me1 modification in the p21
gene promoter region, the gene silencing-associated H3K9Me3
modification was significantly decreased simultaneously, indicating
that the transcriptional regulation of 12 (S)-HETE towards the p21
gene was realized through comprehensive chemical modification of
histones. Due to the complexity of the methylation regulation of
histones and the numerous factors affecting this, the specific
pathogenesis requires further investigation.
In conclusion, the present study demonstrated the
following: The acetylation and methylation of histones were closely
associated with the 12 (S)-HETE-induced transcriptional regulation
towards the gene expression of p21; 12 (S)-HETE inhibited the
expression of LSD1 and its binding in the p21 gene promoter region,
thus inducing the modification of H3K4Me1 in the p21 gene promoter
region. Increasing the binding of p300 in the p21 gene promoter
region induced the modification of H3K9Ac and 12 (S)-HETE
stimulation inhibited the level of H3K9Me3 in the p21 promoter
region. These changes resulted in the p21 gene promoter and
peri-TSS chromatin being in the relaxed transcription activation
state, thus transcriptional activity was increased and the
expression of p21 gene was promoted.
In conclusion, associated epigenetic changes,
including increased histone H3K9Ac and H3K4Me1, and reduced H3K9Me3
may involved in 12(S)-HETE mediated p21 gene regulation, and thus,
be important in the pathogenesis of renal hypertrophy in
diabetes.
Acknowledgments
This study was supported by the Youth Foundation of
national natural Science foundation of China (grant no. 81000300);
The National Natural Science Foundation of China (grant nos.
81370830, 81170669, 81070578 and 81270809), the Jilin Provincial
Department of Science and Technology (grant no. 20130521006JH), the
Doctor Foundation, Ministry of Education of P.R. China (grant no.
2010006120024) and the Item of Bethune Project B, Jilin University
(grant no, 2012226).
References
|
1
|
Hostetter TH: Progression of renal disease
and renal hypertrophy. Annu Rev Physiol. 57:263–278. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Al-Douahji M, Brugarolas J, Brown PA,
Stehman-Breen CO, Alpers CE and Shankland SJ: The cyclin kinase
inhibitor p21WAF/CIP1 is required for glomerular hypertrophy in
experimental diabetic nephropathy. Kidney Int. 56:1691–1699. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bleich D, Chen S, Zipser B, Sun D, Funk CD
and Nadler JL: Resistance to the type 1 diabetes induction in
12-lipoxygenase knockout mice. J Clin Invest. 103:1431–1436. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yuan H, Lanting L, Xu ZG, Li SL, Swiderski
P, Putta S, Jonnalagadda M, Kato M and Natarajan R: Effects of
cholesterol tagged small interfering RNA targeting
12/15-lipoxygenase on parameters of diabetic nephropathy in mouse
model of type 1 diabetes. Am J Physiol Renal Physiol.
295:F605–F617. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma J, Natarajan R, LaPage J, Lanting L,
Kim N, Becerra D, Clemmons B, Nast CC, Surya Prakash GK, Mandal M
and Adler SG: 12/15-lipoxygenase inhibitors in diabetic nephropathy
in the rat. Prostaglandins Leukot Essent Fatty Acids. 72:13–20.
2005. View Article : Google Scholar
|
|
6
|
Kim YS, Xu ZG, Reddy MA, Li SL, Lanting L,
Sharma K, Adler SG and Natarajan R: Novel interactions between
TGF-{beta}1 actions and the 12/15-lipoxygenase pathway in mesangial
cells. J Am Soc Nephrol. 16:352–362. 2005. View Article : Google Scholar
|
|
7
|
Kato M and Natarajan R: Diabetic
nephropathy-emerging epigenetic mechanisms. Nat Rev Nephrol.
10:517–530. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reddy MA, Tak Park J and Natarajan R:
Epigenetic modifications in the pathogenesis of diabetic
nephropathy. Semin Nephrol. 33:341–353. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reddy MA, Sumanth P, Lanting L, Yuan H,
Wang M, Mar D, Alpers CE, Bomsztyk K and Natarajan R: Losartan
reverses permissive epigenetic changes in renal glomeruli of
diabetic db/db mice. Kidney Int. 85:362–373. 2014. View Article : Google Scholar :
|
|
10
|
Natarajan R: Drugs targeting epigenetic
histone acetylation in vascular smooth muscle cells for restenosis
and atherosclerosis. Arterioscler Thromb Vasc Biol. 31:725–727.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan H, Reddy MA, Sun G, Lanting L, Wang
M, Kato M and Natarajan R: Involvement of p300/CBP and epigenetic
histone acetylation in TGF-β1-mediated gene transcription in
mesangial cells. Am J Physiol Renal Physiol. 304:F601–F613. 2013.
View Article : Google Scholar
|
|
12
|
Kato M, Dang V, Wang M, Park JT, Deshpande
S, Kadam S, Mardiros A, Zhan Y, Oettgen P, Putta S, et al: TGF-β
induces acetylation of chromatin and of Ets-1 to alleviate
repression of miR-192 in diabetic nephropathy. Sci Signal.
6:ra432013. View Article : Google Scholar
|
|
13
|
Sun G, Reddy MA, Yuan H, Lanting L, Kato M
and Natarajan R: Epigenetic histone methylation modulates fibrotic
gene expression. J Am Soc Nephrol. 21:2069–2080. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Villeneuve LM and Natarajan R: The role of
epigenetics in the pathology of diabetic complications. Am J
Physiol Renal Physiol. 299:F14–F25. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reddy MA, Zhang E and Natarajan R:
Epigenetic mechanisms in diabetic complications and metabolic
memory. Diabetologia. 58:443–455. 2015. View Article : Google Scholar :
|
|
16
|
Miao F, Chen Z, Genuth S, Paterson A,
Zhang L, Wu X, Li SM, Cleary P, Riggs A, Harlan DM, et al:
Evaluating the role of epigenetic histone modifications in the
metabolic memory of type 1 diabetes. Diabetes. 63:1748–1762. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ling C and Groop L: Epigenetics: A
molecular link between environmental factors and type 2 diabetes.
Diabetes. 58:2718–2725. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pirola L, Balcerczyk A, Okabe J and
El-Osta A: Epigenetic phenomena linked to diabetic complications.
Nat Rev Endocrinol. 6:665–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Marmorstein R and Trievel RC: Histone
modifying enzymes: Structures, mechanisms, and specificities.
Biochim Biophys Acta. 1789:58–68. 2009. View Article : Google Scholar
|
|
20
|
Shilatifard A: Chromatin modifications by
methylation and ubiquitination: Implications in the regulation of
gene expression. Annu Rev Biochem. 75:243–269. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun G, Reddy MA, Yuan H, Lanting L, Kato M
and Natarajan R: Epigenetic histone methylation modulates fibrotic
gene expression. J Am Soc Nephrol. 21:2069–2080. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Reed SM and Quelle DE: p53 Acetylation:
Regulation and consequences. Cancers (Basel). 7:30–69. 2014.
View Article : Google Scholar
|
|
23
|
Marouco D, Garabadgiu AV, Melino G and
Barlev NA: Lysine-specific modifications of p53: A matter of life
and death? Oncotarget. 4:1556–1571. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee JT and Gu W: SIRT1: Regulator of p53
deacetylation. Genes Cancer. 4:112–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi Y, Sawada J, Sui G, Affar el B,
Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y and Shi Y:
Coordinated histone modifications mediated by a CtBP co-repressor
complex. Nature. 422:735–738. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shi Y, Lan F, Matson C, Mulligan P,
Whetstine JR, Cole PA, Casero RA and Shi Y: Histone demethylation
mediated by the nuclear amine oxidase homolog LSD1. Cell.
119:941–953. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang H, Cao R, Xia L, Erdjument-Bromage H,
Borchers C, Tempst P and Zhang Y: Purification and functional
characterization of a histone H3-lysine 4-specific
methyltransferase. Mol Cell. 8:1207–1217. 2001. View Article : Google Scholar
|
|
28
|
Nishioka K, Chuikov S, Sarma K,
Erdjument-Bromage H, Allis CD, Tempst P and Reinberg D: Set9, a
novel histone H3 methyltransferase that facilitates transcription
by precluding histone tail modifications required for
heterochromatin formation. Genes Dev. 16:479–489. 2002. View Article : Google Scholar : PubMed/NCBI
|