Introduction
Stem cells are a major focus of modern scientific
research into the treatment of heart diseases, as their plasticity
may help overcome the limited self-repair capacity of
cardiomyocytes. Human bone marrow-derived mesenchymal stem cells
(hBMSCs) have been widely used in numerous studies and clinical
trials due to their unique properties, including ease of isolation,
expansion in vitro, multipotency and immunological tolerance
(1–4). Previous studies have revealed that
5-azacytidine (5-aza) induces MSC differentiation into myocardial
cells (5–7). It has also been demonstrated that
hBMSCs can differentiate into cardiac troponin T (cTnT)-expressing
cardiomyocytes (2,8–11);
however, the precise mechanisms controlling this process currently
remain unclear.
The mechanism of differentiation is complex and
requires various signaling pathways. Notch signaling, which is
important in cell fate specification during embryogenesis, has
previously been implicated in regulating differentiation of stem
cells in adults (12). Numerous
studies have demonstrated that Notch signaling regulates a wide
variety of processes during embryonic and post-natal development,
including proliferation, apoptosis and cell fate decisions
(13–15). However, the association between
Notch signalling and the differentiation of hBMSCs to
cardiomyocytes remains unclear. The aim of the present study was to
investigate the function of Jagged1-Notch1 signaling during hBMSCs
differentiation into cardiomyocytes. As a result of its widespread
expression, it was hypothesized that Notch signaling may be
involved in the differentiation of hBMSCs.
Materials and methods
Isolation and culture of hBMSCs
hBMSCs were isolated by bone marrow aspiration from
the sternums of 20 male patients (age, 23–48 years) with heart
valve diseases awaiting cardiac surgery, but were healthy with
regard to the circulatory system. Informed consent was obtained
from all patients prior to inclusion in the study. The study used a
protocol approved by the Research Ethics Committee of the First
Affiliated Hospital, Zhejiang University (Hangzhou, China). Bone
marrow mononuclear cells were purified by Ficoll-Paque
density-gradient centrifugation as previously described (16). The purified mononuclear cells were
allowed to adhere to culture flasks in Dulbecco's modified Eagle's
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher
Scientific, Inc.) overnight at 37°C in 5% CO2. Cells from passages
3–6 were used for subsequent experiments.
Cell viability assay
hBMSCs (~5,000 cells/well) were seeded into 96-well
plates. Following overnight incubation, the medium was removed and
Cell Counting Kit 8 (CCK8; Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan) solution was added to each well and incubated at
37°C for 1 h. The absorbance of the solution was measured
spectrophotometrically at 450 nm with a MRX II absorbance reader
(Dynex Technologies, Inc., Chantilly, VA, USA). The growth assay
was performed each day for 5 days consecutively.
Analysis of hBMSCs by flow
cytometry
Culture-expanded cells (passages 3–6) were washed
with phosphate-buffered saline (PBS) containing 0.5% (w/v) bovine
serum albumin (BSA), their concentration adjusted to
1×106 cells/100 µl, and phenotypic analyses performed
via flow cytometry. The hBMSCs were blocked with 1% BSA and
incubated with phycoerythrin- or fluorescein
isothiocyanate-conjugated mouse monoclonal antibodies against human
CD34 (catalog no. sc-19587; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA), CD54 (catalog no. ab27582; Abcam, Cambridge, UK), CD45
(catalog no. 560976; BD Biosciences, San Jose, CA, USA), CD44
(catalog no. 560977; BD Biosciences), CD29 (catalog no. sc-59829;
Santa Cruz Biotechnology, Inc.), human leukocyte antigen-antigen D
related (HLA-DR; catalog no. 560944; BD Biosciences) and CD90
(catalog no. 561969; BD Biosciences) for 60 min in the dark at 4°C,
at dilutions recommended by the manufacturers. Subsequently, cells
were washed with PBS and fixed with 2% paraformaldehyde.
Immunoglobulin isotype incubation was performed as a negative
control. Flow cytometry was performed with a FACSCalibur system
(FC500; Beckman Coulter, Inc., Brea, CA, USA) and analysed using
FlowJo software version 7.6.5 (FlowJo, LLC, Ashland, OR, USA).
Multilineage differentiation
assays
To induce osteogenic differentiation, hBMSCs were
cultured in a commercially available osteogenic differentiation
medium (catalog no. HUXMA-90021; Cyagen Biotechnology Co. Ltd.,
Taicang, China). On day 21, cells were stained with Alizarin Red in
accordance with the manufacturer's protocol. To induce adipogenic
differentiation, hBMSCs were cultured in a commercially available
adipogenic differentiation medium (catalog no. HUXMA-90031; Cyagen
Biotechnology Co. Ltd.). On day 21, cells were stained for 30 min
with Oil Red O, diluted 3:2 with distilled water and filtered. To
induce chondrogenic differentiation, hBMSCs were cultured in a
commercially available chondrogenic differentiation medium
purchased from Cyagen (catalog no. HUXMA-90041; Cyagen
Biotechnology Co. Ltd.). On day 28, cells were stained with 1 mg/ml
Alcian blue for 30 min. Cells were observed under a Nikon Eclipse
E200 light microscope (Nikon Corporation, Tokyo, Japan).
hBMSC5-aza-induced differentiation to
cardiomyocytes
Following the third passage, the cells were washed
twice with PBS, adjusted to a density of 2×104 cells/ml,
and seeded in a 6-well plate. Following 24 h incubation, 10 µmol/l
5-aza was added (for control group, 0 µmol/l 5-aza was added), and
cells were incubated for a further 24 h. The culture medium
containing 5-aza was then removed and complete culture medium was
added. The cells went through the same treatment process course
following each cell passage. The cells were maintained in culture
for 4 weeks following treatment. Samples were taken for detection
of morphological changes by transmission electron microscopy (TEM)
on day 28, for α-actin and cTnT expression by immunocytochemistry
and western blot analysis on day 28, and for analysis of mRNA
expression by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) for GATA binding protein-4 (GATA-4), and NK2
homeobox 5 (Nkx2.5) on day 28, and Jagged1 and Notch1 on days 1, 4
and 7.
TEM analysis
Cultured cells were rinsed with PBS and immersed in
2.5% glutaraldehyde for 4 h, rinsed in 0.1 mol/l sodium cacodylate
buffer (pH 7.3), and postfixed for 1 h in 1% OsO4. The cells were
embedded in Epon resin and cut into 60-nm thick sections with a
Sorvall MTB2 ultramicrotome (Thermo Fisher Scientific, Inc.).
Sections were stained for 20 min at room temperature with uranyl
acetate and lead citrate and examined under a Hitachi H-600
electron microscope (Hitachi, Ltd., Tokyo, Japan).
RNA isolation and RT-qPCR
Total RNA was extracted using TRIzol (Invitrogen;
Thermo Fisher Scientific, Inc.) following the manufacturer's
protocol and reverse transcribed into cDNA using the PrimeScript RT
reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). The
resulting cDNA was quantified by qPCR using SYBR Green (Takara
Biotechnology Co., Ltd.) and an ABI 7500 fast real-time PCR System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). An initial
denaturation step was performed at 95°C for 30 sec, followed by 40
cycles of denaturation at 95°C for 30 sec and annealing at 60°C for
30 sec. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was
used as an internal control. The mRNA and miRNA expression levels
were normalized to GAPDH mRNA. The quantification cycle (Cq) value
of mRNA was calculated using the 2−ΔΔCq method (17). The qPCR primers were provided by
Sangon Biotech Co., Ltd. (Shanghai, China) and the sequences are
presented in Table I.
 | Table I.Primers used for quantitative
polymerase chain reaction. |
Table I.
Primers used for quantitative
polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) |
|---|
| GAPDH | F
AAGGTGAAGGTCGGAGTCA |
|
| R
GGAAGATGGTGATGGGATTT |
| Jagged1 | F
AGCTATTTGCCGACAAGGCT |
|
| R
CACTGCCAGGGCTCATTACA |
| Notch-1 | F
AACGCCTACCTCTGCTTCTG |
|
| R
CTCACAGGCACACTCGTAGC |
| GATA-4 | F
GGAAGCCCAAGAACCTGAAT |
|
| R
TGCCCGTAGTGAGATGACAG |
| NKx2.5 | F
CTACCAGGCTCGGATACCAT |
|
| R
GCCAACAACAACTTCGTGAAC |
Protein extraction and western
blotting
The cells were lysed in cell lysis buffer (Sangon
Biotech Co., Ltd.). A bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.) was used to calculate the total
protein concentration in every lysate. Equivalent amounts of
protein samples (100 µg) were separated on 10% gels by sodium
dodecyl sulphate-polyacrylamide gel electrophoresis and transferred
to polyvinylidene difluoride membranes. Membranes were blocked in
10% skimmed milk for 1 h at room temperature and then incubated
overnight at 4°C with the following primary antibodies: Rabbit
anti-α-actin (catalog no. ab137346; Abcam) and rabbit anti-cTnT
(catalog no. ab45932; Abcam), used at dilutions recommended by the
manufacturer. Following washing, membranes were incubated with the
corresponding goat anti-rabbit secondary antibody (catalog no.
D111018; Sangon Biotech Co., Ltd.) for 1 h at a dilution
recommended by the manufacturer. Protein bands were visualized
using RapidStep™ Enhanced Chemiluminescence reagent (EMD Millipore,
Billerica, MA, USA).
Immunocytochemistry analysis
For immunocytochemical analyses, 1–5×106
cells/ml were seeded onto glass slides and allowed to adhere
overnight. The following day the cells were washed in PBS and fixed
for 10–15 min with 3% H2O2. Non-specific binding was prevented by
blocking with 5% normal goat serum (Sangon Biotech Co., Ltd.). The
cells were then incubated for 2–3 h at 37°C with the α-actin and
cTnT primary antibodies described previously, washed with PBS, and
incubated with the secondary antibody, biotinylated anti-rabbit IgG
(catalog no. D111053; Sangon Biotech Co., Ltd.), at a dilution
recommended by the manufacturer. Horseradish peroxidase was used as
detection reagent and finally diaminobenzidine was used to
visualize antibody binding using a Nikon Eclipse E200 light
microscope (Nikon Corporation).
Statistical analysis
All data are expressed as the mean ± standard
deviation of three independent experiments. Differences between
samples were analyzed by t-tests using SPSS version 17.0 software
(SPSS, Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Isolation and characterization of
hBMSCs in culture
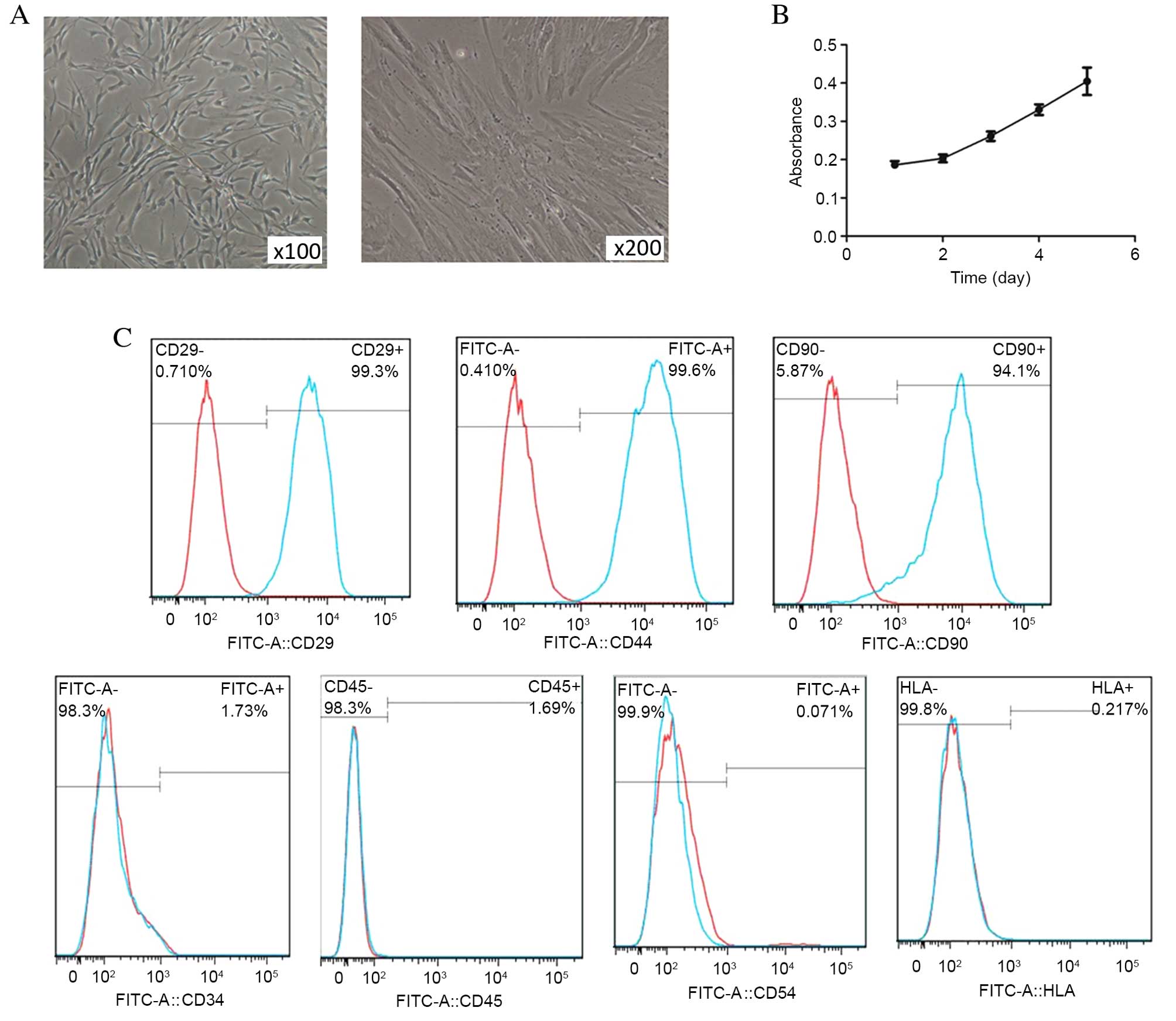
hBMSCs grew as adherent monolayers with a tendency
to grow in clusters, and had the morphological appearance of
spindle-shaped cells (Fig. 1A).
The proliferation ability of the cells was confirmed using CCK-8
(Fig. 1B). Expression of CD105,
CD73 and CD90, and lack of expression of CD45, CD34, CD14 or CD11b,
CD79a or CD19, and HLA-DR are used as markers to define and detect
MSCs. Additionally, MSCs typically differentiate to osteoblasts and
adipocytes in vitro (18).
Flow cytometry data revealed positive staining for CD29, CD44 and
CD90, and negative staining for CD34, CD45, CD54 and HLA-DR,
indicating that the isolated hBMSCs were of mesenchymal origin and
of high purity (Fig. 1C).
Confirmation of the differentiation
capacity of hBMSCs in vitro
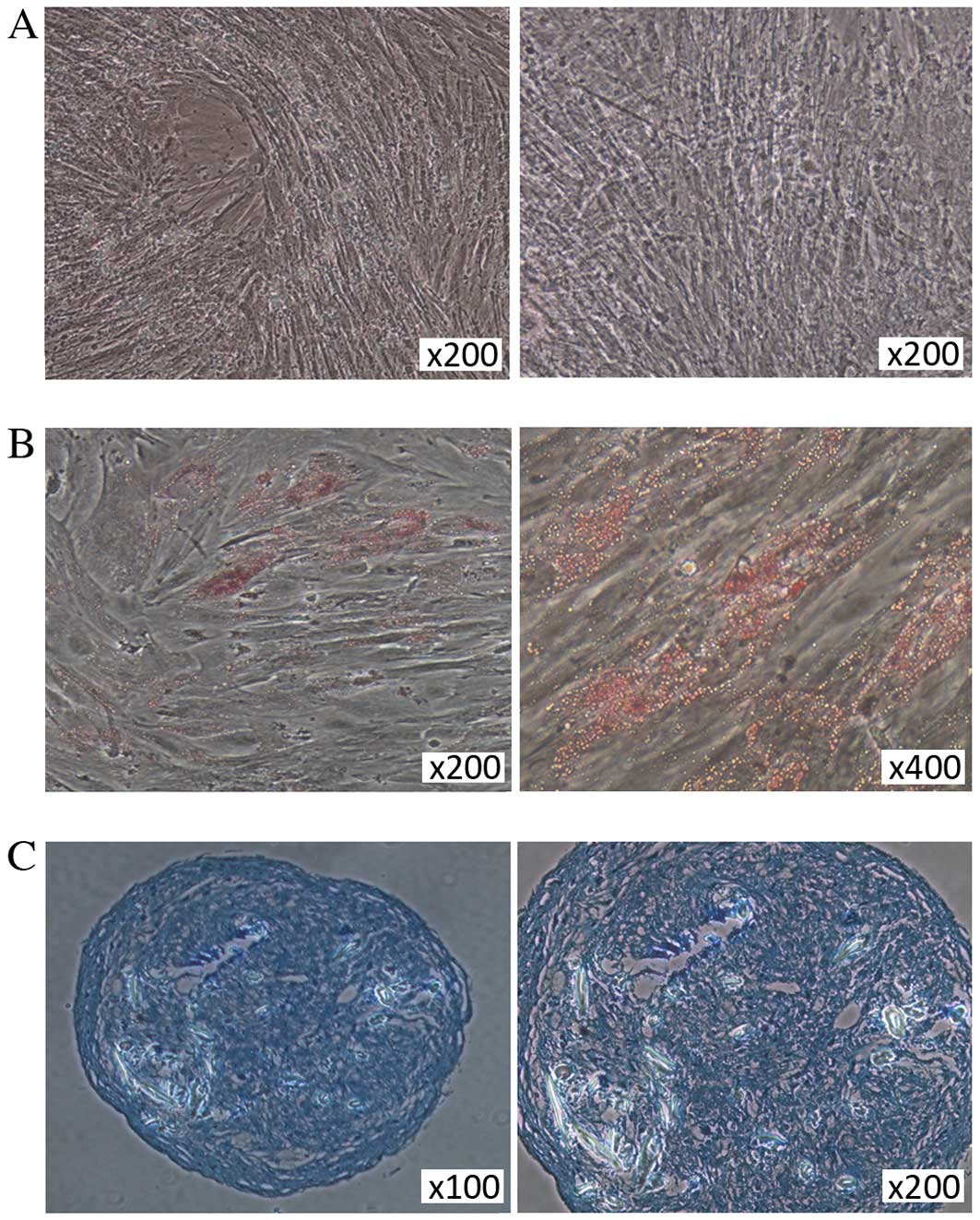
The differentiation capacity of hBMSCs into
mesodermal lineages (osteocytes, adipocytes and chondrocytes) was
assessed in cells cultured in commercially available
differentiation media. Alizarin Red staining demonstrated
mineralization during osteogenic differentiation in hBMSCs on day
21 (Fig. 2A). Adipogenic
differentiation of hBMSCs was characterized by Oil Red O staining,
with lipid droplets visible in the differentiated adipocytes on day
21 following the induction of differentiation (Fig. 2B). Positive Alcian blue staining of
sections from hBMSC pellets following culture in chondrogenic
medium demonstrated the chondrogenic differentiation capabilities
of the adherent cells (Fig.
2C).
Morphological changes of hBMSCs in
response to treatment with 5-aza
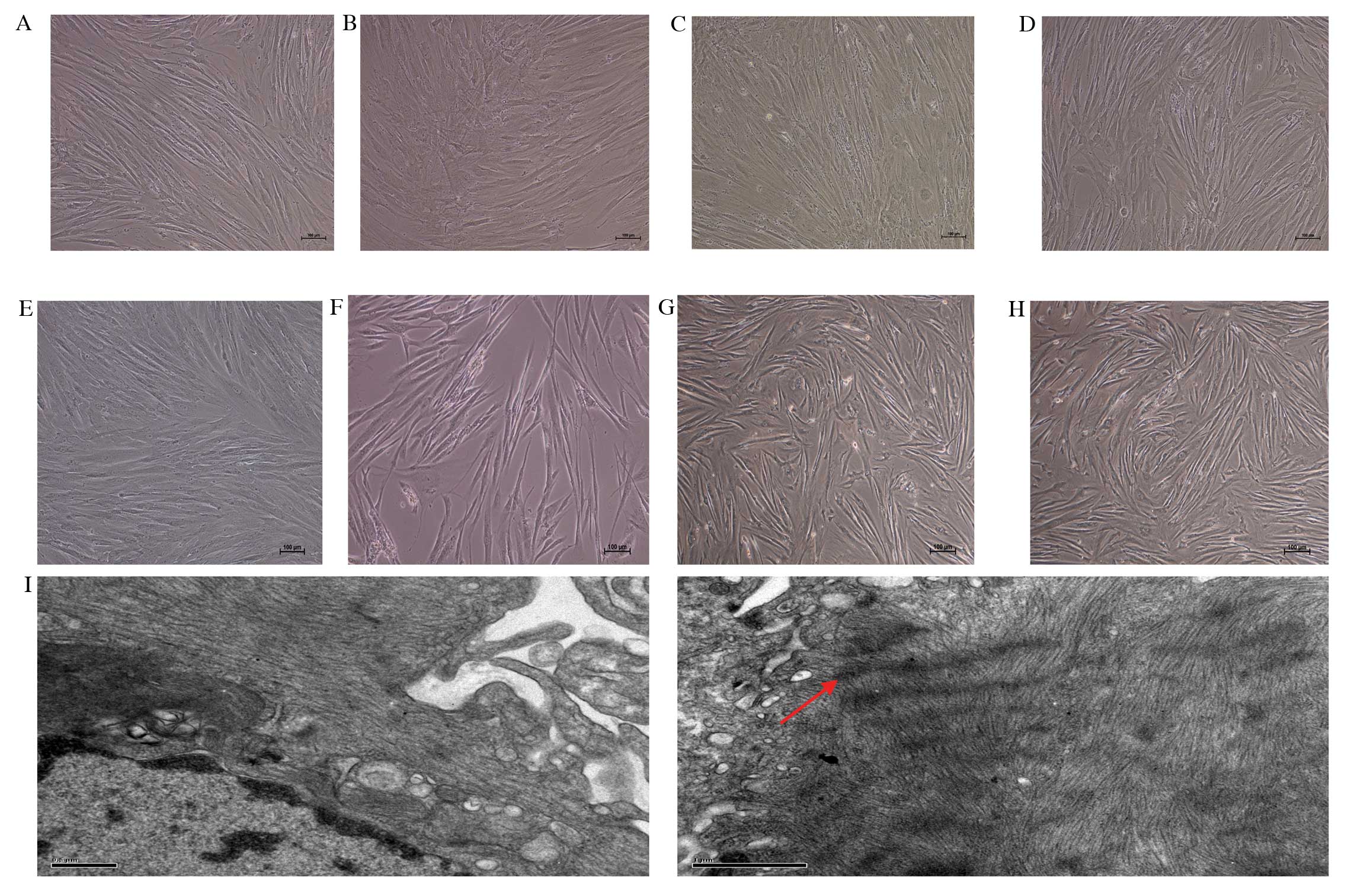
Phase contrast microscopy was used to determine the
morphological changes of 5-aza-treated hBMSCs following 7, 14, 21
and 28 days of treatment. Images of untreated cells on days 7, 14,
21 and 28 are presented in Fig.
3A-D. In the experimental group, certain adherent cells died,
whereas the surviving cells proliferated and differentiated.
Following 7 and 14 days of treatment, cell morphology did not
appear to change (Fig. 3E and F,
respectively), remaining comparable to the spindle-shaped
morphology of undifferentiated cells demonstrated in Fig. 1A. Following 21 and 28 days of
treatment, the appearance of spindle-shaped cells was reduced, with
cells developing a broadened and flattened shape (Fig. 3G and H, respectively). TEM on day
28 also revealed a cardiomyocyte-like ultrastructure of sarcomeres,
suggesting differentiation of hBMSCs into cardiomyocytes (Fig. 3I). No sarcomeres were observed in
undifferentiated cells.
Expression of α-actin and cTnT is
increased in 5-aza-treated cells
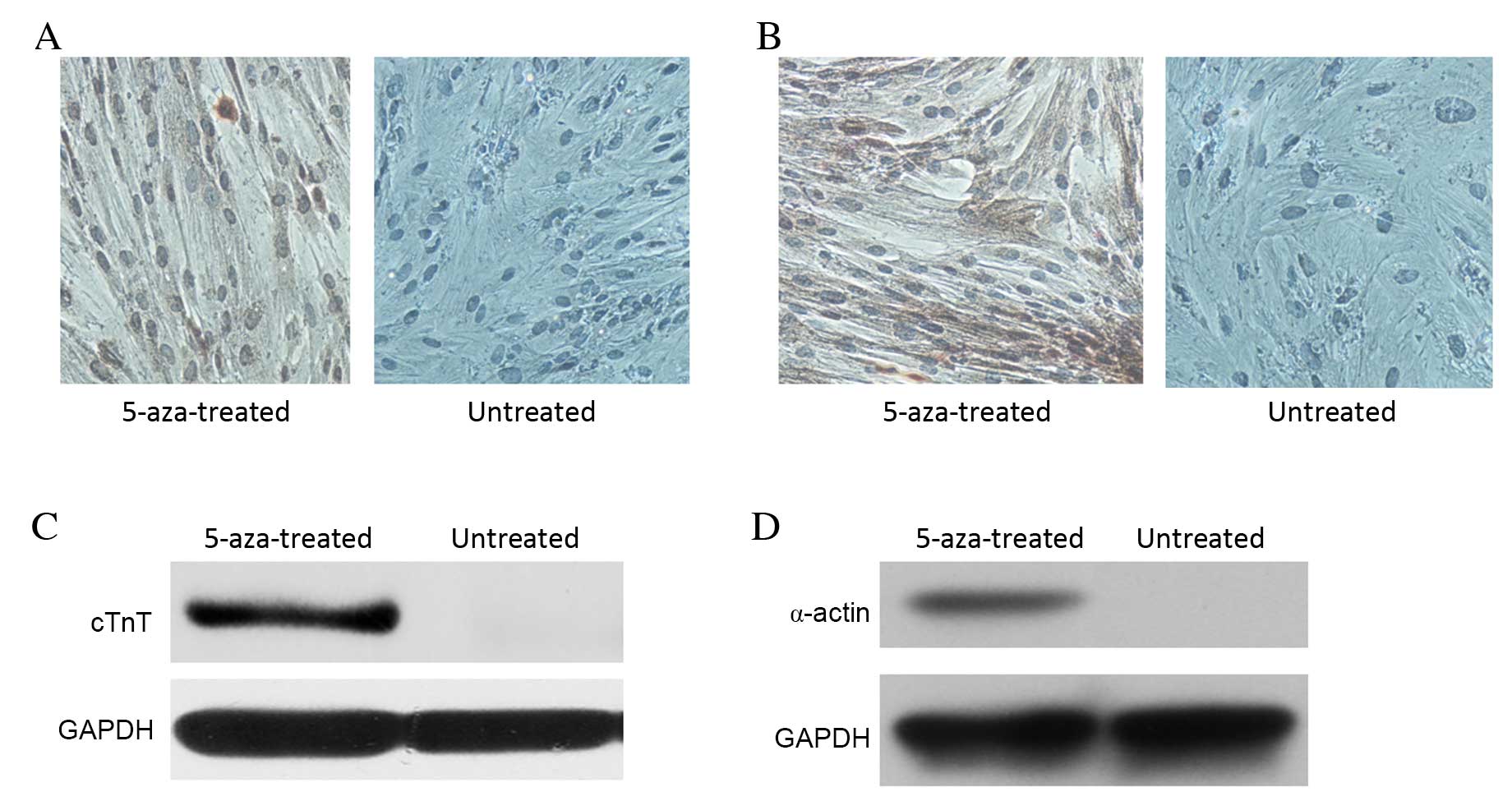
Immunocytochemistry demonstrated that 5-aza
treatment for 28 days induced cTnT and α-actin expression in
hBMSCs, whereas no cTnT or α-actin expression was observed in the
untreated control group (Fig. 4A and
B, respectively). Western blot analysis also demonstrated
increased protein expression levels of cTnT and α-actin in the
5-aza-treated cells, with no cTnT and α-actin detectable in the
untreated control group (Fig. 4C and
D, respectively). These results indicated that certain hBMSCs
in the 5-aza-treated group had undergone differentiation, giving
rise to the expression of molecular markers of cardiomyocytes.
RT-PCR analysis for expression of
transcription factors and Notch signals in hBMSCs
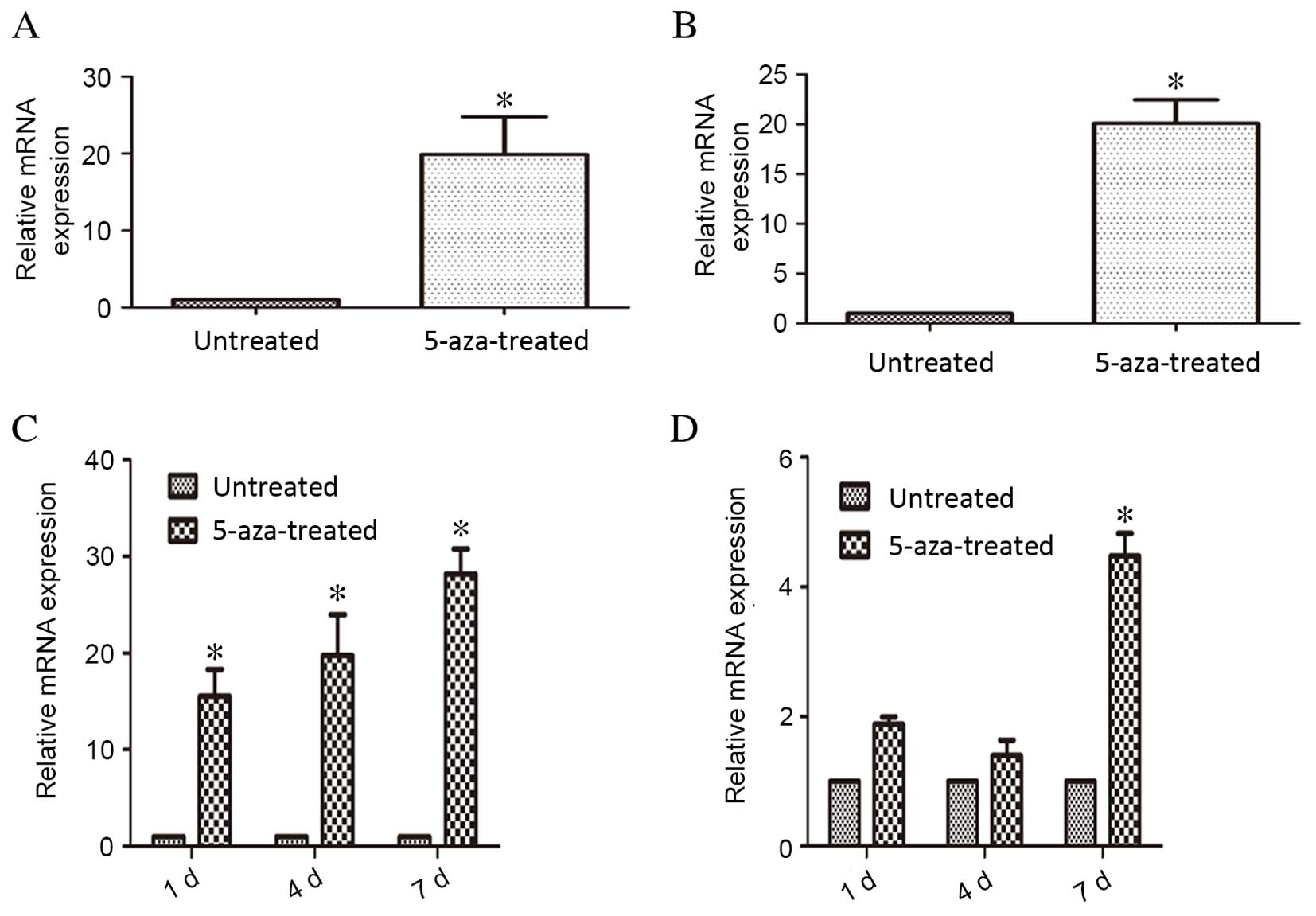
RT-qPCR analysis indicated that mRNA expression
levels of GATA-4 and Nkx2.5 were significantly higher in the
5-aza-treated compared with the untreated control group following
28 days of treatment (P=0.012 and P=0.018, respectively; Fig. 5A and B, respectively). During the
process of 5-aza-induced hBMSC differentiation, RT-qPCR analysis
revealed that Notch1 mRNA expression levels were significantly
upregulated compared with untreated control cells on days 1, 4 and
7 of treatment (P=0.021, P=0.016 and P=0.008, respectively;
Fig. 5C). Jagged1 mRNA expression
levels in cells treated with 5-aza did not differ significantly
compared with untreated control cells on days 1 and 4, but 5-aza
treatment did result in significant upregulation on day 7 compared
with untreated control cells (P=0.035; Fig. 5D). These results suggested that the
Notch1 signaling pathway and Jagged1 ligand contributed to the
differentiation process, during which GATA-4 and Nkx2.5 were
crucial.
Discussion
MSCs possess characteristics which make them a
promising source for cell therapy in numerous diseases (19), with BMSCs currently serving as the
primary source of harvested MSCs. The present study revealed that
hBMSCs have a doubling time of ~5 days, which may be due to samples
having been obtained from patients awaiting surgery. However, a
population of cells, isolated from bone marrow and expanded in
culture, is considered to represent an acceptable source of
undifferentiated MSCs for investigation of MSC differentiation
(20–22). The present study revealed that
hBMSCs have adipogenic, osteogenic and chondrogenic differentiation
potential in vitro, providing further evidence of the
multilineage differentiation potential of hBMSCs that has
previously been demonstrated (23,24).
In the present study, hBMSCs exhibited cardiomyocyte
characteristics when treated with 5-aza, as evidenced by increased
expression of α-actin and cTnT by immunocytochemistry and western
blotting. The ultramicroscopic details observed by TEM in the
5-aza-treated hBMSCs indicated myocardial differentiation.
In mammals, there are four Notch genes (Notch1-4)
and five Notch ligands, Jagged1 and 2, and Delta-like ligands 1, 3
and 4. Upon ligand binding to Notch at the cell membrane, the
cytoplasmic domain is cleaved and released from the cell-surface by
Presenilin/γ-secretase-dependent proteolysis. The cleaved
intracellular Notch protein is translocated to the nucleus, where
it converts the protein recombination signal binding protein for
immunoglobulin κJ region from a transcriptional repressor to a
transcriptional activator (14,25,26).
Notch signalling is part of an evolutionarily
ancient mechanism of cell interaction, with previous studies having
demonstrated Notch signaling at all stages of development,
influencing differentiation, proliferation, and apoptotic events
(27–29). Notch family proteins have been
demonstrated to be expressed in a wide range of mammalian cells and
tissues, and are particularly involved in the formation and
development of the heart (3,30–32).
The present study also examined mRNA expression
levels of important genes of the Notch pathway during the process
of differentiation, and demonstrated that mRNA expression levels of
Notch1, but not Jagged1, increased significantly on days 1, 4 and 7
of 5-aza treatment, compared with untreated cells. The mRNA
expression levels of GATA-4 and Nkx2.5 were also increased
significantly on day 28 in the 5-aza-treated cells compared with
untreated cells. However, the lack of significant difference in
Jagged1 mRNA expression between the 5-aza-treated cells and
untreated control cells on days 1 and 4, despite the significant
difference on day 7, indicated that Jagged1 may be not the
predominant ligand involved in the Notch signalling pathway during
the 5-aza-induceddifferentiation of hBMSCs into cardiomyocytes.
However, the upregulation of Notch1 mRNA expression levels and the
associated transcription factors, GATA-4 and Nkx2.5, suggest that
Notch1 signaling is involved in the differentiation of hBMSCs into
cardiomyocytes. Further studies are required to evaluate the degree
to whichNotch1 promotes this process.
Certain previous studies have demonstrated that the
expression of Nkx2.5 and GATA-4, both of which are involved in the
development of the heart (33), is
enhanced during 5-aza-induced differentiation of hBMSCs into
myocardial cells. This is consistent with the results of the
present study.
In conclusion, the results of the present study
demonstrated that Notch1, GATA-4 and Nkx2.5 are upregulated during
the differentiation process of hBMSCs induced by 5-aza. hBMSC
differentiation may be partially regulated through the
transcription factors, GATA-4 and Nkx2.5. As an important signaling
pathway, Notch1 may exert a regulatory effect on the
differentiation process. By identifying the functions of Notch1, it
may be possible to influence the Notch1 signaling pathway to
regulate hBMSC differentiation induced by 5-aza.
Acknowledgements
We thank all members of State Key Laboratory for
Diagnosis and Treatment of Infectious Diseases for providing us
with laboratory apparatus and technical support. This work was
supported by National Natural Science Funding of China (grant no.
81070264).
References
|
1
|
Kopen GC, Prockop DJ and Phinney DG:
Marrow stromal cells migrate throughout forebrain, and cerebellum
and they differentiate into astrocytes after injection into
neonatal mouse brains. Proc Natl Acad Sci USA. 96:10711–10716.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Saito T, Dennis JE, Lennon DP, Young RG
and Caplan AI: Myogenic Expression of mesenchymal stem cells within
myotubes of mdx mice in vitro and in vivo. Tissue Eng. 1:327–343.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Williams AR and Hare JM: Mesenchymal stem
cells: Biology, pathophysiology, translational findings, and
therapeutic implications for cardiac disease. Circ Res.
109:923–940. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang S, Huang S, Feng C and Fu X:
Umbilical cord-derived mesenchymal stem cells: Strategies,
challenges, and potential for cutaneous regeneration. Front Med.
6:41–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu CX, Zhang EY, Yang SY, Ma JY, An Q and
Shi YK: The expression of GATA-4 and Nkx2.5 gene in the
transformation of Rattus mesenchymal stem cells into
cardiomyocytes. Sichuan Da Xue Xue Bao Yi Xue Ban. 39:882–885.
2008.(In Chinese). PubMed/NCBI
|
|
7
|
Cho J, Rameshwar P and Sadoshima J:
Distinct roles of glycogen synthase kinase (GSK)-3alpha and
GSK-3beta in mediating cardiomyocyte differentiation in murine bone
marrow-derived mesenchymal stem cells. J Biol Chem.
284:36647–36658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rangappa S, Entwistle JW, Wechsler AS and
Kresh JY: Cardiomyocyte-mediated contact programs human mesenchymal
stem cells to express cardiogenic phenotype. J Thorac Cardiovasc
Surg. 126:124–132. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liechty KW, MacKenzie TC, Shaaban AF, Radu
A, Moseley AM, Deans R, Marshak DR and Flake AW: Human mesenchymal
stem cells engraft and demonstrate site-specific differentiation
after in utero transplantation in sheep. Nat Med. 6:1282–1286.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Orlic D, Kajstura J, Chimenti S, Limana F,
Jakoniuk I, Quaini F, Nadal-Ginard B, Bodine DM, Leri A and Anversa
P: Mobilized bone marrow cells repair the infarcted heart,
improving function and survival. Proc Natl Acad Sci USA.
98:10344–10349. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kudo M, Wang Y, Wani MA, Xu M, Ayub A and
Ashraf M: Implantation of bone marrow stem cells reduces the
infarction and fibrosis in ischemic mouse heart. J Mol Cell
Cardiol. 35:1113–1119. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yuan TM and Yu HM: Notch signaling: Key
role in intrauterine infection/inflammation, embryonic development,
and white matter damage? J Neurosci Res. 88:461–468.
2010.PubMed/NCBI
|
|
13
|
Reddy BV, Rauskolb C and Irvine KD:
Influence of fat-hippo and notch signaling on the proliferation and
differentiation of Drosophila optic neuroepithelia. Development.
137:2397–2408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weinmaster G: The ins and outs of notch
signaling. Mol Cell Neurosci. 9:91–102. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gridley T: Notch signaling in vertebrate
development and disease. Mol Cell Neurosci. 9:103–108. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li J, Tao R, Wu W, Cao H, Xin J, Li J, Guo
J, Jiang L, Gao C, Demetriou AA, et al: 3D PLGA scaffolds improve
differentiation and function of bone marrow mesenchymal stem
cell-derived hepatocytes. Stem Cells Dev. 19:1427–1436. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Horwitz EM, Le Blanc K, Dominici M,
Mueller I, Slaper-Cortenbach I, Marini FC, Deans RJ, Krause DS and
Keating A: International Society for Cellular Therapy:
Clarification of the nomenclature for MSC: The international
society for cellular therapy position statement. Cytotherapy.
7:393–395. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Si YL, Zhao YL, Hao HJ, Fu XB and Han WD:
MSCs: Biological characteristics, clinical applications and their
outstanding concerns. Ageing Res Rev. 10:93–103. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tosh D and Slack JM: How cells change
their phenotype. Nat Rev Mol Cell Biol. 3:187–194. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Badorff C, Brandes RP, Popp R, Rupp S,
Urbich C, Aicher A, Fleming I, Busse R, Zeiher AM and Dimmeler S:
Transdifferentiation of blood-derived human adult endothelial
progenitor cells into functionally active cardiomyocytes.
Circulation. 107:1024–1032. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Spees JL, Olson SD, Ylostalo J, Lynch PJ,
Smith J, Perry A, Peister A, Wang MY and Prockop DJ:
Differentiation, cell fusion, and nuclear fusion during ex vivo
repair of epithelium by human adult stem cells from bone marrow
stroma. Proc Natl Acad Sci USA. 100:2397–2402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou M, Yang KM, Zhang H, Zhu WQ, Duan FJ,
Wang H, Song YH, Wei YJ and Hu SS: Transplantation of mesenchymal
stem cells from human bone marrow improves damaged heart function
in rats. Int J Cardiol. 115:220–228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schilling T, Nöth U, Klein-Hitpass L,
Jakob F and Schütze N: Plasticity in adipogenesis and osteogenesis
of human mesenchymal stem cells. Mol Cell Endocrinol. 271:1–17.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ward EJ, Shcherbata HR, Reynolds SH,
Fischer KA, Hatfield SD and Ruohola-Baker H: Stem cells signal to
the niche through the Notch pathway in the Drosophila ovary. Curr
Biol. 16:2352–2358. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weinmaster G: Notch signal transduction: A
real rip and more. Curr Opin Genet Dev. 10:363–369. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brenner M: To be or notch to be. Nat Med.
6:1210–1211. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Artavanis-Tsakonas S, Rand MD and Lake RJ:
Notch signaling: Cell fate control and signal integration in
development. Science. 284:770–776. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kojika S and Griffin JD: Notch receptors
and hematopoiesis. Exp Hematol. 29:1041–1052. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Artavanis-Tsakonas S, Matsuno K and
Fortini ME: Notch signaling. Science. 268:225–232. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li L, Milner LA, Deng Y, Iwata M, Banta A,
Graf L, Marcovina S, Friedman C, Trask BJ, Hood L and Torok-Storb
B: The human homolog of rat Jagged1 expressed by marrow stroma
inhibits differentiation of 32D cells through interaction with
Notch1. Immunity. 8:43–55. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Perdigoto CN and Bardin AJ: Sending the
right signal: Notch and stem cells. Biochim Biophys Acta.
1830:2307–2322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Armiñán A, Gandía C, Bartual M,
García-Verdugo JM, Lledó E, Mirabet V, Llop M, Barea J, Montero JA
and Sepúlveda P: Cardiac differentiation is driven by NKX2.5 and
GATA4 nuclear translocation in tissue-specific mesenchymal stem
cells. Stem Cells Dev. 18:907–918. 2009. View Article : Google Scholar : PubMed/NCBI
|