Introduction
Clear cell renal cell carcinoma (ccRCC), a major
pathological form of kidney cancer, accounts for ~90% of kidney
malignancies (1). There are
>200,000 cases diagnosed and >100,000 deaths from kidney
cancers annually. Approximately one third of patients display
metastasis at diagnosis and thus a poor prognosis, with a 5-year
survival rate of <20% for patients with metastasized ccRCC
(2). Therefore, understanding the
mechanisms of invasion and metastasis in ccRCC is of great
importance to public health.
Metastasis is a major cause of mortality in multiple
cancers, including ccRCC (3–5). The
first stage of tumor metastasis is the detachment of tumor cells
from the basement membrane and extracellular matrix (ECM).
Normally, the growth of tumor cells is reliant on adherence with
the basement membrane and ECM. When the basement membrane and ECM
are damaged or degraded, tumor cells detach and enter the
circulatory system. Most cells entering the circulatory system face
anoikis, a special form of programmed cell death induced by
disengagement from the surrounding ECM or adjacent cells and the
resultant loss of normal cell-matrix interactions. Anoikis is an
important contributor to development, disease and tumor metastasis
(6,7). Resistance to anoikis is a critical
characteristic of metastatic tumor cells, but the mechanisms
underlying this remain largely unknown, particularly in ccRCC.
Tim-3 is an important member of the T cell
immunoglobulin and mucin domain-containing molecule family
(8). It has been reported to
impact multiple diseases, especially human tumor biology: Tim-3 was
reported to participate in colon cancer tumorigenesis and its
expression was hypothesized to be an independent prognostic factor
for patients with colorectal cancer (9). Tim-3 also affected the development
and progression of prostate cancer (10). Increasing evidence suggests that
Tim-3 is involved in maintaining the malignant phenotype of ccRCC
progression, but the mechanisms behind this require further
investigation (11). Previous
studies have demonstrated that anoikis impairs RCC (12), but to the best of our knowledge, no
study has yet examined the effect of Tim-3 on anoikis sensitivity
in ccRCC.
The present study investigated the involvement of
Tim-3 in anoikis and its influence on the invasion of the ccRCC
cell lines, 786-O and Caki-2. Detachment from the ECM was induced
by polyhydroxylethylmethacrylate (poly-HEMA) treatment, and anoikis
was measured by morphological observation, flow cytometry and a
CytoSelect™ 24-well Anoikis Assay kit in the presence and absence
of Tim-3 small interfering RNA (siRNA). Interference with Tim-3
expression increased ECM detachment-induced anoikis and reduced the
invasive ability of ccRCC cells. Therefore, interference with Tim-3
expression may attenuate ccRCC invasion, and therefore metastasis,
through increasing sensitivity to anoikis.
Materials and methods
Materials
Caki-2 and 786-O human ccRCC cell lines were
purchased from the American Tissue Type Collection (Manassas, VA,
USA). RPMI-1640 medium and fetal bovine serum (FBS) were obtained
from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA).
Anti-Tim-3 (cat. no. sc-30326), E-cadherin (cat. no. sc-71007),
N-cadherin (cat. no. sc-59987) and β-actin (cat. no. sc-47778)
antibodies were purchased from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA). Tim-3 specific siRNA (cat. no. sc-72034) and a
negative control siRNA (cat. no. sc-108060) were purchased from
Santa Cruz Biotechnology, Inc. The cell invasion assay kit used was
from Chemicon (Merck Millipore) and the CytoSelectTM 24-well
Anoikis Assay kit (cat. no. CBA-080) was from Cell Biolabs, Inc.
(San Diego, CA, USA). The kit provides an MTT colorimetric system
to detect the growth and viability of cells detached from
extracellular matrix. Cells in each group were seeded (1×106/well)
in the 24-well cell culture plate pre-coated with poly-HEMA
solution (0.5 ml/well), and each group was treated as described
above. MTT Reagent (50 µl) was added into each well, and incubated
for 2–4 h at 37°C. Then, detergent solution (500 µl) was added to
each well, and was mixed gently by pipetting. The wells were
incubated for another 2–4 h in the dark at room temperature. A
total of 200 µl of the incubated solution was added to a 96-well
plate, and the absorbance was measured at 570 nm using an ELISA
reader (Multiskan GO, Thermo Fisher Scientific, Inc.). The
absorbance value reflected the relative viability of each well
under anoikis.
Cell culture
Caki-2 and 786-O cells were cultured in RPMI-1640
medium supplemented with 10% FBS, 100 U/ml penicillin and 100 mg
streptomycin at 37°C in a humidified atmosphere with 5%
CO2. Passage digestion was conducted by 0.25%
trypsin.
Preparation of poly-HEMA-coated
plate
Poly-HEMA powder was dissolved in 95% ethanol in a
65°C water bath. The poly-HEMA solution (1 ml, 52.2 mg/ml) was then
sterilized under a 0.22 µm membrane and added to each culture well
(12-well/plate), and incubated overnight at room temperature. The
plates were washed 2–3 times with sterile double distilled
H2O prior to use.
Interference of Tim-3 expression with
Tim-3 siRNA
As described previously, the target sequences used
for Tim-3 interference were Tim-3 siRNA #1 (SASI_Hs01_00114252)
(8). 786-O and Caki-2 cells were
cultured to 70% confluence. Cells were transfected with 100 pmol
siRNA using Lipofectamine RNAimax (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. Total protein was
extracted 48 h post-transfection and western blots were performed
as described below to analyze protein levels of target genes.
Cell groups
Cells were divided into three groups: Control group
(786-O or Caki-2 cells seeded into normal wells (1×105/well) in the
absence of poly-HEMA pre-coating), the poly-HEMA treated group
(786-O cells or Caki-2 cells cultured in poly-HEMA pre-coated
wells, 1×105/well) and the poly-HEMA + siRNA combined group (786-O
cells or Caki-2 cells (1×105/well) cultured in the poly-HEMA
pre-coated wells and transfected with Tim-3 siRNA for 24 h. The
negative control group (786-O cells or Caki-2 cells cultured in the
poly-HEMA pre-coated wells (1×105/well) were transfected with
negative control (NC) siRNA for 24 h).
Morphological observation
Hoechst 33342 (Ho33342)/propidium iodide (PI) double
staining and acridine orange staining were conducted to observe
morphological changes in anoikitic cells. Prior to staining cells
were washed twice with PBS, fixed with 10% methanol for 15 min, and
stained by Ho33342 (10 µg/ml) and PI (50 µg/ml) at 37°C for 30 min,
or acridine orange (1 mg/ml) at room temperature for 15 min.
Morphological changes were examined by fluorescence microscope at
×200 magnification. In Ho3342/PI staining, cells displaying blue
nuclear fragmentation were interpreted as apoptotic cells, while
cells with red were defined as necrotic. In acridine orange
staining, bright green fluorescence and orange-red fluorescence is
seen in DNA and RNA, respectively, based on the membrane
permeability of the cells.
Flow cytometry
786-O and Caki-2 cells were seeded and treated as
described above (1×105/well). Prior to detection, 786-O and Caki-2
cells were washed with PBS without fixation and resuspended in 200
µl binding buffer containing Annexin V FITC (5 µl) and PI (10 µl)
for 15 min at room temperature. Further binding buffer (300 µl) was
added prior to analysis with a FACScan flow cytometer (BD
Biosciencies, Franklin Lakes, NJ, USA). The FlowJo software
(version, 7.6; FlowJo, LLC, Stanford, CA, USA) was used for further
data analysis. Flow cytometry analysis was used for cell apoptosis
determination.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA extraction and RT-qPCR were performed as
described previously (8). β-actin
was used as a normalization control. All experiments were performed
in triplicate. The primers used were as follows: Tim-3, forward
5′-GCTACTACTTACAAGGTCCTCAG-3′ and reverse,
5′-ATTCACATCCCTTTCATCAGTC-3′; β-actin, forward
5′-TGGCACCCAGCACAATGAA-3 and reverse
5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′.
Western blot analysis
Cells were harvested 48–72 h following transfection
in each group. Cells were lysed under radioimmunoprecipitation
lysis buffer (Beyotime Institute of Biotechnology, Haimen, China)
on ice for 30–40 min, vortexing every 5–10 min. Then, cells were
centrifuged at 4°C for 15 min (13,225 × g). The supernatant was the
protein extraction. Protein concentration was determined using a
bicinchoninic acid assay. Subsequently, 40 µg protein was run on
10% SDS-PAGE gel and transferred to polyvinylidene fluoride (PVDF)
membranes. PVDF membranes were blocked with 10% skim milk in TBST
for 1 h at room temperature and incubated overnight at 4°C with
primary antibodies (all purchased from Santa Cruz Biotechnology,
Inc.), including Tim-3 (1:1,000, cat. no. sc-30326), E-cadherin
(1:200, cat. no. sc-71007), N-cadherin (1:200, cat. no. sc-59987),
and β-actin (1:500, cat. no. sc-47778). Secondary antibodies goat
anti-mouse IgG conjugated to horseradish peroxidase (HRP); cat. no.
sc-2055) and goat anti-rabbit IgG-HRP (cat. no. sc-2004) were added
for further incubation with the membrane of 1 h at room temperature
(1:5,000) and Western Blot Luminal Reagent (CWbio Co., Ltd.,
Beijing, China) were used to visualize the protein bands, and
protein levels were quantified by densitometric analysis using
Image J software (version, 1.42; National Institutes of Health,
Bethesda, MA, USA). All experiments were repeated at least three
times.
Detections of invasive ability
Invasive abilities were detected based on the
Chemicon cell invasion assay kit (8 µm pore size) as described by
Li et al (13). Serum-free
medium (300 µl) was added onto the surface of the insert and
incubated for 2 h to rehydrate. Medium containing 10% FBS (500 µl)
was added to each well beneath the insert as the chemoattractant.
The upper wells were seeded with 100 µl 786-O or Caki-2 cell
suspension, containing ~50,000 cells. Following 24 h incubation at
37°C, cells were fixed with 2% paraformaldehyde at room temperature
for 15 min and stained cells in lower chamber with 0.25% crystal
violet at room temperature for 10 min. The stained cells were
dissolved using 10% acetic acid at room temperature for 10 min, and
the absorbance of the mixture at 560 nm was assayed under a
microplate reader. All experiments were performed in
triplicate.
Statistical analysis
Data are presented as the mean ± standard deviation
and were analyzed using the statistical package SPSS 17.0 (SPSS,
Inc., Chicago, IL, USA). The significance of differences between
groups was determined using two-tailed Student's t-tests. P<0.05
was considered to indicate a statistically significant
difference.
Results
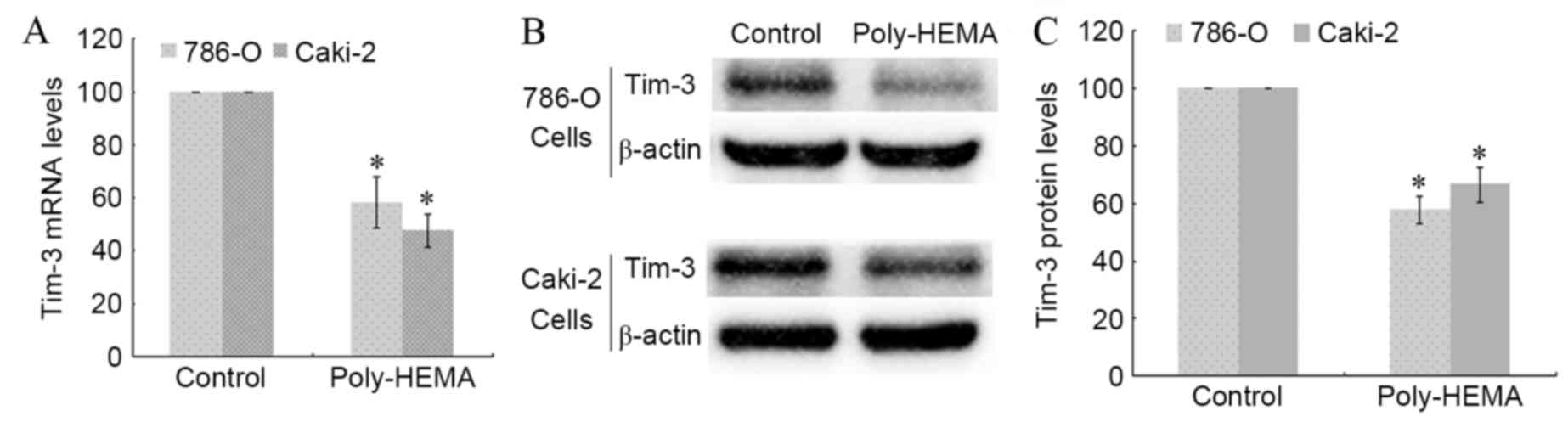
Detachment from the ECM decreases
Tim-3 mRNA and protein expression levels
786-O and Caki-2 cells were seeded in poly-HEMA
pre-coated wells to induce ECM detachment. Total RNA and proteins
were extracted from control and poly-HEMA-treated cells, and the
transcription and expression of Tim-3 were analyzed using RT-qPCR
and western blotting. The Tim-3 transcription level decreased
significantly in poly-HEMA treated 786-O and Caki-2 cells compared
with control cells (P=0.038 and P=0.024, respectively; Fig. 1A). Detachment from the ECM also
significantly decreased Tim-3 protein expression levels in 786-O
and Caki-2 cells compared with control cells (P=0.016 and P=0.035,
respectively; Fig. 1B and C). This
indicates that expression of Tim-3 in ccRCC cells may be associated
with anoikis.
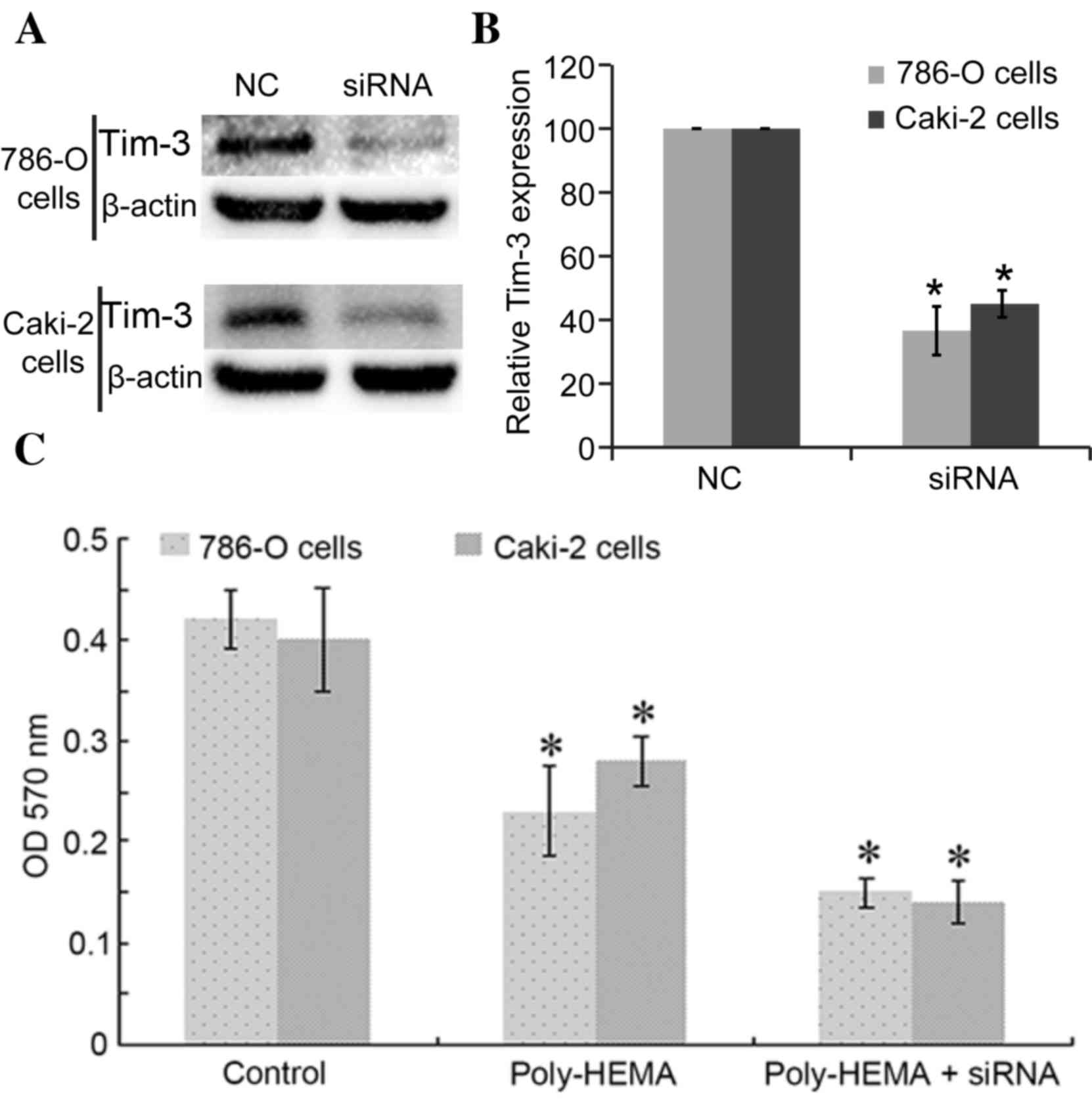
Interference with Tim-3 expression
reduced the cell viabilities of ccRCC cells
Tim-3 expression was blocked with siRNAs, with Tim-3
siRNA successfully inhibiting the expression of Tim-3 in 786-O
cells (36.58±7.65%, P=0.009 vs. control) and in Caki-2 cells
(45.04±4.23%, P=0.024) (Fig. 2A and
B). Cell ability was determined using the CytoSelect™ 24-well
Anoikis Assay kit. Detachment from the ECM (poly-HEMA group cells)
significantly reduced the activities of 786-O cells and Caki-2
cells compared with control cells, with OD570 nm values as follows:
Control 786-O cells, 0.42±0.03, poly-HEMA treated 786-O cells,
0.23±0.044 (P=0.037); control Caki-2 cells, 0.4±0.051, poly-HEMA
treated Caki-2 cells, 0.28±0.025 (P=0.018; Fig. 2C). Knockdown of Tim-3 expression
reduced activities further, with recorded OD570
nm values as follows: Poly-HEMA-treated + siRNA 786-O
cells, 0.15±0.015 P=0.021 vs. poly-HEMA treated cells; poly-HEMA
treated + siRNA Caki-2 cells, 0.14±0.021; P=0.038 vs. poly-HEMA
treated cells (Fig. 2C). Cells
under anoikis show reduced activities, so the lower the measured OD
value, the higher anoikis of cells occurred.
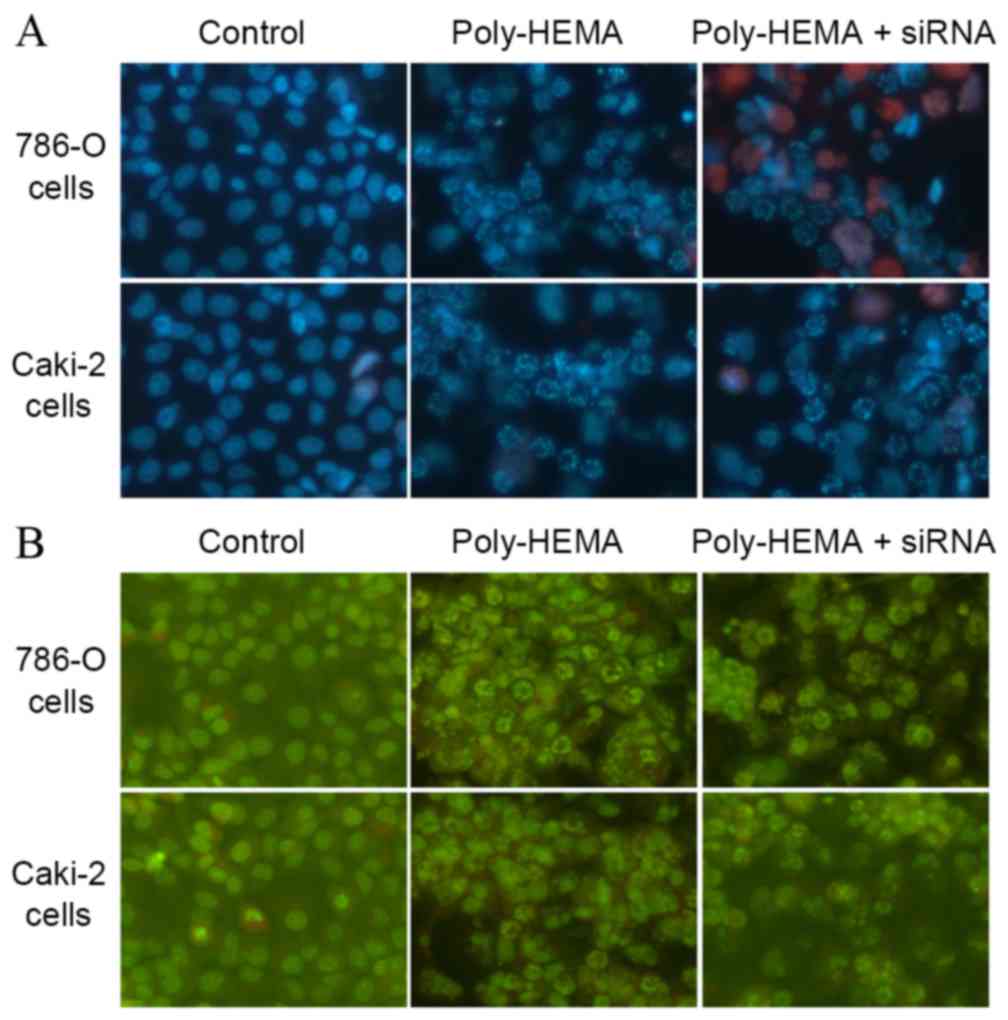
Tim-3 downregulation causes
morphological changes in ccRCC cells
Following Ho33342/PI double staining, control cells
appeared blue with intact nuclei, while poly-HEMA treated cells
demonstrated suspension and aggregation, with non-intact blue
nuclei. Cells transfected with Tim-3 siRNA displayed more damaged
blue nuclei or PI-stained red nuclei (Fig. 3A). In acridine orange staining, the
nuclei of control cells appeared intact, and poly-HEMA treated
cells displayed damaged nuclei (Fig.
3B). This was exacerbated by Tim-3 siRNA transfection (Fig. 3B). Thus, detachment from the ECM
induces anoikis, and knockdown of Tim-3 expression increased
anoikis further.
Knockdown of Tim-3 expression
significantly increased apoptosis
For 786-O cells, apoptosis rate was increased in the
poly-HEMA treatment group (5.37±0.67%) and the poly-HEMA treatment
+ siRNA group (9.83±0.93%) compared with the control group
(1.97±0.21%) (P-=0.041 and P-=0.015, respectively). In addition, a
significant difference was identified between the poly-HEMA
treatment group and the poly-HEMA treatment + siRNA group
(P=0.025). For Caki-2 cells, apoptosis rates were also increased in
the poly-HEMA treatment group (6.07±0.7%) and the poly-HEMA
treatment + siRNA group (10.17±1.19%) compared with the control
group (2.30±0.2%; P=0.001 and P=0.003, respectively). A significant
difference was identified between the poly-HEMA treatment group and
the poly-HEMA treatment + siRNA group (P=0.014; Fig. 4).
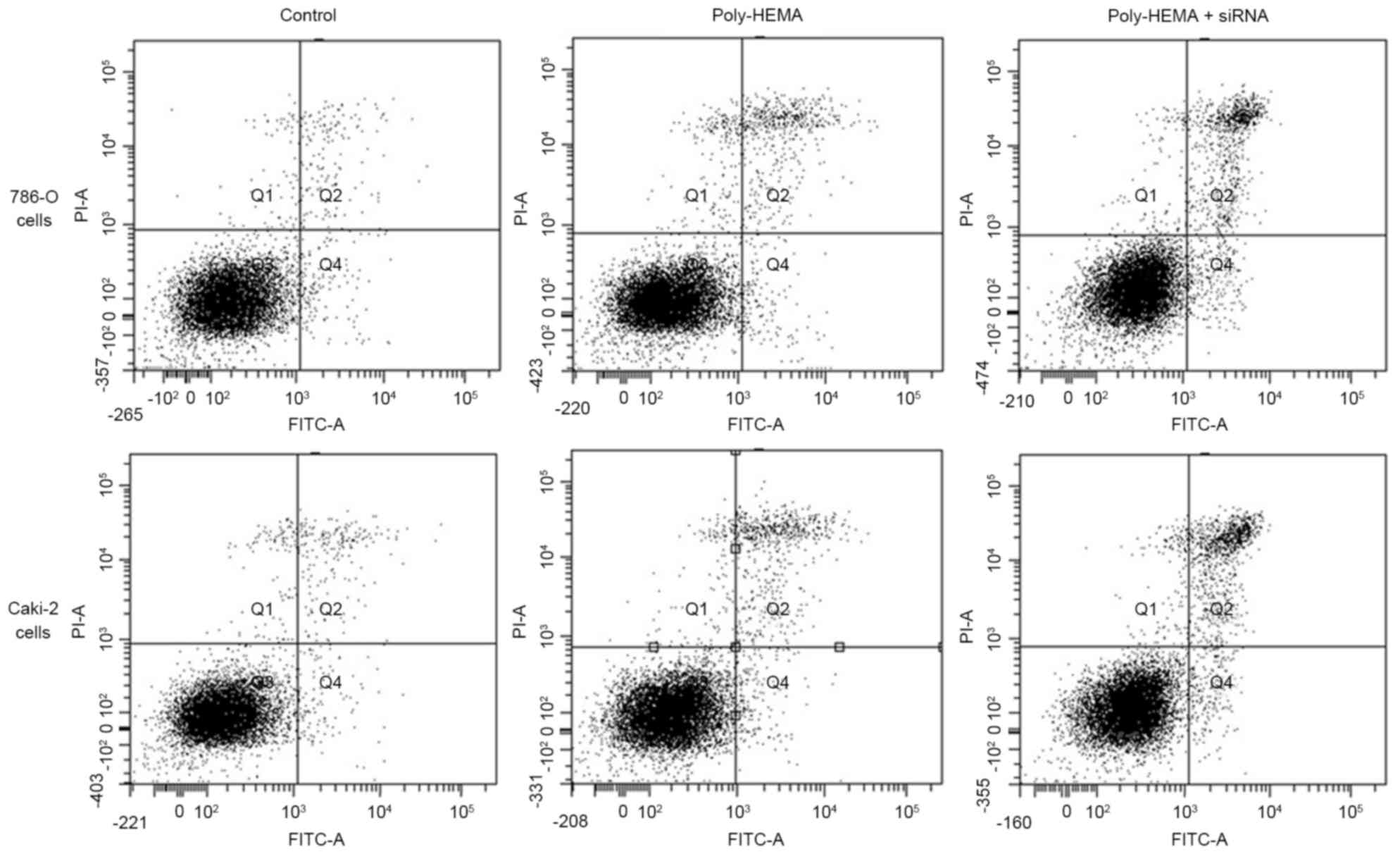 | Figure 4.Flow cytometry analyses of 786-O and
Caki-2 cell apoptosis. A total of 30,000 cells were collected in
each group for flow cytometry analyses. The number of cells in Q4
and Q2 quadrant reflects early and late apoptotic cells,
respectively. Both the Q2 and Q4 cells were counted as apoptosis
here. The number of apoptotic 786-O cells in control, Poly-HEMA,
and Poly-HEMA + siRNA groups were 591±63, 1,611±201 and 2,949±279,
respectively. Meanwhile, the number of apoptotic Caki-2 cells in
control, Poly-HEMA, and Poly-HEMA + siRNA groups were 690±60,
1,821±210 and 3,051±357, respectively. Interference with Tim-3
expression using a small interfering RNA significantly increased
anoikis in 786-O (P=0.015) and Caki-2 (P=0.003). Poly-HEMA,
polyhydroxylethylmethacrylate; siRNA, small interfering RNA. |
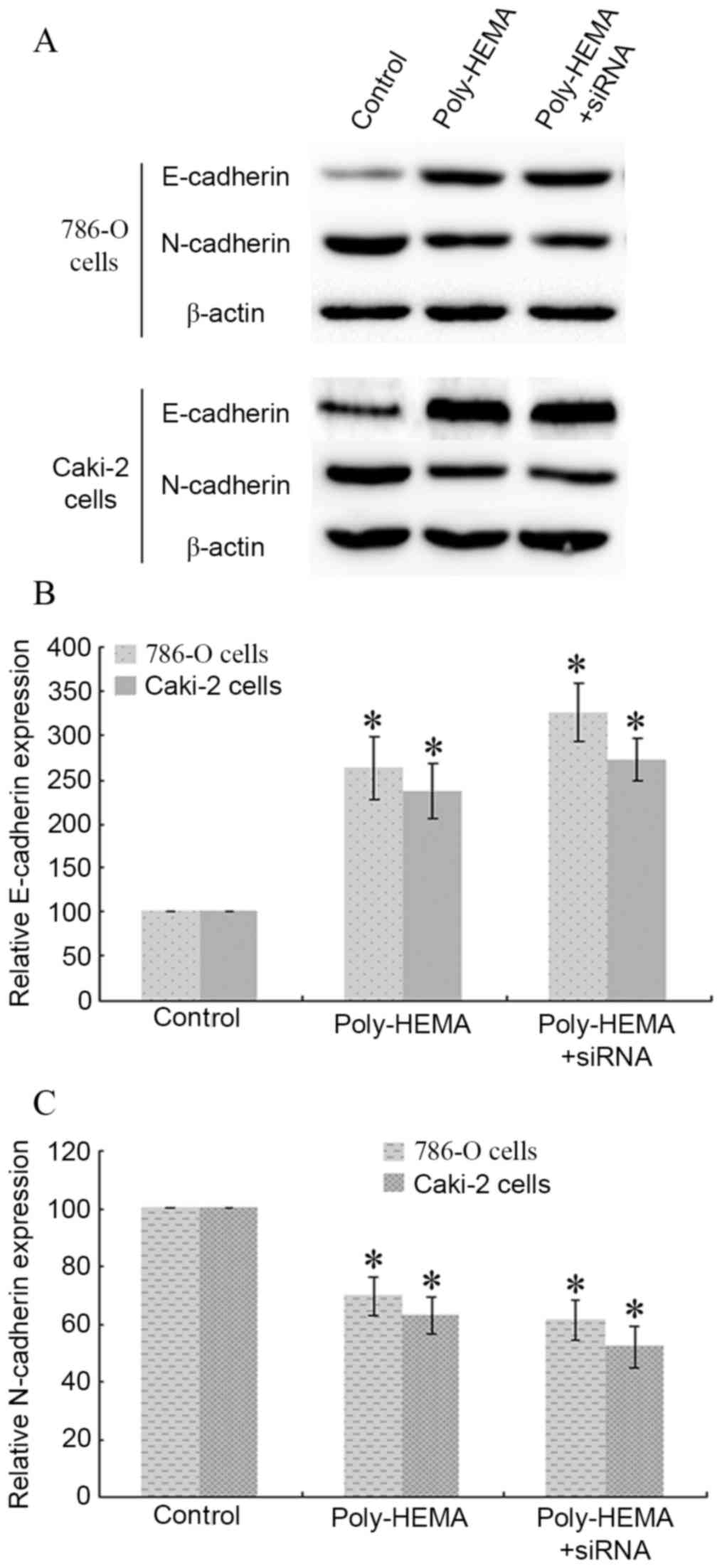
Knockdown of Tim-3 expression
attenuates ECM detachment-induced E-cadherin upregulation and
N-cadherin downregulation in ccRCC cells
786-O cells and Caki-2 cells seeded in poly-HEMA
coated plates were transfected with Tim-3 siRNA, and protein
expression levels of E-cadherin and N-cadherin were analyzed by
western blotting (Fig. 5A).
Detachment from the ECM increased the expression of E-cadherin in
poly-HEMA-treated 786-O and Caki-2 cells compared with control
cells (P=0.037 and P=0.015, respectively; Fig. 5B) and inhibited the expression of
N-cadherin in poly-HEMA treated 786-O and Caki-2 cells compared
with control cells (P=0.019 and P=0.008, respectively; Fig. 5). Tim-3 siRNA transfection enhanced
these changes further in 786-O cells (P=0.023 vs. control group,
P=0.004 vs. poly-HEMA treatment group; Fig. 5) and in Caki-2 cells (P=0.016 vs.
control group, P=0.008 vs. poly-HEMA treatment group; Fig. 5). Therefore, interference with
Tim-3 expression enhanced the ECM detachment-induced E-cadherin
upregulation and N-cadherin downregulation.
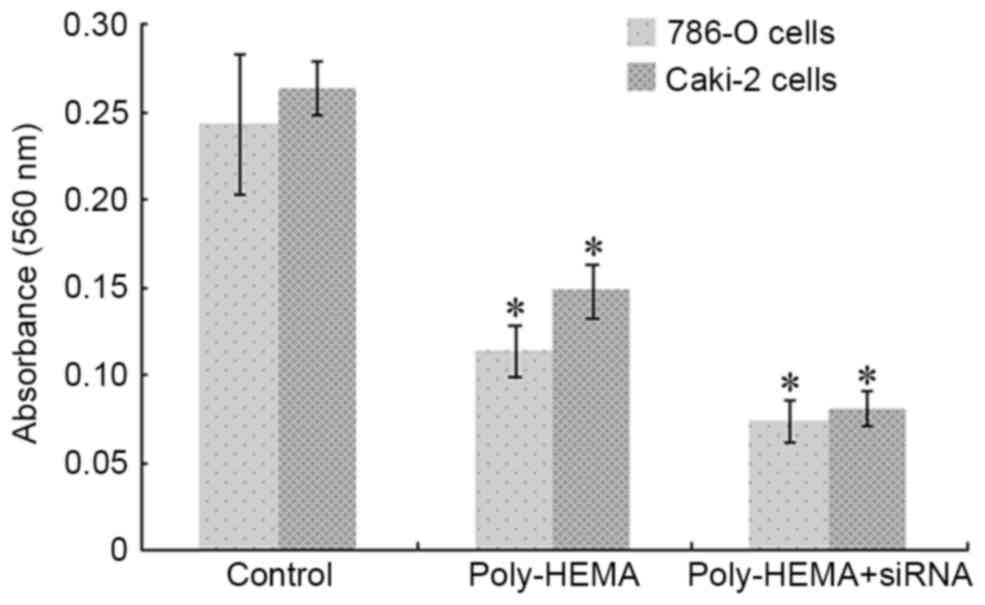
Knockdown of Tim-3 expression
decreases invasion of ccRCC cells
Poly-HEMA treated 786-O cells demonstrated
significantly decreased invasive abilities compared with control
cells, with 560 nm absorbance values of 0.113±0.015 in poly-HEMA
calls and 0.243±0.04 in control cells (P=0.022; Fig. 6). RNAi-mediated silencing of Tim-3
aggravated this reduction, with a 560 nm absorbance value of
0.073±0.012 (P=0.003 vs. control group; P=0.011 vs. poly-HEMA
treatment group; Fig. 6). Similar
results were observed in Caki-2 cells, with 560 nm absorbance
values recorded as 0.263±0.015 for the control group and
0.147±0.015 for the poly-HEMA treatment group (P=0.005, Fig. 6), and 0.08±0.01 for the poly-HEMA
treatment + siRNA group (P=0.016 vs. control group, P=0.018 vs.
poly-HEMA treatment group; Fig.
6).
Discussion
The results of the present study suggest that Tim-3
expression is involved in the process of anoikis in ccRCCs, which
may influence their invasion ability and, thus, their metastatic
potential. Knockdown of Tim-3 expression exacerbated anoikis in
786-O and Caki-2 cells, which displayed obvious morphological
changes and increased apoptosis ratios as measured by flow
cytometry. In addition, interference with Tim-3 expression
attenuated the invasion of 786-O and Caki-2 cells, increased
E-cadherin expression and decreased N-cadherin expression. To the
best of our knowledge, few studies have investigated the role of
Tim-3 in anoikis, especially in ccRCC. These results provide novel
insights regarding the function of Tim-3 in tumor biology.
ccRCC is the most frequently occurring kidney
cancer, with high rate of metastasis through the blood circulation.
It has previously been reported that ~33% of patients with ccRCC
displayed metastasis at diagnosis, resulting in poor prognosis and
a low 5-year survival rate (14).
However, previous studies have demonstrated the prognosis of RCC
improves when the expression of a number of genes is targeted,
including Tim-3 (15,16).
Tim-3 expression levels are increased in ccRCC
tissue compared with adjacent normal renal tissue (17) and high levels of Tim-3 expression
are considered as an independent predictor of ccRCC-specific
survival and progression-free survival (11). Previous research has indicated that
Tim-3 is involved in the invasive potential of ccRCC cells, by
either activating or inhibiting GATA binding protein 3 (8). The results of the present study
further confirm the involvement of Tim-3 in the invasion ability of
ccRCC cells, indicating Tim-3 as a potential therapeutic target for
treating ccRCC. This is consistent with the work of Yuan et
al (11).
Anoikis-resistant carcinoma cells are a cause of
metastasis in cancers, including ccRCC. Cancer cell growth relies
on adherence with an intact basement membrane and ECM, which are
essential for their growth. Damage to the basement membrane or ECM
results in an unfavorable environment for cell growth. Cells
detached from the basement membrane or ECM undergo anoikis, and
only cells that escape anoikis enter the blood circulation and
ultimately form metastases. Therefore, anoikis resistance is
critical for tumor metastasis. Previous research has reported
certain factors that modulate anoikis sensitivity and resistance in
ccRCC, including histone deacetylase inhibitors (18), tumor progression locus 2 kinase
(19), and HMGA1 (20). Further mechanisms of anoikis
sensitivity and resistance in ccRCC remain unknown. The present
study demonstrated morphological changes to cancer cells using
Ho33342/PI double staining and acridine orange staining, assayed
the apoptosis rate by flow cytometry analysis, and measured the
rate of anoikis using the CytoSelect™ 24-well assay kit. It was
revealed that knockdown of Tim-3 enhanced the rate of anoikis in
786-O and Caki-2 cells, indicating that Tim-3 is an anoikis
inhibitory molecule. To the best of our knowledge, this is the
first time Tim-3 has been demonstrated to modulate anoikis in
ccRCC.
The present results provided novel insights into the
role of Tim-3 in ccRCC anoikis. However, little has been examined
regarding the molecular mechanism involved in Tim-3-mediated
anoikis. Neither of the core cell apoptosis pathways, including the
extracellular pathway (death receptor pathway) and intracellular
pathway (mitochondrial pathway), were involved in the present
study. Future studies concerning the cell apoptosis pathway
involved in Tim-3-mediated anoikis will be of great value to the
field.
To summarize, the present study demonstrated that
interference with Tim-3 expression using siRNA may attenuate ccRCC
invasion by aggravating anoikis following cell detachment,
indicating Tim-3 might be a potential target for treating
ccRCC.
References
|
1
|
la Rosa AH, Acker M, Swain S and Manoharan
M: The role of epigenetics in kidney malignancies. Cent European J
Urol. 68:157–164. 2015.PubMed/NCBI
|
|
2
|
Klatte T, Pantuck AJ, Riggs SB, Kleid MD,
Shuch B, Zomorodian N, Kabbinavar FF and Belldegrun AS: Prognostic
factors for renal cell carcinoma with tumor thrombus extension. J
Urol. 178:1189–1195. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu B, Wen X and Cheng Y: Survival or
death: Disequilibrating the oncogenic and tumor suppressive
autophagy in cancer. Cell Death Dis. 4:e8922013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Criscitiello C, Esposito A and Curigliano
G: Tumor-stroma crosstalk: Targeting stroma in breast cancer. Curr
Opin Oncol. 26:551–555. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sato H, Hagiwara H, Ohde Y, Senba H,
Virgona N and Yano T: Regulation of renal cell carcinoma cell
proliferation, invasion and metastasis by connexin 32 gene. J Membr
Biol. 216:17–21. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Paoli P, Giannoni E and Chiarugi P:
Anoikis molecular pathways and its role in cancer progression.
Biochim Biophys Acta. 1833:3481–3498. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Strauss SJ, Ng T, Mendoza-Naranjo A,
Whelan J and Sorensen PH: Understanding micrometastatic disease and
Anoikis resistance in ewing family of tumors and osteosarcoma.
Oncologist. 15:627–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng H, Guo X, Tian Q, Li H and Zhu Y:
Distinct role of Tim-3 in systemic lupus erythematosus and clear
cell renal cell carcinoma. Int J Clin Exp Med. 8:7029–7038.
2015.PubMed/NCBI
|
|
9
|
Zhou E, Huang Q, Wang J, Fang C, Yang L,
Zhu M, Chen J, Chen L and Dong M: Up-regulation of Tim-3 is
associated with poor prognosis of patients with colon cancer. Int J
Clin Exp Pathol. 8:8018–8027. 2015.PubMed/NCBI
|
|
10
|
Piao YR, Jin ZH, Yuan KC and Jin XS:
Analysis of Tim-3 as a therapeutic target in prostate cancer.
Tumour Biol. 35:11409–1480. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yuan J, Jiang B, Zhao H and Huang Q:
Prognostic implication of TIM-3 in clear cell renal cell carcinoma.
Neoplasma. 61:35–40. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sakamoto S, Schwarze S and Kyprianou N:
Anoikis disruption of focal adhesion-Akt signaling impairs renal
cell carcinoma. Eur Urol. 59:734–744. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Jiang AC, Dong P, Wang H, Xu W and
Xu C: MDR1/P-gp and VEGF synergistically enhance the invasion of
Hep-2 cells with multidrug resistance induced by taxol. Ann Surg
Oncol. 16:1421–1428. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang YQ and Chen J: Predictive role of
vascular endothelial growth factor polymorphisms in the survival of
renal cell carcinoma patients. Genet Mol Res. 13:5011–5017. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai C, Wang L, Wu Z, Li M, Chen W and Sun
Y: T-cell immunoglobulin- and mucin-domain-containing molecule 3
gene polymorphisms and renal cell carcinoma. DNA Cell Biol.
31:1285–1289. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Komohara Y, Morita T, Annan DA, Horlad H,
Ohnishi K, Yamada S, Nakayama T, Kitada S, Suzu S, Kinoshita I, et
al: The coordinated actions of TIM-3 on cancer and myeloid cells in
the regulation of tumorigenicity and clinical prognosis in clear
cell renal cell carcinomas. Cancer Immunol Res. 3:999–1007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dannenmann SR, Thielicke J, Stöckli M,
Matter C, von Boehmer L, Cecconi V, Hermanns T, Hefermehl L,
Schraml P, Moch H, et al: Tumor-associated macrophages subvert
T-cell function and correlate with reduced survival in clear cell
renal cell carcinoma. Oncoimmunology. 2:e235622013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hamed HA, Das SK, Sokhi UK, Park MA,
Cruickshanks N, Archer K, Ogretmen B, Grant S, Sarkar D, Fisher PB
and Dent P: Combining histone deacetylase inhibitors with
MDA-7/IL-24 enhances killing of renal carcinoma cells. Cancer Biol
Ther. 14:1039–1049. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lee HW, Joo KM, Lim JE, Cho HJ, Cho HJ,
Park MC, Seol HJ, Seo SI, Lee JI, Kim S, et al: Tpl2 kinase impacts
tumor growth and metastasis of clear cell renal cell carcinoma. Mol
Cancer Res. 11:1375–1386. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takaha N, Sowa Y, Takeuchi I, Hongo F,
Kawauchi A and Miki T: Expression and role of HMGA1 in renal cell
carcinoma. J Urol. 187:2215–2222. 2012. View Article : Google Scholar : PubMed/NCBI
|