Introduction
Rheumatoid arthritis (RA) is an inflammatory and
autoimmune disease that affects ~1% of the world's population. RA
is characterized by chronic and invasive synovitis, damage to
cartilage and bone, and cartilage destruction (1,2).
Despite the advances in RA treatment, including aggressive immune
suppression with biologics and modifying anti-rheumatic drugs, the
general efficacy of treatment remains unsatisfactory (3). Accumulating evidence suggests that
fibroblast-like synoviocytes (FLSs) serve critical roles in the
initiation and progression of RA (4). The abnormal proliferation of
fibroblast-like synoviocytes (FLSs) leads to destructive invasion
of the cartilage and bones through secreting digestive proteases
and inflammatory cytokines (5,6).
Furthermore, FLSs exhibit resistance to apoptosis induced by
several apoptotic stimuli (7,8).
Therefore, understanding the molecular mechanism of FLS
proliferation and developing pro-apoptosis drugs for FLS is
important.
The human transcriptome contains a large number of
protein-coding mRNAs and noncoding transcripts (9). Among these, long noncoding RNA
(lncRNA) is defined as a molecule of RNA >200 nucleotides in
length (10). Accumulating studies
have reported that numerous lncRNAs have important biological
functions, including chromatin remodeling, transcriptional control
and posttranscriptional modification (11,12).
Dysregulation of lncRNA has been identified to be associated with
human diseases, including inflammation and cancer (13–15).
The abnormal expression of lncRNA may contribute to disease
initiation and progression through regulating expression of the
gene products (16,17). A recent study has reported that
differentiation and activation of immune cells are associated with
lncRNAs, which have an essential role in autoimmune diseases,
including RA (18). However, the
role of lncRNA in FLSs has yet to be elucidated.
Long noncoding-interleukin-7 receptor (lnc-IL7R) has
been recently identified, and it overlaps with the 3′ untranslated
region (3′UTR) of the interleukin-7 receptor α-subunit gene (IL7R).
Lnc-IL7R is activated by LPS stimulation, and reduces the
LPS-induced inflammatory response (19). A previous study (20) demonstrated the differential
expression of lncRNAs between RA patients and non-RA patients. From
the different expression profiles, it was identified that lnc-IL7R
is one of these lncRNAs. However, the role of lnc-IL7R in FLSs has
yet to be elucidated. The present study demonstrates that lnc-IL7R
promotes cell proliferation, cell cycle progression and inhibits
apoptosis in FLSs. Further investigation identified that lnc-IL7
interacts with enhancer of zeste homolog 2 (EZH2), and is required
for polycomb repressive complex 2 (PRC2)-mediated suppression.
Materials and methods
Isolation and culture of FLSs
Isolation and culture of FLSs was performed as
previously described (21). In
brief, synovial tissues were obtained during surgery from RA
patients diagnosed according to American Rheumatism Association
criteria. The present study was approved by the ethics committee of
Cangzhou Central Hospital (Cangzhou, China) and written informed
consent was provided by all patients. The synovial tissues were
digested with 2.5 g/l trypsin for 2 h, and cells were centrifuged
at 2,500 × g for 10 min at 4°C and cultured in Dulbecco's minimum
essential medium in 5% CO2 at 37°C. Cells within the
3rd-8th passages were used for all experiments.
MTT assays
A total of 1×103 cells per well were
seeded in 96-well plates and the viability of the cells was
ascertained from three replicates in three independent experiments
using an MTT assay [Vazyme Biotech (Nanjing) Co., Ltd., Nanjing,
Jiangsu, China] after 3 days.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated using Trizol reagent
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
1 µg complementary DNAS (cDNAs) were synthesized using a Prime
Script RT Reagent kit (Takara Bio, Inc., Otsu, Japan) according to
the manufacturer's protocol. Quantification of gene expression was
determined using FastStart Universal SYBR Green Master mix in the
StepOne Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The relative expression of RNAs was calculated
using the comparative Cq method (or the 2−ΔΔCq method)
(22). Gene-specific primers were
as follows: lnc-IL7R: 5′-CCAGCCTTTGCCTCTTCCTTCAAT-3′ (forward),
5′-CCGTACCAAGTCTCTTAGCCCCTC-3′ (reverse); p16:
5′-GCCCAACGCACCGAATAGTTA-3′ (forward), 5′-ACGGGTCGGGTGAGAGT-3′
(reverse); p21: 5′-CCTCATCCCGTGTTCTCCTTT-3′ (forward),
5′-GTACCACCCAGCGGACAAGT-3′ (reverse).
Transfection
The cDNA encoding lnc-IL7R was subcloned into
pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.). Cells
were transfected with the plasmids pcDNA3.1 or pcDNA-lnc-IL7R using
Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Cells were transfected
and grown for 48 h prior to the subsequent assays. The small
interfering RNA (siRNA) against lnc-IL7R was purchased from
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The target sequence
was as follows: AAGAGATATATTTCATCGAGA. Cells were transfected with
10 0 nM siRNA for 48 h prior to subsequent assays.
Cell-cycle analysis
Cell-cycle analysis was performed as previously
described (23). In brief, cells
were permeabilized with 75% ethanol, and DNA was stained with 50
µg/ml propidium iodide. Cellular DNA content was measured using
flow cytometric analysis using a FACScan flow cytometer, and the
data were analyzed with Kaluza software (version 1.20; Beckman
Coulter, Inc., Brea, CA, USA).
Apoptosis analysis
Cells were trypsinized, resuspended and stained with
fluorescein isothiocyanate-conjugated annexin V and
7-aminoactinomycin D (7-AAD; Nanjing KeyGen Biotech Co., Ltd.,
Nanjing, China) according to the manufacturer's protocol. Cells
were then measured by flow cytometric analysis using the FACScan
flow cytometer with the data analyzed with Kaluza software.
Western blot analysis
Cells were lysed in RIPA lysis buffer with cocktail
protease inhibitor (Selleck Chemicals, Houston, TX, USA). Samples
were separated by SDS-PAGE and transferred onto polyvinylidene
fluoride (PVDF) membranes (Merck Millipore, Darmstadt, Germany).
The PVDF membranes were blocked with 5% non-fat milk for 1 h at
room temperature and incubated with primary antibodies (1:1,000)
raised against CDK4 (cat. no. 12790), CDK6 (cat. no. 13331),
caspase 3 (cat. no. 9661), poly (ADP-ribose) polymerase (PARP; cat.
no. 5625) or GAPDH (cat. no. 2118) overnight at 4°C, followed by
incubation with anti-rabbit secondary antibodies (1:10,000; cat.
no. 7074) coupled to horseradish peroxidase (all antibodies
purchased from Cell Signaling Technology, Danvers, MA, USA) for 1 h
at room temperature. Signals were visualized with an ECL
chemiluminescence system (Pierce ECL Western Blotting Substrate;
Thermo Fisher Scientific, Inc.).
RNA immunoprecipitation (RIP) and
chromatin immunoprecipitation (ChIP) assays
RIP and ChIP assays were performed using an EZ-Magna
RIP RNA-binding protein immunoprecipitation kit and an EZ-Magna
ChIP kit, according to the manufacturer's protocol (Merck
Millipore). RIP and ChIP products were analyzed by RT-qPCR.
ChIP-enriched promoter of target genes was quantified by RT-qPCR
using the following primers: p16: Forward:
5′-CGCTAAGTGCTCGGAGTTAATA-3′, reverse: 5′-CGACCCTGTCCCTCAAATC-3′;
p21: Forward: 5′-GTGTCCAGCGCACCAAC-3′, reverse:
5′-TGAGCCTGGCCGAGTTC-3′.
RNA pull-down assays
RNA pull-down assays were performed as described
previously (24). Briefly, RNAs
were biotin-labeled with the Biotin RNA Labeling mix (Roche
Diagnostics, Basel, Switzerland) and transcribed in vitro
using T7 RNA polymerase (Roche Diagnostics, Switzerland).
Biotinylated RNAs were mixed and incubated with cell lysates for 2
h at 4°C. Streptavidin-agarose beads (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) were added to each binding reaction, and
incubated for 1 h at room temperature. The beads were then boiled
in sodium dodecyl sulfate (SDS) buffer. The eluted proteins were
detected by western blot analysis.
Statistical analysis
All statistical analyses were performed by using
SPSS 19.0 software (IBM SPSS, Armonk, NY, USA). Student's t-test
was performed for the results from all assays. In all cases,
P<0.05 was considered to indicate a statistically significant
difference.
Results
Overexpression of lnc-IL7R promotes FLS
proliferation, induces cell cycle progression and decreases cell
apoptosis. To determine the role of lnc-IL7R in FLSs,
gain-of-function studies were performed. The cell proliferation of
FLSs with empty vector or lnc-IL7R overexpression was detected by
performing an MTT assay. It was identified that lnc-IL7R
overexpression increased the proliferation of FLSs compared with
control cells (Fig. 1A and B). To
understand the mechanism by which lnc-IL7R promotes cell
proliferation, the apoptosis and cell cycle distribution of
lnc-IL7R-overexpressed FLSs was detected by fluorescence-activated
cell sorting (FACS). It was identified that lnc-IL7R overexpression
significantly promoted G1/S transition in FLSs (Fig. 1C). Consistent with the FACS data,
the expression of G1/S-phase checkpoint markers, including CDK4 and
CDK6, were upregulated in lnc-IL7R-overexpressed FLSs (Fig. 1D). However, lnc-IL7R-overexpressed
FLSs showed a significantly higher percentage of annexin V-positive
cells than did control cells (Fig.
1E). Consistent with the FACS results, apoptosis markers,
including cleaved caspase 3 and PARP, markedly decreased in FLSs
with lnc-IL7R overexpression (Fig.
1F). Taken together, the results demonstrated that lnc-IL7R
promotes FLS proliferation, via driving cell cycle progression and
decreasing cell apoptosis.
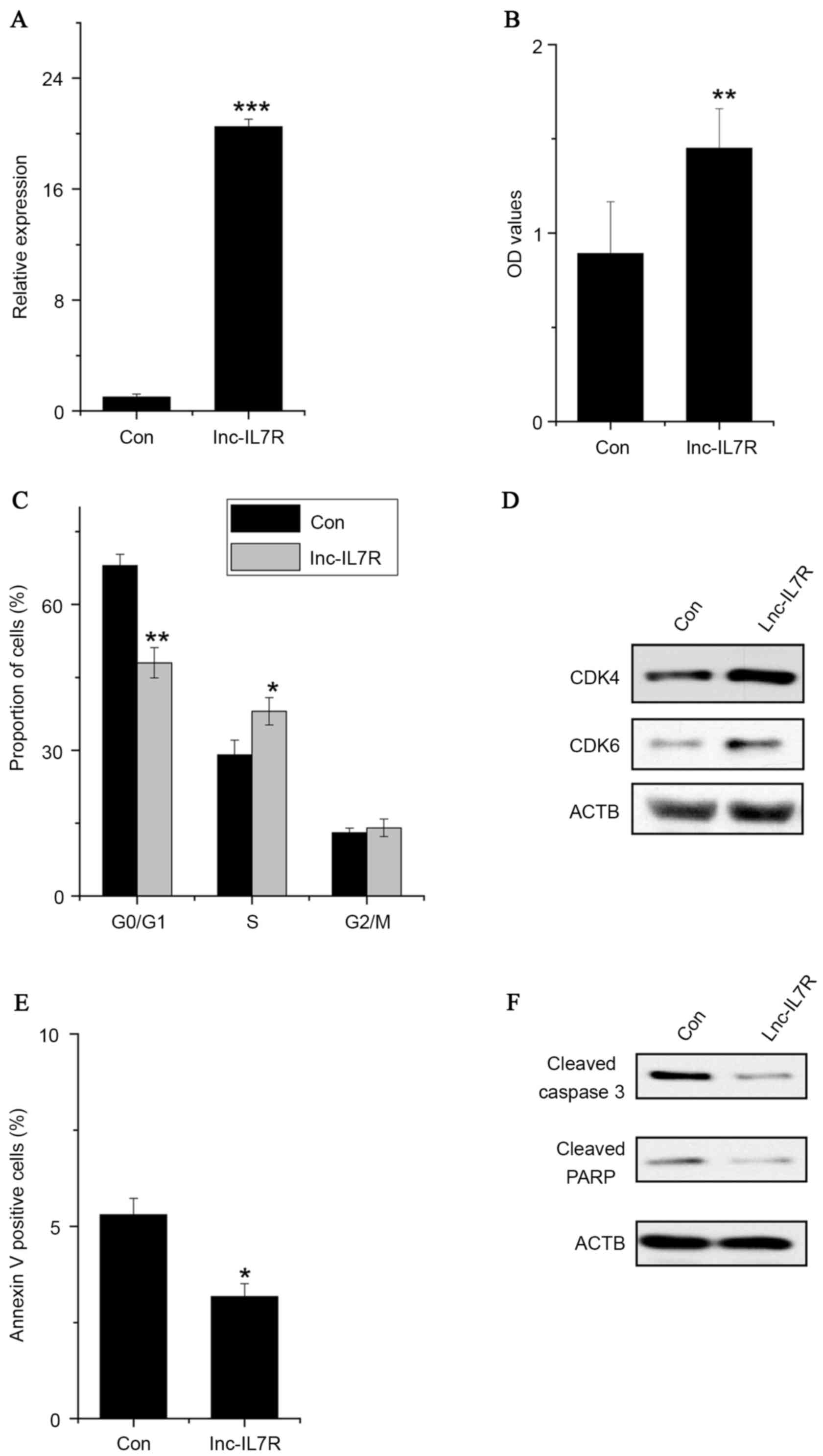 | Figure 1.Overexpression of lnc-IL7R promotes
FLS proliferation, induces cell cycle progression and decreases
cell apoptosis. (A) Relative expression of lnc-IL7R in FLSs
expressing control and lnc-IL7R. (B) Cell growth determined by MTT
proliferation assays. (C) Data derived from the FACS analysis,
illustrating increases or decreases of the cells in S or G1 phases,
respectively, in FLSs with lnc-IL7R overexpression. (D) CDK4 and
CDK6 expression levels detected by western blot analysis following
lnc-IL7R overexpression. (E) Cells with lnc-IL7R overexpression
stained with a combination of annexin V and 7-AAD and analyzed
using FACS; cells positive for annexin V staining were counted as
apoptotic cells. (F) Caspase 3 and PARP expression levels were
detected by western blot analysis following lnc-IL7R
overexpression. Data are shown as the mean ± standard deviation
(n=3). *P<0.05, **P<0.01, ***P<0.001 (Student's t-test).
Lnc-IL7R, long noncoding-interleukin-7 receptor; FLSs,
fibroblast-like synoviocytes; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; FACS,
fluorescence-activated cell sorting; CDK4, cyclin-dependent kinase
4; CDK6, cyclin-dependent kinase 6; 7-AAD, 7-amino-actinomycin D;
PARP, poly(ADP) ribose polymerase; Con, control. |
Knockdown of lnc-IL7R inhibits FLS
proliferation, induces cell cycle arrest and promotes cell
apoptosis
Lnc-IL7R expression in FLSs was silenced (Fig. 2A) and an MTT assay was performed to
detect the cell proliferation of FLSs with lnc-IL7R small hairpin
RNA (shRNA). It was identified that knockdown of lnc-IL7R decreased
the proliferative capacity of FLSs compared with control cells
(Fig. 2B). Similarly, a FACS assay
demonstrated that lnc-IL7R knockdown significantly inhibited G1/S
transition and promoted cell apoptosis (Fig. 2C and D). Consistent with the FACS
data, the expression of G1/S phase checkpoint markers were markedly
downregulated, while the apoptosis markers were upregulated in
lnc-IL7R knockdown FLSs (Fig. 2E and
F). Collectively, these data indicate that lnc-IL7R serves an
important role in FLS proliferation, cell cycle and apoptosis.
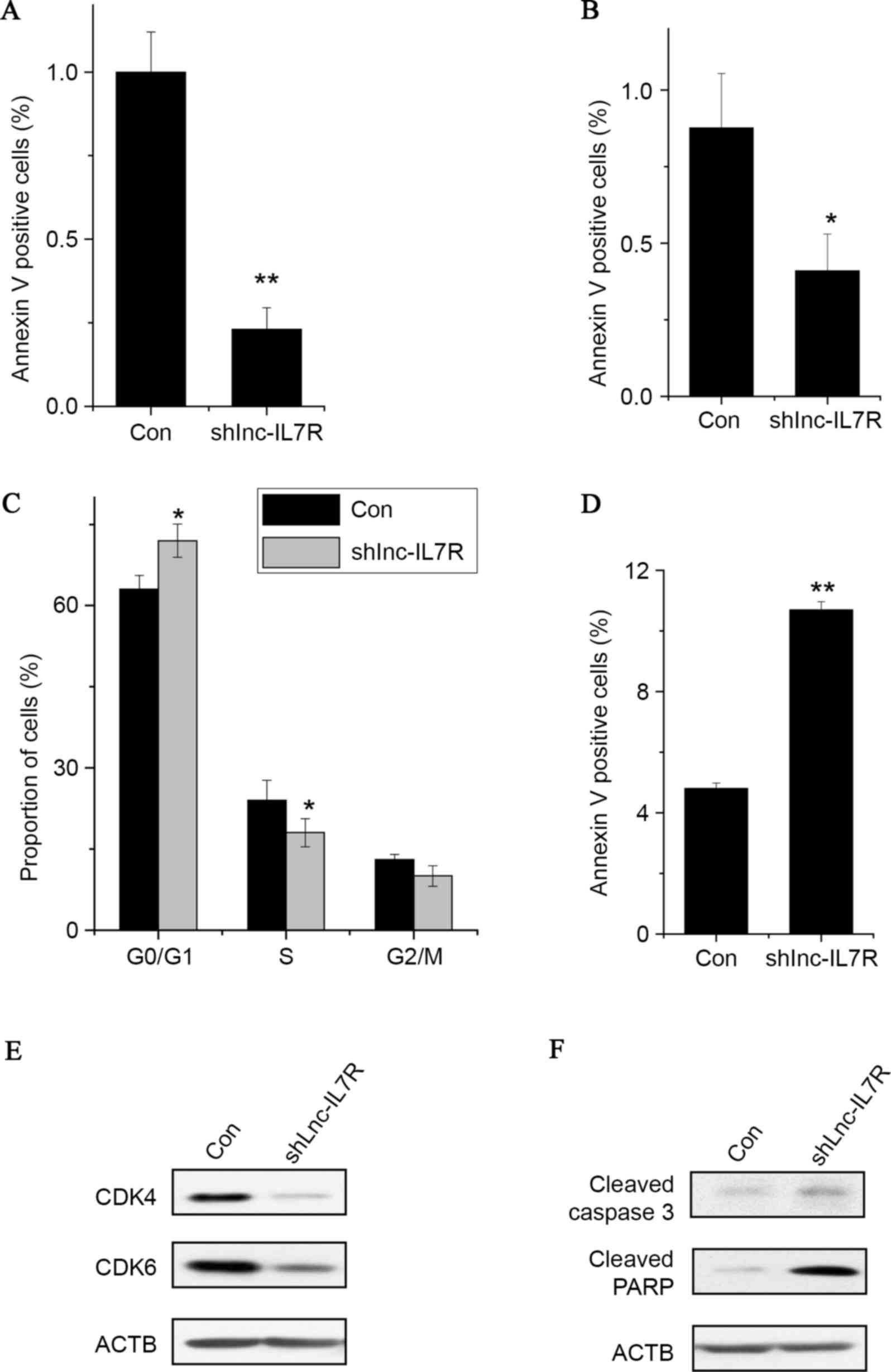 | Figure 2.Knockdown of lnc-IL7R inhibits FLS
proliferation, induces cell cycle arrest and promotes cell
apoptosis. (A) Relative expression of lnc-IL7R in FLSs expressing
Con and lnc-IL7R shRNA. (B) Cell growth determined by MTT
proliferation assay. (C) FACS analysis showed a marked decrease or
increase of cells in the S or G1 phase, respectively, in FLSs with
lnc-IL7R shRNA. (D) Cells with lnc-IL7R shRNA stained with a
combination of annexin V and 7-AAD and analyzed by FACS. Cells
positive for annexin V staining were counted as apoptotic cells.
(E) CDK4 and CDK6 expression levels detected by western blot
analysis following lnc-IL7R silencing. (F) Caspase 3 and PARP
expression levels detected by western blot analysis following
lnc-IL7R silencing. Data are shown as the mean ± standard deviation
(n=3). *P<0.05, **P<0.01 (Student's t-test). Lnc-IL7R, long
noncoding-interleukin-7 receptor; FLS, fibroblast-like synoviocyte;
shRNA, small hairpin RNA; MTT,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; Con,
control; FACS, fluorescence-activated cell sorting; CDK4,
cyclin-dependent kinase 4; CDK6, cyclin-dependent kinase 6; 7-AAD,
7-amino-actinomycin D; PARP, poly(ADP) ribose polymerase. |
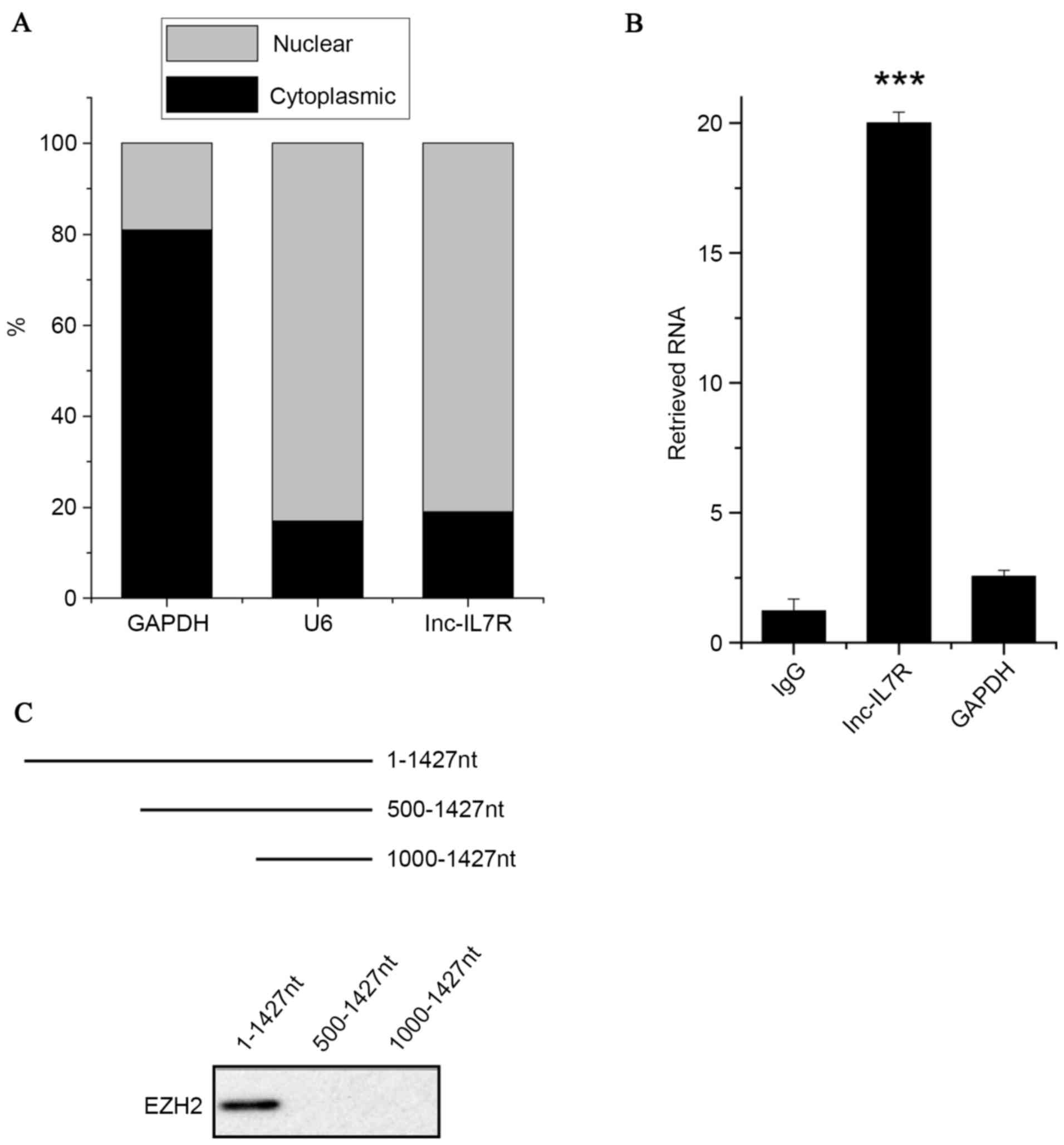
Inc-IL7R interacts with EZH2
The molecular mechanisms by which lnc-IL7R promotes
cell proliferation were subsequently investigated. First, cellular
fractionation assays were performed to analyze the subcellular
localization of lnc-IL7R. It was identified that lnc-IL7R was
localized in the nuclei of FLSs (Fig.
3A). This suggested that lnc-IL7R may be involved in
transcriptional regulation.
A previous study (25) reported that 20% of human lncRNs are
associated with PRC2. It was hypothesized that lnc-IL7R may
interact with EZH2, a core component of PRC2, and regulate gene
expression. To confirm this, an RIP assay was performed with an
EZH2 antibody from nuclear extracts of FLSs. A marked enrichment of
lnc-IL7R by the EZH2 antibody was identified, but not any
enrichment of GAPDH compared with the IgG control antibody
(Fig. 3B). Consistent with the RIP
assay, the association between EZH2 and lnc-IL7R was validated
through a RNA pull-down assay (Fig.
3C). The deletion-mapping assay showed that 500 nucleotides at
the 5′ end of lnc-IL7R are required for its interaction with EZH2.
Collectively, the results demonstrated that there is an interaction
between lnc-IL7R and EZH2.
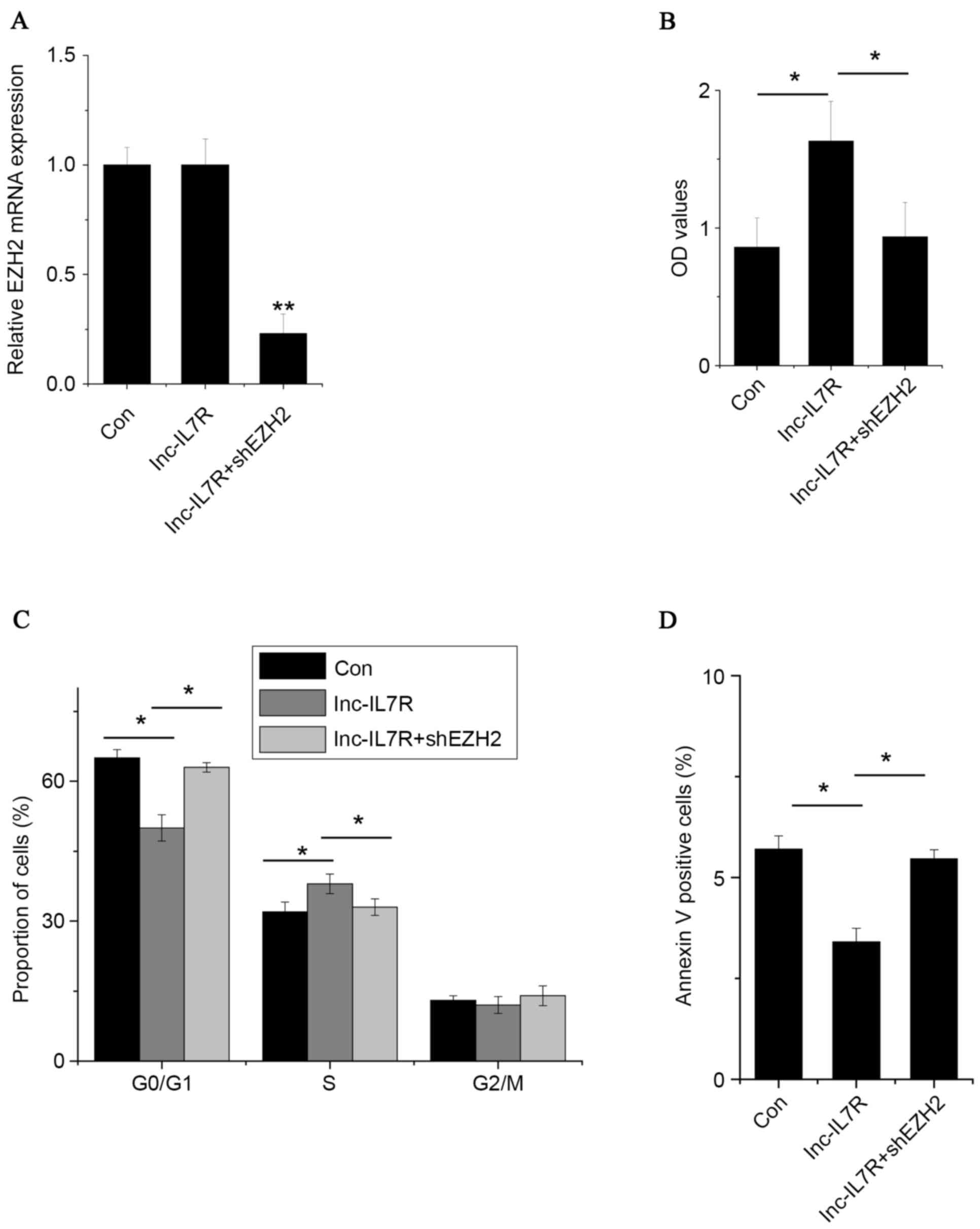
Inc-IL7R promotes FLS proliferation,
induces cell cycle progression and decreases cell apoptosis via
association with EZH2
To investigate the practical relevance of the
interaction of lnc-IL7R and EZH2, EZH2 was silenced in
lnc-IL7R-overexpressed FLSs (Fig.
4A). EZH2 knockdown reversed the effects of promotion of cell
proliferation and cell cycle progression, and the anti-apoptosis
effects, induced by lnc-IL7R (Fig.
4B-D). These data suggest that lnc-IL7R exerts its function, at
least in part, through interaction with EZH2.
Lnc-IL7R regulate target gene expression
via epigenetic mechanisms
To further corroborate the practical relevance of
the association between lnc-IL7R and EZH2, PRC2 target genes, which
regulate the cell cycle, were detected, including cyclin dependent
kinase inhibitor 2A (p16) and cyclin dependent kinase inhibitor 1A
(p21) (Fig. 5A). p16 and p21
expression levels were decreased by lnc-IL7R overexpression, and
reversed by knockdown of EZH2. By contrast, both p16 and p21
expression levels were increased by lnc-IL7R knockdown (Fig. 5B). A ChIP assay in
lnc-IL7R-overexpressed FLSs was performed to test whether lnc-IL7R
is involved in transcriptional regulation by recruiting EZH2 to
target genes. The results demonstrated that lnc-IL7R increased the
binding of EZH2 and H3K27me3 levels across the p16 and p21 promoter
regions (Fig. 5C and D).
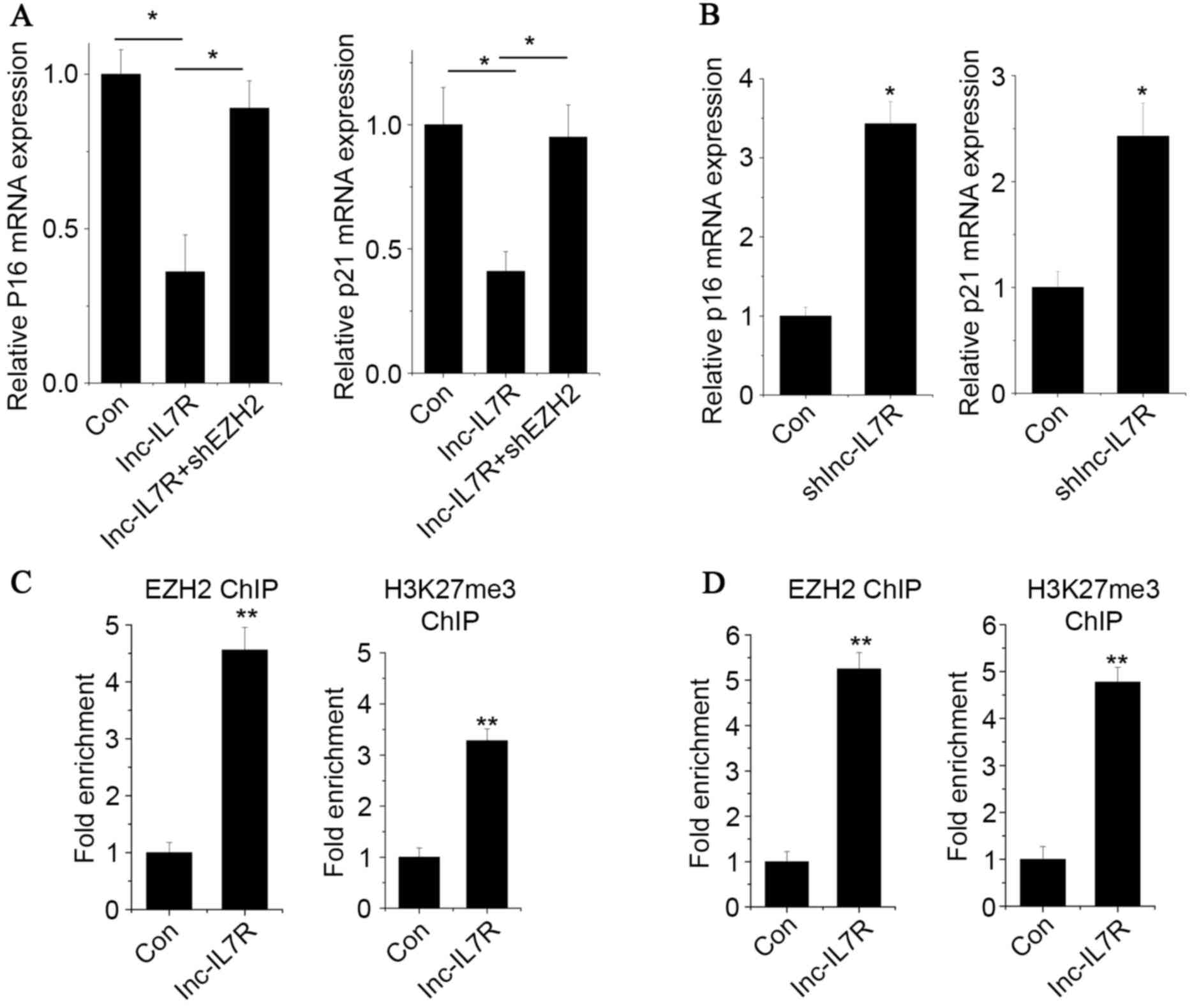 | Figure 5.Lnc-IL7R regulate target gene
expression via epigenetic mechanisms. The (A) p16 and (B) p21
expression levels of the groups as detected by RT-qPCR. (C) The
occupancy of EZH2 and H3K27me3 in the promoter regions of p16
measured by EZH2 and H3K27me3 ChIP assay followed by RT-qPCR. (D)
The occupancy of EZH2 and H3K27me3 in the promoter regions of p21
was measured by EZH2 and H3K27me3 ChIP assay followed by RT-qPCR.
Data are shown as mean ± standard deviation; n=3. *P<0.05,
**P<0.01, ***P<0.001 (Student's t-test). Lnc-IL7R, long
noncoding-interleukin-7 receptor; p16, cyclin-dependent kinase
inhibitor 2A; p21, cyclin-dependent kinase inhibitor 1A; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; EZH2,
enhancer of zeste homolog 2; H3K27me3, histone 3 lysine 27
trimethylation; ChIP, chromatin immunoprecipitation; Con,
control. |
Discussion
Development of RA is closely associated with
characteristic changes of the synovium, including FLS hyperplasia,
abnormal angiogenesis and infiltration of immune cells (26,27).
Previous studies (28,29) have reported that the excessive
proliferation of FLSs is associated with aberrant activation of the
inflammatory signal pathway. Noncoding RNA (ncRNA) is also involved
in regulating FLS proliferation. For example, microRNA (miR)-152
overexpression significantly decreases FLS proliferation through
the upregulation of SFRP4, a negative regulator of the Wnt
signaling pathway (30). However,
the function of lncRNA in FLSs has yet to be elucidated. The
present study demonstrated that lnc-IL7R promotes cell
proliferation and cell cycle progression, and inhibits apoptosis in
FLSs. Although the present study has demonstrated that lnc-IL7R
diminishes the LPS-induced inflammatory response in human
monocytes, this phenomenon was not observed in FLSs (data not
shown). Lnc-IL7R may therefore exert its function in a
tissue-specific manner.
Aberrant epigenetic modifications have been
thoroughly elucidated in human diseases. Epigenetic modifications,
including DNA methylation and histone modification, regulate gene
expression (31,32). Previous studies (33,34)
have shown an extensive aberrant methylation of gene promoters in
FLSs, including C-X-C motif chemokine ligand 12, interleukin 10 and
interleukin 6. Many of these genes are involved in the immune
response and in inflammation (35,36).
However, histone modifications involved in RA have yet to be
elucidated (37,38). A previous study (39) demonstrated that EZH2 is
overexpressed in FLSs, and inhibits the expression of secreted
frizzled-related protein 1. However, none of the PRC2 core
components are responsible for the sequence-specific DNA-binding
activity. Previous studies (40,41)
have reported that PRC2 may be recruited to target genes by lncRNA.
In the present study, it was demonstrated that lnc-IL7R associates
with EZH2. Lnc-IL7R depletion leads to upregulation of genes which
are suppressed by EZH2, including p16 and p21. Additionally,
lnc-IL7R overexpression increases the binding of EZH2 and H3K27me3
levels across p16 and p21 promoters. Overall, the present study
proposes a model in which lncRNA regulates proliferation via
association with EZH2. Whether lnc-IL7R is associated with other
proteins requires further investigation. Understanding the precise
molecular mechanisms by which lnc-IL7R functions in FLSs will aid
the exploration of new therapies for RA.
Acknowledgements
This study was supported by a grant from the Support
Project of Hebei province science and technology (grant no.
1123038ZD).
References
|
1
|
Gibofsky A: Epidemiology, pathophysiology,
and diagnosis of rheumatoid arthritis: A Synopsis. Am J Manag Care.
20:S128–S135. 2014.PubMed/NCBI
|
|
2
|
Huber LC, Distler O, Tarner I, Gay RE, Gay
S and Pap T: Synovial fibroblasts: Key players in rheumatoid
arthritis. Rheumatology (Oxford). 45:669–675. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang CL, Or TC, Ho MH and Lau AS:
Scientific basis of botanical medicine as alternative remedies for
rheumatoid arthritis. Clin Rev Allergy Immunol. 44:284–300. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mor A, Abramson SB and Pillinger MH: The
fibroblast-like synovial cell in rheumatoid arthritis: A key player
in inflammation and joint destruction. Clin Immunol. 115:118–128.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bottini N and Firestein GS: Duality of
fibroblast-like synoviocytes in RA: Passive responders and
imprinted aggressors. Nat Rev Rheumatol. 9:24–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartok B and Firestein GS: Fibroblast-like
synoviocytes: Key effector cells in rheumatoid arthritis. Immunol
Rev. 233:233–255. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Smith MD and Walker JG: Apoptosis a
relevant therapeutic target in rheumatoid arthritis? Rheumatology
(Oxford). 43:405–407. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bai S, Liu H, Chen KH, Eksarko P, Perlman
H, Moore TL and Pope RM: NF-kappaB-regulated expression of cellular
FLIP protects rheumatoid arthritis synovial fibroblasts from tumor
necrosis factor alpha-mediated apoptosis. Arthritis Rheum.
50:3844–3855. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wilusz JE, Sunwoo H and Spector DL: Long
noncoding RNAs: Functional surprises from the RNA world. Genes Dev.
23:1494–1504. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu G, Mattick JS and Taft RJ: A
meta-analysis of the genomic and transcriptomic composition of
complex life. Cell Cycle. 12:2061–2072. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang JL, Zheng L, Hu YW and Wang Q:
Characteristics of long non-coding RNA and its relation to
hepatocellular carcinoma. Carcinogenesis. 35:507–514. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lam MT, Li W, Rosenfeld MG and Glass CK:
Enhancer RNAs and regulated transcriptional programs. Trends
Biochem Sci. 39:170–182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y, Chen H, Pan T, Jiang C, Zhao Z, Wang
Z, Zhang J, Xu J and Li X: LncRNA ontology: Inferring lncRNA
functions based on chromatin states and expression patterns.
Oncotarget. 24:39793–39805. 2015.
|
|
14
|
Keenan CR, Schuliga MJ and Stewart AG:
Pro-inflammatory mediators increase levels of the noncoding RNA
GAS5 in airway smooth muscle and epithelial cells. Can J Physiol
Pharmacol. 93:203–206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheng HS, Njock MS, Khyzha N, Dang LT and
Fish JE: Noncoding RNAs regulate NF-κB signaling to modulate blood
vessel inflammation. Front Genet. 5:4222014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang H, Modise T, Helm R, Jensen RV and
Good DJ: Characterization of the hypothalamic transcriptome in
response to food deprivation reveals global changes in long
noncoding RNA and cell cycle response genes. Genes Nutr. 10:482015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng
J, Xiao W, Yu G, Yao W, Zhou H, et al: LncRNA MALAT1 functions as a
competing endogenous RNA to regulate ZEB2 expression by sponging
miR-200s in clear cell kidney carcinoma. Oncotarget. 6:38005–38015.
2015.PubMed/NCBI
|
|
18
|
Spurlock CF III, Tossberg JT, Matlock BK,
Olsen NJ and Aune TM: Methotrexate inhibits NF-κB activity via long
intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol.
66:2947–2957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui H, Xie N, Tan Z, Banerjee S,
Thannickal VJ, Abraham E and Liu G: The human long noncoding RNA
lnc-IL7R regulates the inflammatory response. Eur J Immunol.
44:2085–2095. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Messemaker TC, Frank-Bertoncelj M, Marques
RB, Adriaans A, Bakker AM, Daha N, Gay S, Huizinga TW, Toes RE,
Mikkers HM and Kurreeman F: A novel long non-coding RNA in the
rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels.
Genes Immun. 17:85–92. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun J, Yan P, Chen Y, Chen Y, Yang J, Xu
G, Mao H and Qiu Y: MicroRNA-26b inhibits cell proliferation and
cytokine secretion in human RASF cells via the Wnt/GSK-3β/β-catenin
pathway. Diagn Pathol. 10:722015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Braconi C, Valeri N, Kogure T, Gasparini
P, Huang N, Nuovo GJ, Terracciano L, Croce CM and Patel T:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cao C, Sun J, Zhang D, Guo X, Xie L, Li X,
Wu D and Liu L: The long intergenic noncoding RNA UFC1, a target of
MicroRNA 34a, interacts with the mRNA stabilizing protein HuR to
increase levels of β-catenin in HCC cells. Gastroenterology.
148:415–426, e18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Morales Rivea D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:11667–11672. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sweeney SE and Firestein GS: Rheumatoid
arthritis: Regulation of synovial inflammation. Int J Biochem Cell
Biol. 36:372–378. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Umar S, Hedaya O, Singh AK and Ahmed S:
Thymoquinone inhibits TNF-α-induced inflammation and cell adhesion
in rheumatoid arthritis synovial fibroblasts by ASK1 regulation.
Toxicol Appl Pharmacol. 287:299–305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Fang Y, Wang Y, Wang Z, Zou Q, Shi
Y, Chen J and Peng D: Inhibitory effect of curcumol on Jak2-STAT
signal pathway molecules of fibroblast-like synoviocytes in
patients with rheumatoid arthritis. Evid Based Complement Alternat
Med. 2012:7464262012.PubMed/NCBI
|
|
29
|
Carrion M, Juarranz Y, Martínez C,
González-Álvaro I, Pablos JL, Gutiérrez-Cañas I and Gomariz RP:
IL-22/IL-22R1 axis and S100A8/A9 alarmins in human osteoarthritic
and rheumatoid arthritis synovial fibroblasts. Rheumatology
(Oxford). 52:2177–2186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miao CG, Yang YY, He X, Huang C, Huang Y,
Qin D, Du CL and Li J: MicroRNA-152 modulates the canonical Wnt
pathway activation by targeting DNA methyltransferase 1 in
arthritic rat model. Biochimie. 106:149–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Picascia A, Grimaldi V, Pignalosa O, De
Pascale MR, Schiano C and Napoli C: Epigenetic control of
autoimmune diseases: From bench to bedside. Clin Immunol. 157:1–15.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gonzalez S, Aguilera S, Urzúa U, Quest AF,
Molina C, Alliende C, Hermoso M and González MJ:
Mechanotransduction and epigenetic control in autoimmune diseases.
Autoimmun Rev. 10:175–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karouzakis E, Rengel Y, Jüngel A, Kolling
C, Gay RE, Michel BA, Tak PP, Gay S, Neidhart M and Ospelt C: DNA
methylation regulates the expression of CXCL12 in rheumatoid
arthritis synovial fibroblasts. Genes Immun. 12:643–652. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin SY, Hsieh SC, Lin YC, Lee CN, Tsai MH,
Lai LC, Chuang EY, Chen PC, Hung CC, Chen LY, et al: A whole genome
methylation analysis of systemic lupus erythematosus:
Hypomethylation of the IL10 and IL1R2 promoters is associated with
disease activity. Genes Immun. 13:214–220. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wilkinson J: Study reveals a DNA methylome
signature in rheumatoid arthritis. Epigenomics.
4:4812012.PubMed/NCBI
|
|
36
|
Nakano K, Whitaker JW, Boyle DL, Wang W
and Firestein GS: DNA methylome signature in rheumatoid arthritis.
Ann Rheum Dis. 72:110–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Viatte S, Plant D and Raychaudhuri S:
Genetics and epigenetics of rheumatoid arthritis. Nat Rev
Rheumatol. 9:141–153. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huber LC, Brock M, Hemmatazad H, Giger OT,
Moritz F, Trenkmann M, Distler JH, Gay RE, Kolling C, Moch H, et
al: Histone deacetylase/acetylase activity in total synovial tissue
derived from rheumatoid arthritis and osteoarthritis patients.
Arthritis Rheum. 56:1087–1093. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Trenkmann M, Brock M, Gay RE, Kolling C,
Speich R, Michel BA, Gay S and Huber LC: Expression and function of
EZH2 in synovial fibroblasts: Epigenetic repression of the Wnt
inhibitor SFRP1 in rheumatoid arthritis. Ann Rheum Dis.
70:1482–1488. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|