Introduction
Staphylococcus aureus and Pseudomonas
aeruginosa are the main bacteria that opportunistically infect
patients with burns (1). However,
recent reports (2–5) indicate that the proportion of
infections caused by Acinetobacter baumannii is gradually
increasing and, in some instances, already exceeds the number of
infections caused by P. aeruginosa. A. baumannii is the most
commonly detected Gram-negative organism infecting patients with
burns (2–5). However, the emergence of
multidrug-resistant A. baumannii complicates the clinical
treatment of these infections (6–8).
Quorum sensing is a form of cell-cell communication
that bacteria use to coordinate the expression of genes involved in
certain behaviours, such as flagellar movement (9,10),
virulence factor production (11,12),
and secondary metabolite and biofilm production (9,13).
Various quorum-sensing signalling molecules have been identified,
including oligopeptides in Gram-positive bacteria and N-acyl
homoserine lactones (AHLs) in some Gram-negative bacteria (14). Acinetobacter spp. also
produce AHLs that possess quorum-sensing activity (15), and the A. baumannii AHL,
N-3-hydroxy-dodecanoyl-homoserine lactone
(N-3-OH-C12-HSL), is known to affect its motility and
biofilm formation (16,17).
Since quorum sensing allows bacteria to respond to
environmental changes as a colony and thereby boosts survival,
disrupting the quorum-sensing system may be a promising new
strategy for the treatment of infections (18–20).
It is important to investigate novel strategies for the inhibition
of A. baumannii by targeting AHLs (16,17,21–23),
but little is currently known about the types and functions of AHLs
produced by A. baumannii.
All AHLs share a common homoserine moiety but can
contain acyl side-chains of various lengths and degrees of
saturation and with various groups at the third carbon position.
AHLs generate characteristic fragment ions on electrospray
ionization (ESI) at a mass-to-charge ratio (m/z) of 102, and
the acyl side-chains generate the corresponding fragment ions at
m/z [M+H-101]+ (15,24–33).
The most common methods for identifying AHLs involve
a combination of thin-layer chromatography and biosensors (34,35).
These methods are simple and inexpensive but are limited by the
sensitivity of the biosensor and the use of standard substances as
references. In the present study, a clinical isolate of A.
baumannii strain S (AbS) was collected from the wound of a burn
patient and cultured. AHLs produced by AbS were subsequently
analysed by high-performance liquid chromatography (HPLC) and
either tandem quadrupole (TQ) or quadrupole time-of-flight (Q-TOF)
high-resolution mass spectrometry (HRMS). The present study adds to
the growing body of research on the quorum-sensing system of A.
baumannii and may contribute to the development of novel
antibacterial therapies that target AHLs for treating
multidrug-resistant A. baumannii infection.
Materials and methods
Bacterial strains and growth
conditions
A single nosocomial specimen of AbS was collected
from the wound surface exudates of a patient admitted to the
Department of Burns and Plastic Surgery at Ruijin Hospital
(Shanghai Jiaotong University School of Medicine, Shanghai, China)
in 2008. Antibiotic sensitivity was assessed according to the
guidelines provided by the Clinical and Laboratory Standards
Institute (CLSI) (36), which
included using ATB test strips (BioMérieux, Marcy l'Etoile, France)
and the Kirby-Bauer disk diffusion method with antibiotic discs
from Oxoid, Ltd. (Thermo Fisher Scientific, Inc., Waltham, MA,
USA). Agrobacterium tumefaciens strain KYC55 was used as a
biosensor, and was kindly provided by Professor Jun Zhu (College of
Life Sciences, Nanjing Agricultural University, Nanjing, China).
A. baumannii was cultured statically in Luria-Bertani (LB)
medium or Mueller-Hinton (MH) medium (Oxoid, Basingstoke, UK) at
37°C; A. tumefaciens KYC55 was cultured statically in LB
medium at 28°C.
Preparation of AHL extract
A. baumannii and A. tumefaciens were
stored at −80°C in bacteria stock solution (Beyotime Biotechnology,
Shanghai, China). A. baumannii were inoculated on LB agarose
plates and incubated overnight at 37°C. Individual colonies
(1×108 colony-forming units (CFU)/ml) were selected and
cultured in 15 ml LB medium at 37°C.
For HPLC-MS, bacteria were cultured in 500 ml LB
medium from the overnight LB agarose plates, and 500 ml bacterial
liquid was collected at 8 h, as determined by the AHL activity
curve. Bacterial samples were centrifuged (4,500 × g for 20 min)
and supernatants were passed through a 0.22 µm filter. An equal
volume of 100% ethyl acetate was added to the filtrate, and the
ethyl acetate phase was collected for AHL extraction and dried in a
vacuum centrifuge. The residue was the AHL extract and was then
re-dissolved in 50 µl ethyl acetate.
Analysis of AHL activity
AHL activity was measured at 4, 8, 16, 24, 32, 40
and 48 h after seeding. At each time point, 3 bacterial liquid were
collected and the optical density (OD) 600 was measured. AHL
extracts in 50 µl ethyl acetate from 4, 8, 16, 24, 32, 40 and 48 h
were added to cultures of A. tumefaciens KYC55 cultures, and
β-galactosidase activity was measured to indirectly indicate AHL
activity, as described previously (15,37).
Following overnight incubation, the OD600 was measured
and 0.8 ml Z buffer (in each litre containing: 16.1 g
(Na2HPO4) 7H2O, 5.5 g
(NaH2PO4) H2O, 0.75 g KCl, 0.245 g
(MgSO4) 7H2O, 2.7 ml 2-mercaptoethanol,
adjusted to pH 7.0 with HCl), 10 µl 0.05% sodium dodecyl sulphate,
15 µl chloroform and 0.1 ml ortho-nitrophenyl-β-galactoside (4
mg/ml) were added, with a final sample volume of 0.2 ml. The time
(T) taken for the solution to turn yellow was recorded, and 0.6 ml
1 M Na2CO3 was added to terminate the
reaction. The OD420 of supernatants was determined, and
relative AHL activity was calculated as follows: Activity in Miller
units=(1,000xOD420)/(OD600xTx0.2).
Identification of AHLs using
HPLC-MS
AHLs of different structures contain the same
homoserine lactone (HSL) ring, and this moiety generates
characteristic fragment ions at m/z 102 (25). Based on this principle, AbS AHLs
were identified by HPLC combined with either TQ or Q-TOF HRMS using
a 1200 HPLC-6140 TQ MS or a 1260 HPLC-6538 Q-TOF HRMS (Agilent
Technologies, Inc., Santa Clara, CA, USA), respectively. Resultant
chromatograms were compared with those of standard substances to
elucidate the structure of AbS AHLs. The test conditions were as
follows: An Agilent Poroshell 120 SB-C18 chromatographic column
(2.7 µm, 2.1×100 mm; Agilent Technologies, Inc.) was used with
acetonitrile and water as the mobile phase. The initial
acetonitrile concentration was 40%, which was increased to 100%
after 30 min, with a z-flow rate of 0.3 ml/min and sample injection
volume of 10 µl. Positive-ion ESI was conducted with the ion source
at 350°C. The dry N2 flow rate was 8 l/min, and the air
pressure of atomizing N2 was 40 psi. The capillary
voltage was 4,000 V. HPLC-TQ HRMS was performed with a precursor
ion scan and daughter ion scan (collision energy, 15–30 units),
while HPLC-Q-TOF HRMS involved MS1 and MS2 full scans (collision
energy, 15–30 units).
Establishment of an AHL-deficient AbS
mutant (AbS-M)
An AbS mutant that is unable to produce AHLs was
established using pKNG101.abaI::Km, as previously described
(38); the pKNG101.abaI::Km
plasmid was provided by Professor Philip N. Rather (Department of
Microbiology and Immunology, Emory University School of Medicine,
Atlanta, GA, USA). This vector was transformed into Escherichia
coli strain SM10, and the resultant SM10/pKNG101.abaI::Km was
cultured with AbS in a filter-mating system in LB medium at 37°C
without antibiotics for 24 h, after which it was cultured and
screened on LB agarose plates containing 10% sucrose without NaCl.
Sucrose resistance indicates that the bacteria have lost the
integrated pKNG101 plasmid and therefore streptomycin sensitivity.
AbS-M was screened for kanamycin resistance, and Southern blotting
was used to confirm that colonies with this phenotype had abaI::Km
disruption, as previously described (38).
Antibacterial sensitivity of AbS and
AbS-M
The minimum inhibitory concentration (MIC) of common
antibacterial drugs (including, meropenem, piperacillin,
ceftazidime, ciprofloxacin, sulfamethoxazole/trimethoprim and
minocycline) against AbS, AbS-M and AbS-M supplemented with 10 µmol
N-3-OH-C12-HSL (AbS-M+HSL; #53727; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) was assessed using the
broth-micro-dilution method, according to the CLSI protocol.
Briefly, overnight bacterial cultures were inoculated at
5×105 CFU/ml in 1 ml MH medium containing a range of
concentrations (128, 64, 32, 16, 8, 4, 2, 1, 0.5, 0.25, 0.125,
0.0625, 0.03125 and 0 µg/ml) of the antibacterial drugs. Following
24 h incubation at 37°C, the MIC against bacterial growth was
assessed by visual examination. Each antibiotic concentration was
tested three times.
Expression of drug-resistance genes in
AbS, AbS-M and AbS-M+HSL treated with 0.125 μg/ml meropenem or AbS
untreated with meropenem (AbS-U) for 24 h
A total of 45 µl 0.5 McFarland bacterial liquid
(AbS, AbS-M and AbS-M+AHL) was added to 3 ml LB medium containing
0.125 µg/ml meropenem, with a final concentration of 10 µM AHL
(N-3-OH-C12-HSL). Alternatively, 45 µl 0.5 McFarland
bacterial liquid was added into 3 ml LB medium without meropenem
(AbS-U). The cultures were incubated at 37°C. After 24 h, 1 ml
bacterial liquid was centrifuged (10,621 × g for 1 min) and
supernatants were discarded. Total RNA was extracted using TRIzol
Reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol; concentration and
purity were determined using an ultraviolet spectrophotometer. RNA
was reverse transcribed into cDNA using the AMV First Strand cDNA
Synthesis kit (New England Biolabs, Inc., Ipswich, MA, USA),
according to the manufacturer's protocol. AbS-U, AbS, AbS-M and
AbS-M+HSL cultures were incubated for 24 h and the expression
levels of 16S rRNA, Oxacillinase (OXA)-51, AmpC type
β-lactamase (AmpC), oxacillinase (OXA)-23, IMP type
metallo-β-lactamase (IMP)-4, verona integron-mediated
metallo-β-lactamase (VIM)-2, Acinetobacter drug
efflux (Ade) A, AdeB and AdeC were assessed by
quantitative polymerase chain reaction (qPCR) using a StepOnePlus
Real-Time PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) and SYBR-Green Master Mix (Thermo Fisher Scientific, Inc.);
primers used are listed in Table
I. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression
was measured as a reference, and gene expressions were calculated
in terms of fold change using the comparative Cq method; relative
mRNA expression was calculated using the 2−ΔΔCq method
(39). The experiments were
repeated 3 times.
 | Table I.Primer sequences used for
quantitative polymerase chain reaction. |
Table I.
Primer sequences used for
quantitative polymerase chain reaction.
| Gene | Primer sequence
(5′-3′) | Product length
(bp) |
|---|
| 16S rRNA | F:
ACGGTCGCAAGACTAAAACTCA | 108 |
|
| R:
GTATGTCAAGGCCAGGTAAGGT |
|
| OXA-51 | F:
CTATGGTAATGATCTTGCTCGTG | 104 |
|
| R:
TGGTGGTTGCCTTATGGTG |
|
| AmpC | F:
TTATGCGGGCAATACACCA | 207 |
|
| R:
CTGACAGAACCTAGCTCAAAAATG |
|
| OXA-23 | F:
AAGGGCGAGAAAAGGTCATT | 89 |
|
| R:
TCCTGATAGACTGGGACTGCA |
|
| IMP-4 | F:
ATTCTCAATCCATCCCCACG | 185 |
|
| R:
CCTTTCAGGCAGCCAAACTAC |
|
| VIM-2 | F:
AACTCTTCTATCCTGGTGCTGC | 105 |
|
| R:
TGCGTGACAACTCATAAATCG |
|
| AdeA | F:
AGTCGGAGGTATCATTGAAAAGG | 162 |
|
| R:
TGAACTTTGAGTCTTGCCACCT |
|
| AdeB | F:
ATGCGTGAAATGGAACAACTG | 145 |
|
| R:
CCAAGACAAGGAAGACAACTAACA |
|
| AdeC | F:
GCCATTCAATCAGCTTTTCGT | 117 |
|
| R:
GAGTTTATAGGTTGCAGCAGTCG |
|
| GAPDH | F:
ACCACAGTCCATGCCATCAC | 440 |
|
| R:
TCCACCACCCTGTTGCTGTA |
|
Statistical analyses
Data were presented as the mean ± standard deviation
and analysed using Student's t-test, analysis of variance and least
significant difference pot hoc test with SPSS version 19.0 (IBM
SPSS, Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
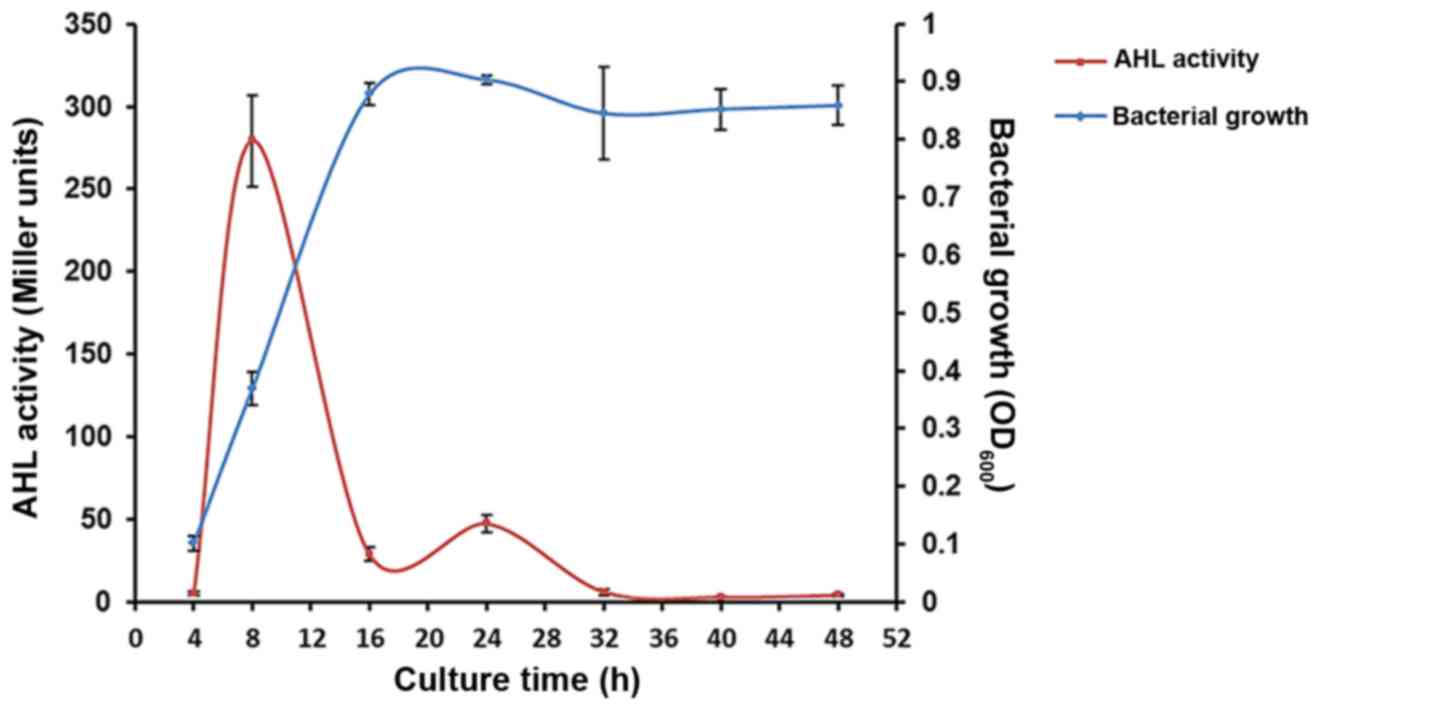
Changes in AHL activity of AbS
AbS growth rate and AHL activity were measured
periodically between 4 and 48 h incubation (Fig. 1). AHL activity increased from
5.00±1.00 Miller units at 4 h culture to a maximum of 279.33±27.59
Miller units at 8 h. Subsequently, the activity decreased to
28.67±4.16 Miller units at 16 h and plateaued. The AbS growth curve
did not correlate with AHL activity after bacterial growth reached
the log phase; the OD600 (bacterial growth) peaked at
0.90±0.01 after 24 h of culture and then plateaued.
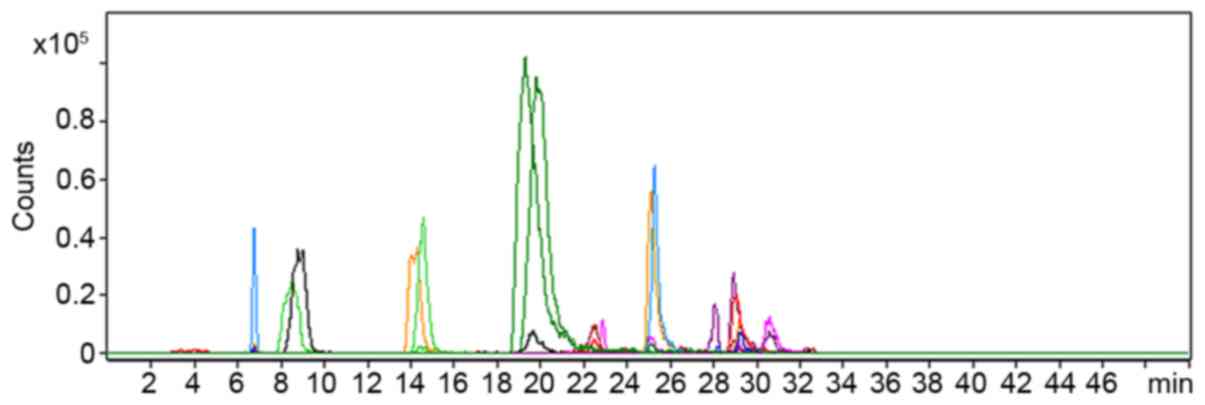
AHLs produced by AbS
AHLs that were extracted from AbS culture
supernatants using ethyl acetate were screened using HPLC-TQ MS. As
presented in Fig. 2, the precursor
ion scan at m/z 102 detected 30 precursor ions, including
those at m/z 282, 284 and 300 (each precursor ion represents
one compound). MS2 spectrum analysis of these 30 ions confirmed
that they could generate fragment ions at m/z 102,
suggesting that they may be AHLs.
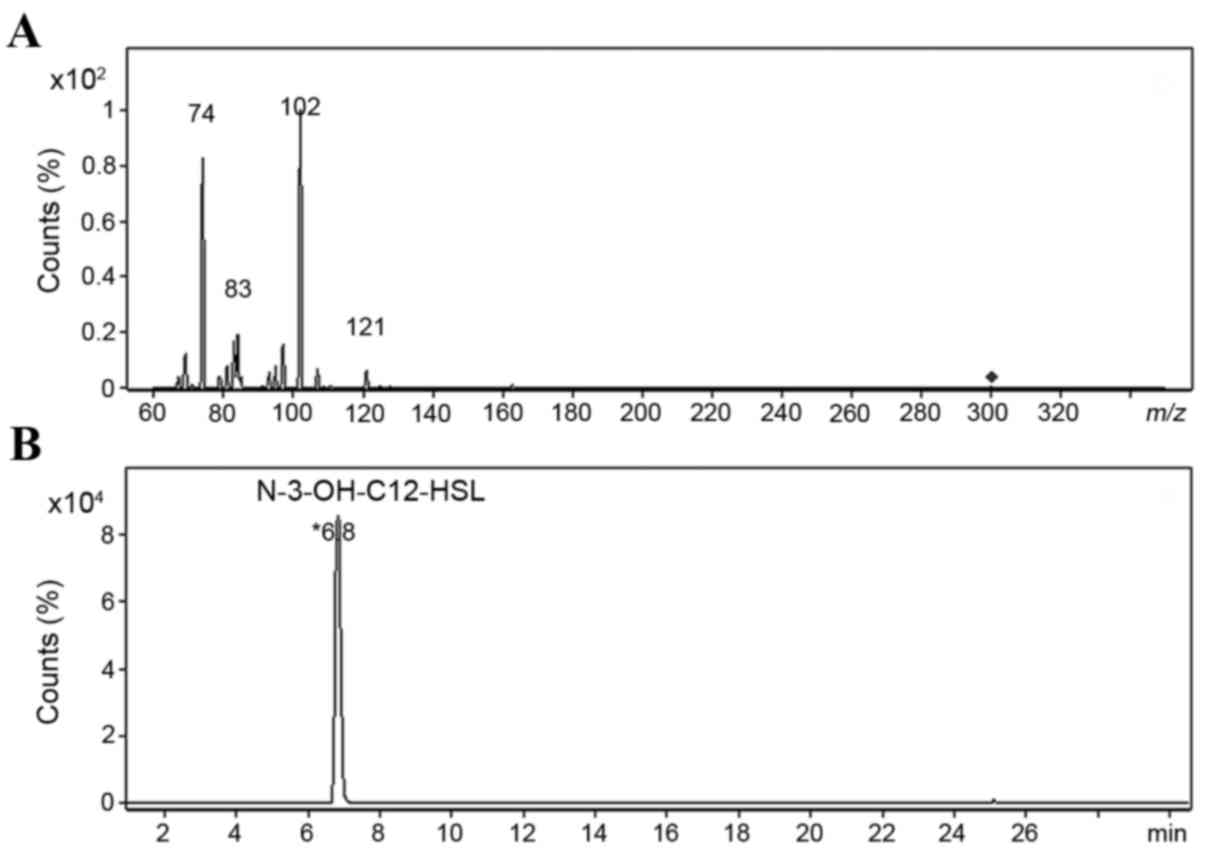
In the positive ionization mode of ESI, extracted
AHLs generated a quasi-molecular ion at m/z 300 and major
fragment ions at m/z 102 and m/z 74 (Fig. 3A). The ion at m/z 102 was
the most abundant, and in order to determine its structure,
HPLC-Q-TOF HRMS was used to examine the elemental composition of
this ion and related daughter fragment ions. The elemental
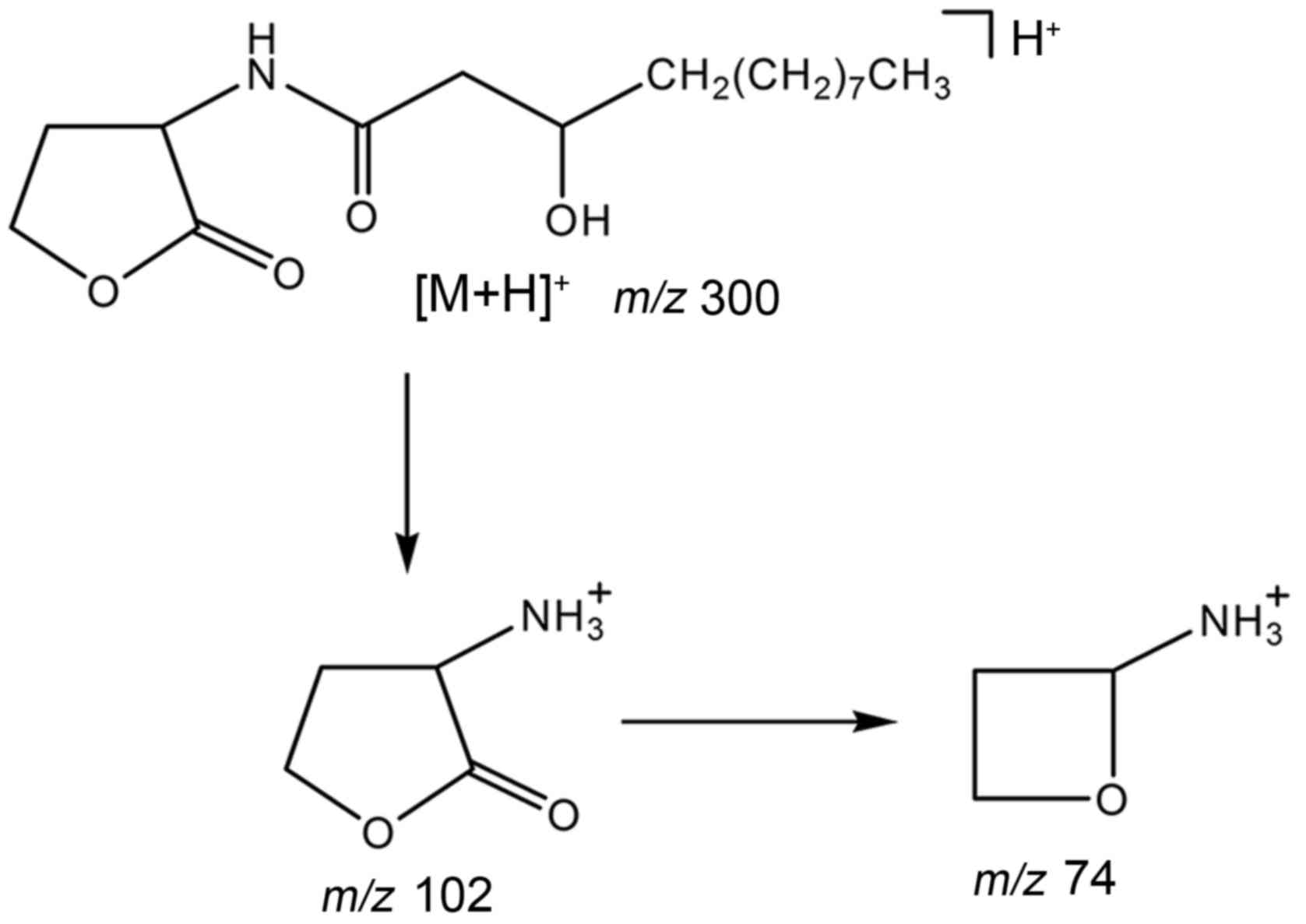
composition at m/z 300 was
C16H30NO4, representing the [M+H]
+ ions of N-3-OH-C12-HSL. The two major
fragment ions were C4H8NO (m/z 102)
and C3H8NO (m/z 74), both of which
were derived from the HSL ring of N-3-OH-C12-HSL, which
was the only AHL molecule identified (Fig. 3B). The major fragmentation pathway
is shown in Fig. 4. In addition,
some low-abundance ions were detected in the MS2 spectrum
(m/z 121, 97 and 83), and they contained only two elements,
H and C. We hypothesized that these were derived from the
fragmentation of carbon chains near the acyl group.
Since components from the culture media may
interfere with AHL detection, HPLC-Q-TOF HRMS and tandem MS were
used to scan for AHL molecules identified in previous screens. The
present study also determined the elemental compositions of the 30
precursor ions and their corresponding daughter ions at m/z
102. The results revealed that only ions detected at m/z
300≥102 met the structural requirement for AHLs. The daughter ions
at m/z 102 were derived from 29 candidate molecules that
contained C5H12NO and therefore could not be
AHLs.
According to the composition and degree of
unsaturation, the signal molecule at m/z 300 was inferred to
be N-3-OH-C12-HSL. HPLC-MS was then used to examine a
commercially available N-3-OH-C12-HSL, and the results
confirmed that it was structurally identical to the
N-3-OH-C12-HSL detected in the present study.
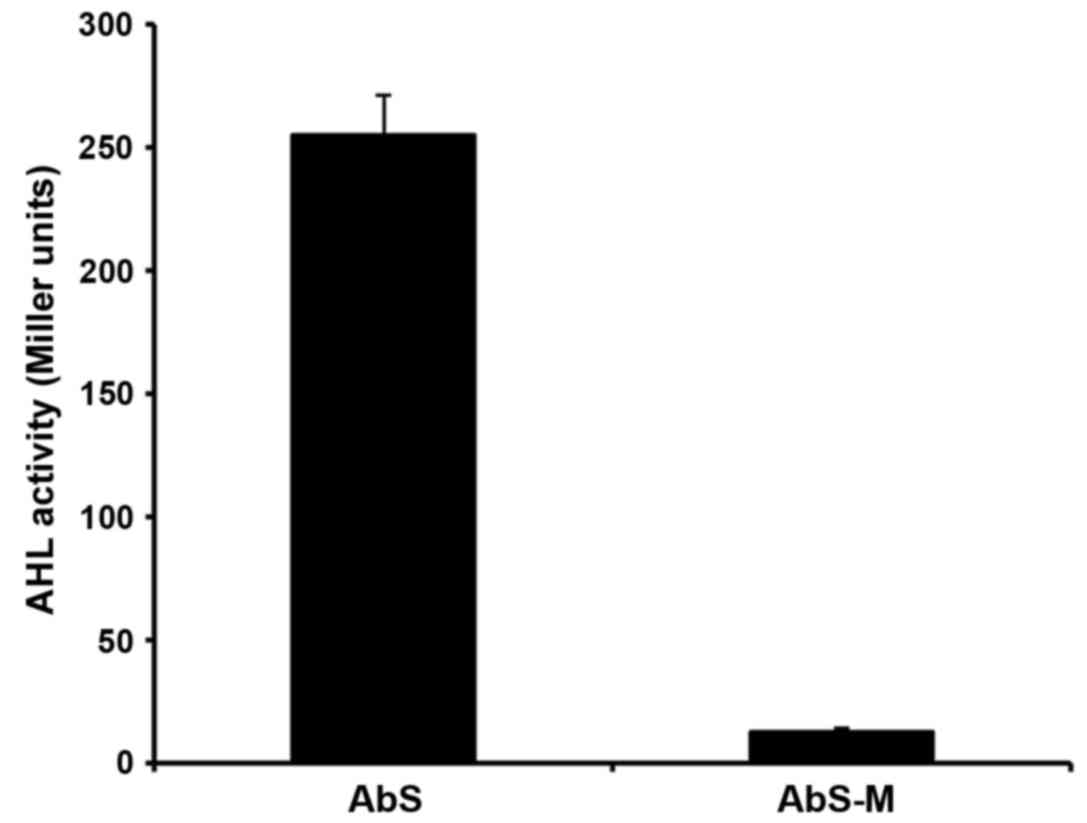
Activity of mutant AHL
AHLs were isolated from the supernatant of AbS and
AbS-M cultures incubated for 8 h at 37°C and the activity levels
were analysed. AHL activity was significantly lower in AbS-M
(12.67±1.53 Miller units) compared with wild-type AbS (255.67±16.01
Miller units) (P<0.01; Fig.
5).
Antibiotic sensitivity of AbS, AbS-M
and AbS-M+HSL
AbS was sensitive to amikacin, cefuroxime,
ceftazidime, imipenem, gentamicin, ciprofloxacin,
sulfamethoxazole/trimethoprim, sulperazone, tazocin, cefepime,
panipenem, meropenem, ampicillin, Unasyn (which is a combination of
ampicillin and sulbactam) and piperacillin. Then the MIC of AbS,
AbS-M and AbS-M + HSL cultures to meropenem, piperacillin,
ceftazidime, ciprofloxacin, sulfamethoxazole/trimethoprim and
minocycline was assessed (Table
II). The MICs of meropenem and piperacillin against AbS-M (0.25
and 1 µg/ml, respectively) were lower than the MICs of these
antibiotics against wild-type AbS (0.5 and 2 µg/ml, respectively).
However, the addition of HSL to the AbS-M culture raised the MICs
of meropenem and piperacillin to similar levels as wild-type AbS
(0.5 and 2 µg/ml, respectively). By contrast, the MICs of
ceftazidime, ciprofloxacin, sulfamethoxazole/trimethoprim and
minocycline were similar for AbS, AbS-M and AbS-M + HSL.
 | Table II.Antibiotic sensitivity. |
Table II.
Antibiotic sensitivity.
|
| Minimum inhibitory
concentration (µg/ml) |
|---|
|
|
|
|---|
| Antibiotic | AbS | AbS-M | AbS-M + HSL |
|---|
| Meropenem | 0.5 | 0.25 | 0.5 |
| Piperacillin | 2.0 | 1.0 | 2.0 |
| Ceftazidime | 0.25 | 0.25 | 0.25 |
| Ciprofloxacin | 0.5 | 0.5 | 0.5 |
|
Sulfamethoxazole/trimethoprim | 0.25/4.75 | 0.25/4.75 | 0.25/4.75 |
| Minocycline | 0.5 | 0.5 | 0.5 |
Expression of drug-resistance genes in
AbS-U, AbS, AbS-M and AbS-M + HSL treated with meropenem for 24
h
AbS, AbS-M and AbS-M + HSL were cultured for 24 h in
LB medium supplemented with meropenem (0.125 µg/ml), and AbS
untreated with meropenem (AbS-U) was additionally cultured. The
mRNA expression levels of drug-resistance genes were assessed by
qPCR (Table III). Meropenem
treatment increased the expression of OXA-51, AmpC, AdeA and
AdeB in all three bacterial cultures. The mRNA expression
levels of these four genes were significantly lower in AbS-M
compared with wild-type AbS; however, supplementation of AbS-M
cultures with N-3-OH-C12-HSL increased the mRNA
expression of these four drug-resistance genes to higher levels
compared with wild-type AbS and untreated AbS-M. The expression of
OXA-23, IMP-4, VIM-2 and AdeC could not be detected
in any of the three strains.
 | Table III.mRNA expression levels of
multidrug-resistance genes in meropenem-treated cultures or
untreated cultures. |
Table III.
mRNA expression levels of
multidrug-resistance genes in meropenem-treated cultures or
untreated cultures.
| Gene | AbS-U | AbS | AbS-M | AbS-M +HSL |
|---|
| OXA-51 | 0.13±0.02 | 1.09±0.13 |
0.68±0.04a | 1.74±0.04 |
| AmpC | 0.12±0.03 | 0.94±0.11 |
0.60±0.04a | 1.55±0.04 |
| OXA-23 | ND | ND | ND | ND |
| IMP-4 | ND | ND | ND | ND |
| VIM-2 | ND | ND | ND | ND |
| AdeA | 0.08±0.04 | 1.17±0.17 |
0.59±0.08a | 1.66±0.25 |
| AdeB | 0.09±0.08 | 1.08±0.16 |
0.51±0.09a | 1.31±0.11 |
| AdeC | ND | ND | ND | ND |
Discussion
Quorum sensing affects bacterial biofilm formation
(27,28), antibacterial drug sensitivity
(29) and bacterial virulence
(30), suggesting that inhibition
of this system may be a useful therapeutic strategy in combating
the emergence of antibiotic-resistant strains of pathogenic
bacteria. The present study aimed to contribute to the growing body
of literature on AHLs produced by clinical isolates of A.
baumannii.
The present study found that although the activity
of AHLs produced by AbS was positively correlated with bacterial
density in the log phase of growth, AHL activity reduced as growth
plateaued; this trend has been previously reported for other
bacteria (31,32). N-(3-oxohexanoyl)-L-HSL produced by
Erwinia carotovora was revealed to be unstable at pH
>7–8, which is the pH of the stationary phase of bacterial
growth (40). Another study
demonstrated that, during growth plateauing, A. tumefaciens
produces abundant levels of acyl-homoserine lactonases, which
reduce AHL activity (32). Thus,
AHL activity seems to be regulated by the growth rate, which allows
bacteria to respond to their changing density.
In the present study, HPLC-MS with TQ and Q-TOF was
used to successfully identify AHLs. This method is advantageous
because it does not depend on biosensor sensitivity and reference
substances, and thus may be preferable to the conventional methods
used for AHL identification, which combine thin-layer
chromatography with biosensors.
AHLs produced by A. baumannii have been
proposed to vary depending on culture conditions (41). One previous study identified
3-OH-C12-HSL and other AHLs of unknown structure in
cultures of A. baumannii strain M2 (38), whereas another study identified
C6-HSL and C8-HSL in cultures of A.
baumannii strain 4KT (15).
Furthermore, P. aeruginosa infections in patients with
cystic fibrosis have been reported to produce different AHLs in
vivo and in vitro (42). Thus, the AHL identified in the
present study may differ from those identified previously from
A. baumannii, owing to the particular strain and culture
conditions used. Additional experiments are required to identify
the range of AHLs produced by this organism.
The present study established an AHL-deficient AbS
mutant that was used to determine whether AHLs affected
antibacterial drug sensitivity of AbS. Antibacterial
drug-sensitivity assays revealed that the MICs of meropenem and
piperacillin were lower in AbS-M compared with wild-type AbS;
however, the MICs returned to wild-type AbS levels when AbS-M
cultures were treated HSL. Although this AHL-mediated increase in
MICs was not substantial, this finding is promising in that it
confirms the association between AHLs and antibiotic resistance in
A. baumannii.
A previous report regarding the influence of AHLs on
bacterial drug resistance mainly focused on their influence on
biofilm formation (9). Previous
studies have also described multiple mechanisms of drug resistance
in A. baumannii, including the production of β-lactamases
(43), which can be divided into
four categories: Extended spectrum β-lactamases (44–46),
metallo-β-lactamases (47,48), AmpC enzyme (49) and oxacillinases (50,51).
However, the present study sought to determine the influence of
AHLs on the expression of drug-resistance genes and revealed that
in the presence of meropenem AbS expressed OXA-51 and
AmpC, but not OXA-23, IMP-4, or VIM-2.
OXA-51 was previously demonstrated to be strongly expressed
in Acinetobacter spp. and may be the main drug-resistance
gene (52,53), whereas AmpC is often found
in A. baumannii strains from China (54,55).
The present study found that the mRNA expression levels of
OXA-51 and AmpC were significantly lower in AbS-M
compared with wild-type AbS, but the levels recovered upon
supplementation of the AbS-M culture with an AHL extract. These
results indicate that AHLs may strengthen drug resistance by
moderating the expression of drug-resistance genes.
In addition to producing β-lactamases, A.
baumannii expresses efflux pump genes AdeA, AdeB and
AdeC, which confer resistance to β-lactam antibiotics,
aminoglycosides, erythromycins, quinolones, tetracyclines,
chloramphenicol and trimethoprim (43,56–62).
The present study found that AdeA and AdeB were
expressed by AbS in the presence of meropenem. It was not
unexpected that AdeC was not detected, since this gene is
not essential for efflux pump activity (59). The mRNA expression levels of
AdeA and AdeB were significantly lower in AbS-M than
in wild-type AbS, and the expression of both AdeA and
AdeB was recovered with AHL supplementation. Results from
the present study indicated that AbS AHLs promote the expression of
OXA-51, AmpC, AdeA and AdeB in the presence of
meropenem, suggesting that AbS produces AHLs to enhance antibiotic
resistance. Furthermore, upregulation of AdeB expression has
been reported to be associated with the emergence of pan-resistant
A. baumannii (57). Thus,
AHLs may promote the emergence of meropenem-induced multidrug- and
pan-resistance.
The present study has some limitations that should
be noted. Although a mutant strain AbS was designed to be deficient
in AHL, subsequent experiments with this mutant may have been
influenced by the presence of abaI homologues; AbaI is
similar to the LuxI family of autoinducer synthases (37). In addition, it is well known that
AHLs can be degraded by N-acylhomoserine lactone-lactonase
(32). Therefore, we cannot rule
out the possibility of AHL degradation due to lactonolysis. Lastly,
the present study did not determine whether the mutation in AbS-M
specifically reduces the transcription of abaI or whether it
causes a generalized reduction in transcription.
In Gram-negative bacteria, AHL receptor systems
include the cytoplasmic LuxR receptor and the transmembrane LuxN
receptor (19). Inactivation of
suppressor of division inhibition (SdiA), a bacterial homolog of
LuxR, hampers the expression of the efflux pump drug-resistance
genes acrA and acrB, which are responsible for
bacterial multidrug resistance, and AHLs may interact with SdiA to
enhance the expression of acrA and acrB (63). Similar systems may exist in A.
baumannii, and the interaction of AHLs with such systems may be
able to induce the expression of drug-resistance genes. However,
the MIC of the antibiotics ceftazidime, ciprofloxacin,
sulfamethoxazole/trimethoprim and minocycline did not differ
between the presence and absence of AHLs, suggesting that
drug-resistant phenotypes may be produced by a diverse range of
factors and genes. Conversely, exposure to meropenem for 24 h was
perhaps insufficient to induce significant phenotypic alterations,
and additional experiments are required to rule out longer-term
changes to genes encoding resistance to these antibiotics. However,
results from the present study are notable, since to the best of
our knowledge no previous study has addressed the mechanisms
underlying the influence of AbS AHLs on the expression of
drug-resistance genes.
In the present study, the quorum-sensing system of
AbS was demonstrated to involve N-3-OH-C12-HSL, which
induced the expression of drug-resistance genes OXA-51,
AmpC, AdeA and AdeB in the presence of meropenem.
Loss of AHL production in AbS-M resulted in reduced mRNA expression
of these four drug-resistance genes, while treatment with
N-3-OH-C12-HSL restored their expression. Thus,
AHL-mediated induction of AdeA and AdeB expression
could in turn lead to multidrug resistance in A. baumannii.
These results highlight a new direction for the development of
drugs targeting A. baumannii, particularly pan-resistant
strains.
Acknowledgements
The authors thank Professor Jun Zhu (College of
Life Sciences, Nanjing Agricultural University, Nanjing, China) for
his kind gift of A. tumefaciens KYC55 and Professor Philip
N. Rather (Department of Microbiology and Immunology, Emory
University School of Medicine, Atlanta, GA, USA) for his kind gift
of the pKNG101.abaI::Km plasmid. The authors are also grateful to
the Shanghai Ninth Peoples Hospital, Shanghai Jiaotong University
School of Medicine, Shanghai Research Institute of Stomatology and
Shanghai Key Laboratory of Stomatology (Shanghai, China) for
experimental support.
References
|
1
|
Dou Y, Zhang Q and Liao ZJ: Investigation
on the drug resistance of Pseudomonas aeruginosa in our burn ward
in the past 11 years. Zhonghua Shao Shang Za Zhi. 20:6–9. 2004.(In
Chinese).
|
|
2
|
Dou Y, Zhang X, Zhang Q and Shi Y:
Analysis of the drug-resistance of Pseudomonas aeruginosa and the
use of antibiotics in burn wards. Zhonghua Shao Shang Za Zhi.
27:109–113. 2011.(In Chinese).
|
|
3
|
Essayagh T, Zohoun A, Essayagh M, Elameri
A, Zouhdi M, Ihrai H and Elhamzaoui S: Bacterial epidemiology in
the burns unit at military teaching hospital Mohamed V of Rabat.
Ann Biol Clin (Paris). 69:71–76. 2011.(In French).
|
|
4
|
Chong SJ, Ahmed S, Tay JM, Song C and Tan
TT: 5 year analysis of bacteriology culture in a tropical burns
ICU. Burns. 37:1349–1353. 2011. View Article : Google Scholar
|
|
5
|
Glik J, Kawecki M, Gázdzik T and Nowak M:
The impact of the types of microorganisms isolated from blood and
wounds on the results of treatment in burn patients with sepsis.
Pol Przegl Chir. 84:6–16. 2012.
|
|
6
|
Dijkshoorn L, Nemec A and Seifert H: An
increasing threat in hospitals: Multidrug-resistant Acinetobacter
baumannii. Nat Rev Microbiol. 5:939–951. 2007. View Article : Google Scholar
|
|
7
|
Lin MF and Lan CY: Antimicrobial
resistance in Acinetobacter baumannii From bench to bedside. World
J Clin Cases. 2:787–814. 2014. View Article : Google Scholar :
|
|
8
|
Simor AE, Lee M, Vearncombe M, Jones-Paul
L, Barry C, Gomez M, Fish JS, Cartotto RC, Palmer R and Louie M: An
outbreak due to multiresistant Acinetobacter baumannii in a burn
unit: Risk factors for acquisition and management. Infect Control
Hosp Epidemiol. 23:261–267. 2002. View
Article : Google Scholar
|
|
9
|
Li YH and Tian X: Quorum sensing and
bacterial social interactions in biofilms. Sensors (Basel).
12:2519–2538. 2012. View Article : Google Scholar :
|
|
10
|
Zan J, Heindl JE, Liu Y, Fuqua C and Hill
RT: The CckA-ChpT-CtrA phosphorelay system is regulated by quorum
sensing and controls flagellar motility in the marine sponge
symbiont Ruegeria sp. KLH11. PLoS One. 8:e663462013. View Article : Google Scholar :
|
|
11
|
Smith RS, Harris SG, Phipps R and Iglewski
B: The Pseudomonas aeruginosa quorum-sensing molecule
N-(3-oxododecanoyl)homoserine lactone contributes to virulence and
induces inflammation in vivo. J Bacteriol. 184:1132–1139. 2002.
View Article : Google Scholar :
|
|
12
|
Antunes LC, Ferreira RB, Buckner MM and
Finlay BB: Quorum sensing in bacterial virulence. Microbiology.
156:2271–2282. 2010. View Article : Google Scholar
|
|
13
|
Bhargava N, Sharma P and Capalash N:
Quorum sensing in Acinetobacter: An emerging pathogen. Crit Rev
Microbiol. 36:349–360. 2010. View Article : Google Scholar
|
|
14
|
Miller MB and Bassler BL: Quorum sensing
in bacteria. Annu Rev Microbiol. 55:165–199. 2001. View Article : Google Scholar
|
|
15
|
Chan KG, Cheng HJ, Chen JW, Yin WF and
Ngeow YF: Tandem mass spectrometry detection of quorum sensing
activity in multidrug resistant clinical isolate Acinetobacter
baumannii. ScientificWorldJournal. 2014:8910412014. View Article : Google Scholar :
|
|
16
|
Stacy DM, Welsh MA, Rather PN and
Blackwell HE: Attenuation of quorum sensing in the pathogen
Acinetobacter baumannii using non-native N-Acyl homoserine
lactones. ACS Chem Biol. 7:1719–1728. 2012. View Article : Google Scholar :
|
|
17
|
Chow JY, Yang Y, Tay SB, Chua KL and Yew
WS: Disruption of biofilm formation by the human pathogen
Acinetobacter baumannii using engineered quorum-quenching
lactonases. Antimicrob Agents Chemother. 58:1802–1805. 2014.
View Article : Google Scholar :
|
|
18
|
Roy V, Adams BL and Bentley WE: Developing
next generation antimicrobials by intercepting AI-2 mediated quorum
sensing. Enzyme Microb Technol. 49:113–123. 2011. View Article : Google Scholar
|
|
19
|
Chen G, Swem LR, Swem DL, Stauff DL,
O'Loughlin CT, Jeffrey PD, Bassler BL and Hughson FM: A strategy
for antagonizing quorum sensing. Mol Cell. 42:199–209. 2011.
View Article : Google Scholar :
|
|
20
|
Kalia VC: Quorum sensing inhibitors: An
overview. Biotechnol Adv. 31:224–245. 2013. View Article : Google Scholar
|
|
21
|
Berger M, Neumann A, Schulz S, Simon M and
Brinkhoff T: Tropodithietic acid production in Phaeobacter
gallaeciensis is regulated by N-acyl homoserine lactone-mediated
quorum sensing. J Bacteriol. 193:6576–6585. 2011. View Article : Google Scholar :
|
|
22
|
Churchill ME and Chen L: Structural basis
of acyl-homoserine lactone-dependent signaling. Chem Rev.
111:68–85. 2011. View Article : Google Scholar
|
|
23
|
Van Mooy BA, Hmelo LR, Sofen LE, Campagna
SR, May AL, Dyhrman ST, Heithoff A, Webb EA, Momper L and Mincer
TJ: Quorum sensing control of phosphorus acquisition in
Trichodesmium consortia. ISME J. 6:422–429. 2012. View Article : Google Scholar
|
|
24
|
Bruhn JB, Christensen AB, Flodgaard LR,
Nielsen KF, Larsen TO, Givskov M and Gram L: Presence of acylated
homoserine lactones (AHLs) and AHL-producing bacteria in meat and
potential role of AHL in spoilage of meat. Appl Environ Microbiol.
70:4293–4302. 2004. View Article : Google Scholar :
|
|
25
|
Gould TA, Herman J, Krank J, Murphy RC and
Churchill ME: Specificity of acyl-homoserine lactone synthases
examined by mass spectrometry. J Bacteriol. 188:773–783. 2006.
View Article : Google Scholar :
|
|
26
|
Wayne P: Clinical and Laboratory Standards
Institute: Methods for Dilution Antimicrobial Susceptibility Tests
for bacteria that grow aerobicallyApproved Standard. seventh. pp.
M7–A7. CLSI; 2006
|
|
27
|
Cady NC, McKean KA, Behnke J, Kubec R,
Mosier AP, Kasper SH, Burz DS and Musah RA: Inhibition of biofilm
formation, quorum sensing and infection in Pseudomonas aeruginosa
by natural products-inspired organosulfur compounds. PLoS One.
7:e384922012. View Article : Google Scholar :
|
|
28
|
Jakobsen TH, Bragason SK, Phipps RK,
Christensen LD, Van Gennip M, Alhede M, Skindersoe M, Larsen TO,
Høiby N, Bjarnsholt T and Givskov M: Food as a source for quorum
sensing inhibitors: Iberin from horseradish revealed as a quorum
sensing inhibitor of Pseudomonas aeruginosa. Appl Environ
Microbiol. 78:2410–2421. 2012. View Article : Google Scholar :
|
|
29
|
Brackman G, Cos P, Maes L, Nelis HJ and
Coenye T: Quorum sensing inhibitors increase the susceptibility of
bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob
Agents Chemother. 55:2655–2661. 2011. View Article : Google Scholar :
|
|
30
|
Koh KH and Tham FY: Screening of
traditional Chinese medicinal plants for quorum-sensing inhibitors
activity. J Microbiol Immunol Infect. 44:144–148. 2011. View Article : Google Scholar
|
|
31
|
Byers JT, Lucas C, Salmond GP and Welch M:
Nonenzymatic turnover of an Erwinia carotovora quorum-sensing
signaling molecule. J Bacteriol. 184:1163–1171. 2002. View Article : Google Scholar :
|
|
32
|
Zhang HB, Wang LH and Zhang LH: Genetic
control of quorum-sensing signal turnover in Agrobacterium
tumefaciens. Proc Natl Acad Sci USA. 99:pp. 4638–4643. 2002;
View Article : Google Scholar :
|
|
33
|
Ortori CA, Atkinson S, Chhabra SR, Camara
M, Williams P and Barrett DA: Comprehensive profiling of
N-acylhomoserine lactones produced by Yersinia pseudotuberculosis
using liquid chromatography coupled to hybrid quadrupole-linear ion
trap mass spectrometry. Anal Bioanal Chem. 387:497–511. 2007.
View Article : Google Scholar
|
|
34
|
Shaw PD, Ping G, Daly SL, Cha C, JE Jr,
Rinehart KL Cronan and Farrand SK: Detecting and characterizing
N-acyl-homoserine lactone signal molecules by thin-layer
chromatography. Proc Natl Acad Sci USA. 94:pp. 6036–6041. 1997;
View Article : Google Scholar :
|
|
35
|
Cha C, Gao P, Chen YC, Shaw PD and Farrand
SK: Production of acyl-homoserine lactone quorum-sensing signals by
gram-negative plant-associated bacteria. Mol Plant Microbe
Interact. 11:1119–1129. 1998. View Article : Google Scholar
|
|
36
|
Clinical and Laboratory Standards
Institute, . Performance standards for antimicrobial susceptibility
testing: Twenty-forth Informational Supplement M100-S24. CLSI;
Wayne, PA, USA: 2014
|
|
37
|
Zhu J, Chai Y, Zhong Z, Li S and Winans
SC: Agrobacterium bioassay strain for ultrasensitive detection of
N-acylhomoserine lactone-type quorum-sensing molecules: Detection
of autoinducers in Mesorhizobium huakuii. Appl Environ Microbiol.
69:6949–6953. 2003. View Article : Google Scholar :
|
|
38
|
Niu C, Clemmer KM, Bonomo RA and Rather
PN: Isolation and characterization of an autoinducer synthase from
Acinetobacter baumannii. J Bacteriol. 190:3386–3392. 2008.
View Article : Google Scholar :
|
|
39
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
40
|
Byers JT, Lucas C, Salmond GP and Welch M:
Nonezymatic turnover of an Erwinia carotovora quorum-sensing signal
molecule. J Bacteriol. 184:1163–1171. 2002. View Article : Google Scholar :
|
|
41
|
González RH, Nusblat A and Nudel BC:
Detection and characterization of quorum sensing signal molecules
in Acinetobacter strains. Microbiol Res. 155:271–277. 2001.
View Article : Google Scholar
|
|
42
|
Middleton B, Rodgers HC, Cámara M, Knox
AJ, Williams P and Hardman A: Direct detection of N-acylhomoserine
lactones in cystic fibrosis sputum. FEMS Microbiol Lett. 207:1–7.
2002. View Article : Google Scholar
|
|
43
|
Peleg AY, Seifert H and Paterson DL:
Acinetobacter baumannii: Emergence of a successful pathogen. Clin
Microbiol Rev. 21:538–582. 2008. View Article : Google Scholar :
|
|
44
|
Poirel L, Mugnier PD, Toleman MA, Walsh
TR, Rapoport MJ, Petroni A and Nordmann P: ISCR2, another vehicle
for bla(VEB) gene acquisition. Antimicrob Agents Chemother.
53:4940–4943. 2009. View Article : Google Scholar :
|
|
45
|
Naas T, Bogaerts P, Bauraing C, Degheldre
Y, Glupczynski Y and Nordmann P: Emergence of PER and VEB
extended-spectrum beta-lactamases in Acinetobacter baumannii in
Belgium. J Antimicrob Chemother. 58:178–182. 2006. View Article : Google Scholar
|
|
46
|
Nagano N, Nagano Y, Cordevant C, Shibata N
and Arakawa Y: Nosocomial transmission of CTX-M-2
beta-lactamase-producing Acinetobacter baumannii in a neurosurgery
ward. J Clin Microbiol. 42:3978–3984. 2004. View Article : Google Scholar :
|
|
47
|
Yum JH, Yi K, Lee H, Yong D, Lee K, Kim
JM, Rossolini GM and Chong Y: Molecular characterization of
metallo-beta-lactamase-producing Acinetobacter baumannii and
Acinetobacter genomospecies 3 from Korea: Identification of two new
integrons carrying the bla(VIM-2) gene cassettes. J Antimicrob
Chemother. 49:837–840. 2002. View Article : Google Scholar
|
|
48
|
Houang ET, Chu YW, Lo WS, Chu KY and Cheng
AF: Epidemiology of rifampin ADP-ribosyltransferase (arr-2) and
metallo-btea-lactamase (blaIMP-4) gene cassettes in
class 1 integrons in Acinetobacter strains isolated from blood
cultures in 1997 to 2000. Antimicrob Agents Chemother.
47:1382–1390. 2003. View Article : Google Scholar :
|
|
49
|
Tian GB, Adams-Haduch JM, Taracila M,
Bonomo RA, Wang HN and Doi Y: Extended-spectrum AmpC
cephalosporinase in Acinetobacter baumannii ADC-56 confers
resistance to cefepime. Antimicrob Agents Chemother. 55:4922–4925.
2011. View Article : Google Scholar :
|
|
50
|
Mendes RE, Bell JM, Turnidge JD,
Castanheira M and Jones RN: Emergence and widespread dissemination
of OXA-23, −24/40 and −58 carbapenemases among Acinetobacter spp.
in Asia-Pacific nations: Report from the SENTRY surveillance
program. J Antimicrob Chemother. 63:55–59. 2009. View Article : Google Scholar
|
|
51
|
Zong Z, Lu X, Valenzuela JK, Partridge SR
and Iredell J: An outbreak of carbapenem-resistant Acinetobacter
baumannii producing OXA-23 carbapenemase in western China. Int J
Antimicrob Agents. 31:50–54. 2008. View Article : Google Scholar
|
|
52
|
Feizabadi MM, Fathollahzadeh B,
Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, Soroush
S and Mohammadi-Yegane S: Antimicrobial susceptibility patterns and
distribution of blaOXA genes among Acinetobacter spp. isolated from
patients at Tehran hospitals. Jpn J Infect Dis. 61:274–278.
2008.
|
|
53
|
Wang H, Guo P, Sun H, Wang H, Yang Q, Chen
M, Xu Y and Zhu Y: Molecular epidemiology of clinical isolates of
carbapenem-resistant Acinetobacter spp. from Chinese hospitals.
Antimicrob Agents Chemother. 51:4022–4028. 2007. View Article : Google Scholar :
|
|
54
|
Wei LH, Zhang J, Deng JJ, Zou FM, Liu G
and Si XQ: The isolation of Acinetobacter strain from burn wound
and the analysis of its antibiotic resistance. Zhonghua Shao Shang
Za Zhi. 20:17–19. 2004.(In Chinese).
|
|
55
|
Wang H, Liu YM, Chen MJ, Sun HL, Xie XL
and Xu YC: Mechanism of carbapenems resistance in Acinetobacter
baumannii. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 25:567–572.
2003.(In Chinese).
|
|
56
|
Héritier C, Poirel L, Lambert T and
Nordmann P: Contribution of acquired carbapenem-hydrolyzing
oxacillinases to carbapenem resistance in Acinetobacter baumannii.
Antimicrob Agents Chemother. 49:3198–3202. 2005. View Article : Google Scholar :
|
|
57
|
Higgins PG, Wisplinghoff H, Stefanik D and
Seifert H: Selection of topoisomerase mutations and overexpression
of adeB mRNA transcripts during an outbreak of Acinetobacter
baumannii. J Antimicrob Chemother. 54:821–823. 2004. View Article : Google Scholar
|
|
58
|
Magnet S, Courvalin P and Lambert T:
Resistance-nodulation-cell division-type efflux pump involved in
aminoglycoside resistance in Acinetobacter baumannii strain BM4454.
Antimicrob Agents Chemother. 45:3375–3380. 2001. View Article : Google Scholar :
|
|
59
|
Marchand I, Damier-Piolle L, Courvalin P
and Lambert T: Expression of the RND-type efflux pump AdeABC in
Acinetobacter baumannii is regulated by the AdeRS two-component
system. Antimicrob Agents Chemother. 48:3298–3304. 2004. View Article : Google Scholar :
|
|
60
|
Nemec A, Maixnerova M, van der Reijden TJ,
van den Broek PJ and Dijkshoorn L: Relationship between the AdeABC
efflux system gene content, netilmicin susceptibility and multidrug
resistance in a genotypically diverse collection of Acinetobacter
baumannii strains. J Antimicrob Chemother. 60:483–489. 2007.
View Article : Google Scholar
|
|
61
|
Peleg AY, Adams J and Paterson DL:
Tigecycline efflux as a mechanism for nonsusceptibility in
Acinetobacter baumannii. Antimicrob Agents Chemother. 51:2065–2069.
2007. View Article : Google Scholar :
|
|
62
|
Ruzin A, Keeney D and Bradford PA: AdeABC
multidrug efflux pump is associated with decreased susceptibility
to tigecycline in Acinetobacter calcoaceticusAcinetobacter
baumannii complex. J Antimicrob Chemother. 59:1001–1004. 2007.
View Article : Google Scholar
|
|
63
|
Rahmati S, Yang S, Davidson AL and
Zechiedrich EL: Control of the AcrAB multidrug efflux pump by
quorum-sensing regulator SdiA. Mol Microbiol. 43:677–685. 2002.
View Article : Google Scholar
|