Introduction
Colorectal cancer (CRC), the most common
gastrointestinal malignant tumor, is one of the most common
malignant tumors and the third leading cause of cancer-associated
mortality worldwide (1,2). Despite the advances in diagnostic
methods and therapy, the prognosis of patients with CRC remains
poor (3,4). Therefore, it is urgent to investigate
novel diagnostic, prognostic and treatment strategies for CRC.
MicroRNAs (miRs) are a family of short noncoding
RNAs (18–25 nucleotides long) that regulate diverse biological
processes, including cell proliferation, cell migration,
metastasis, invasion and apoptosis (5–8). It
has been previously reported that miRs can act as tumor oncogenes
or suppressors during cancer development, dependent on the role of
their target genes (9–11). miR-598 was reported to be a
biomarker for bile duct cancer (12). Zhao et al (13) indicated that miR-598 acted as a
prognostic biomarker in esophageal cancer. However, the function
and molecular mechanisms of miR-598 in human CRC remain to be fully
elucidated. In the present study, miR-598 expression in CRC was
investigated through analyzing data from two public datasets: The
Caner Genome Atlas project (TCGA) and Gene Expression Omnibus (GEO;
accession number GSE30454). In addition, in the present study
miR-598 expression was demonstrated to be increased in CRC tissues
and cell lines, and miR-598 may contribute to cell proliferation
and cell cycle progression by targeting inositol
polyphosphate-5-phosphatase E (INPP5E). The results of the present
study indicated that miR-598 was frequently upregulated in CRC, may
act as a tumor promoter and has potential as a prognostic biomarker
for patients with CRC.
Materials and methods
Clinical specimens
Human CRC tissues and the matched adjacent non-tumor
tissues were obtained from eight patients, including four males and
four females, between 38 and 76 years old, with CRC and
histopathologically diagnosed at the Department of General Surgery,
Huizhou First Hospital (Huizhou, China) between 1 June 2015 and 31
December 2015. The present study was approved by the Ethics
Committee of the Department of General Surgery, Huizhou First
Hospital. Written informed consent was obtained from all patients.
Tissue samples were collected during surgery, immediately frozen in
liquid nitrogen and stored until total RNA or proteins were
extracted.
Cell culture
Human CRC cell lines SW620, COLO320DM, SW403, SW480,
HT-29 and COLO205 and normal colonic cell line FHC were purchased
from the National Rodent Laboratory Animal Resources (Shanghai,
China). All CRC cell lines were grown in Dulbecco's modified Eagle
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich,
Merck KGaA, Darmstadt, Germany) and normal colon FHC cells were
grown in DMEM/F-12 medium with 10% FBS, 10 ng/ml cholera toxin
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 5 µg/ml
transferrin, 5 µg/ml insulin, 100 ng/ml hydrocortisone and 10 mM
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES). Cell
lines were cultured in a humidified 37°C incubator with 5%
CO2.
Plasmids, small interfering RNA
(siRNA) and transfection
SW480 cells (5×105) were transfected with
2.5 µg each of scrambled miRs as negative controls (NCs), miR-598
mimic and miR-598-in (miR-598-inhibitor; GeneCopoeia, Inc.,
Rockville, MD, USA), Mutant miR-598 was constructed by GeneCopoeia,
Inc. INPP5E siRNAs and NCs (stQ0004501-1; http://www.ribobio.com/sitecn/product_info.aspx?id=338920)
were purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
Transfection of plasmids and siRNAs was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from culture cells and
patient samples using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol, then cDNA was synthesized with using reverse
transcription reagent kit (Takara Biotechnology Co., Ltd., Dalian,
China) at 37°C for 15 min, followed at 85°C for 5 sec, and then
cooling to 4°C. RT-qPCR was carried out using an ABI 7900HT Fast
Real-time PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using the SYBR-Green PCR kit (Takara Biotechnology Co.,
Ltd.). The primers selected by GeneCopoeia, Inc., were miR-598
(cat. no. HmiRQP0702), U6 (cat. no. HmiRQP9001), cyclin D1 (cat.
no. HQP016204), cyclin-dependent kinase inhibitor 1B (p27; cat. no.
MQP028863) and GAPDH (cat. no. HQP006940). Thermocycling conditions
were 95°C for 30 sec, followed by 40 cycles of amplification at
95°C for 5 sec, 59°C for 30 sec and then 72°C for 30 sec.
Relative miR-598 or cyclin D1 and p27 mRNA
expression were normalized to U6 or GAPDH, respectively. Relative
quantification was calculated as 2−∆∆Cq (14) and was used to calculate
fold-changes.
Western blotting
Protein lysates were lysed with
radioimmunoprecipitation buffer (Beyotime Institute of
Biotechnology, Haimen, China), equal quantities (50 µg) of protein
were separated on a 10% SDS-PAGE and transferred onto
polyvinylidene difluoride membranes. Membranes were incubated with
primary antibodies as follows: Rabbit anti-INPP5E monoclonal
antibody (ab191520; 1:1,000); rabbit anti-cyclin D1 monoclonal
antibody (ab134175; 1:1,000); rabbit anti-p27 monoclonal antibody
(ab32034; 1:1,000) overnight at 4°C, and anti-α-tubulin antibody
(ab7291; 1:5,000; all from Abcam, Cambridge, MA, USA) was used as a
reference protein. The next day, membranes were incubated with the
appropriate horseradish peroxidase-conjugated secondary antibodies
(ab205718; 1:10,000; Abcam) for 2 h at room temperature and bound
antibodies were visualized using an enhanced chemiluminesence kit
(Beyotime Institute of Biotechnology) with the SuperSignal West
PICO chemiluminescent detection system (Pierce; Thermo Fisher
Scientific, Inc.).
MTT and colony formation assays
Cell growth was measured by MTT assay
(Sigma-Aldrich; Merck KGaA) according to the manufacturers'
protocol. Transfected SW480 cells were seeded in 96-well plates at
5×103 cells/well. Cells were allowed to grow for 0–5
days. A total of 20 µl, 5 mg/ml MTT solution was then added to each
well and incubated for 4 h, and then 150 µl DMSO (Sigma-Aldrich;
Merck KGaA) was added. Absorbance was measured at 490 nm using a
microplate reader (Molecular Devices, LLC, Sunnyvale, CA, USA).
Anchorage-independent growth
assay
The soft agar colony-forming assay was performed
using transfected SW480 (2,000 cells/well), which were seeded into
each well of a 6-well plate with 0.5% agar (Sigma-Aldrich; Merck
KGaA). After 14 days, three fields were randomly selected and cell
colonies >0.1 mm2 were counted with an inverted
microscope at a magnification of ×200.
Cell cycle analysis
Following 48 h of incubation post-transfection,
SW480 cells were removed from culture plates by trypsinization and
were washed with PBS. Cells were fixed with 75% ice-cold ethanol at
4°C. Following overnight incubation at 4°C and washing with PBS,
the cells were treated with RNase A (300 µg/ml) at 37°C for 30 min
and stained with propidium iodide (PI) at 4°C for 30 min in the
dark, and the cell cycle distribution was analyzed using a flow
cytometer (BD FACSCalibur™; BD Biosciences, Franklin
Lakes, NJ, USA). The percentages of cells in
G0/G1, S and G2/M phases were
counted by BD CellQuest™ Pro Software version 3.3 and compared.
Experiments were repeated three times.
Luciferase assays
The INPP5E open reading frame with the
3′-untranslated region (UTR) was cloned into pGL3 vectors were
purchased from GeneCopoeia, Inc. (Guangzhou, China). SW480 cells
(5×104/well) were cultured in 24-well plates. Transient
co-transfection with either the INPP5E 3′-UTR wild type and miR-598
or miR-598- in or control mimics were performed using Lipofectamine
2000 reagent. A total of 48 h following transfection, firefly and
Renilla luciferase activities were measured using the
Dual-Luciferase Reporter Assay System (Promega Corporation,
Madison, WI, USA) according to manufacturer's protocol.
Bromodeoxyuridine (BrdU) labeling and
immunofluorescence
Following transfection cells were grown on cover
slips (Thermo Fisher Scientific, Inc.) and were incubated with BrdU
for 1 h and then stained with an anti-BrdU antibody (20-BS17;
1:500; Upstate Biotechnology, Inc., Lake Placid, NY, USA) according
to the manufacturer's protocol. Gray level images were acquired
using a laser-scanning microscope (Axioskop 2 plus; Carl Zeiss AG,
Oberkochen, Germany).
Statistical analysis
All statistical analyses were performed using SPSS
software (version 17.0; SPSS, Inc., Chicago, IL, USA). All data are
expressed as the mean ± standard deviation. Student's t-test was
used to evaluate the significance of the differences between two
groups of data and one-way analysis of variance followed by Tukey's
post hoc test was used for multiple comparisons. P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-598 expression was elevated in CRC
tissues and cell lines
To investigate the potential roles of miR-598 in
CRC, the expression data downloaded from TCGA and GEO (accession
number GSE30454) was analyzed and indicated that miR-598 expression
was notably upregulated in CRC tissues (n=218) compared with normal
colorectal tissue (n=8; Fig. 1A).
The results of GEO analysis demonstrated that compared with normal
colorectal tissue (n=20), the expression of miR-598 was
significantly upregulated in CRC samples (n=54; P<0.05; Fig. 1B). In addition, RT-qPCR was
performed to analyze the expression of miR-598 in CRC and
demonstrated that compared with the matched tumor adjacent tissues,
miR-598 expression was upregulated in the CRC tissues (Fig. 1C) and in all six tested CRC cell
lines (SW620, COLO320DM, SW403, SW480, HT-29 and COLO205) had
significantly upregulated expression of miR-598 compared with the
normal colonic cell line FHC (P<0.05; Fig. 1D). These results indicated that
miR-598 was upregulated in CRC cell lines and tissues. miR-598
expression in SW480 cells was decreased compared with COLO320DM,
HT-29, SW403 and COLO205, but it was increased compared with SW620.
SW480, therefore appeared to be a suitable cellular model for
processing up and downregulation of miR-598.
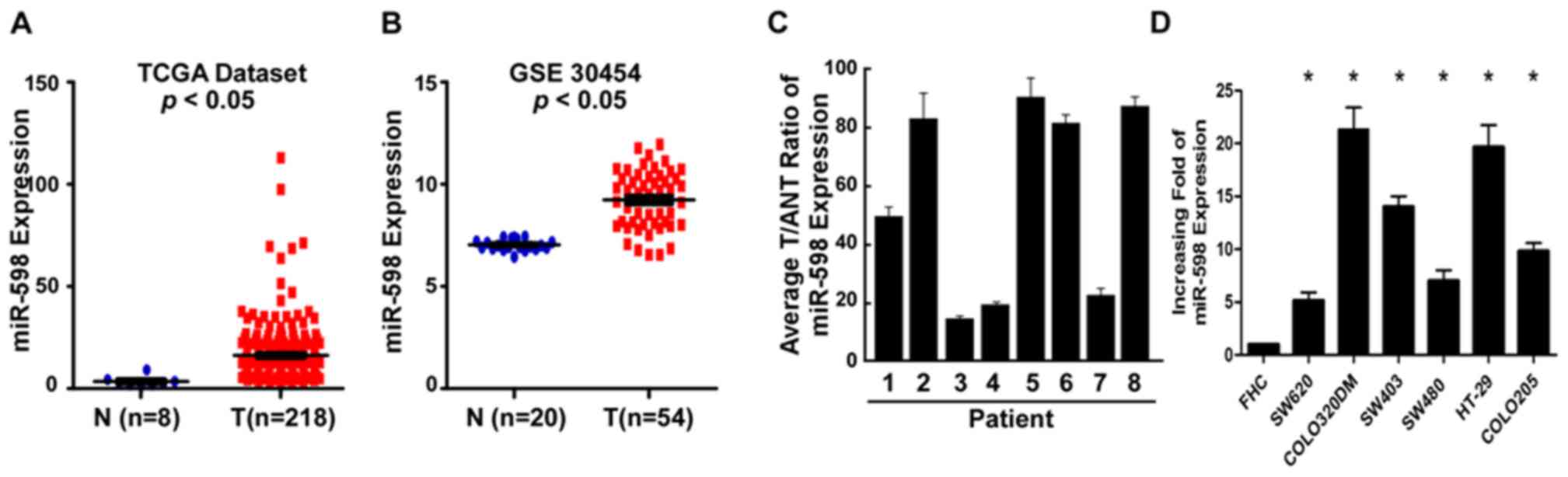 | Figure 1.Expression of miR-598 in human CRC
tissues and cell lines. (A) The expression levels of miR-598 in CRC
tissues from the TCGA dataset. (B) Analysis of miR-598 data from
the Gene Expression Omnibus database. (C) Relative miR-598 mRNA
expression levels in eight paired primary CRC tissues and the
matched ANT from the same patients were analyzed by PCR. (D)
Reverse transcription-quantitative PCR analysis of miR-598
expression in FHC cells and CRC cell lines, including SW620,
COLO320DM, SW403, SW480, HT-29 and COLO205. *P<0.05 vs. FHC.
CRC, colorectal cancer; PCR, polymerase chain reaction; miR,
microRNA; TCGA, The Cancer Genome Atlas; ANT, adjacent non-tumor
tissues; GSE30454; Gene Expression Omnibus dataset; T, primary CRC
tissues. |
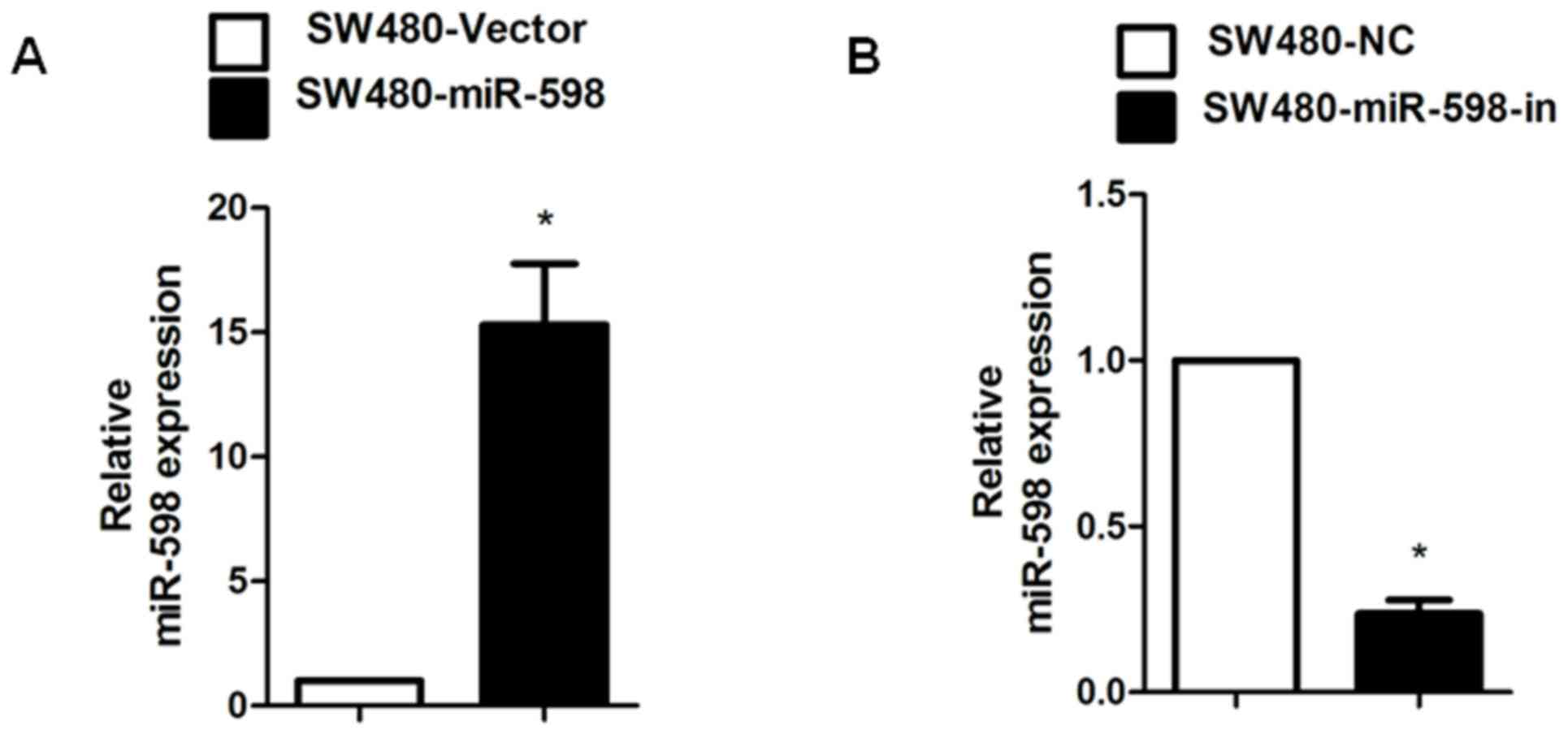
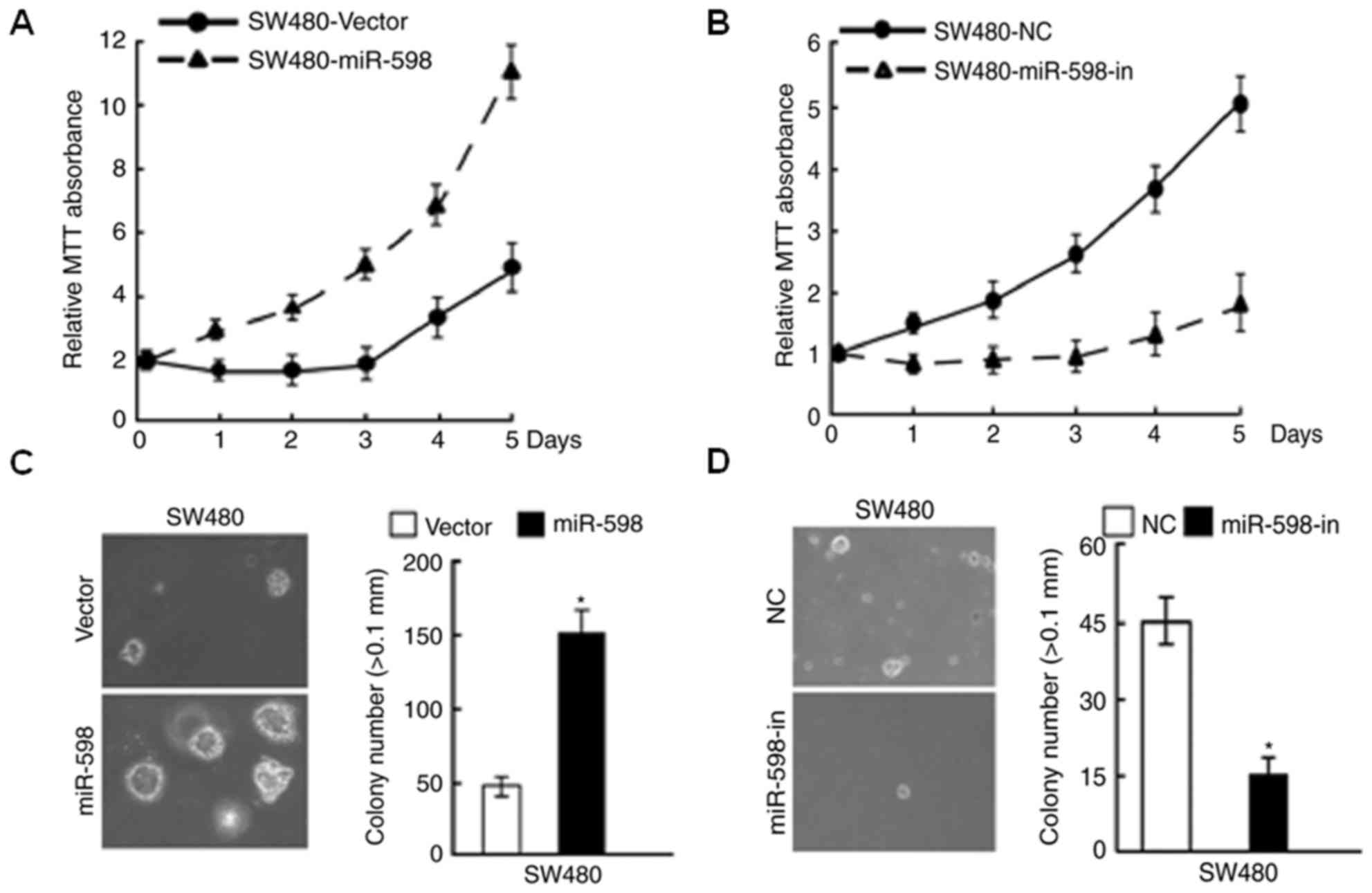
miR-598 promoted CRC cell
proliferation whereas miR-598-in inhibited proliferation
In order to investigate the role of miR-598 in CRC,
an miR-598 mimic, miR-598-in or the relative controls were
transiently transfected into SW480 cells. Relative miR-598
expression in SW480 cells was confirmed using RT-qPCR (Fig. 2A and B). Using MTT and
anchorage-independent growth assays, it was observed that miR-598
promoted cell proliferation (Fig. 3A
and C), while the growth rate of SW480 cells following
transfection with miR-598-in was decreased (Fig. 3B and D), compared with respective
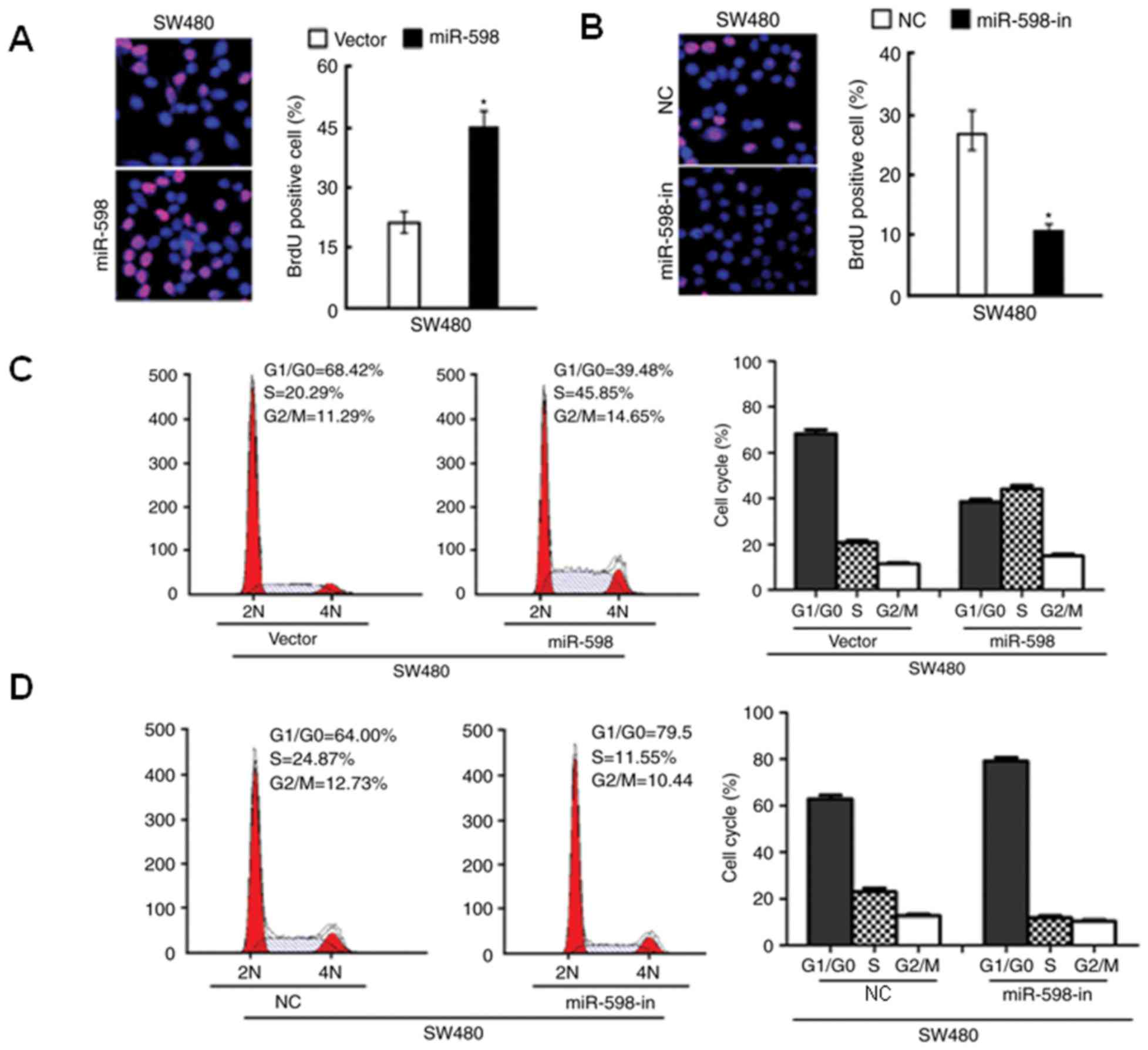
NC-transfected cells. The results of the BrdU analysis indicated
that miR-598 overexpression significantly increased the percentage
of BrdU positive cells (P<0.05; Fig. 4A), while BrdU positive cells were
significantly reduced in SW480 cells with miR-598-in (P<0.05;
Fig. 4B). Additionally, flow
cytometry indicated an increase in the percentage of cells in the S
phase and a decrease in the percentage of cells in
G1/G0 phase in miR-598 transfected SW480
cells (Fig. 4C). In SW480 cells
transfected with miR-598-in, the number of cells in the S phase of
the cell cycle decreased and the number in
G1/G0 phase increased (Fig. 4D). These results suggested that
miR-598 serves an important role in cell proliferation and cell
cycle progression in CRC cells.
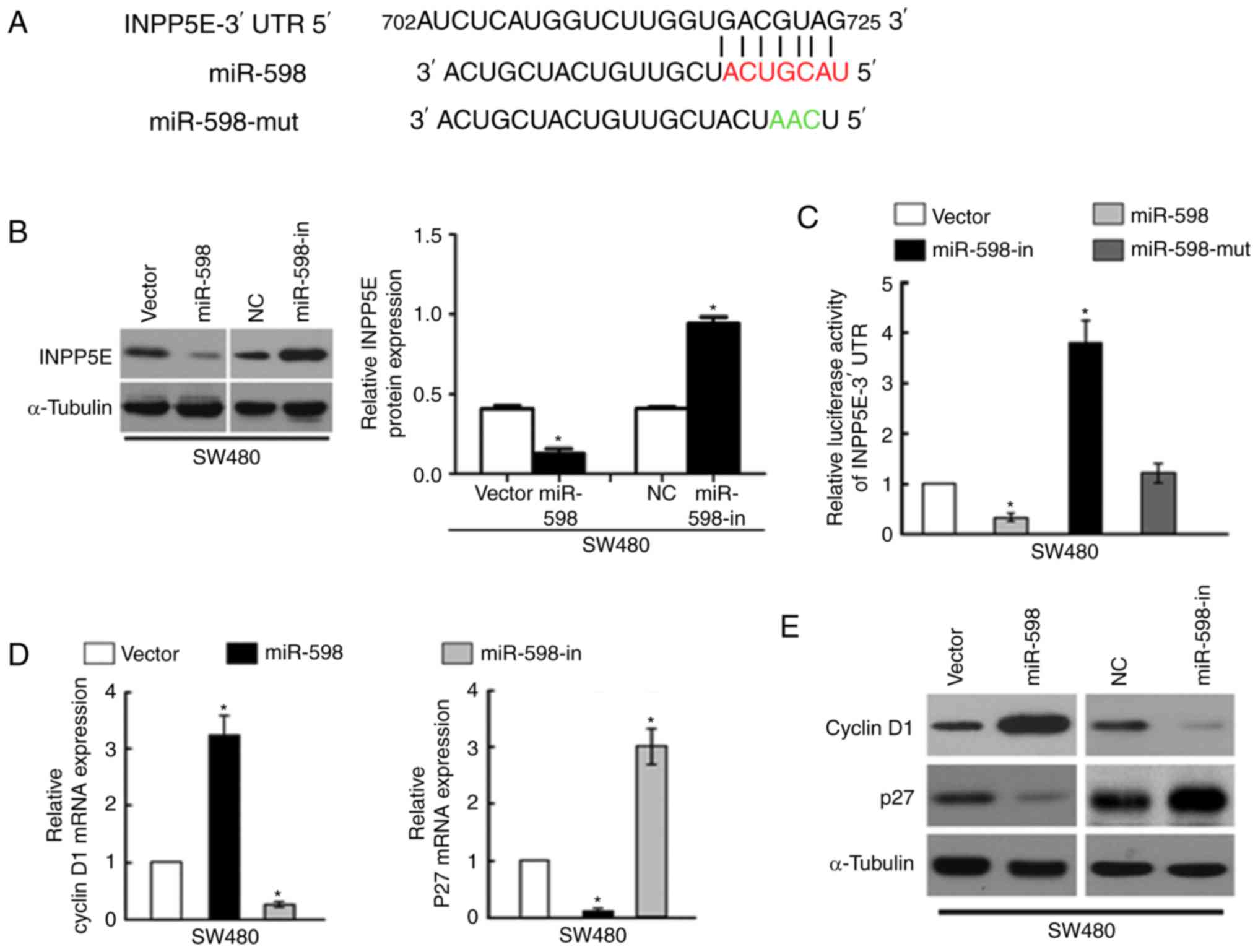
miR-598 directly targets INPP5E by
binding to its 3′-UTR and alters levels of proteins associated with
cell proliferation and cell cycle in CRC cells
Using the miR target prediction website (www.targetscan.org), INPP5E was demonstrated to act as
the potential target of miR-598. The wild type INPP5E 3-UTR mRNA
contained a length of conserved sequence with miR-598 and a mutant
miR-598 was constructed (Fig. 5A).
Western blot analysis of the INPP5E expression levels demonstrated
that INPP5E levels in the miR-598 mimic transfection group were
significantly lower than those in the NC transfection group,
whereas cells in the miR-598-in group demonstrated significantly
increased INPP5E levels compared with the NC (P<0.05; Fig. 5B).
To confirm that INPP5E is directly targeted by
miR-598, the present study investigated whether miR-598 recognizes
the 3′UTR of INPP5E mRNA using a dual-luciferase reporter assay.
The relative luciferase activity of the reporter containing the
wild-type INPP5E 3′-UTR was significantly decreased following
co-transfection with miR-598; however, this was notably increased
following co-transfection with miR-598-in. The luciferase activity
was not significantly affected by co-transfection with the
miR-598-mut (P<0.05; Fig.
5C).
To analyze the effect of miR-598 on the expression
levels of mRNA and proteins associated with cell proliferation and
cell cycle progression in CRC cells, miR-598 or miR-598-in were
transfected into SW480 cells and RT-qPCR and western blot analysis
were performed. The results indicated that mRNA and protein
expression of cyclin D1 expression was significantly upregulated
and p27 was downregulated by miR-598 (P<0.05; Fig. 5D). In contrast, the expression
level of cyclin D1 was downregulated and p27 was upregulated in
SW480 cells transfected with the miR-598-in (Fig. 5E). These results confirmed that
INPP5E is a target of miR-598.
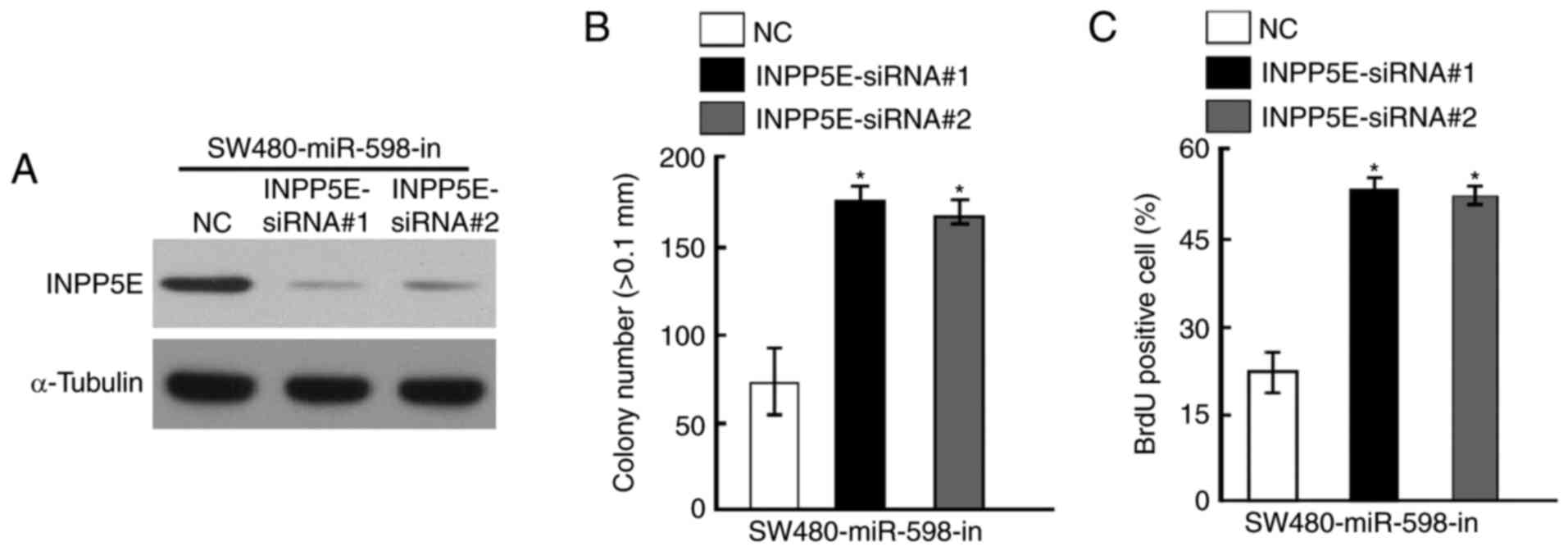
Silencing of INPP5E counteracted the
cell growth arrest caused by an miR-598-in
To further investigate the role of INPP5E in CRC,
specific siRNAs against INPP5E were designed and synthesized.
Western blot analysis indicated that INPP5E protein level was
decreased following treatment with siRNAs in SW480 cells
transfected with miR-598-in (Fig.
6A). The results of anchorage-independent growth assays
demonstrated that knockdown of INPP5E counteracted the growth
arrest by miR-598-in (Fig. 6B). In
addition, silencing of INPP5E also increased the percentage of BrdU
positive cells in SW480 cells transfected with miR-598-in compared
with the NC (Fig. 6C).
Discussion
In the present study, miR-598 was demonstrated to be
upregulated in CRC tissues in comparison with matched non-tumor
tissues. Additional experiments using RT-qPCR indicated that the
expression of miR-598 was upregulated in CRC tissues and cells
compared with the matched tumor adjacent tissues and the normal
colonic cell line FHC. Biological functions of miR-598 in CRC were
investigated using gain or loss of function studies. miR-598 was
determined to act as a tumor promoter by targeting INPP5E.
Recently, a number of studies have indicated
dysfunction of miRs may lead to a variety of disorders, as a result
of the impact on the target genes regulated by the miRs (15–17).
It has been previously reported that a number of miRs serve
essential roles in CRC aggression (18–20).
The discovery of miRs provided novel insight into understanding the
molecular mechanisms and treatment of cancer. MiR-219-5p was
reported to serve a tumor suppressive role in colon cancer by
targeting oncogene Sal-like protein 4 (21). miR-320b was demonstrated to
suppress cell proliferation of human CRC by targeting c-Myc
(22). Cai et al (23) indicated that miR-211 expression
promoted CRC cell growth by targeting CHD5. Zhang et al
(24) demonstrated that miR-638
suppresses cell proliferation, invasion and regulated cell cycle by
targeting tetraspanin-1 in human colorectal carcinoma. Liang et
al (25) demonstrated that
miR-892a promoted cell proliferation in human CRC by regulating
serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B
α isoform expression. In the present study, the expression of
miR-598 in CRC samples from TCGA and GEO was analyzed, which
demonstrated that miR-598 expression was upregulated in CRC. In
addition, significantly higher miR-598 expression was demonstrated
in CRC tissues in comparison with matched non-tumor tissues. To
investigate the association of the miR-598 upregulation and cell
proliferation in CRC, miR-598 gain or loss of-function studies were
performed, the results indicated that overexpression of miR-598
promoted proliferation, whereas miR-598-in CRC inhibited cell
proliferation.
To further investigate the mechanisms via which
miR-598 to promoted cell proliferation and cell cycle progression
in CRC, INPP5E was identified as a potential target gene of miR-598
using bioinformatics analysis. INPP5E is an inositol polyphosphate
5-phosphatase that hydrolyzes the 5-phosphate of
phosphatidylinositol 3,4,5 phosphatase 3 [PtdIns(3,4,5)P3] and
PtdIns(4,5)P2 and serves an essential role in cancer, diabetes and
inflammation (26–28). INPP5E was reported to act as an
essential inhibitor of the phosphoinositide 3-kinase/protein kinase
B/mammalian target of rapamycin C1 signaling axis and regulated the
development of renal epithelial cells (29). In the present study, bioinformatics
analysis predicted INPP5E as a target of miR-598. The
bioinformatics prediction was verified and the association between
INPP5E and miR-598 investigated using western blotting that
demonstrated INPP5E protein expression was downregulated by
miR-598. In addition, miR-598 was demonstrated to recognize the
3′-UTR of INPP5E, as investigated using a dual luciferase reporter
gene assay. Further functional experiments indicated that silencing
of INPP5E counteracted the arrest of cell growth caused by an
miR-598-in in CRC. The results of the present study suggested that
miR-598 promoted CRC cell proliferation by suppressing INPP5E.
In conclusion, the present study indicated that
miR-598 was frequently upregulated in CRC. miR-598 promoted CRC
cell proliferation and cell cycle progression by silencing INPP5E,
providing novel insight into the pathogenesis of CRC.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Guangdong Province (grant no. 2014Y062).
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lieberman DA: Clinical practice. Screening
for colorectal cancer. N Engl J Med. 361:1179–1187. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Haggar FA and Boushey RP: Colorectal
cancer epidemiology: Incidence, mortality, survival, and risk
factors. Clin Colon Rectal Surg. 22:191–197. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang ZS, Zhong M, Bian YH, Mu YF, Qin SL,
Yu MH and Qin J: MicroRNA-187 inhibits tumor growth and invasion by
directly targeting CD276 in colorectal cancer. Oncotarget.
7:44266–44276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Minna E, Romeo P, Dugo M, De Cecco L,
Todoerti K, Pilotti S, Perrone F, Seregni E, Agnelli L, Neri A, et
al: miR-451a is underexpressed and targets AKT/mTOR pathway in
papillary thyroid carcinoma. Oncotarget. 7:12731–12747. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li N, Zhao X, Wang L, Zhang S, Cui M and
He J: miR-494 suppresses tumor growth of epithelial ovarian
carcinoma by targeting IGF1R. Tumour Biol. 37:7767–7776. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tian J, Hu L, Li X, Geng J, Dai M and Bai
X: MicroRNA-130b promotes lung cancer progression via PPAR
γ/VEGF-A/BCL-2-mediated suppression of apoptosis. J Exp Clin Cancer
Res. 35:1052016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao Z, Qin L and Li S: miR-411
contributes the cell proliferation of lung cancer by targeting
FOXO1. Tumour Biol. 37:5551–5560. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Xu G, Liu G, Ye Y, Zhang C, Fan
C, Wang H, Cai H, Xiao R, Huang Z and Luo Q: miR-411-5p inhibits
proliferation and metastasis of breast cancer cell via targeting
GRB2. Biochem Biophys Res Commun. 476:607–613. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xia K, Zhang Y, Cao S, Wu Y, Guo W, Yuan W
and Zhang S: miR-411 regulated ITCH expression and promoted cell
proliferation in human hepatocellular carcinoma cells. Biomed
Pharmacother. 70:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang M, Wen TF, He LH, Li C, Zhu WJ and
Trishul NM: A six-microRNA set as prognostic indicators for bile
duct cancer. Int J Clin Exp Med. 8:17261–17270. 2015.PubMed/NCBI
|
|
13
|
Zhao BS, Liu SG, Wang TY, Ji YH, Qi B, Tao
YP, Li HC and Wu XN: Screening of microRNA in patients with
esophageal cancer at same tumor node metastasis stage with
different prognoses. Asian Pac J Cancer Prev. 14:139–143. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhu X, Li D, Yu F, Jia C, Xie J, Ma Y, Fan
S, Cai H, Luo Q, Lv Z and Fan L: miR-194 inhibits the
proliferation, invasion, migration, and enhances the
chemosensitivity of non-small cell lung cancer cells by targeting
forkhead box A1 protein. Oncotarget. 7:13139–13152. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xue L, Xu Z, Wang K, Wang N, Zhang X and
Wang S: Network analysis of microRNAs, transcription factors,
target genes and host genes in human anaplastic astrocytoma. Exp
Ther Med. 12:437–444. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang L, Yang C, Song Y, Liu W, Wang K, Li
S and Zhang Y: MicroRNA-23a-3p promotes the development of
osteoarthritis by directly targeting SMAD3 in chondrocytes. Biochem
Biophys Res Commun. 478:467–473. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang PY, Chen CC, Chang YS, Tsai WS, You
JF, Lin GP, Chen TW, Chen JS and Chan EC: MicroRNA-223 and
microRNA-92a in stool and plasma samples act as complementary
biomarkers to increase colorectal cancer detection. Oncotarget.
7:10663–10675. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deng B, Wang B, Fang J, Zhu X, Cao Z, Lin
Q, Zhou L and Sun X: MiRNA-203 suppresses cell proliferation,
migration and invasion in colorectal cancer via targeting of
EIF5A2. Sci Rep. 6:283012016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang F, Xie YQ, Tang SQ, Wu XB and Zhu HY:
miR-143 regulates proliferation and apoptosis of colorectal cancer
cells and exhibits altered expression in colorectal cancer tissue.
Int J Clin Exp Med. 8:15308–15312. 2015.PubMed/NCBI
|
|
21
|
Cheng J, Deng R, Zhang P, Wu C, Wu K, Shi
L, Liu X, Bai J, Deng M, Shuai X, et al: miR-219-5p plays a tumor
suppressive role in colon cancer by targeting oncogene Sall4. Oncol
Rep. 34:1923–1932. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang H, Cao F, Li X, Miao H, E J, Xing J
and Fu CG: miR-320b suppresses cell proliferation by targeting
c-Myc in human colorectal cancer cells. BMC Cancer. 15:7482015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cai C, Ashktorab H, Pang X, Zhao Y, Sha W,
Liu Y and Gu X: MicroRNA-211 expression promotes colorectal cancer
cell growth in vitro and in vivo by targeting tumor suppressor
CHD5. PLoS One. 7:e297502012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Fei B, Wang Q, Song M, Yin Y,
Zhang B, Ni S, Guo W, Bian Z, Quan C, et al: MicroRNA-638 inhibits
cell proliferation, invasion and regulates cell cycle by targeting
tetraspanin 1 in human colorectal carcinoma. Oncotarget.
5:12083–12096. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liang WL, Cao J, Xu B, Yang P, Shen F, Sun
Z, Li WL, Wang Q and Liu F: miR-892a regulated PPP2R2A expression
and promoted cell proliferation of human colorectal cancer cells.
Biomed Pharmacother. 72:119–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ooms LM, Horan KA, Rahman P, Seaton G,
Gurung R, Kethesparan DS and Mitchell CA: The role of the inositol
polyphosphate 5-phosphatases in cellular function and human
disease. Biochem J. 419:29–49. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bielas SL, Silhavy JL, Brancati F,
Kisseleva MV, Al-Gazali L, Sztriha L, Bayoumi RA, Zaki MS,
Abdel-Aleem A, Rosti RO, et al: Mutations in INPP5E, encoding
inositol polyphosphate-5-phosphatase E, link phosphatidyl inositol
signaling to the ciliopathies. Nat Genet. 41:1032–1036. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hakim S, Bertucci MC, Conduit SE, Vuong DL
and Mitchell CA: Inositol polyphosphate phosphatases in human
disease. Curr Top Microbiol Immunol. 362:247–314. 2012.PubMed/NCBI
|
|
29
|
Hakim S, Dyson JM, Feeney SJ, Davies EM,
Sriratana A, Koenig MN, Plotnikova OV, Smyth IM, Ricardo SD, Hobbs
RM and Mitchell CA: Inpp5e suppresses polycystic kidney disease via
inhibition of PI3K/Akt-dependent mTORC1 signaling. Hum Mol Genet.
25:2295–2313. 2016. View Article : Google Scholar : PubMed/NCBI
|