Introduction
Lung cancer is one of the most common type of cancer
and the leading cause of cancer-associated death worldwide, which
accounts for >1.6 million cases and >1.3 million mortalities
due to this disease (1). Non-small
cell lung cancer (NSCLC), which accounts for >80% of lung cancer
cases, is comprised of three histological subtypes: Squamous-cell
carcinoma, adenocarcinoma and large-cell carcinoma. At present, the
primary therapeutic treatments for NSCLC patients are
comprehensive, including surgery resection, chemotherapy and
radiotherapy (2). Unfortunately,
despite recent progress in the diagnosis and treatment of cancer,
the five-year survival rate for patients with NSCLC remains at 11%
(3). Metastasis is a major cause
of mortality among patients with NSCLC (4). Therefore, fully understanding the
mechanisms underlying the carcinogenesis and progression of NSCLC
would facilitate the development of novel therapeutic targets and
improve the prognosis.
Emerging evidences has suggested that microRNAs
(miRNAs) are deregulated in NSCLC and serve important roles in
NSCLC initiation and development (5–7).
miRNAs represent a large family of non-coding and small RNA
molecules of ~20–23 nucleotides in length (8). They can negative regulate the
expression of their target messenger RNAs (mRNAs) via interacting
with the 3′ untranslated regions (3′UTRs) of genes, and therefore
enhancing mRNA degradation or translational suppression (9,10).
There are ~1881 precursor and 2588 mature miRNAs in the human
genome which are estimated to regulate the expression of >60% of
coding-genes (11). Through these
mechanisms, miRNAs are involved in regulation of diverse
bio-behaviors including cell proliferation, cell cycle, apoptosis,
metastasis and chemotherapy resistance, which are important in
developmental processes and tumorigenesis of cancers (12–14).
It has previously been reported that miRNAs whose expression are
dysregulated in human cancers can function as oncogenes or tumor
suppressors, primarily depending on the characteristic of their
target genes (15). For example,
the expression level of miR-370 is reduced in NSCLC, and
upregulation of miR-370 inhibits cell proliferation and induces
apoptosis via directly targeting oncogene tumor necrosis factor
receptor associated factor 4 (16). In contract, miR-575 is upregulated
in NSCLC, and targets the tumor suppressor gene BH3-like
motif-containing cell death inducer to promote cell proliferation,
migration and invasion in vitro (17).
The present study revealed that miR-454 was
downregulated in NSCLC tissues and cell lines. Low miR-454
expression was significantly associated with aggressive
clinicopathological features in NSCLC. Functional experiments
indicated that miR-454 overexpression suppressed proliferation,
migration and invasion of NSCLC cells in vitro. Furthermore,
signal transducer and activator of transcription-3 (STAT3) was
demonstrated to be a direct target gene of miR-454. These results
suggested that miR-454 served as a tumor suppressor in NSCLC via
directly targeting STAT3.
Materials and methods
Tissue samples
Tissue samples, including NSCLC and adjacent normal
tissues, were obtained from 67 NSCLC patients who underwent surgery
at Department of Thoracic Surgery, Yantaishan Hospital (Yantai,
China) between 2012 and 2014. The adjacent normal tissues were
obtained 5 cm from the edge of tumor tissues. None of these NSCLC
patients received chemotherapy or radiotherapy prior to surgery.
The Ethical Committee of Yantaishan Hospital approved this study,
and written informed consent was obtained from all subjects.
Patient characteristics are presented in Table I.
 | Table I.Correlations between miR-454
expression and clinicopathological features in NSCLC. |
Table I.
Correlations between miR-454
expression and clinicopathological features in NSCLC.
|
|
| miR-454
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | Cases | High | Low | P-value |
|---|
| Gender |
|
|
| 0.376 |
|
Male | 32 | 13 | 19 |
|
|
Female | 35 | 18 | 17 |
|
| Age (years) |
|
|
| 0.434 |
|
<55 | 29 | 15 | 14 |
|
|
≥55 | 38 | 16 | 22 |
|
| Smoke |
|
|
| 0.318 |
|
Yes | 45 | 19 | 26 |
|
| No | 22 | 12 | 10 |
|
|
Differentiation |
|
|
| 0.773 |
|
I–II | 30 | 15 | 15 |
|
|
III–IV | 37 | 16 | 21 |
|
| TNM stage |
|
|
| 0.023 |
|
I–II | 29 | 18 | 11 |
|
|
III–IV | 38 | 13 | 25 |
|
| Lymph node
metastasis |
|
|
| 0.001 |
| No | 29 | 20 | 9 |
|
|
Yes | 38 | 11 | 27 |
|
Cell lines and culture conditions
The A549, SPC-A-1, H520 and H1299 NSCLC cell lines
and the BEAS-2B human lung epithelial cell were obtained from the
China Center for Type Culture Collection (Wuhan, China). All cells
were cultured in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS), 2 µM glutamine, and
1% penicillin and streptomycin (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) in humidified air with 5%
CO2 at 37°C.
Transient transfection
Both oligonucleotide miR-454 mimics, negative
control (NC), STAT3 small interfering (si)RNA and NC siRNA were
synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
STAT3 siRNA sequence was 5′-CAUCUGCCUAGAUCGGCUA-3′ and the NC siRNA
sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′. NSCLC cells were seeded
into 6-well plates (5×105 cells/well) and transfected
with oligonucleotides using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The transfection efficiency was assessed
by using reverse transcription-quantitative polymerase chain
reaction (RT-qPCR).
RT-qPCR
Total RNA was extracted using TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. RNA concentration was determined using a
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific,
Inc.). For miR-454 expression, reverse transcription was performed
using TaqMan MicroRNA Reserve Transcription kit, followed by qPCR
with a Taqman MicroRNA Assay kit (both from Applied Biosystems;
Thermo Fisher Scientific, Inc.). The thermocycling conditions were
as follows: 40 cycles of denaturation at 95°C for 15 sec and
annealing/extension at 60°C for 60 sec. To quantify STAT3 mRNA
expression, cDNA was synthesized using a PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd., Dalian, China) and qPCR was
performed with a SYBR-Green PCR Master Mix (Takara Biotechnology
Co., Ltd). The thermocycling conditions were as follows: 95°C for
10 min, then 40 cycles of 95°C for 15 sec and 60°C for 1 min. PCR
was performed on an Applied Biosystems Real-Time 7500 Sequence
Detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.). U6 and GADPH served as internal controls for miR-454 and
STAT3 mRNA expression, respectively. The primers were designed as
follows: miR-454 forward, 5′-ACCCTATCAATATTGTCTCTGC-3′ and reverse,
5′-GCGAGCACAGAATTAATACGAC-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; STAT3 forward,
5′-TTGCCAGTTGTGGTGATC-3′ and reverse, 5′-AGACCCAGAAGGAGCCGC-3′; and
GAPDH forward, 5′-CATCTTCTTTTGCGTCGCCA-3′ and reverse,
5′-TCGCCCCACTTGATTTTGG-3′. The relative fold expressions were
determined with the 2−ΔΔCq method (18).
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was measured using a CCK-8
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan).
Transfected (miRNA/siRNA) cells were cultured into 96-well plates
at a density of 2,000 cells/well. CCK-8 assay was performed every
24 h for 96 h following the manufacturer's protocol. CCK-8 (10 µl)
solution was added and cells were cultured for 2 h in humidified
air with 5% CO2 at 37°C. Absorbance was then measured at
a wavelength of 450 nm using a microplate reader. All experiments
were performed in triple.
In vitro migration and invasion
assay
In vitro migration and invasion assays were
performed with 24-well Transwell chambers (8-mm pore size; BD
Biosciences). For the invasion assay, Transwell chambers were
pre-coated with Matrigel (BD Biosciences). For both assays,
5×104 transfected cells in 100 µl FBS-free culture
medium were injected into the upper chamber, while 500 µl culture
medium containing 20% FBS was added into the lower chambers as a
chemoattractant. A total of 48 h after transfection in humidified
air with 5% CO2 at 37°C, the Transwell chambers were
fixed with 95% methanol and stained with 0.1% crystal violet. After
washing, migrated and invaded cells were counted and imaged under
an inverted microscope at ×200 magnification (IX71; Olympus
Corporation).
Bioinformatics analysis
Bioinformatics analysis was performed to predict the
putative targets of miR-454 using TargetScan (version 6.0;
www.targetscan.org).
Luciferase reporter assay
Luciferase reporter plasmids, pmirGLO-STAT3-3′UTR
wild-type (Wt) and pmirGLO-STAT3-3′UTR mutant (Mut), were
synthesized by Shanghai GenePharma. For the luciferase reporter
assay, HEK293T cells (China Center for Type Culture Collection)
were transfected with miR-454 mimics or NC together with
pmirGLO-STAT3-3′UTR Wt or pmirGLO-STAT3-3′UTR Mut using
Lipofectamine 2000. A total of 48 h after co-transfection,
luciferase activities were measured using the Dual-Luciferase
Reporter Assay system (Promega Corporation, Madison, WI, USA).
Renilla luciferase activity was used for normalization. This
assay was performed in triplicate and repeated three times.
Western blotting
A total of 72 h after transfection, cells were
washed with PBS (Gibco; Thermo Fisher Scientific, Inc.) and lysed
in radioimmunoprecipitation assay buffer supplemented with freshly
added protease inhibitors (Roche Diagnostics GmbH, Mannheim,
Germany). Following centrifugation at 16,000 × g and 4°C for 10
min, protein concentration was determined using a Bicinchoninic
Acid assay kit (Pierce; Thermo Fisher Scientific, Inc.). Equal
amounts of protein (30 µg) were loaded onto a 10% SDS-PAGE gel and
transferred onto polyvinylidene fluoride membranes (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and subsequently blocked
with 5% non-fat milk in TBS containing 0.1% Tween-20 (TBST). After
incubation with primary antibodies at 4°C overnight, the membranes
were washed with TBST three times and then probed with a
corresponding horseradish peroxidase (HRP)-conjugated secondary
antibody (1:10,000; cat no. sc-2005; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA) for 2 h at room temperature. The protein
bands were visualized using an Enhanced Chemiluminescence kit (EMD
Millipore, Billerica, MA, USA), and the relative expression of
STAT3 was normalized to the level of GADPH. The primary antibodies
used in this study were mouse anti-human STAT3 monoclonal (1:1,000;
cat no. sc-77441) and mouse anti-human monoclonal GADPH (1:1,000;
cat no. sc-166574) (both from Santa Cruz Biotechnology, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation.
SPSS version 19.0 (SPSS Inc., Chicago, IL, USA) was used for
statistical analysis and a Student's t-test or one-way analysis of
variance, with a Student-Newman-Keuls post hoc test for multiple
comparisons, were performed. P<0.05 was considered to indicate a
statistically significant difference.
Results
Low levels of miR-454 in NSCLC tissues
and cell lines
To investigate the potential roles of miR-454 in
NSCLC progression, miR-454 expression in NSCLC and adjacent normal
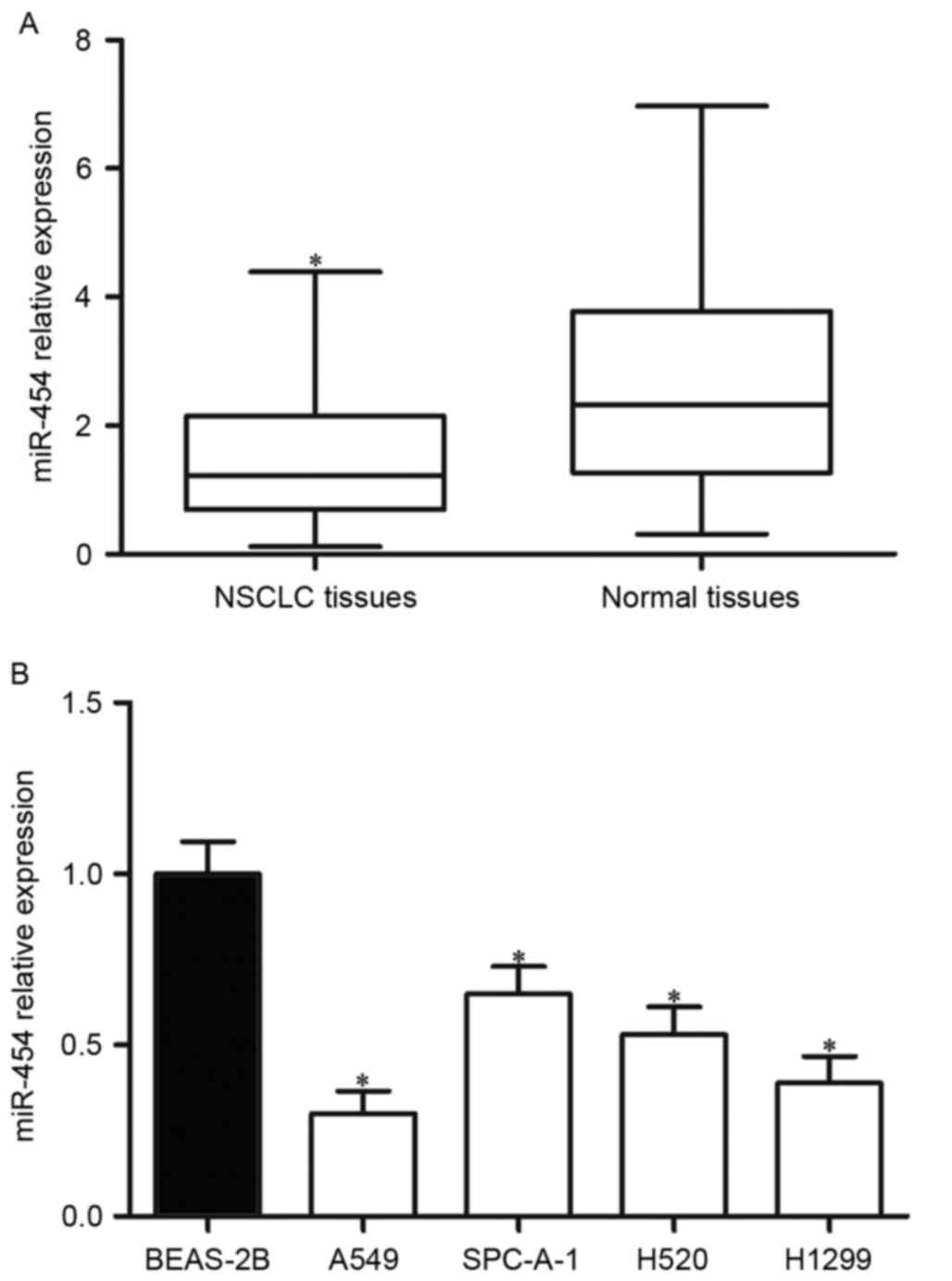
tissues was assessed. As presented in Fig. 1A, miR-454 was downregulated in
NSCLC tissues compared with adjacent normal tissues. The expression
of miR-454 in NSCLC cell lines (A549, SPC-A-1, H520 and H1299) and
the human lung epithelial cell (BEAS-2B) was also detected; the
expression levels of miR-454 were significantly lower in NSCLC cell
lines than in the BEAS-2B cell line (Fig. 1B).
Correlations between miR-454
expression and clinicopathological features in NSCLC
The correlations between miR-454 expression and
clinicopathological features in NSCLC are presented in Table I. The results of the statistical
analysis revealed that miR-454 expression in NSCLC was
significantly correlated with TNM stage (P=0.023) and lymph node
metastasis (P=0.001), but no correlation was observed between
miR-454 expression and gender (P=0.376), age (P=0.434), smoking
(P=0.318) and differentiation (P=0.773).
miR-454 inhibits proliferation,
migration and invasion of NSCLC cells
A549 and H1299 cells, which express relatively low
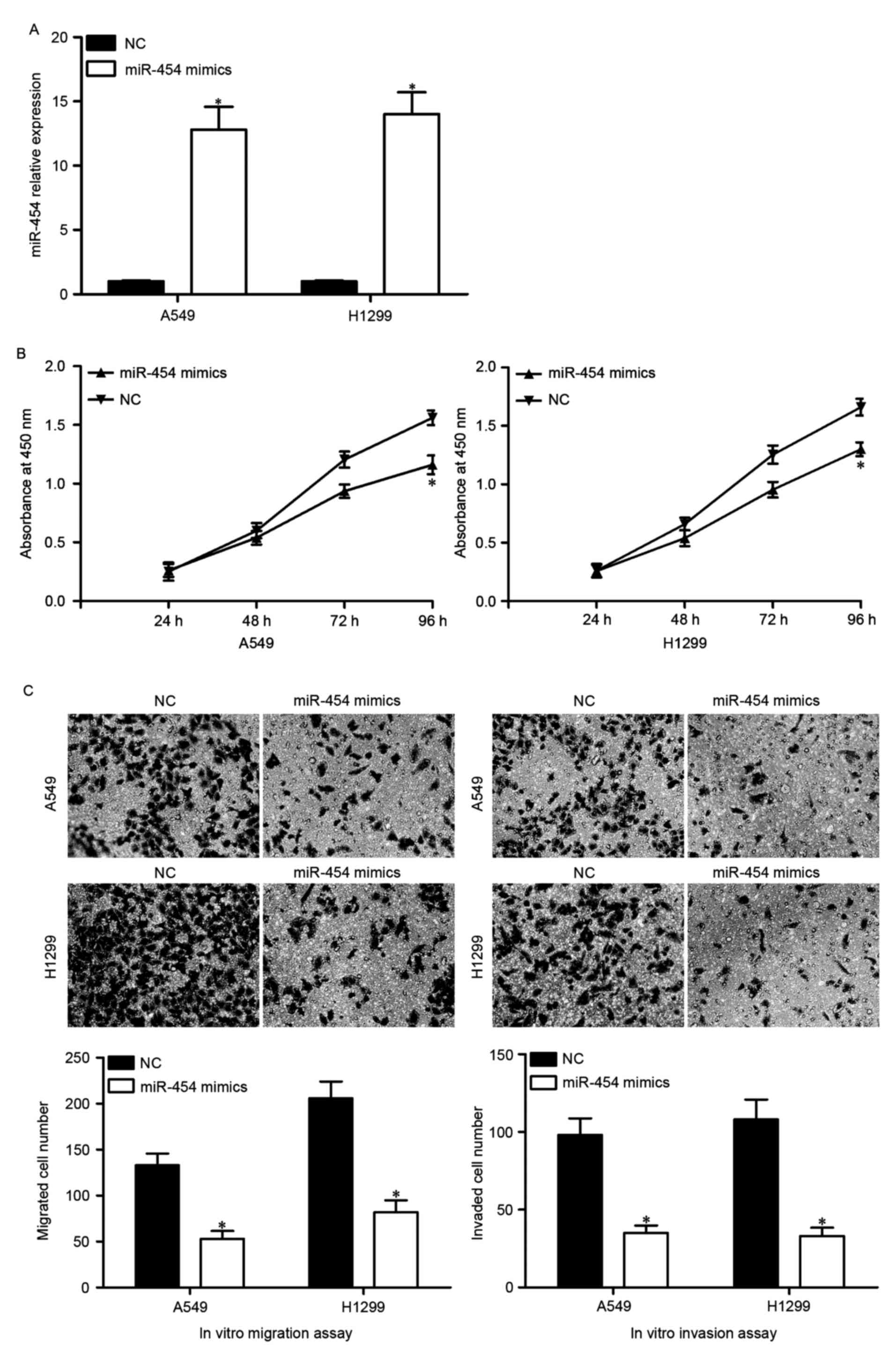
levels of endogenous miR-454, were transfected with miR-454 mimics
or NC. The transfection efficiency was assessed by RT-qPCR, and
miR-454 was demonstrated to be markedly overexpressed in both A549
and H1299 cells following transfection with miR-454 mimics
(Fig. 2A).
Following this, the effects of miR-454
overexpression on the proliferation, migration and migration of
A549 and H1299 cells was assessed using CCK-8 and in vitro
migration and invasion assays, respectively. As presented in
Fig. 2B, A549 and H1299 cells
transfected with miR-454 mimics exhibited reduced proliferation.
Furthermore, the number of migrated and invaded cells was
significantly lower in A549 and H1299 cells transfected with
miR-454 mimics (Fig. 2C).
Collectively, these results indicated that miR-454 serves as a
tumor suppressor in NSCLC via inhibiting tumor cell proliferation,
migration and invasion.
STAT3 is a direct target of miR-454 in
NSCLC
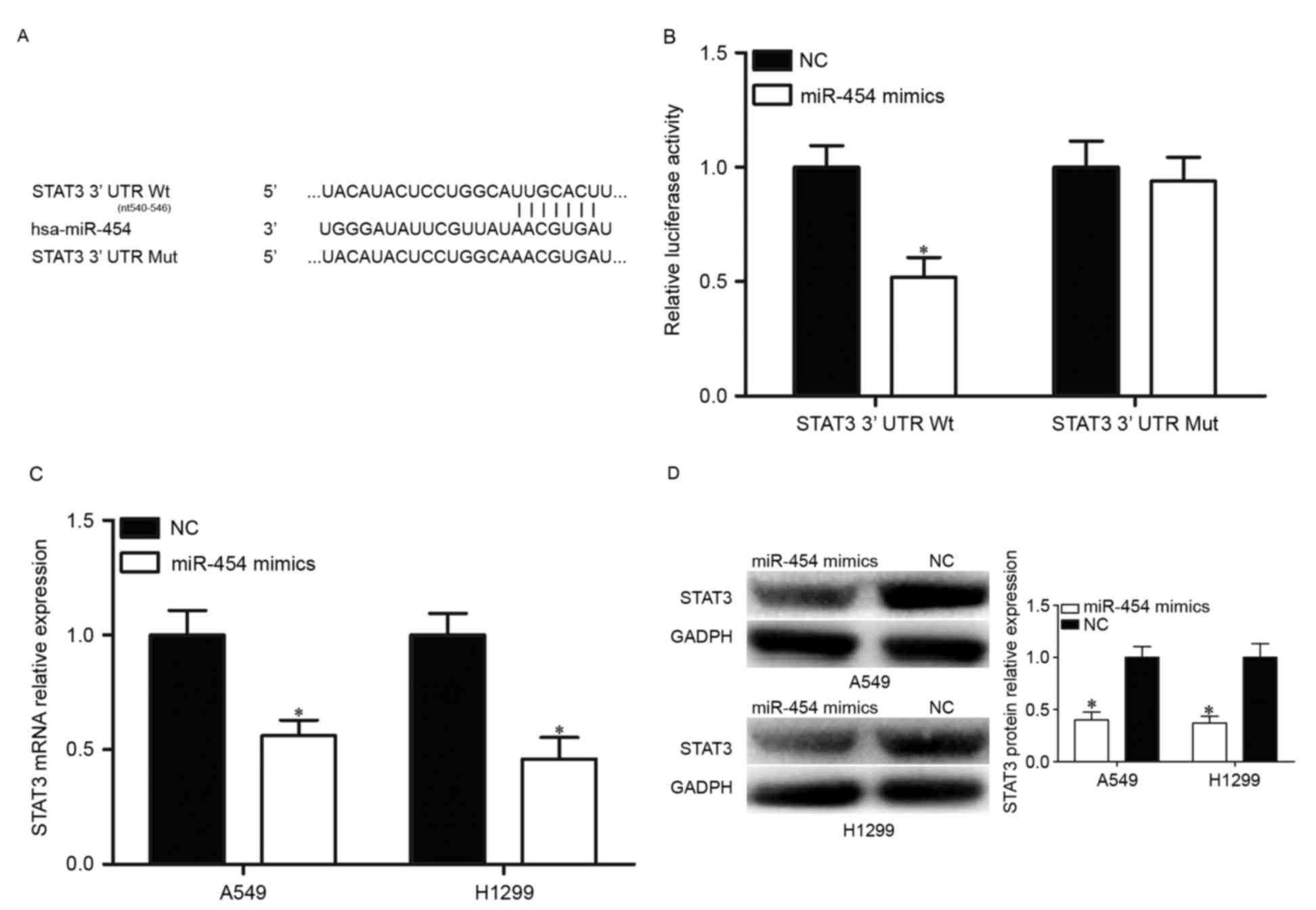
To identify potential target genes of miR-454,
bioinformatics analysis was performed with TargetScan (http://www.targetscan.org/). The analysis of
TargetScan indicated that STAT3 is a putative target of miR-454
(Fig. 3A). Following this, a
luciferase reporter assay was performed to investigate whether the
3′UTR of STAT3 could be directly targeted by miR-454. HEK293T cells
were co-transfected with miR-454 mimics or NC, and
pmirGLO-STAT3-3′UTR Wt or pmirGLO-STAT3-3′UTR Mut. The results
demonstrated that overexpression of miR-454 resulted in decreased
luciferase activities in pmirGLO-STAT3-3′UTR Wt, but not
pmirGLO-STAT3-3′UTR Mut (Fig.
3B).
Following this, the regulatory effects of miR-454
overexpression on STAT3 expression were investigated. RT-qPCR and
western blotting were performed to detect STAT3 mRNA and protein
expression levels in A549 and H1299 transfected with miR-454 mimics
or NC, respectively. As presented in Fig. 3C, upregulation of miR-454
attenuated STAT3 mRNA expression in A549 and H1299 cells. The
results of western blot analysis revealed that STAT3 protein
expression was reduced by miR-454 overexpression in A549 and H1299
cells (Fig. 3D). These results
demonstrated that STAT3 is a direct target of miR-454 in NSCLC.
Downregulation of STAT3 may simulate
the miR-454-induced tumor suppressive roles in NSCLC
To investigate the roles of STAT3 in NSCLC, A549 and
H1299 cells were transfected with STAT3 siRNA or NC siRNA. The
transfection efficiency was determined using RT-qPCR and western
blotting, and it was demonstrated that STAT3 was significantly
downregulated in A549 and H1299 cells at both the mRNA (Fig. 4A) and protein (Fig. 4B) level. In addition, decreased
cell proliferation (Fig. 4C) was
accompanied by decreased cell migration and invasion (Fig. 4D) in A549 and H1299 cells
transfected with STAT3 siRNA. Therefore, it was concluded that the
tumor suppressive roles of miR-454 in NSCLC cells was a consequence
of reduced STAT3 expression in NSCLC.
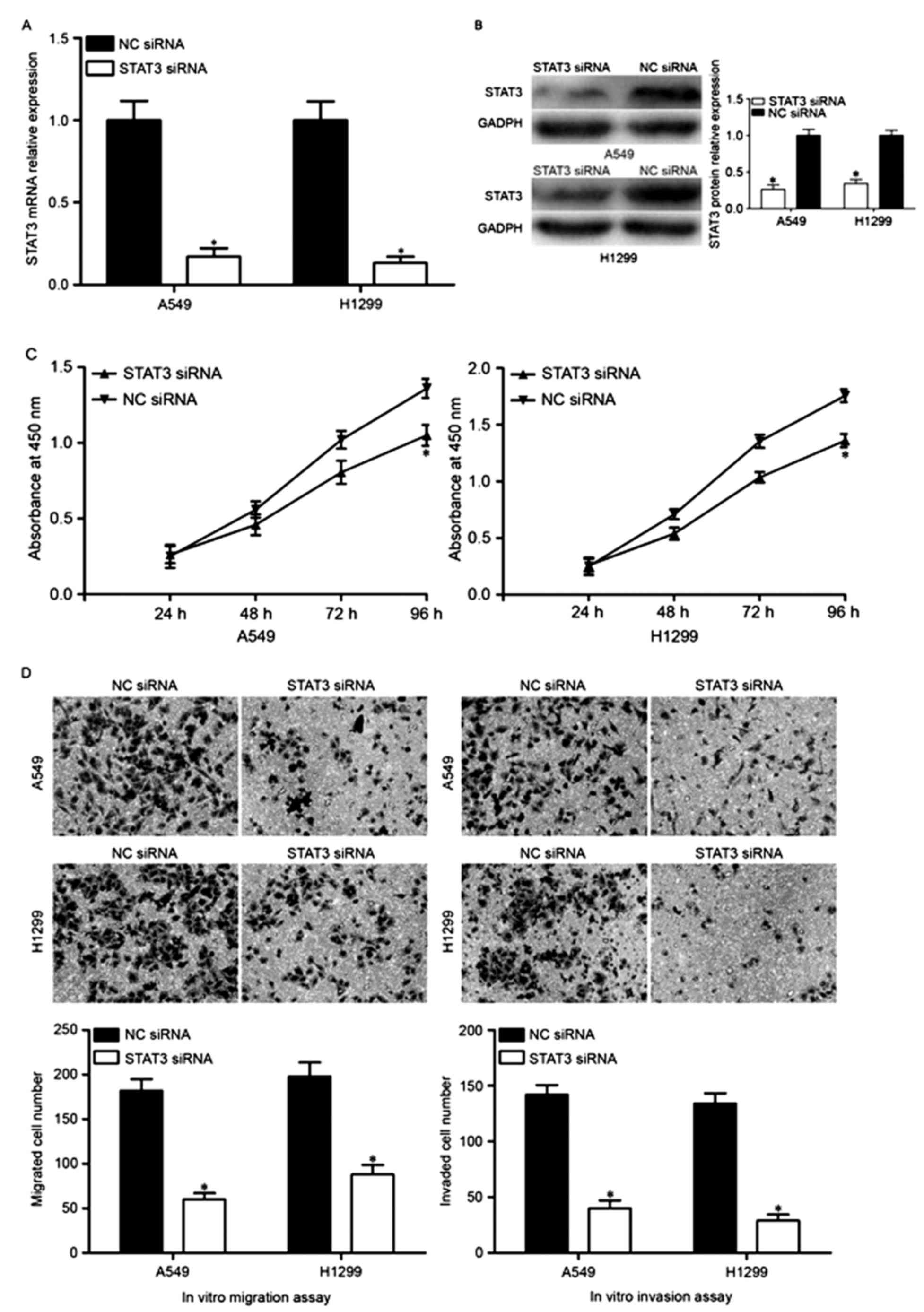 | Figure 4.Downregulation of STAT3 inhibits A549
and H1299 cell proliferation, migration and invasion. A549 and
H1299 cells were injected with STAT3 siRNA or NC siRNA, and the (A)
mRNA and (B) protein expression levels of STAT3 were determined by
reverse transcription-quantitative polymerase chain reaction and
western blot analysis, respectively. The effects of STAT3
underexpression on cell proliferation, migration and invasion were
evaluated by (C) Cell Counting kit-8 and (D) in vitro
migration and invasion assays, respectively. Data are expressed as
the mean ± standard deviation. NSCLC, non-small cell lung cancer;
miR-454, microRNA-454; NC, negative control; STAT3, signal
transducers and activators of transcription 3; siRNA, small
interfering RNA. |
Discussion
Previous studies have revealed that a great deal of
miRNAs are dysregulated in NSCLC, and part of these miRNAs have
been demonstrated to be correlated with particular
clinicopathological factors of NSCLC (19–21).
Notably, accumulated evidence has verified that miRNAs are involved
in NSCLC carcinogenesis and progression via regulation of cell
growth, apoptosis, angiogenesis, migration and invasion (22–24).
Therefore, it is beneficial to investigate the expression,
functions and corresponding targets of deregulated miRNAs in NSCLC,
which could provide therapeutic targets to improve the prognosis of
this disease. In the present study, it was demonstrated that
miR-454 is downregulated in NSCLC. In addition, reduced miR-454
expression was significantly correlated with aggressive
clinicopathological features. miR-454 overexpression suppressed
cell proliferation, migration and invasion of NSCLC. In addition,
STAT3 was identified as a direct target of miR-454. To the best of
our knowledge, our current study is the first to investigate the
expression, clinical significance and biological roles of miR-454
in NSCLC.
The aberrant expression of miR-454 has been reported
in various kinds of cancers. Sun et al (25) demonstrated that miR-454 is
obviously upregulated in uveal melanoma. In colorectal cancer, the
expression level of miR-454 was higher in tumor tissues and cell
lines than the matched tumor adjacent tissues and normal colonic
cell line, respectively (26).
Zhou et al (27) reported
that miR-454 was upregulated in hepatocellular carcinoma, and
upregulation of miR-454 was associated with a low 5-year overall
survival. Multivariate analysis indicated that miR-454
overexpression was an independent prognostic factor for 5-year
overall survival and disease-free survival (27). However, a study by Niu et al
(28) demonstrated that expression
of miR-454 was lower in osteosarcoma tissue specimens and cell
lines. Fang et al (29)
revealed that miR-454 was significantly downregulated in
glioblastoma tissue samples and cells. These studies suggested that
miR-454 expression is deregulated in cancers, and may contribute to
tumorigenesis and tumor progression.
The biological functions of miR-454 have been
studied in many kinds of human cancer. For example, upregulation of
miR-454 suppresses osteosarcoma cell proliferation and invasion via
negative regulation of c-Met (28). miR-454 targets stromal cell derived
factor-1 to regulate the growth of pancreatic ductal adenocarcinoma
cells (30). Downregulation of
miR-454 represses hepatocellular carcinoma cell proliferation,
invasion and epithelial mesenchymal transition by directly
targeting chromodomain helicase DNA binding protein 5 (31). miR-454 serves as an oncogene in
uveal melanoma via promotion of cell growth, colony formation,
invasion and induction of the cell cycle (25). Liang et al (26) demonstrated that miR-454 enhances
cell proliferation in colorectal cancer via blockade of ubiquitin
carboxyl-terminal hydrolase CYLD (26). In glioblastoma, miR-454
overexpression suppresses cell growth by downregulation of
3-phosphoinositide-dependent protein kinase 1 (29). These findings suggested that
miR-454 may represent an efficient therapeutic target in human
cancers.
Furthermore, STAT3, an oncogene, was identified as a
direct target of miR-454. STAT3 is a signal mediator that can be
activated by various kinds of cytokines, growth factors and
interferons (32). Mounting
studies have demonstrated that STAT3 is constitutively activated in
dozens of human cancers, including prostate (33), breast (34), head and neck (35) and lung (36). In the present study, STAT3 mRNA and
protein expression levels were upregulated in NSCLC. Furthermore,
high STAT3 expression was associated with tumor differentiation,
clinical stage and lymph node metastasis of NSCLC patients. NSCLC
patients with high STAT3 expression had a poorer prognosis compared
with patients with low STAT3 expression. In a previous study,
multivariate analysis revealed that high STAT3 protein expression
was an independent prognostic factor for NSCLC patients (37). A functional study by Zhu et
al (38) revealed that
antisense oligonucleotides targeting STAT3 inhibit NSCLC cell
proliferation and induce apoptosis (37). Siveen et al (39) reported that STAT3 inhibits
apoptosis and induces cell proliferation, angiogenesis, invasion
and metastasis in various human cancers (39). Given the importance and role of
STAT3 in multiple oncogenic signaling pathways, STAT3 may be
validated as a therapeutic target for patients with NSCLC.
In conclusion, the current study provided evidence
that miR-454 may function as a tumor suppressor in NSCLC, partly by
regulating STAT3 expression. Therefore, regulating miR-454
expression represents a potential strategy for the treatment of
NSCLC patients.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang F, Zhou J, Zhang Y, Wang Y, Cheng L,
Bai Y and Ma H: The value of microRNA-155 as a prognostic factor
for survival in non-small cell lung cancer: A meta-analysis. PLoS
One. 10:e01368892015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Verdecchia A, Francisci S, Brenner H,
Gatta G, Micheli A, Mangone L and Kunkler I; EUROCARE-4 Working
Group, : Recent cancer survival in Europe: A 2000–02 period
analysis of EUROCARE-4 data. Lancet Oncol. 8:784–796. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen C, Zhao Z, Liu Y and Mu D:
MicroRNA-99a is downregulated and promotes proliferation, migration
and invasion in non-small cell lung cancer A549 and H1299 cells.
Oncol Lett. 9:1128–1134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Li Y, Liu J, Fan Y, Li X, Dong M,
Liu H and Chen J: Expression levels of microRNA-145 and
microRNA-10b are associated with metastasis in non-small cell lung
cancer. Cancer Biol Ther. 17:272–279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Yang X, Wu H, Zhou W and Liu Z:
MicroRNA-145 inhibits migration and invasion via inhibition of
fascin 1 protein expression in non-small-cell lung cancer cells.
Mol Med Rep. 12:6193–6198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li D, Wei Y, Wang D, Gao H and Liu K:
MicroRNA-26b suppresses the metastasis of non-small cell lung
cancer by targeting MIEN1 via NF-κB/MMP-9/VEGF pathways. Biochem
Biophys Res Commun. 472:465–470. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gee GV, Koestler DC, Christensen BC,
Sugarbaker DJ, Ugolini D, Ivaldi GP, Resnick MB, Houseman EA,
Kelsey KT and Marsit CJ: Downregulated microRNAs in the
differential diagnosis of malignant pleural mesothelioma. Int J
Cancer. 127:2859–2869. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Donadeu FX, Schauer SN and Sontakke SD:
Involvement of miRNAs in ovarian follicular and luteal development.
J Endocrinol. 215:323–334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liwak U, Faye MD and Holcik M: Translation
control in apoptosis. Exp Oncol. 34:218–230. 2012.PubMed/NCBI
|
|
14
|
Rutnam ZJ and Yang BB: The involvement of
microRNAs in malignant transformation. Histol Histopathol.
27:1263–1270. 2012.PubMed/NCBI
|
|
15
|
Joshi P, Middleton J, Jeon YJ and Garofalo
M: MicroRNAs in lung cancer. World J Methodol. 4:59–72. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen T, Gao F, Feng S, Yang T and Chen M:
MicroRNA-370 inhibits the progression of non-small cell lung cancer
by downregulating oncogene TRAF4. Oncol Rep. 34:461–468. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Yan C, Shi X, Zheng J, Deng L,
Yang L, Yu F, Yang Y and Shao Y: MicroRNA-575 targets BLID to
promote growth and invasion of non-small cell lung cancer cells.
FEBS Lett. 589:805–811. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li D, Li DQ, Liu D and Tang XJ: miR-613
induces cell cycle arrest by targeting CDK4 in non-small cell lung
cancer. Cell Oncol (Dordr). 39:139–147. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cortinovis D, Monica V, Pietrantonio F,
Ceresoli GL, La Spina CM and Wannesson L: MicroRNAs in non-small
cell lung cancer: Current status and future therapeutic promises.
Curr Pharm Des. 20:3982–3990. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Boeri M, Pastorino U and Sozzi G: Role of
microRNAs in lung cancer: microRNA signatures in cancer prognosis.
Cancer J. 18:268–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
miR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hou Y, Zhen J, Xu X, Zhen K, Zhu B, Pan R
and Zhao C: miR-215 functions as a tumor suppressor and directly
targets ZEB2 in human non-small cell lung cancer. Oncol Lett.
10:1985–1992. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou YL, Xu YJ and Qiao CW: miR-34c-3p
suppresses the proliferation and invasion of non-small cell lung
cancer (NSCLC) by inhibiting PAC1/MAPK pathway. Int J Clin Exp
Pathol. 8:6312–6322. 2015.PubMed/NCBI
|
|
25
|
Sun L, Wang Q, Gao X, Shi D, Mi S and Han
Q: MicroRNA-454 functions as an oncogene by regulating PTEN in
uveal melanoma. FEBS Lett. 589:2791–2796. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liang HL, Hu AP, Li SL, Xie JP, Ma QZ and
Liu JY: miR-454 prompts cell proliferation of human colorectal
cancer cells by repressing CYLD expression. Asian Pac J Cancer
Prev. 16:2397–2402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou L, Qu YM, Zhao XM and Yue ZD:
Involvement of miR-454 overexpression in the poor prognosis of
hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 20:825–829.
2016.PubMed/NCBI
|
|
28
|
Niu G, Li B, Sun J and Sun L: miR-454 is
down-regulated in osteosarcomas and suppresses cell proliferation
and invasion by directly targeting c-Met. Cell Prolif. 48:348–355.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fang B, Zhu J, Wang Y, Geng F and Li G:
miR-454 inhibited cell proliferation of human glioblastoma cells by
suppressing PDK1 expression. Biomed Pharmacother. 75:148–152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fan Y, Xu LL, Shi CY, Wei W, Wang DS and
Cai DF: MicroRNA-454 regulates stromal cell derived factor-1 in the
control of the growth of pancreatic ductal adenocarcinoma. Sci Rep.
6:227932016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu L, Gong X, Sun L, Yao H, Lu B and Zhu
L: miR-454 functions as an oncogene by inhibiting CHD5 in
hepatocellular carcinoma. Oncotarget. 6:39225–39234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wagner KU and Schmidt JW: The two faces of
Janus kinases and their respective STATs in mammary gland
development and cancer. J Carcinog. 10:322011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mora LB, Buettner R, Seigne J, Diaz J,
Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, et
al: Constitutive activation of Stat3 in human prostate tumors and
cell lines: Direct inhibition of Stat3 signaling induces apoptosis
of prostate cancer cells. Cancer Res. 62:6659–6666. 2002.PubMed/NCBI
|
|
34
|
Dolled-Filhart M, Camp RL, Kowalski DP,
Smith BL and Rimm DL: Tissue microarray analysis of signal
transducers and activators of transcription 3 (Stat3) and
phospho-Stat3 (Tyr705) in node-negative breast cancer shows nuclear
localization is associated with a better prognosis. Clin Cancer
Res. 9:594–600. 2003.PubMed/NCBI
|
|
35
|
Nagpal JK, Mishra R and Das BR: Activation
of Stat-3 as one of the early events in tobacco chewing-mediated
oral carcinogenesis. Cancer. 94:2393–2400. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Song L, Turkson J, Karras JG, Jove R and
Haura EB: Activation of Stat3 by receptor tyrosine kinases and
cytokines regulates survival in human non-small cell carcinoma
cells. Oncogene. 22:4150–4165. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yin Z, Zhang Y, Li Y, Lv T, Liu J and Wang
X: Prognostic significance of STAT3 expression and its correlation
with chemoresistance of non-small cell lung cancer cells. Acta
Histochem. 114:151–158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhu BR, Cai JM, Tang GS, Li BL, Gao F, Cui
JG and Liu HC: Effects of STAT3 antisense oligonucleotide on
proliferation and apoptosis of non-small cell lung cancer cell line
A549. Ai Zheng. 26:820–827. 2007.(In Chinese). PubMed/NCBI
|
|
39
|
Siveen KS, Sikka S, Surana R, Dai X, Zhang
J, Kumar AP, Tan BK, Sethi G and Bishayee A: Targeting the STAT3
signaling pathway in cancer: Role of synthetic and natural
inhibitors. Biochim Biophys Acta. 1845:136–154. 2014.PubMed/NCBI
|