Introduction
Gastric cancer is the fourth most common cancer and
the second most common cause of cancer-associated mortality in the
world (1–3). Gastric cancer remains difficult to
cure, primarily since the majority patients present with advanced
disease or even suffer from gastric cancer metastasis. Therefore,
the mechanisms of gastric cancer invasion and migration remain to
be elucidated.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs,
which are approximately 20 nucleotides (nt) long and are involved
in vast range of cellular processes (4–6).
miRNAs negatively regulate gene expression by repressing
translation or inducing target mRNA degradation (5,6),
processes which are notably involved in cell proliferation,
differentiation and apoptosis (7).
miRNAs have been observed to serve roles in numerous types of
cancer. Increasing evidence has indicated that miRNAs may function
as oncogenes or tumor suppressor genes, which are crucial in the
initiation and progression of human malignancies (8). miR-21 is an extensively-researched
oncogenic miRNA in a number of solid human tumors, including
non-muscle-invasive bladder cancer (9), colorectal cancer (10), non-small cell lung cancer (11) and gastric cancer (12). miR-21 promoted cell growth and
invasion by repressing phosphatidylinositol 3,4,5-trisphosphate
3-phosphatase and dual-specificity protein phosphatase PTEN in
colorectal cancer (13), similar
to the impact of miR-21 on gastric cancer (14). Recently, miR-373 was demonstrated
to be an important gene in numerous human malignant tumors
(15–17), which provoked widespread
attention.
Hsa-miR-373 belongs to the miR-371-3 gene cluster,
which is transcribed into the primary transcript pri-miR-3771-373.
The pri-miR-371-3 is processed into three pre-miRNAs (pre-miR371,
pre-miR372 and pre-miR-373), giving rise to four mature miRNAs
(miR-371, miR-372, miR-373 and miR-373*) (18). Hsa-miR-373, which was first
reported to be a human embryonic stem cell (ESC)-specific miRNA
(18). Subsequently, miR-373,
consistent with miR-372, was identified to be an oncogene in the
tumorigenesis of human testicular germ cell tumors, through the
direct inhibition of large tumor suppressor 1/2 expression
(18). In 2011, Zhang et al
(19) reported that the
downregulation of hsa-miR-373 had the potential to predict
recurrence risk in patients with gastric cancer following surgical
resection. The following year, it was discovered that PR domain
zinc finger protein 4, a recurrence risk biomarker in patients with
gastric cancer, was regulated by miR-373 (20). An additional study revealed that
miR-373 is upregulated in gastric adenocarcinoma tissue and gastric
carcinoma cell lines, and it induces cell proliferation in gastric
adenocarcinoma cells in vitro via the downregulation of
broad-complex, tramtrack and Bric a brac/Poxvirus and zinc finger
domain-containing adapter for cullin3-mediated RhoA degradation
protein 2 (21). Although
oncogenic roles of miR-373 have been demonstrated, the molecular
mechanisms of miR-373 in gastric cancer metastasis remain unclear.
Mmu-miR-294 is one member of the miR-290 family, which has been
demonstrated to be the homologous gene family of the hsa-miR-371-3
gene family (22). Therefore, the
role of mmu-miR-294 in mouse gastric cancer remains to be
elucidated.
In the present study, the effect of
hsa-miR-373/mmu-miR-294 on gastric cancer cellular metastasis was
investigated. The results demonstrated that hsa-miR-373 and
mmu-miR-294 were able to suppress the metastasis of gastric cancer
cells by negatively regulating the expression of vimentin.
Furthermore, it was observed that hsa-miR-373 and mmu-miR-294 were
able to promote gastric cancer cell proliferation. These results
may provide a novel therapeutic strategy for gastric cancer with
metastasis.
Materials and methods
Clinical gastric cancer tissues
The primary malignant gastric cancer tissues and
their matching and adjacent non-tumorous tissues (located >5-10
cm away from the primary site) were collected from 28 patients with
gastric cancer undergoing surgery at the Affiliated Hospital of
Jiangsu University from March 2016 to May 2017. (Zhenjiang, China).
Informed consent was given by all patients examined. All samples
were confirmed by pathological examination. Histological grade was
defined according to the World Health Organization classification
system. The tissues obtained were immediately stored at −80°C. The
present study was approved by the Medical Ethics Committee of
Jiangsu University.
Cell culture
Human gastric cancer cell lines SGC-7901 and HGC-27,
and mouse gastric cancer cell line MFC, were obtained from the
Institute of Biochemistry and Cell Biology at the Chinese Academy
of Sciences (Shanghai, China). These cell lines were cultured in
RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). All medium was supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc.). Cells were incubated
at 37°C in humidified air with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) for mRNA expression
Total RNA from human gastric tissues and gastric
cancer cells was extracted using TRIzol reagent (Life Technologies;
Thermo Fisher Scientific, Inc.). RT was performed using a HiScript
1st Strand cDNA Synthesis kit (Vazyme, Piscataway, NJ, USA) and the
signal-stranded cDNA was obtained. RT was performed at 25°C for 5
min, 50°C for 15 min and 85°C for 5 min. The qPCR reactions were
performed using the QuantiTect SYBR Green PCR kit (Toyobo Co.,
Ltd., Osaka, Japan). Thermal cycling parameters were as follows:
94°C for 5 min; 40 cycles at 94°C for 30 sec, 55°C for 30 sec and
72°C for 30 sec; and 65–95°C for drawing the dissociation curve.
The expression level of mRNA was normalized to the expression of
β-actin (23). The primer
sequences were as follows: Vimentin sense,
5′-ATACTGCTGGCGCACATCAC-3′, and vimentin antisense,
5′-CCCTTTCCCCAGTTTTTAATAGG-3′; β-actin sense,
5′-GACCTGTACGCCAACACAGT-3′, and β-actin antisense,
5′-CTCAGGAGGAGCAATGATCT-3′.
RT-qPCR for miRNA expression
RT was performed using the miScript II RT kit
(Qiagen China Co., Ltd., Shanghai, China) and the signal-stranded
cDNA was obtained. RT was performed at 42°C for 60 min, followed by
an inactivation reaction at 70°C for 15 min. The CFX96 Real Time
Instrument (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and
miScript SYBR Green PCR kit (Qiagen China Co., Ltd.) were used for
quantitative RNA detection. Thermal cycling parameters were as
follows: 95°C for 5 min; 40 cycles at 94°C for 15 sec, 55°C for 15
sec and 72°C for 20 sec; and 65–95°C for drawing the dissociation
curve. The relative expression levels of miRNA were normalized to
the expression of small nuclear RNA U6 (23). PCR primers were as follows: miR-373
sense, 5′-CGCGCGAAGTGCTTCGATTT-3′, and miR-373 antisense,
5′-GTGCAGGGTCCGAGGT-3′; U6 sense, 5′-GCTTCGGCAGCACATATACTAAAAT-3′,
and U6 antisense, 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
Transfection
miRNA mimics were purchased from Shanghai GenePharma
Co., Ltd. (Shanghai, China), and Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection. The concentrations of hsa-miR-373 mimics or
mmu-miR-294 mimics were 10, 12.5 and 25 nM. The same concentrations
of mimic controls (NC) were used. The sequences of the
oligonucleotides were as follows: miR-294 mimics NC sense,
5′-UUCUUCGAACGUGUCACGUTT-3′, miR-294 mimics NC antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-373 mimics NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′; miR-373 mimics NC antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; miR-373 mimics sense,
5′-GAAGUGCUUCGAUUUUGGGGUGU-3′; and miR-373 mimics antisense,
5′-ACCCCAAAAUCGAAGCACUUCUU-3′.
Cell cycle
A total of 1×105 SGC-7901,
1×105 HGC-27 or 1.5×105 MFC cells were seeded
in each well of a 6-well plate 1 day prior to transfection, and
transfected with miRNA mimics. A total of 6 h subsequently, cells
were washed twice with PBS and placed in fresh medium containing
10% FBS for 48 h. Cells were collected and washed twice with PBS,
and stained with 10 µg/ml propidium iodide (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) in 500 µl PBS (containing 100 µg/ml
RNase) for 30 min in the dark at room temperature. A filter screen
was used to isolate suspended cells. The distribution of cells in
the different phases of the cell cycle was detected using a flow
cytometer (FACS Calibur; BD Biosciences, Franklin Lakes, NJ, USA)
and analyzed with a flow cytometry software (FlowJo v7.6.5; BD
Biosciences).
Colony formation assay
A total of 1,000 SGC-7901 or HGC-27 cells were
seeded in each well of a 6-well plate in triplicate and incubated
at 37°C in a 5% CO2 humidified incubator for 8 days. The
medium was changed at 3-day intervals. At the end of the incubation
period, the cultures were fixed with 4% paraformaldehyde for 30 min
at room temperature and stained with crystal violet for 20 min at
room temperature. Images were captured in five random fields
(magnification, ×40) using an inverted fluorescent microscope
(Nikon Corporation, Tokyo, Japan).
Wound healing assay
SGC-7901, HGC-27 and MFC cells were seeded at a
density of 2×105, 3×105 and 6×105
cells/well, respectively, in 6-well plates and incubated at 37°C in
5% CO2 for 24 h to create confluent monolayers. The
monolayers were scratched with a sterile pipette tip. To measure
the mobility of cells, images were captured in five random fields
(magnification, ×40) at 12 h post-scratching using an inverted
fluorescent microscope (Nikon Corporation).
Transwell invasion assay
The abilities of cell migration and invasion were
detected by Transwell assay. All cells were seeded at
1×105 cells/well in each well of a 24-well plate with
FBS-free RPMI-1640 media in the upper chamber, and there was 500 µl
fresh medium containing 10% FBS in the lower chamber. Prior to
staining with crystal violet for 20 min at room temperature, cells
were incubated at 37°C in humidified air with 5% CO2 for
12 h. Images were captured in 5 random fields (magnification, ×40)
using an upright fluorescent microscope (Nikon Corporation).
In silico target prediction of
miRNA
TargetScan 7.1 (www.targetscan.org) target prediction software was
used to indicated the miRNAs that may potentially target the
3′untranslated region of vimentin.
Western blotting
The total protein in gastric cancer cells was
extracted using a lysis buffer containing 50 mM NaCl, 1 mM ethylene
glycol tetra-acetic acid, 0.1% SDS, 1 mM NaF, 1 mM Na3VO4, 1 mg/ml
aprotinin and 1 mg/ml leupeptin in 10 mM Tris (pH 7.4) and
proteinase inhibitors (1 mM phenylmethylsulfonyl fluoride). Protein
concentrations were determined using spectrophotometry (Nanodrop;
Thermo Fisher Scientific, Inc.). A total of 30 ug total protein
were separated on 10% SDS-PAGE gels and transferred to
nitrocellulose membranes (EMD Millipore, Billerica, MA, USA).
Membranes were blocked with 5% milk in PBS for 1 h at 37°C and
incubated with monoclonal antibodies against β-actin (BS6007M),
proliferating cell nuclear antigen (PCNA; BZ02108), vimentin
(MB9006) and E-cadherin (LM0206; all obtained from Bioworld
Technology, Inc., St Louis Park, MN, USA) at 4°C overnight, the
dilutions of which were 1:2,000, 1:4,000, 1:600 and 1:500,
respectively. Following incubation with the secondary antibodies
[1:2,000; goat anti-mouse horseradish peroxidase immunoglobulin G
(IgG; BS12478) and goat anti-rabbit horseradish peroxidase
conjugated IgG (BS10043); all obtained from Bioworld Technology,
Inc.] for 1 h at 37°C, the signals were visualized using a
chemiluminescent and fluorescent imaging system (EMD Millipore).
β-actin was used as the loading control. The software used for
analysis was ImageQuant LT 7.0 (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA).
Statistical analysis
Statistical analysis of the differences in the
expression levels of miR-373 in paired gastric cancer and adjacent
non-tumorous tissues were analyzed using a paired t-test with Prism
version 5. The correlation between miR-373 expression and
clinicopathological factors was estimated using Fisher's exact
test. Potential differences between groups with different
treatments were analyzed using one-way analysis of variance,
followed by a Dunnett t-test. All experiments were performed at
least in triplicate (n=3). Data are presented as the mean ±
standard deviation. All statistical analysis was performed with
GraphPad Prism 5.0 software (GraphPad Software Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Hsa-miR-373 is upregulated in human
gastric cancer
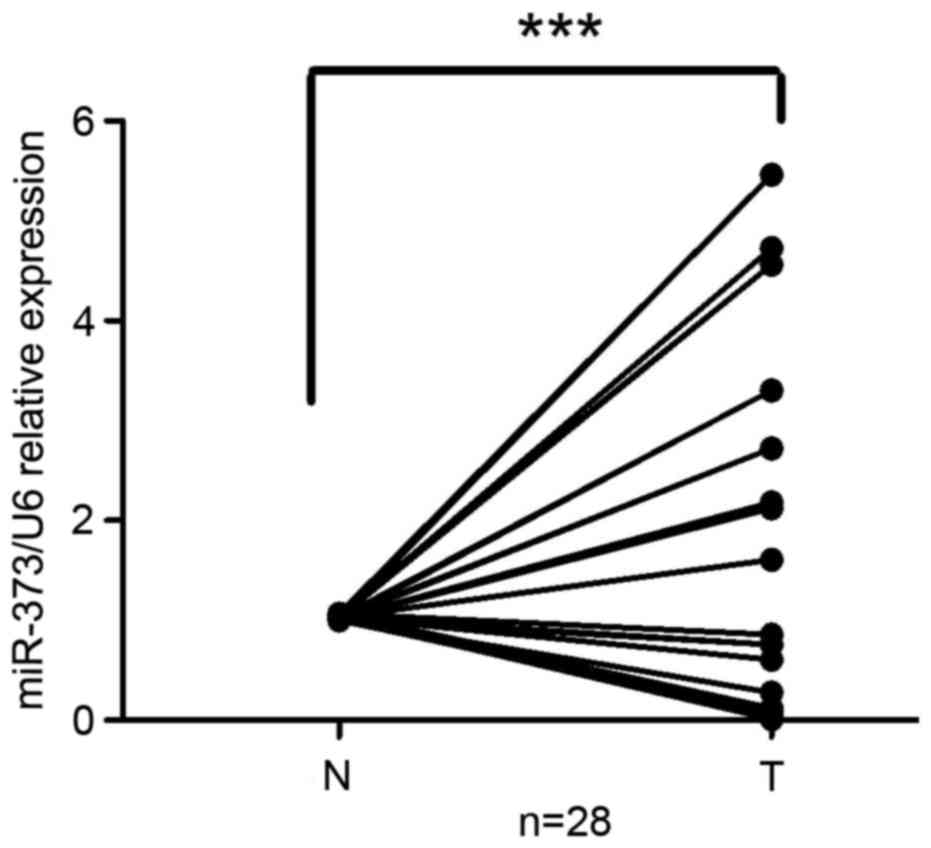
To investigate the clinical significance of miR-373
in gastric cancer, miR-373 expression in 28 paired primary
malignant gastric cancer tissues and adjacent non-tumorous tissues
was compared. The results demonstrated that miR-373 was upregulated
in gastric cancer tissues compared with normal gastric tissues
(n=28, P<0.001; Fig. 1). The
correlation between miR-373 expression and clinical pathological
characteristics was analyzed. Gastric cancer tissues with a more
than two-fold increase in miR-373 (relative to adjacent
non-tumorous tissues) were defined as the high group, while those
with a less than two-fold increase or a decrease in miR-373
(relative to adjacent non-tumorous tissues) were defined as the low
group. The results demonstrated that relative miR-373 expression
was increased in patients with N0/N1 stage lymph node metastasis
(7/8), and a lower expression was observed in patients with N2/N3
stage lymph node metastasis (11/20). miRNA-373 levels were
negatively correlated with lymph node metastasis (Table I). No correlation was observed
between miR-373 expression and age, gender, tumor size,
histological grade or clinical stage (Table I). Thus, it was hypothesized that
miR-373 may be used to distinguish between cases of malignant
primary gastric cancer with or without lymph node metastasis, and
gastric cancer with a lower level of miR-373 may be more
invasive.
 | Table I.Correlation between clinical
pathological factors and miR-373 expression in patients with
gastric cancer. |
Table I.
Correlation between clinical
pathological factors and miR-373 expression in patients with
gastric cancer.
|
|
| miR-373
expression |
|
|---|
|
|
|
|
|
|---|
| Factor | No. (n=28) | Low group | High group | P-value |
|---|
| Age, years |
|
|
| 0.6270 |
|
≥60 | 16 | 8 | 8 |
|
|
<60 | 12 | 4 | 8 |
|
| Sex |
|
|
| 0.4725 |
|
Male | 24 | 12 | 12 |
|
|
Female | 4 | 0 | 4 |
|
| Size, cm |
|
|
| 1.0000 |
| ≥5 | 18 | 8 | 10 |
|
|
<5 | 10 | 4 | 6 |
|
| Histological
grade |
|
|
| 0.4589 |
| Poorly
+ signet | 12 | 4 | 8 |
|
|
Moderately + well | 16 | 8 | 8 |
|
| Stage |
|
|
| 0.4725 |
|
I/II | 4 | 0 | 4 |
|
|
III/IV | 24 | 12 | 12 |
|
| T grade |
|
|
| 0.4725 |
| T1+
T2 | 4 | 0 | 4 |
|
| T3+
T4 | 24 | 12 | 12 |
|
| Lymph node
metastasis |
|
|
|
|
| (N factor) |
|
|
| 0.0882 |
|
N0+N1 | 8 | 1 | 7 |
|
|
N2+N3 | 20 | 11 | 9 |
|
Hsa-miR-373 represses the migration
and invasion of gastric cancer SGC-7901 and HGC-27 cells
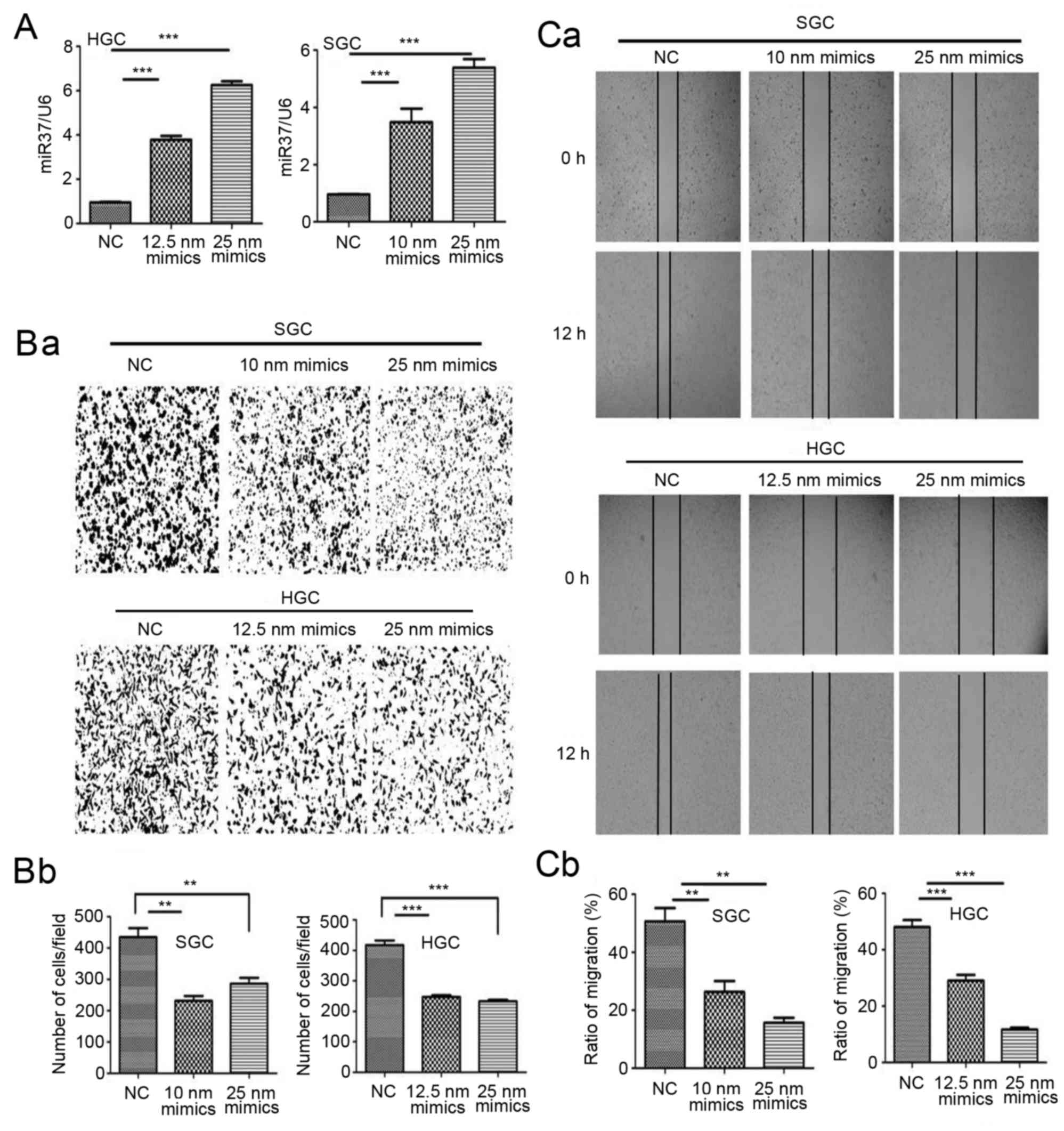
To investigate the role of miR-373 in gastric cancer
cellular metastasis in vitro, SGC-7901 and HGC-27 cells were
transfected with miR-373 mimics in vitro. The effect of
miR-373 on invasion and migration were studied by Transwell
invasion assay and wound healing assay, respectively. Following
treatment with miR-373 mimics, the level of miR-373 in transfected
cells increased (Fig. 2A). The
results of the Transwell invasion assay demonstrated that SGC-7901
and HGC-27 invasion was decreased by miR-373 mimics compared with
control cells (Fig. 2B). The
results of the wound healing assay revealed that the wound size in
miR-373-treated SGC-7901 and HGC-27 cells was larger compared with
control cells (Fig. 2C). These
findings suggested that miR-373 was able to inhibit the migration
and invasion of SGC-7901 and HGC-27 cells.
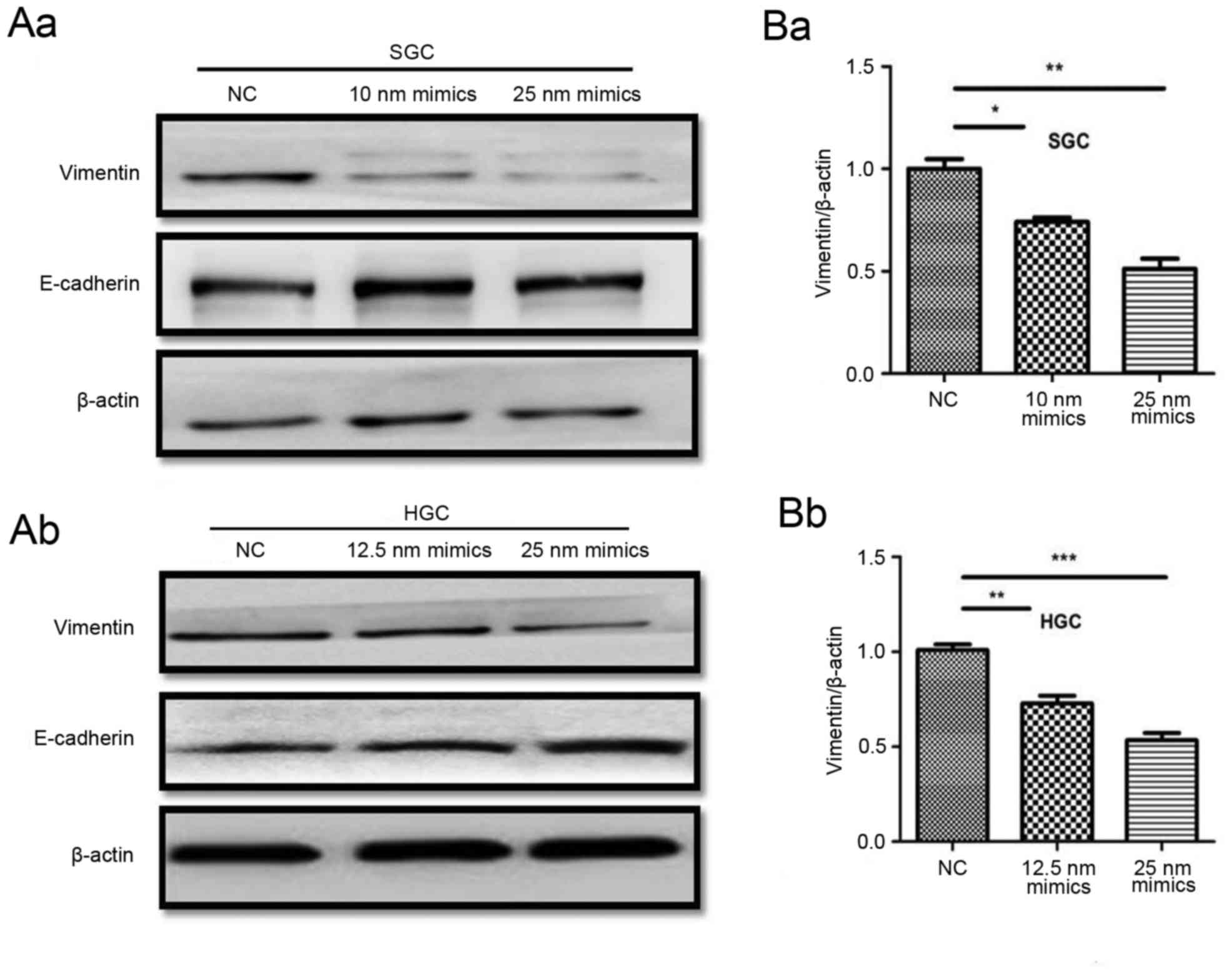
Hsa-miR-373 downregulates vimentin
expression in SGC-790 1 and HGC-27 cells
Previously miR-373 has been reported to be an
oncogene, and to serve a role in tumor metastasis (24). However, the mechanism underlying
the role of miR-373 in tumor metastasis remains unclear.
Epithelial-mesenchymal transition has been demonstrated to serve an
important role in promoting metastasis in gastric cancer (25). Vimentin is an important mesenchymal
marker and E-cadherin is an epithelial marker. The effect of
miR-373 on vimentin and E-cadherin expression was detected by
western blotting. The results demonstrated that vimentin was
downregulated in miR-373 mimics-treated cells compared with the
negative control, and E-cadherin was increased (Fig. 3A). The mRNA expression of vimentin
was additionally decreased in miR-373-treated cells compared with
the negative control (Fig. 3B).
These results revealed that miR-373 was able to downregulate
vimentin expression in SGC-7901 and HGC-27 cells.
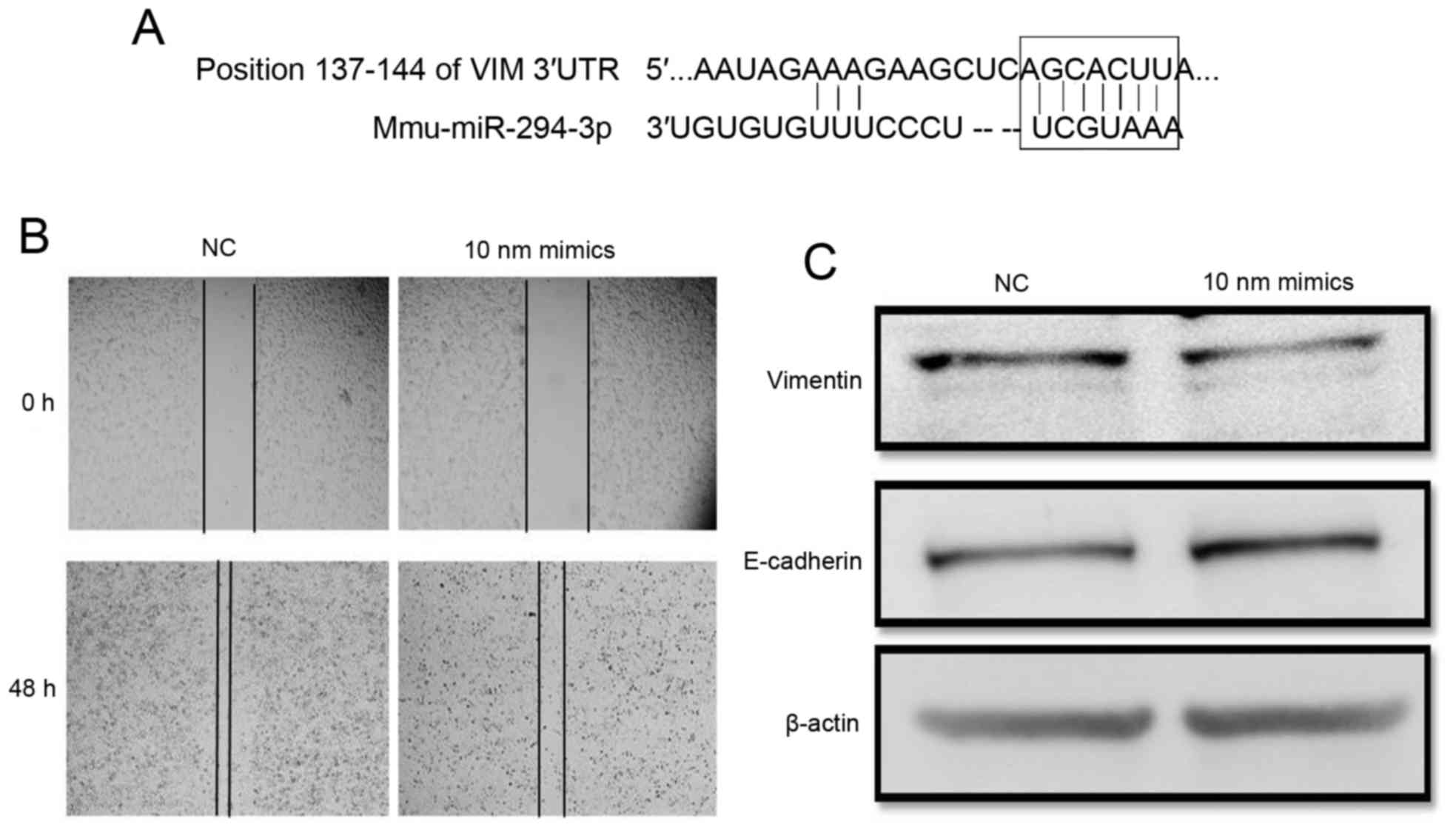
Mmu-miR-294 represses the metastasis
of mouse gastric cancer MFC cells by inhibiting vimentin
Hsa-miR-373 belongs to the miR-371-3 gene cluster,
which has been demonstrated to be the homologous gene family of the
Mmu-miR-294 gene family (22).
Mouse miR-294, a homologous gene of miR-373, was used to further
investigate the function of miR-373 in mouse gastric cancer
metastasis (Fig. 4). Mmu-miR-294
was overexpressed in the mouse gastric cancer MFC cell line. The
wound healing results demonstrated that the wound size in miR-294
mimics-treated cells was larger compared with negative control
cells at 48 h post miRNA transfection (Fig. 4B).
To verify the factors mediating the effect of
miR-294 in gastric cancer, the target of miR-294 was identified
using an online database. The prediction revealed that vimentin
mRNA contained a potential miR-294 seed sequence within its
3′-untranslated region (Fig. 4A).
Western blotting revealed that the alteration in vimentin
expression was inversely associated with miR-294 abundance
(Fig. 4C). These results
demonstrated that mmu-miR-294 was able to repress the migration of
gastric cancer cells by downregulating vimentin expression in
vitro.
Hsa-miR-373 promotes the proliferation
of gastric cancer SGC-7901 and HGC-27 cells in vitro
miR-373 is able to reduce the metastasis of gastric
cancer cells, in addition to promoting tumor cell proliferation
under certain conditions. For example, overexpression of miR-373
inhibits proliferation and tumor growth in colon cancer (26) and pancreatic cancer (24), while it promotes proliferation in
testicular germ cell cancer (18).
It is not clear whether miR-373 is able to promote proliferation of
gastric cancer cell. The present study investigated the role of
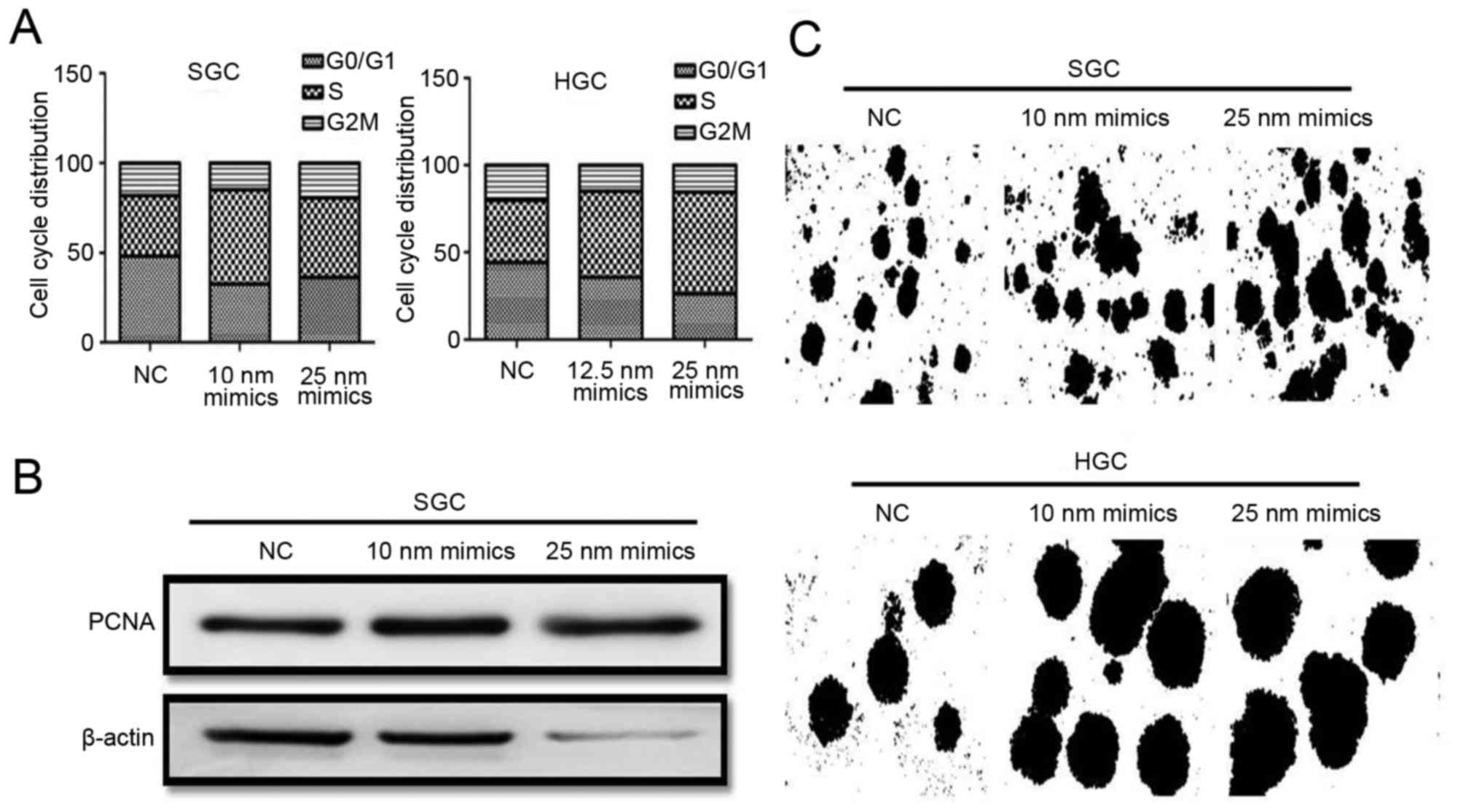
miR-373 in gastric cancer cell proliferation. Cell cycle analysis
demonstrated that the percentage of cells in the S phase was
increased following treatment with mimics (Fig. 5A). The expression of PCNA in
mimics-treated cells, as detected by western blotting, was
upregulated markedly (Fig. 5B).
The results of the colony-formation assay demonstrated that cells
transfected with mimics formed more colonies compared with control
cells, and the diameter of colony-formation units in transfected
cells was larger compared with control cells (Fig. 5C). The results of the present study
demonstrated that miR-373 was able to promote the proliferation of
gastric cancer SGC-7901 and HGC-27 cells in vitro.
Discussion
Gastric cancer is difficult to cure, since patients
with gastric cancer frequently exhibit no obvious symptoms in the
early stages. The majority of patients with gastric cancer are at
the mid-late stages when diagnosed. Over one-half of patients with
gastric cancer undergoing radical surgery suffer metastasis, and
the long-term survival of such patients is shortened (27). Thus, the exact molecular mechanisms
in gastric cancer metastasis require further study.
miRNAs are a type of biomolecule involved in vast
range of cellular processes, including proliferation,
differentiation, senescence and apoptosis (4–6). The
aberrant expression of miRNAs may lead to diseases, including
cancer (28,29). miR-373 was first identified to be
an oncogene in the tumorigenesis of human testicular germ cell
tumors (18), and was upregulated
in seminoma compared with normal testicular tissue. The expression
of miR-373 was additionally upregulated in breast cancer (30) and hepatocellular carcinoma
(31). To elucidate the role of
miR-373 in gastric cancer, the expression of miR-373 was analyzed
in gastric cancer tissues and their paired non-tumorous tissues by
RT-qPCR. The correlation between miR-373 expression and clinical
pathological characteristics was subsequently assessed. The data
showed that miR-373 is up-regulated in gastric cancer tissue than
paired non-tumorous tissues. Furthermore, the miRNA-373 level was
negatively correlated with lymph node metastasis.
Accumulating evidence has indicated that miR-373
participates in tumor metastasis (32), and the role of human miR-373 in
metastasis is dependent the tissue and organ type. miR-373 was
first identified to be a metastasis-promoting miRNA in breast
cancer (30). Cluster of
differentiation 44 was determined to be a functional target of
miR-373 and miR-520c, which was responsible for the migration
phenotype (30). In human
fibrosarcoma HT1080 cells, miR-373 and miR-520c may promote
migration. Serine/threonine-protein kinase mechanistic target of
rapamycin and NAD-dependent protein deacetylase sirtuin-1, which
are negative regulators of matrix metalloproteinase 9 expression,
have been demonstrated to be directly downregulated by miR-373 and
miR-520 (33). These previous
results illustrated the role of human miR-373 in promoting breast
cancer and fibrosarcoma metastasis.
However, miR-373 was additionally demonstrated to
function as a suppressor of cell migration and invasion. Wu et
al (34) reported that the
overexpression of miR-373 in lung cancer A549 cells decreased
migration by targeting E-cadherin. The hepatitis B antigen, which
is involved in HBV-associated hepatocellular carcinoma, was
demonstrated to be able to downregulate the expression of miR-373
which consequently reduces E-cadherin expression, suggesting that
hepatocellular carcinoma with low miR-373 expression may be more
invasive (31). Although these
data demonstrated that miR-373 is involved in the metastasis of
tumors from different tissues and organs, no clear explanation of
the role of miR-373 in gastric cancer metastasis was reported. To
examine the impact of miR-373 on the metastasis of gastric cancer,
the present study used miRNA mimics to perform gain-of-function
experiments. The results demonstrated that miR-373 overexpression
was able to inhibit the migration and invasion of gastric cancer
SGC-7901 and HGC-27 cells in vitro by inhibiting vimentin
expression.
Although it was observed that miR-373 served an
important role in the metastasis of gastric cancer, that gastric
cancer with a lower level of miR-373 may be more invasive, and that
miR-373 overexpression inhibited the migration and invasion of
gastric cancer cells by repressing vimentin, the question of
whether vimentin is the direct target of miR-373 was not examined
in the present study. Therefore, further studies are required to
clarify the direct target of miR-373 in gastric cancer.
In conclusion, these findings suggested that miR-373
may be associated with gastric cancer, and that gastric cancer with
a lower level of miR-373 may be more invasive. miR-373 may inhibit
the metastasis of gastric cancer cells by suppressing the
expression level of vimentin. The results of the present study
demonstrated an oncogenic role of miR-373 in the metastasis of
human gastric cancer, and may provide a novel therapeutic strategy
for gastric cancer with lymph node metastasis.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 31340040, 81272481,
81270214, 81572075, 81602883 and 81670549), the China Postdoctoral
Science Foundation (grant nos. 2015M580403 and 2016T90431), the
China Postdoctoral Science Foundation Funded Project (grant no.
2016M600383), the Special Funded Projects of National Postdoctoral
Fund (grant no. 2017T100337), and the Innovation Project for
Graduate Student Research of Jiangsu Province (grant no.
KYLX15_1096).
References
|
1
|
Brenner H, Rothenbacher D and Arndt V:
Epidemiology of stomach cancer. Methods Mol Biol. 472:467–477.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Desai AM, Pareek M, Nightingale PG and
Fielding JW: Improving outcomes in gastric cancer over 20 years.
Gastric Cancer. 7:196–201. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crew KD and Neugut AI: Epidemiology of
gastric cancer. World J Gastroenterol. 12:354–362. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mukherji S, Ebert MS, Zheng GX, Tsang JS,
Sharp PA and van Oudenaarden A: MicroRNAscan generate thresholds in
target gene expression. Nat Genet. 43:854–859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pichinuk E, Broday L and Wreschner DH:
Endogenous RNA cleavages at the ribosomal SRL site likely reflect
miRNA mediated translational suppression. Biochem Biophys Res
Commun. 414:706–711. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang
X, Tao Y, Zhang L and Xu W: miR-17-5p/20a are important markers for
gastric cancer and murine double minute 2 participates in their
functional regulation. Eur J Cancer. 49:2010–2021. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mitash N, Agnihotri S, Tiwari S, Agrawal V
and Mandhani A: MicroRNA-21 could be a molecular marker to predict
the recurrence of nonmuscle invasive bladder cancer. Indian J Urol.
33:283–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiao W, Leng X, Zhou Q, Wu Y, Sun L, Tan
Y, Ni H, Dong X, Shen T, Liu Y and Li J: Different miR-21-3p
isoforms and their different features in colorectal cancer. Int J
Cancer. 141:2103–2111. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Song Y, Zuo Y, Qian XL, Chen ZP, Wang SK,
Song L and Peng LP: Inhibition of MicroRNA-21-5p promotes the
radiation sensitivity of non-small cell lung cancer through HMSH2.
Cell Physiol Biochem. 43:1258–1272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Motoyama K, Inoue H, Mimori K, Tanaka F,
Kojima K, Uetake H, Sugihara K and Mori M: Clinicopathological and
prognostic significance of PDCD4 and microRNA-21 in human gastric
cancer. Int J Oncol. 36:1089–1095. 2010.PubMed/NCBI
|
|
13
|
Wu Y, Song Y, Xiong Y, Wang X, Xu K, Han
B, Bai Y, Li L, Zhang Y and Zhou L: MicroRNA-21 (Mir-21) promotes
cell growth and invasion by repressing tumor suppressor PTEN in
colorectal cancer. Cell Physiol Biochem. 43:945–958. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang BG, Li JF, Yu BQ, Zhu ZG, Liu BY and
Yan M: microRNA-21 promotes tumor proliferation and invasion in
gastric cancer by targeting PTEN. Oncol Rep. 27:1019–1026. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang L, Qu J, Zhou L, Liao F and Wang J:
MicroRNA-373 inhibits cell proliferation and invasion via targeting
BRF2 in human non-small cell lung cancer A549 cell line. Cancer Res
Treat. Oct 12–2017.(Epub ahead of print). View Article : Google Scholar :
|
|
16
|
Li Y, Zhang D and Wang J: MicroRNA-373
promotes tumorigenesis of renal cell carcinoma in vitro and in
vivo. Mol Med Rep Sep. 16:7048–7055. 2017. View Article : Google Scholar
|
|
17
|
Hua Y, Chen H, Wang L, Wang F, Wang P,
Ning Z, Li Y, Liu L, Chen Z and Meng Z: Low serum miR-373 predicts
poor prognosis in patients with pancreatic cancer. Cancer Biomark.
20:95–100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suh MR, Lee Y, Kim JY, Kim SK, Moon SH,
Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al: Human embryonic
stem cells express a unique set of microRNAs. Dev Biol.
270:488–498. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui
J, Liu Y, Gao Z, Li J, Shen L and Lu Y: Combination of hsa-miR-375
and hsa-miR-142-5p as a predictor for recurrence risk in gastric
cancer patients following surgical resection. Ann Oncol.
22:2257–2266. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yan Z, Xiong Y, Xu W, Li M, Cheng Y, Chen
F, Ding S, Xu H and Zheng G: Identification of recurrence-related
genes by integrating microRNA and gene expression profiling of
gastriccancer. Int J Oncol. 41:2166–2174. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang X, Li X, Tan Z, Liu X, Yang C, Ding
X, Hu X, Zhou J, Xiang S, Zhou C and Zhang J: MicroRNA-373 is
upregulated and targets TNFAIP1 in human gastric cancer,
contributing to tumorigenesis. Oncol Lett. 6:1427–1434. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim KS, Kim JS, Lee MR, Jeong HS and Kim
J: A study of microRNAs in silico and in vivo: Emerging regulators
of embryonic stem cells. FEBS J. 276:2140–2149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nakata K, Ohuchida K, Mizumoto K, Aishima
S, Oda Y, Nagai E and Tanaka M: MicroRNA-373 is down-regulated in
pancreatic cancer and inhibits cancer cell invasion. Ann Surg
Oncol. 21 Suppl 4:S564–S574. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Peng Z, Wang CX, Fang EH, Wang GB and Tong
Q: Role of epithelial-mesenchymal transition in gastric cancer
initiation and progression. World J Gastroenterol. 20:5403–5410.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tanaka T, Arai M, Wu S, Kanda T, Miyauchi
H, Imazeki F, Matsubara H and Yokosuka O: Epigenetic silencing of
microRNA-373 plays an important role in regulating cell
proliferation in colon cancer. Oncol Rep. 26:1329–1335.
2011.PubMed/NCBI
|
|
27
|
Cao Y, DePinho RA, Ernst M and Vousden K:
Cancer research: Past, present and future. Nat Rev Cancer.
11:749–754. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Han C, Shen JK, Hornicek FJ, Kan Q and
Duan Z: Regulation of microRNA-1 (miR-1)expression in human cancer.
Biochim Biophys Acta. 1860:227–232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsuda A, Yan IK, Foye C, Parasramka M
and Patel T: MicroRNAs as paracrine signaling mediators in cancers
and metablic diseases. Best Pract Res Clin Endocrinol Metab.
30:577–590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang Q, Gumireddy K, Schrier M, Le Sage
C, Nagel R, Nair S, Egan DA, Li A, Huang G, Klein-Szanto AJ, et al:
The microRNAs miR-373 and miR-520c promote tumour invasion and
metastasis. Nat Cell Bio. 10:202–210. 2008. View Article : Google Scholar
|
|
31
|
Arzumanyan A, Friedman T, Kotei E, Ng IO,
Lian Z and Feitelson MA: Epigeneticrepression of E-cadherin
expression by hepatitis B virus × antigen in liver cancer.
Oncogene. 31:563–572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Stacy AJ, Craig MP, Sakaram S and Kadakia
M: ΔNp63α and microRNAs: Leveraging the epithelial-mesenchymal
transition. Oncotarget. 8:2114–2129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu P and Wilson MJ: miR-520c and miR-373
upregulate MMP9 expression by targeting mTOR and SIRT1 and activate
the Ras/Raf/MEK/Erk signaling pathway and NF-κB factor in human
fibrosarcoma cells. J Cell Physiol. 227:867–876. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu W, He X, Kong J and Ye B: Mir-373
affects human lung cancer cells' growth and its E-cadherin
expression. Oncol Res. 20:163–170. 2012. View Article : Google Scholar : PubMed/NCBI
|