Introduction
Intervertebral disc degeneration (IDD) is a major
contributor of low back pain (LBP) and has been recognized to be
associated with various etiological factors, including aging,
smoking, genetic predisposition, trauma, infection and abnormal
mechanical loading (1–7). IDD is a disc cell-mediated
pathological process (8). The
primary cellular events in the process of IDD have been identified
as disorder of extracellular matrix (ECM) metabolism in disc cells,
aberrant production of matrix proteases, inflammatory cytokines and
chemokines, cell senescence, apoptosis and cell death (9–12).
It is of note that these cellular events have been identified to be
strongly associated with a reduced nutrient supply in
intervertebral disc (IVD) (13,14).
The nutrient supply of disc cells primarily depends
on the diffusion of nutrient solutes from blood vessels through
cartilage endplate (CEP) (15,16).
However, the reduced permeability of CEP due to calcification
impedes the availability of nutrients to disc cells and suppresses
the clearance of metabolites. Therefore, the microenvironment of
the disc becomes harsh, impairing the viability and activity of
disc cells and consequently contributing to IDD (13,17).
The causes of CEP calcification remain to be elucidated. The
underlying molecular mechanism is not well understood. Previous
studies have determined the association between CEP calcification
and abnormal mechanical loading of the spine (18,19).
Excessive mechanical loading has been identified to lead to the
damage of chondrocytes and ECM in CEP (8). CEP chondrocytes, the basic units of
CEP, have an important function in sensing and responding the
mechanical stress loaded on CEP. Therefore, investigating the
responses of CEP chondrocytes to mechanical loading may provide a
novel insight into the mechanism of CEP calcification and the
pathogenesis of IDD.
Cyclic mechanical tension (CMT) generated by the
Flexcell system in vitro was widely used to simulate the
mechanical strain which CEP chondrocytes undergo in vivo
(20–24). The effects of CMT on the
calcification of CEP chondrocytes have been discussed intensively
in previous studies (20–26). It varies with the types of CMT:
Intermittent CMT (ICMT, stimulation for several h each day in
consecutive days) significantly upregulated the expression of
calcification-associated genes, such as collagen type I, type X,
osteocalcin and osteopontin and downregulated the expression of
anti-calcification genes, including progressive ankylosis (ANK)
gene, extracellular nucleotide phosphatase/phosphodiesterase 1
(ENPP1) and transforming growth factor-β1 (TGF-β1) leading to
ICMT-induced calcification of CEP chondrocytes (21,22).
Conversely, continuous CMT (CCMT, continuous stimulation for
several days) upregulated the expression of progressive ankylosis
(ANK) and TGF-β1 and protected CEP chondrocytes from calcification
(20,23). Additionally, CMT has been
identified to regulate the cartilage matrix metabolism, autophagy
and cytoskeleton arrangement in CEP chondrocytes (22,24–26).
In summary, the response of CEP chondrocytes to CMT was
complicated, and involved in complex biological processes and
signaling pathways. However, the regulatory mechanism underlying
the response of CEP chondrocytes to CMT remains to be
elucidated.
MicroRNAs (miRNAs) are members of the family of
small non-coding RNAs and are 20–22 nucleotides in length. They
bind to their target mRNAs in order to trigger degradation or to
suppress their translation (27).
MiRNAs have been determined to regulate various biological
processes, such as cell proliferation, differentiation, apoptosis,
senescence and development (27).
Additionally, the involvement of miRNAs in the development and
progression of various diseases has been discussed in previous
studies, including carcinoma, neurodegenerative disorders,
inflammatory diseases and reproductive disorders (28–34).
The importance of miRNAs in musculoskeletal disorders, such as IDD,
bone tumors, osteoarthritis and osteonecrosis of the femoral head,
have been previously reported (35–40).
However, the miRNA expression profile of CEP chondrocytes under CMT
stimulation remains to be elucidated. Elucidating the altered miRNA
expression profile of CEP chondrocytes under CMT stimulation helps
us understand the mechanism underlying the regulatory effects of
CMT on the biological processes and signaling pathways in CEP
chondrocytes.
The present study aimed to determine the
differentially expressed miRNAs of CEP chondrocytes under ICMT
stimulation. CEP chondrocytes were isolated from human CEP
specimens. Subsequently, ICMT stimulation was applied and the total
RNA of CEP chondrocytes was extracted for miRNA microarray.
Bioinformatics analysis was performed to determine the
significantly differentially expressed miRNAs and their target
genes. The signaling pathways and biological processes in which
these target genes were involved were predicted. The findings of
the present study identified the miRNA expression profile
associated with the biological responses of CEP under ICMT
stimulation, which may aid in identifying the mechanism underlying
the mechanical force-induced changes of CEP. Additionally, the
present study improved the clinical understanding of the
pathogenesis of IDD.
Materials and methods
Ethical statement
All experimental procedures for human IVD samples
were approved by the Ethical Committee of Xinqiao Hospital
(Chongqing, China) and were in consistent with the Declaration of
Helsinki. Signed informed consent was obtained from the
participating patients.
Human CEP specimens
CEP specimens used in this study were obtained from
patients who underwent posterior discectomy and a fusion procedure
for IDD at the orthopedics department of Xinqiao Hospital between
September 2015 and November 2015. The degenerative changes of CEP
were evaluated using the method previously described by Thompson
et al (41). In order to
reduce the bias resulting from age and different CEP degenerative
degrees, the present study selected the patients between the ages
of 40 and 50. Additionally, the modic types of the selected
patients were type 0, normal. The specimens from 6 patients were
used in the present study. Detailed information about the 6
patients and samples is presented in Table I.
 | Table I.Patient information and the usage of
cartilage endplate specimens. |
Table I.
Patient information and the usage of
cartilage endplate specimens.
| Case no. | Age | Gender | Level | Modic type | Validation
type |
|---|
| 1 | 43 | Female | L4/L5 | 0 | Microarray |
| 2 | 45 | Male | L3/L4 | 0 | Microarray |
| 3 | 49 | Male | L5/S1 | 0 | Microarray |
| 4 | 47 | Male | L5/S1 | 0 | PCR |
| 5 | 44 | Female | L5/S1 | 0 | PCR |
| 6 | 48 | Male | L5/S1 | 0 | PCR |
CEP chondrocyte culture
CEP chondrocytes were isolated from IVD specimens
according to the protocol described in previous studies (42,43).
IVD specimens were placed in a sterilized culture dish and washed
with phosphate buffered saline (PBS). CEP tissues were carefully
separated from IVD specimens under a dissecting microscope.
Subsequently, CEP tissues were minced into 1-mm3 tissue
blocks followed by digestion with Dulbecco's modified Eagle's
medium (DMEM)/F-12 medium (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 0.2% type II collagenase
(Sigma-Aldrich; Merck Millipore, Darmstadt, Germany) for 2 h at
37°C. The cell suspension was filtered with a 70 µm cell strainer
and subsequently centrifuged for 10 min at 100 × g and then
re-suspended in DMEM/F12 containing 10% fetal bovine serum (FBS)
and 1% penicillin/streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.). The isolated CEP chondrocytes were cultured at
37°C and 5% CO2. The medium was replaced twice a week.
When the cells reached about 80% confluence, they were
sub-cultivated. The cells at the second passage were used in the
experiments.
ICMT treatment
Following trypsinization, CEP chondrocytes were
seeded on 6-well flexible silicone rubber BioFlex plates coated
with collagen type I (Flexcell International Corporation,
Burlington, NC, USA) at a density of 1.0×105 cells/well.
When the cells reached 80–90% confluence, ICMT at 0.5 Hz sinusoidal
curve at of 10% elongation was applied for 4 h/day in 14
consecutive days using a FX-5000T Flexcell Tension Plus system
(Flexcell International Corporation). The culture medium was
replaced every 2 days. The cells were cultured in a humidified
atmosphere at 37°C and 5% CO2 ICMT treatment. The
morphology of the cells was observed and imaged using a phase
contrast microscope (Olympus Corporation, Tokyo, Japan). The cells
were harvested immediately after following ICMT treatment. The CEP
chondrocytes from the same patients without ICMT stimulation were
used as the control.
RNA isolation and microRNA
microarray
CEPs isolated from 3 patients (case no. 1, 2 and 3)
were treated with ICMT and then termed the ICMT samples (n=3). The
CEPs from the same patient without ICMT treatment were used as the
control samples (n=3). Total RNA of all samples (n=6) lysed with 1
ml TRIzol reagent (Takara Bio Inc., Otsu, Japan) were sent to
Kangchen Biotech Co., Ltd. (Shanghai, China) for microRNA
microarray analysis. RNA quantity and quality were determined by a
NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Inc.).
All samples met the quality control standards. RNA was labeled
using a miRCURY Array Power Labeling kit (Exiqon, Woburn, MA, USA)
and then was hybridized to a one-color miRCURY Array (Exiqon,
Woburn). The slides were scanned with the Axon GenePix 4000B
microarray scanner (Molecular Devices, LLC, Sunnyvale, CA, USA).
After image scanning, GenePix Pro version 6.0 was used to read the
raw intensity of the image. The intensity of green signal was
calculated after background subtraction and four replicated spots
of each probe on the same slide have been averaged. The median
normalization method was used to obtain the ‘Normalized Data’, as
follows: Normalized Data=(Foreground-Background)/median, the median
is 50 percent quantile of microRNA intensity that is >30 in all
samples after background correction. The difference in the
normalized data of miRNAs between the ICMT group (n=3) and the
control group (n=3) was statistically tested using a paired sample
Student's t-test. The fold-change of the upregulated miRNAs was
calculated using the following equation: Fold-change=ICMT
normalized data/Control normalized data. The fold-change of the
downregulated miRNAs was calculated as following:
Fold-change=−(Control normalized data/the normalized data of the
ICMT groups). The differentially expressed miRNAs were identified
according to their fold-change (fold-change ≥2 or ≤-2). The
significantly differentially expressed miRNAs were selected
according to the following thresholds P<0.05 and fold-change ≥2
or ≤-2.
Reversed transcription-quantitative
polymerase chain reaction (RT-qPCR)
A total of 10 significantly differentially expressed
miRNAs with relatively high fold-changes (fold-change ≥4 or ≤-4)
were selected for validation using the RT-qPCR method, including 5
upregulated miRNAs (hsa-let-7a, hsa-miR-29c, hsa-miR-142,
hsa-miR-181a, hsa-miR-19b) and 5 downregulated miRNAs
(hsa-miR-548q, hsa-miR-637, hsa-miR-614, hsa-miR-581,
hsv1-miR-H14). RNA from the CEPs of the patients (case no. 1, 2, 3,
4, 5 and 6) was reverse transcribed using a microRNA First Strand
cDNA Synthesis kit (Sangon Biotech Co., Ltd., Shanghai, China)
according to the manufacturer's protocol. The reaction was
performed at 37°C for 60 min followed by 85°C for 5 min. qPCR was
performed in triplicate on a StepOnePlus Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) with SYBR
Premix Ex Taq™ II (Takara Bio) according to the protocol provided
by the manufacturer. The following thermocycling conditions were
used for the PCR: 95°C for 30 sec; 40 cycles of 95°C for 5 sec and
60°C for 34 sec. PCR products were subjected to melting curve
analysis. Relative expression was calculated using the
2−ΔΔCq method (44). U6
was used as the internal reference. Mean Cq values were normalized
to U6. The forward primers of the miRNAs investigated in the
present study were listed in Table
II. The reverse primers used for the miRNAs were universal PCR
primer R.
 | Table II.Forward primers used in reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II.
Forward primers used in reverse
transcription-quantitative polymerase chain reaction analysis.
| microRNA | Forward |
|---|
| hsa-miR-548q |
GGCCGCTGGTGCAAAAGTAA |
| hsa-miR-637 |
ACTGGGGGCTTTCGGGC |
| hsa-miR-614 |
GAACGCCTGTTCTTGCCAG |
| hsv1-miR-H14 |
AGTCGCACTCGTCCCTGG |
| hsa-miR-581 |
GGTCTTGTGTTCTCTAGATCAG |
| hsa-miR-142-3p |
GTGTAGTGTTTCCTACTTTATGGA |
| hsa-let-7a-5p |
GCTGAGGTAGTAGGTTGTATAG |
|
hsa-miR-181a-5p |
AACATTCAACGCTGTCGGTGA |
| hsa-miR-19b-3p |
GGCTGTGCAAATCCATGCAAA |
| hsa-miR-29c-3p |
GCTAGCACCATTTGAAATCGGT |
Bioinformatics analysis
The present study used three popular databases
MiRBase (www.mirbase.org) (45), TargetScan (www.targetscan.org) (46) and MiRanda (www.microrna.org) (47) to predict the target genes of the
significantly differentially expressed miRNAs. Genes that
overlapped all three databases were selected as the target genes
for further function annotation analysis. The Gene Ontology
analysis (GO; www.geneontology.org) was used to annotate the
functions of the target genes. The Kyoto Encyclopedia of Genes and
Genomes pathway analysis (KEGG; www.genome.jp/kegg) was performed to identify the
enriched signaling pathways by the target genes. P<0.05 was
considered to indicate statistically significant difference.
Statistical analysis
Data were presented as the mean ± standard error of
the mean. Data from the RT-qPCR were statistically tested using
Kruskal-Wallis non-parametric analysis and Mann-Whitney U post-hoc
tests as previously described (48). The remaining data were analyzed
using SPSS version 22.0 (IBM Corporation, Armonk, NY, USA) and
GraphPad Prism version 6 (GraphPad Software Inc., La Jolla, CA,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
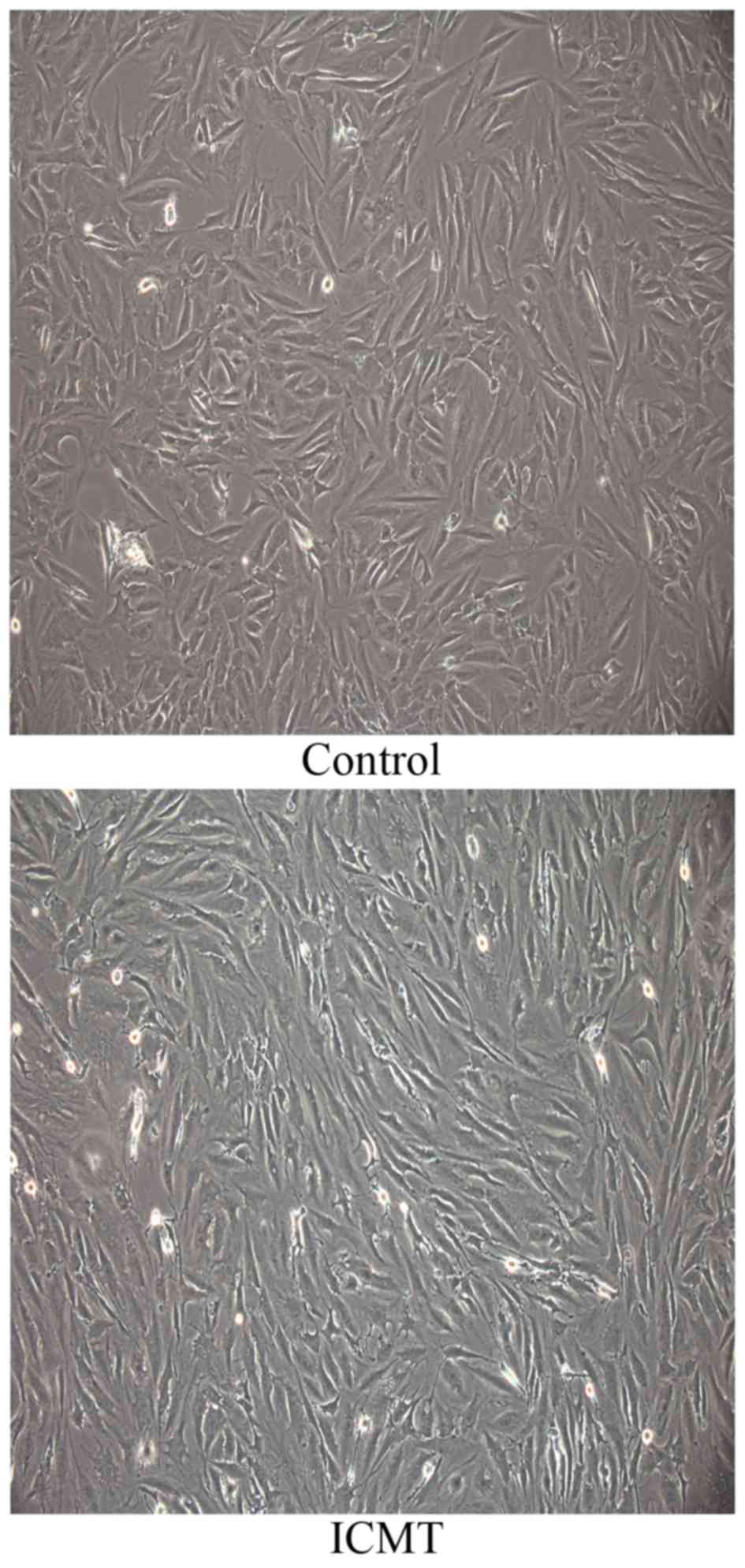
Morphological changes of CEP
chondrocytes subjected to ICMT
The morphology of CEP chondrocytes subjected to ICMT
changed from a polygonal morphology into a spindle morphology.
Additionally, ICMT induced an orderly alignment of CEP chondrocytes
with a certain direction (Fig.
1).
Differentially expressed miRNAs of CEP
chondrocytes induced by ICMT
All differentially expressed miRNAs were filtered
according to their fold-changes (fold-change ≥2 or ≤-2). There were
86 upregulated miRNAs and 188 downregulated miRNAs compared with
the control. From these, 83 miRNAs (21 upregulated and 62
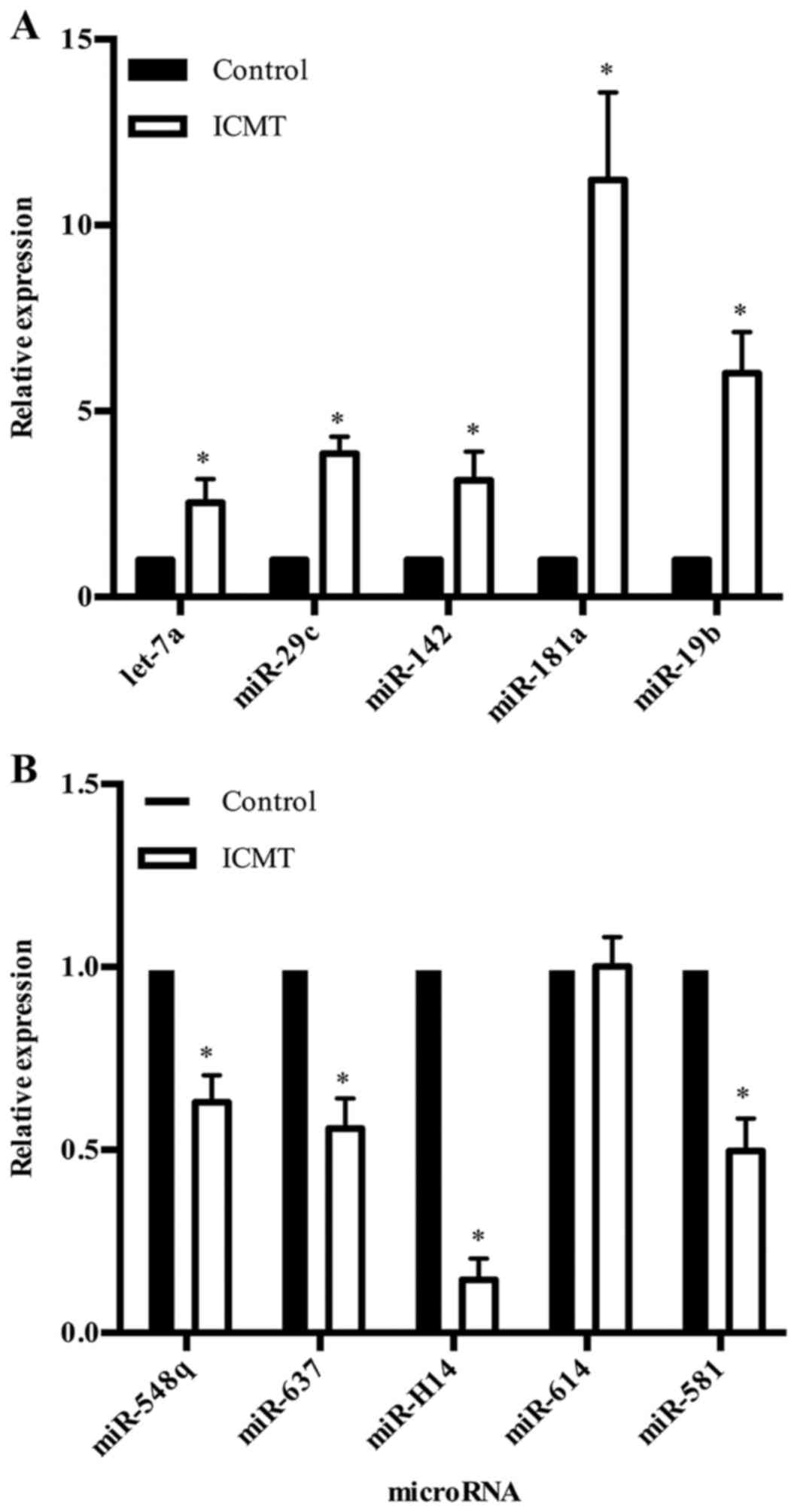
downregulated) were statistically significant (P<0.05; Tables III and IV). To validate the microarray results,
10 significantly differentially expressed miRNAs with high
fold-changes (fold-change ≥4 or ≤-4), including 5 upregulated
miRNAs (hsa-let-7a, hsa-miR-29c, hsa-miR-142, hsa-miR-181a,
hsa-miR-19b) and 5 downregulated miRNAs (hsa-miR-548q, hsa-miR-637,
hsa-miR-614, hsa-miR-581, hsv1-miR-H14) were selected as
representative miRNAs for RT-qPCR assays. With the exception of
has-miR-614, these miRNAs were significantly regulated by ICMT,
which was consistent with the microarray results (Fig. 2).
 | Table III.Significantly upregulated microRNAs
in cartilage endplate chondrocytes under cyclic mechanical
tension. |
Table III.
Significantly upregulated microRNAs
in cartilage endplate chondrocytes under cyclic mechanical
tension.
| ID | Name | Fold-change | P-value |
|---|
| 42,865 |
hsa-miR-181a-5p | 15.84 |
3.29×10−2 |
| 10,947 | hsa-miR-142-3p | 14.65 |
2.29×10−2 |
| 11,041 | hsa-miR-29c-3p | 10.53 |
2.00×10−2 |
| 10,998 | hsa-miR-19b-3p | 10.07 |
3.11×10−2 |
| 11,013 |
hsa-miR-181a-3p |
9.22 |
4.65×10−3 |
| 4610 | hsa-miR-126-3p |
8.47 |
1.94×10−2 |
| 9938 | hsa-let-7i-5p |
6.00 |
3.82×10−2 |
| 145,693 | hsa-miR-92a-3p |
5.19 |
4.85×10−2 |
| 10,990 |
hsa-miR-196a-5p |
5.04 |
8.43×10−3 |
| 17,506 | hsa-miR-24-3p |
4.90 |
1.11×10−2 |
| 32,884 | hsa-miR-342-3p |
4.76 |
2.17×10−2 |
| 147,162 | hsa-let-7a-5p |
4.44 |
3.52×10−2 |
| 19,588 | hsa-miR-17-3p |
3.63 |
2.25×10−2 |
| 146,049 | hsa-miR-28-5p |
3.15 |
2.17×10−2 |
| 33,902 | hsa-miR-128-3p |
2.88 |
1.30×10−2 |
| 10,925 | hsa-miR-10b-5p |
2.64 |
3.42×10−2 |
| 17,935 | hsa-miR-101-5p |
2.52 |
2.21×10−2 |
| 42,783 | hsa-miR-197-3p |
2.47 |
9.19×10−2 |
| 42,829 | hsa-miR-127-3p |
2.25 |
1.96×10−2 |
| 168,925 |
hsa-miR-1273g-3p |
2.18 |
1.72×10−2 |
| 169,385 | hsa-miR-4500 |
2.02 |
3.12×10−2 |
 | Table IV.Significantly downregulated microRNAs
in cartilage endplate chondrocytes under cyclic mechanical
tension. |
Table IV.
Significantly downregulated microRNAs
in cartilage endplate chondrocytes under cyclic mechanical
tension.
| ID | Name | Fold-change | P-value |
|---|
| 145,750 | hsa-miR-614 | −15.33 |
1.40×10−2 |
| 147,815 |
hsv1-miR-H14-5p |
−7.84 |
1.47×10−2 |
| 14,962 | hsa-miR-581 |
−5.81 |
5.64×10−2 |
| 146,148 | hsa-miR-548q |
−5.60 |
4.05×10−2 |
| 169,118 |
hsa-miR-5009-3p |
−5.55 |
4.19×10−2 |
| 148,677 | hsa-miR-637 |
−5.29 |
1.54×10−2 |
| 147,623 | hsa-miR-4304 |
−5.17 |
1.33×10−2 |
| 168,895 | hsa-miR-548ag |
−4.40 |
1.58×10−2 |
| 46,606 |
hsa-miR-1288-3p |
−4.15 |
3.34×10−2 |
| 17,402 |
ebv-miR-BART2-5p |
−4.12 |
3.10×10−2 |
| 168,667 |
hsa-miR-4999-3p |
−3.87 |
1.65×10−2 |
| 45,775 | hsa-miR-1279 |
−3.84 |
2.98×10−2 |
| 148,140 |
hsa-miR-181d-3p |
−3.84 |
1.75×10−2 |
| 169,131 |
hsa-miR-4724-3p |
−3.82 |
3.26×10−2 |
| 46,235 | hsa-miR-524-5p |
−3.79 |
8.34×10−3 |
| 42,493 | hsa-miR-892b |
−3.58 |
1.87×10−2 |
| 169,281 | hsa-miR-4752 |
−3.27 |
4.72×10−2 |
| 169,084 | hsa-miR-5708 |
−3.17 |
3.51×10−3 |
| 147,866 | hsa-miR-3134 |
−3.12 |
4.62×10−2 |
| 42,786 | hsa-miR-188-3p |
−3.11 |
2.14×10−2 |
| 11,154 |
hsa-miR-517c-3p |
−3.07 |
4.48×10−2 |
| 148,474 |
hsa-miR-3622a-5p |
−3.00 |
2.71×10−2 |
| 169,318 |
hsa-miR-4649-3p |
−2.95 |
3.40×10−2 |
| 169,164 | hsa-miR-4705 |
−2.92 |
2.64×10−2 |
| 42,773 |
ebv-miR-BART17-3p |
−2.90 |
4.87×10−2 |
| 17,851 |
hsa-miR-200c-5p |
−2.88 |
3.22×10−2 |
| 168,698 |
hsa-miR-3127-3p |
−2.73 |
4.35×10−2 |
| 168,886 |
hsa-miR-4745-5p |
−2.71 |
2.72×10−2 |
| 148,215 |
hsa-miR-3591-3p |
−2.71 |
3.65×10−2 |
| 147,979 |
hsa-miR-3150a-3p |
−2.68 |
1.33×10−2 |
| 46,634 | hsa-miR-1281 |
−2.65 |
4.57×10−2 |
| 148,102 |
hsa-miR-3619-5p |
−2.60 |
4.67×10−2 |
| 147,977 |
hsa-miR-3190-3p |
−2.48 |
1.02×10−2 |
| 46,800 |
hsa-miR-1224-3p |
−2.40 |
3.47×10−4 |
| 168,745 |
hsa-miR-4667-3p |
−2.40 |
3.02×10−2 |
| 146,006 | hsa-miR-670-5p |
−2.40 |
3.03×10−2 |
| 169,080 |
hsa-miR-4684-5p |
−2.36 |
2.83×10−2 |
| 146,042 | hsv1-miR-H8-3p |
−2.33 |
4.63×10−2 |
| 42,613 |
ebv-miR-BART19-5p |
−2.33 |
3.63×10−2 |
| 46,363 | hsa-miR-1272 |
−2.32 |
2.82×10−2 |
| 46,556 | hsa-miR-623 |
−2.28 |
2.30×10−2 |
| 46,705 | hsa-miR-548k |
−2.28 |
5.75×10−3 |
| 17,336 | hsa-miR-618 |
−2.28 |
2.53×10−3 |
| 146,169 | mcv-miR-M1-3p |
−2.22 |
4.99×10−2 |
| 46,210 |
hsa-miR-1249-3p |
−2.20 |
2.25×10−2 |
| 21,498 | hsa-miR-654-3p |
−2.19 |
3.65×10−2 |
| 169,166 | hsa-miR-4683 |
−2.17 |
1.80×10−2 |
| 17,306 | ebv-miR-BART12 |
−2.15 |
2.81×10−2 |
| 42,584 | hsa-miR-432-3p |
−2.15 |
2.51×10−3 |
| 168,904 | hsa-miR-4473 |
−2.12 |
5.84×10−3 |
| 17,299 |
hcmv-miR-UL22A-3p |
−2.09 |
4.27×10−2 |
| 148,063 | hsa-miR-3713 |
−2.09 |
2.82×10−2 |
| 169,038 | hsa-miR-488-3p |
−2.09 |
3.31×10−3 |
| 168,603 |
hsa-miR-4664-5p |
−2.07 |
2.58×10−3 |
| 148,398 | hsa-miR-3908 |
−2.07 |
1.93×10−3 |
| 147,837 | hsa-miR-3119 |
−2.07 |
4.05×10−2 |
| 168,850 |
hsa-miR-3191-5p |
−2.07 |
2.24×10−2 |
| 169,185 |
hsa-miR-5187-3p |
−2.07 |
2.71×10−3 |
| 46,408 | hsa-miR-1322 |
−2.06 |
1.04×10−2 |
| 42,457 |
hsa-miR-323a-5p |
−2.05 |
1.68×10−2 |
| 168,841 |
hsa-miR-5588-3p |
−2.04 |
9.37×10−3 |
| 148,643 |
hsa-miR-642a-5p |
−2.04 |
3.38×10−2 |
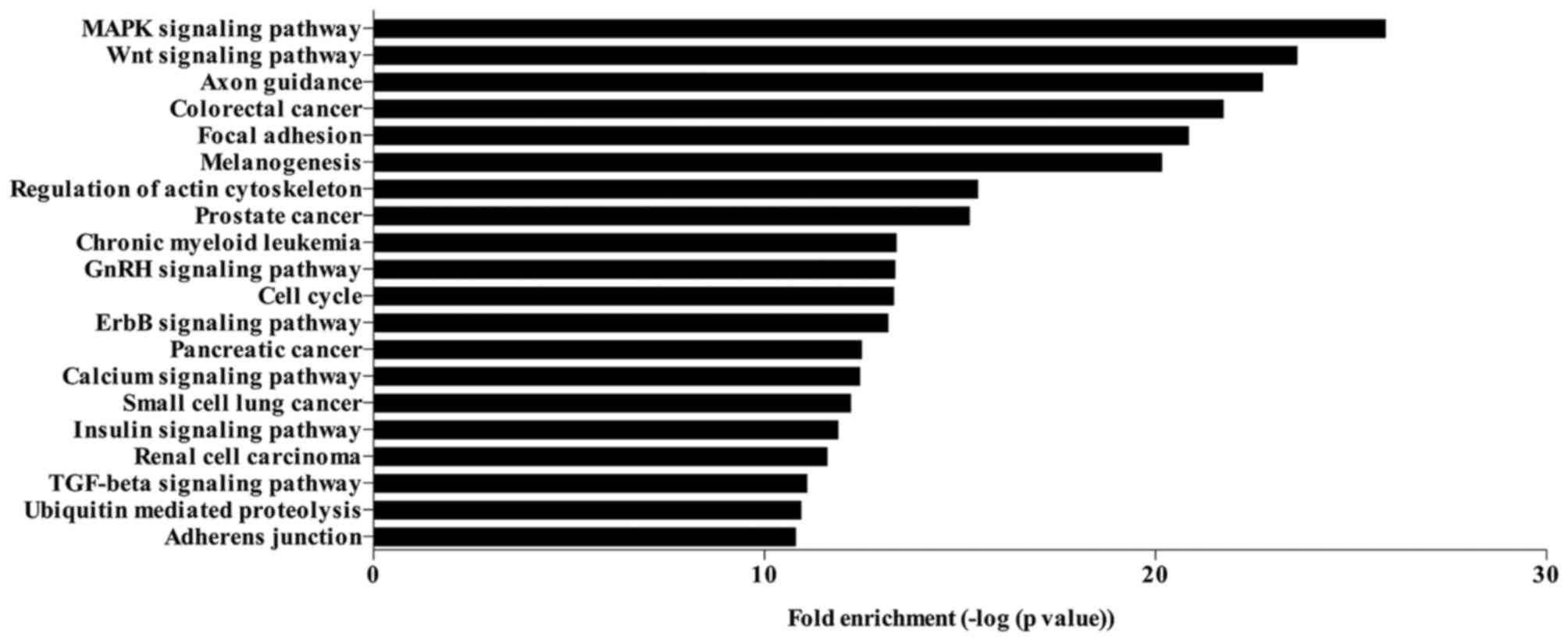
KEGG pathway analysis
The target genes of the 83 significantly
differentially expressed miRNAs were identified based on the
overlapping of 3 databases (MiRBase, MiRanda and TargetScan). A
total of 1,836 genes were identified to be targeted by the
significantly regulated miRNAs. The KEGG pathways analysis of these
target genes revealed that the genes targeted by significantly
differentially expressed miRNAs were enriched in various signaling
pathways. The top 20 signaling pathways regulated by the
significantly differentially expressed miRNAs are presented in
Fig. 3, including the
mitogen-activated protein kinase (MAPK) and Wnt signaling pathways,
focal adhesion, regulation of actin cytoskeleton, GnRH signaling
pathway, cell cycle, ErbB signaling pathway, insulin signaling
pathway, TGF-β signaling pathway and adherens junction.
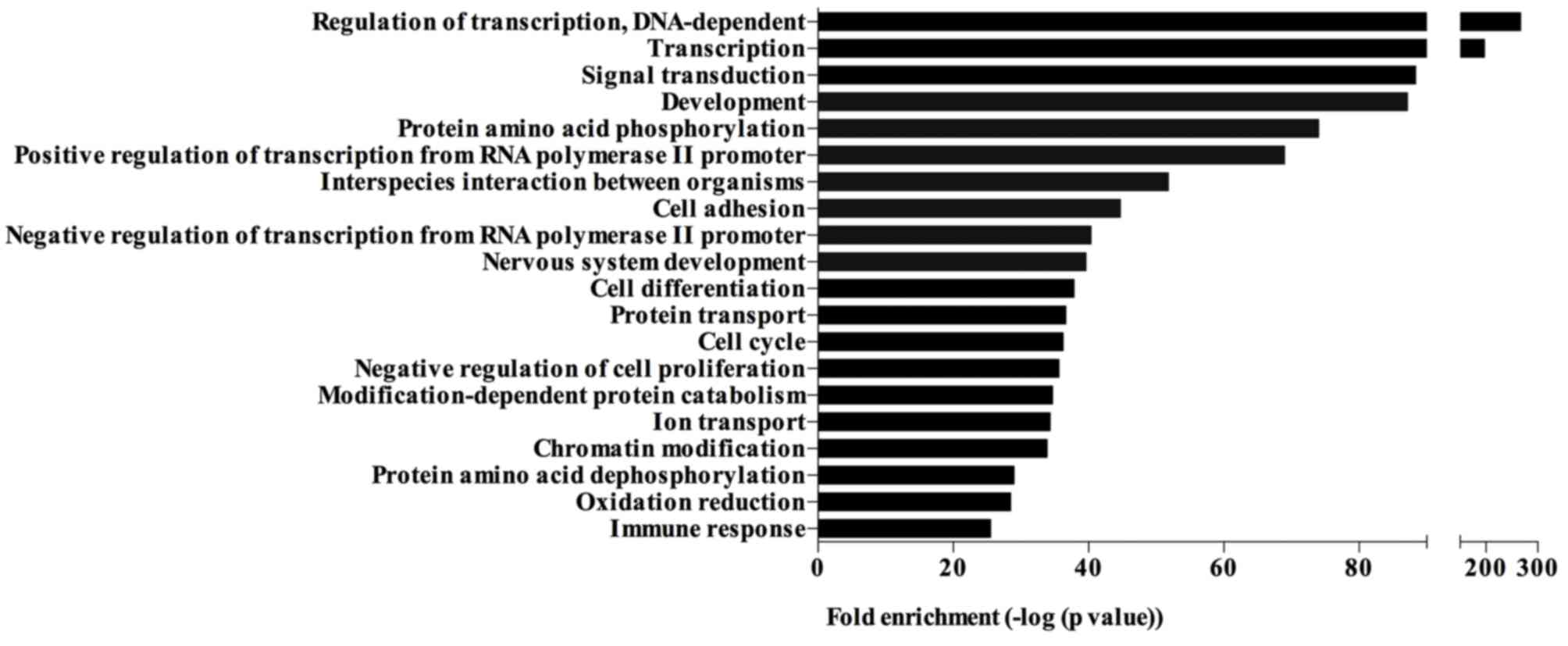
GO analysis
The significantly differentially expressed miRNAs
were involved in different biological processes, including
regulation of transcription, signal transduction, development, cell
adhesion, cell cycle, negative regulation of cell proliferation and
chromatin modification (Fig. 4).
The top 20 biological processes enriched by the target genes of
significantly differentially expressed miRNAs are presented in
Fig. 4.
Discussion
Previous studies have reported that ICMT markedly
induced the calcification of CEP chondrocytes (21,22).
Furthermore, CMT has been identified to regulate the matrix
metabolism, autophagy and cytoskeleton arrangement of CEP
chondrocytes through MAPK and Wnt/β-catenin pathways (22,24–26).
Briefly, CMT is involved in the regulation of complex biological
processes and signaling pathways of CEP chondrocytes. It is of note
that CEP calcification is a major pathological characteristic of
degenerative discs, as it impedes the availability of nutrients to
disc cells and suppresses the clearance of metabolites (13,17).
The presence of disc cell autophagy and the disturbance of the
balance between matrix anabolism and catabolism in human
degenerative disc cells has also been identified as causal factor
of IDD (49–52). Therefore, mechanical tension is a
widely recognized as a primary contributor of IDD. However, the
mechanism underlying the effect of ICMT on the viability and
function of CEP chondrocytes remains to be fully elucidated. The
present study determined that 83 miRNAs were significantly
differentially expressed in human CEP chondrocytes in response to
ICMT. Additionally, GO and KEGG pathway analyses revealed that the
differentially expressed miRNAs were widely involved in the
regulation of various cellular processes and signaling pathways of
human CEP chondrocytes, suggesting that they are crucial
intermediators of the effects of ICMT on the viability and function
of CEP chondrocytes. More importantly, the present study suggests
that miRNA dysregulation has an important function in the
establishment and progression of IDD through mediation of the
effect of mechanical loadings on the structural and functional
homeostasis of IVDs.
The target genes of the significantly differentially
expressed miRNAs were enriched in various biological processes
according to the GO analysis. It is of note that several GO terms
were associated with regulation of cell transcription, including
regulation of transcription (DNA-dependent), positive regulation of
transcription from RNA polymerase II promoter, negative regulation
of transcription from RNA polymerase II promoter and chromatin
modification. The present findings suggest that there is a
regulatory effect of ICMT on the transcriptome of human CEPs. This
is consistent with the findings of previous studies (21,24).
ICMT has been identified to regulate the transcription of
ECM-associated genes, including TGF-β gene and the genes associated
with calcification, including collagen type I, type X, osteocalcin,
osteopontin, ANK and ENPP1. Subsequently, the CEP chondrocytes
undergo a phenotype shift from a cartilage-specific to
calcification-associated, which also promotes IDD progression.
Based on the current findings, miRNAs are crucial regulators of CEP
chondrocyte transcription under ICMT stimulation. Therefore, it is
possible that this ICMT-induced phenotype shift is mediated by
miRNAs, indicating that miRNA dysregulation contributes to the
initiation and progression of CEP calcification. The effect of
miRNAs on the transcriptome of human CEP chondrocytes should be
discussed in further studies. In addition, there were some GO terms
identified to be associated with cell signal transduction,
including signal transduction, protein amino acid phosphorylation,
modification-dependent protein catabolism and protein amino acid
dephosphorylation. Considering the essential function of signal
transduction in the regulation of cell viability and function, the
current findings indicate that miRNAs affect the biological
responses of CEP chondrocytes to mechanical stimulation by
regulation of signal transduction pathways in CEP chondrocytes.
However, the function of miRNAs in regulating mechanical
force-induced signal transduction remains unclear and should be
investigated in future research.
The KEGG pathway analysis was used to annotate the
functional roles of significantly differentially expressed miRNAs
in regulating the biological responses of CEP chondrocytes to ICMT.
When exposed to ICMT, human CEP chondrocytes changed from a
randomly distributed polygonal morphology into a spindle morphology
with a specific directional alignment, which was consistent with
the findings of previous studies (20,26).
Multiple mechanotransduction pathways, including focal adhesion
pathway, action cytoskeleton regulation pathway and adherens
junction pathway were previously identified to be responsible for
these changes (53–55). The findings of the present study
predicted that these signaling pathways were regulated by the
significantly differentially expressed miRNAs under ICMT
stimulation, suggesting that miRNAs may regulate the migration and
morphology of CEP cells via these signaling pathways.
The proliferation of CEP chondrocytes is crucial for
the maintenance of the number of functional cells in CEP, which is
regulated by cell cycle progression. In the current study, some GO
terms and KEGG pathways enriched by the target genes of miRNAs were
associated with cell cycle and negative regulation of cell
proliferation, suggesting that ICMT regulates cell cycle
progression to affect the proliferation of CEP chondrocytes via
miRNAs. ICMT has been previously reported to promote the
proliferation of CEP chondrocytes (26). However, the mechanism underlying
this effect had not been previously investigated. The present study
investigated miRNA regulation in order to elucidate the effect of
mechanical stimulation on CEP chondrocyte proliferation.
TGF-β1 is an established regulator of cell
proliferation, differentiation and ECM metabolism. Previous studies
have demonstrated the function of TGF-β1 in the ICMT-induced
calcification of CEP chondrocytes (21,22).
TGF-β1 induced the expression of ANK and ENPP1 to slow down the
process of CEP calcification. However, ICMT reduced the expression
of TGF-β1 and in turn reduced the expression of ANK and ENPP1
leading to calcification of the CEP cells (21,22).
Additionally, TGF-β1 has been demonstrated to promote the ECM
anabolism of disc cells (56).
Previous studies have determined that the downregulation of
collagen type II and aggrecan was accompanied by the downregulation
of TGF-regulationnion of collagen type he ECM anabolis (21,26).
It is of note that the present study identified an association
between the significantly differentially expressed miRNAs and the
TGF-β signaling pathway, indicating the roles of differentially
expressed miRNAs in the pathogenesis of IDD through regulation of
the matrix metabolism and calcification of CEP chondrocytes via the
TGF-β signaling pathway.
The activation of Wnt pathway has been identified in
human degenerative CEP specimens (26). ICMT has been identified to activate
the Wnt signaling pathway in order to induce the loss of the
chondrogenic phenotype of human CEP chondrocytes (26). Furthermore, the activation of the
Wnt signaling pathway has been found to promote IDD by inducing
nucleus pulposus (NP) cell senescence and upregulating the
expression of matrix metalloproteinase-3 (MMP-3), MMP-7, MMP-9 and
MMP-10 in NP cells (48).
Therefore, the Wnt signaling pathway is involved in the occurrence
and development of IDD. Additionally, the KEGG pathway analysis
performed in the present study suggested that the Wnt signaling
pathway is associated with the significantly differentially
expressed miRNAs under ICMT stimulation. The function of the Wnt
pathway in regulating biological responses of CEP chondrocytes
under ICMT provided novel insights into the involvement of miRNA
dysregulation in the pathogenesis of IDD.
The MAPK signaling pathway has been previously
revealed to regulate the expression of ANK and be responsible for
mechanotransduction (57–59). CMT has been previously reported to
activate the p38-MAPK signaling pathway to regulate the expression
of ANK in CEP chondrocytes (23),
indicating the crucial role of the MAPK signaling pathway in the
regulation of ICMT-induced calcification of CEP chondrocytes.
According the findings of the current study, the MAPK signaling
pathway may be regulated by the differentially expressed miRNA
under ICMT stimulation. The mechanism underlying the regulatory
effects of miRNAs on the MAPK signaling pathway will be discussed
in future studies.
There are two major limitations to the current
study. First one is that the sample size of miRNA microarray
analysis was relatively small. Therefore, more RT-qPCR validation
is required to confirm the effect of ICMT on the expression of
miRNAs in human CEP chondrocytes. Secondly, further studies using
dysregulated miRNAs are necessary to elucidate the effects of
miRNAs on the biological functions of CEP chondrocytes in
detail.
In conclusion, the present study identified the
differentially expressed miRNAs of human CEP chondrocytes under
ICMT stimulation. The bioinformatics analyses suggest that miRNAs
are essential for the regulation of the biological responses of CEP
chondrocytes to ICMT, particularly in terms of the ICMT-induced
calcification of CEP chondrocytes. The present study improved the
current understanding of the involvement of miRNAs in the
pathogenesis of CEP calcification, suggesting the possible function
of miRNA dysregulation in the pathogenesis of IDD. Determining the
detailed regulatory functions of these differentially expressed
miRNAs contributes to identifying novel approaches to protect CEP
cells from calcification under abnormal mechanical stress and
subsequently suppressing the process of IDD.
Acknowledgements
The current study was supported by the National
Natural Science Foundation of China (grant nos. 81271982, 81472076
and 81572186).
Glossary
Abbreviations
Abbreviations:
|
IVD
|
intervertebral disc
|
|
IDD
|
intervertebral disc degeneration
|
|
NP
|
nucleus pulposus
|
|
AF
|
annulus fibrosus
|
|
ICMT
|
intermittent cyclic mechanical
tension
|
|
ECM
|
extracellular matrix
|
|
ANK
|
progressive ankylosis
|
|
ENPP1
|
extracellular nucleotide
phosphatase/phosphodiesterase 1
|
|
TGF-β1
|
transforming growth factor-β1
|
|
CMT
|
cyclic mechanical tension
|
|
LBP
|
lower back pain
|
|
CEP
|
cartilage endplate
|
|
miRNA
|
microRNA
|
|
MMP
|
matrix metalloproteinase
|
|
FBS
|
fetal bovine serum
|
|
PBS
|
phosphate-buffered saline
|
References
|
1
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88 Suppl 2:S10–S14. 2006. View Article : Google Scholar
|
|
2
|
Kanayama M, Togawa D, Takahashi C, Terai T
and Hashimoto T: Cross-sectional magnetic resonance imaging study
of lumbar disc degeneration in 200 healthy individuals. J Neurosurg
Spine. 11:501–507. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheung KM, Karppinen J, Chan D, Ho DW,
Song YQ, Sham P, Cheah KS, Leong JC and Luk KD: Prevalence and
pattern of lumbar magnetic resonance imaging changes in a
population study of one thousand forty-three individuals. Spine
(Phila Pa 1976). 34:934–940. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Battie MC, Videman T, Kaprio J, Gibbons
LE, Gill K, Manninen H, Saarela J and Peltonen L: The twin spine
study: Contributions to a changing view of disc degeneration. Spine
J. 9:47–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vergroesen PP, Kingma I, Emanuel KS,
Hoogendoorn RJ, Welting TJ, van Royen BJ, van Dieën JH and Smit TH:
Mechanics and biology in intervertebral disc degeneration: A
vicious circle. Osteoarthritis Cartilage. 23:1057–1070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang D, Nasto LA, Roughley P, Leme AS,
Houghton AM, Usas A, Sowa G, Lee J, Niedernhofer L, Shapiro S, et
al: Spine degeneration in a murine model of chronic human tobacco
smokers. Osteoarthritis Cartilage. 20:896–905. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stirling A, Worthington T, Rafiq M,
Lambert PA and Elliott TS: Association between sciatica and
Propionibacterium acnes. Lancet. 357:2024–2025. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration and what causes it? Spine (Phila
Pa 1976). 31:2151–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vo NV, Hartman RA, Yurube T, Jacobs LJ,
Sowa GA and Kang JD: Expression and regulation of
metalloproteinases and their inhibitors in intervertebral disc
aging and degeneration. Spine J. 13:331–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang L, Zhang X, Zheng X, Ru A, Ni X, Wu
Y, Tian N, Huang Y, Xue E, Wang X and Xu H: Apoptosis, senescence
and autophagy in rat nucleus pulposus cells: Implications for
diabetic intervertebral disc degeneration. J Orthop Res.
31:692–702. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gruber HE, Ingram JA, Norton HJ, Hanley EN
and Hanley EN Jr: Senescence in cells of the aging and degenerating
intervertebral disc: Immunolocalization of senescence-associated
beta-galactosidase in human and sand rat discs. Spine (Phila Pa
1976). 32:321–327. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang YC, Urban JP and Luk KD:
Intervertebral disc regeneration: Do nutrients lead the way? Nat
Rev Rheumatol. 10:561–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vo NV, Hartman RA, Patil PR, Risbud MV,
Kletsas D, Iatridis JC, Hoyland JA, Le Maitre CL, Sowa GA and Kang
JD: Molecular mechanisms of biological aging in intervertebral
discs. J Orthop Res. 34:1289–1306. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Urban JP, Smith S and Fairbank JC:
Nutrition of the intervertebral disc. Spine (Phila Pa 1976).
29:2700–2709. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grunhagen T, Shirazi-Adl A, Fairbank JC
and Urban JP: Intervertebral disk nutrition: A review of factors
influencing concentrations of nutrients and metabolites. Orthop
Clin North Am. 42:465–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bibby SR and Urban JP: Effect of nutrient
deprivation on the viability of intervertebral disc cells. Eur
Spine J. 13:695–701. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bian Q, Liang QQ, Wan C, Hou W, Li CG,
Zhao YJ, Lu S, Shi Q and Wang YJ: Prolonged upright posture induces
calcified hypertrophy in the cartilage end plate in rat lumbar
spine. Spine (Phila Pa 1976). 36:2011–2020. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Peng B, Hou S, Shi Q and Jia L: The
relationship between cartilage end-plate calcification and disc
degeneration: An experimental study. Chin Med J (Engl).
114:308–312. 2001.PubMed/NCBI
|
|
20
|
Xu HG, Hu CJ, Wang H, Liu P, Yang XM,
Zhang Y and Wang LT: Effects of mechanical strain on ANK, ENPP1 and
TGF-β1 expression in rat endplate chondrocytes in vitro. Mol Med
Rep. 4:831–835. 2011.PubMed/NCBI
|
|
21
|
Xu HG, Zhang XH, Wang H, Liu P, Wang LT,
Zuo CJ, Tong WX and Zhang XL: Intermiåttent cyclic mechanical
tension-induced calcification and downregulation of ankh gene
expression of end plate chondrocytes. Spine (Phila Pa 1976).
37:1192–1197. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu HG, Li ZR, Wang H, Liu P, Wang LT, Zuo
CJ Tong WX and Zhang XL: Intermittent cyclic mechanical
tension-induced down-regulation of ectonucleotide pyrophosphatase
phosphodiesterase 1 gene expression is mainly dependent on TGF-β1
in end-plate chondrocytes. Orthop Surg. 5:40–45. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu H and Zhang X, Wang H, Zhang Y, Shi Y
and Zhang X: Continuous cyclic mechanical tension increases ank
expression in endplate chondrocytes through the TGF-β1 and p38
pathway. Eur J Histochem. 57:e282013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu HG, Yu YF, Zheng Q, Zhang W, Wang CD,
Zhao XY, Tong WX, Wang H, Liu P and Zhang XL: Autophagy protects
end plate chondrocytes from intermittent cyclic mechanical tension
induced calcification. Bone. 66:232–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu HG, Ma MM, Zheng Q, Shen X, Wang H,
Zhang SF, Xu JJ, Wang CD and Zhang XL: P120-catenin protects
endplate chondrocytes from intermittent cyclic mechanical tension
induced degeneration by inhibiting the expression of RhoA/ROCK-1
signaling pathway. Spine (Phila Pa 1976). 41:1261–1271. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu HG, Zheng Q, Song JX, Li J, Wang H, Liu
P, Wang J, Wang CD and Zhang XL: Intermittent cyclic mechanical
tension promotes endplate cartilage degeneration via canonical Wnt
signaling pathway and E-cadherin/β-catenin complex cross-talk.
Osteoarthritis Cartilage. 24:158–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Teague EM, Print CG and Hull ML: The role
of microRNAs in endometriosis and associated reproductive
conditions. Hum Reprod Update. 16:142–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Alexandrov PN, Dua P and Lukiw WJ:
Up-Regulation of miRNA-146a in progressive, age-related
inflammatory neurodegenerative disorders of the human CNS. Front
Neurol. 5:1812014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lv Z, Shi Q, Huang W, Xing C, Hao Y, Feng
X, Yang Y, Zhang A, Kong Q, Yuki N and Wang Y: MicroRNA expression
profiling in Guillain-Barre syndrome. J Neuroimmunol. 301:12–15.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Men Y, Fan Y, Shen Y, Lu L and Kallen AN:
The Steroidogenic acute regulatory protein (StAR) is regulated by
the H19/let-7 Axis. Endocrinology. 158:402–409. 2016.
|
|
33
|
Lu L, Katsaros D, Canuto EM, Biglia N,
Risch HA and Yu H: LIN-28B/let-7a/IGF-II axis molecular subtypes
are associated with epithelial ovarian cancer prognosis. Gynecol
Oncol. 141:121–127. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu L, Katsaros D, Risch HA, Canuto EM,
Biglia N and Yu H: MicroRNA let-7a modifies the effect of
self-renewal gene HIWI on patient survival of epithelial ovarian
cancer. Mol Carcinog. 55:357–365. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ohrt-Nissen S, Døssing KB, Rossing M,
Lajer C, Vikeså J, Nielsen FC, Friis-Hansen L and Dahl B:
Characterization of miRNA expression in human degenerative lumbar
disks. Connect Tissue Res. 54:197–203. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao B, Yu Q, Li H, Guo X and He X:
Characterization of microRNA expression profiles in patients with
intervertebral disc degeneration. Int J Mol Med. 33:43–50. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yuan HF, Von Roemeling C, Gao HD, Zhang J,
Guo CA and Yan ZQ: Analysis of altered microRNA expression profile
in the reparative interface of the femoral head with osteonecrosis.
Exp Mol Pathol. 98:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Miyaki S and Asahara H: Macro view of
microRNA function in osteoarthritis. Nat Rev Rheumatol. 8:543–552.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang X, Qian W, Wu Z, Bian Y and Weng X:
Preliminary screening of differentially expressed circulating
microRNAs in patients with steroidinduced osteonecrosis of the
femoral head. Mol Med Rep. 10:3118–3124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu X, Che L, Xie YK, Hu QJ, Ma CJ, Pei
YJ, Wu ZG, Liu ZH, Fan LY and Wang HQ: Noncoding RNAs in human
intervertebral disc degeneration: An integrated microarray study.
Genom Data. 5:80–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thompson KJ, Dagher AP, Eckel TS, Clark M
and Reinig JW: Modic changes on MR images as studied with
provocative diskography: Clinical relevance-a retrospective study
of 2457 disks. Radiology. 250:849–855. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiong CJ, Huang B, Zhou Y, Cun YP, Liu LT,
Wang J, Li CQ, Pan Y and Wang H: Macrophage migration inhibitory
factor inhibits the migration of cartilage end plate-derived stem
cells by reacting with CD74. PLoS One. 7:e439842012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu LT, Huang B, Li CQ, Zhuang Y, Wang J
and Zhou Y: Characteristics of stem cells derived from the
degenerated human intervertebral disc cartilage endplate. PLoS One.
6:e262852011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: microRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Hiyama A, Sakai D, Risbud MV, Tanaka M,
Arai F, Abe K and Mochida J: Enhancement of intervertebral disc
cell senescence by WNT/β-catenin signaling-induced matrix
metalloproteinase expression. Arthritis Rheum. 62:3036–3047. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ye W, Xu K, Huang D, Liang A, Peng Y, Zhu
W and Li C: Age-related increases of macroautophagy and
chaperone-mediated autophagy in rat nucleus pulposus. Connect
Tissue Res. 52:472–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Jiang W, Zhang X, Hao J, Shen J, Fang J,
Dong W, Wang D, Zhang X, Shui W, Luo Y, et al: SIRT1 protects
against apoptosis by promoting autophagy in degenerative human disc
nucleus pulposus cells. Sci Rep. 4:74562014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gruber HE, Hoelscher GL, Ingram JA, Bethea
S and Hanley EN Jr.: Autophagy in the degenerating human
intervertebral disc: In vivo molecular and morphological evidence
and induction of autophagy in cultured annulus cells exposed to
proinflammatory cytokines-implications for disc degeneration. Spine
(Phila Pa 1976). 40:773–782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Engl W, Arasi B, Yap LL, Thiery JP and
Viasnoff V: Actin dynamics modulate mechanosensitive immobilization
of E-cadherin at adherens junctions. Nat Cell Biol. 16:587–594.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Xu Y, Wang Q, Li Y, Gan Y, Li P, Li S,
Zhou Y and Zhou Q: Cyclic tensile strain induces tenogenic
differentiation of tendon-derived stem cells in bioreactor culture.
Biomed Res Int. 2015:7908042015. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tamiello C, Buskermolen ABC, Baaijens FPT,
Broers JLV and Bouten CVC: Heading in the Right Direction:
Understanding Cellular Orientation Responses to Complex Biophysical
Environments. Cell Mol Bioeng. 9:12–37. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Walsh AJ, Bradford DS and Lotz JC: In vivo
growth factor treatment of degenerated intervertebral discs. Spine
(Phila Pa 1976). 29:156–163. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ding L, Heying E, Nicholson N, Stroud NJ,
Homandberg GA, Buckwalter JA, Guo D and Martin JA: Mechanical
impact induces cartilage degradation via mitogen activated protein
kinases. Osteoarthritis Cartilage. 18:1509–1517. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tetsunaga T, Nishida K, Furumatsu T,
Naruse K, Hirohata S, Yoshida A, Saito T and Ozaki T: Regulation of
mechanical stress-induced MMP-13 and ADAMTS-5 expression by RUNX-2
transcriptional factor in SW1353 chondrocyte-like cells.
Osteoarthritis Cartilage. 19:222–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cailotto F, Bianchi A, Sebillaud S,
Venkatesan N, Moulin D, Jouzeau JY and Netter P: Inorganic
pyrophosphate generation by transforming growth factor-beta-1 is
mainly dependent on ANK induction by Ras/Raf-1/extracellular
signal-regulated kinase pathways in chondrocytes. Arthritis Res
Ther. 9:R1222007. View
Article : Google Scholar : PubMed/NCBI
|