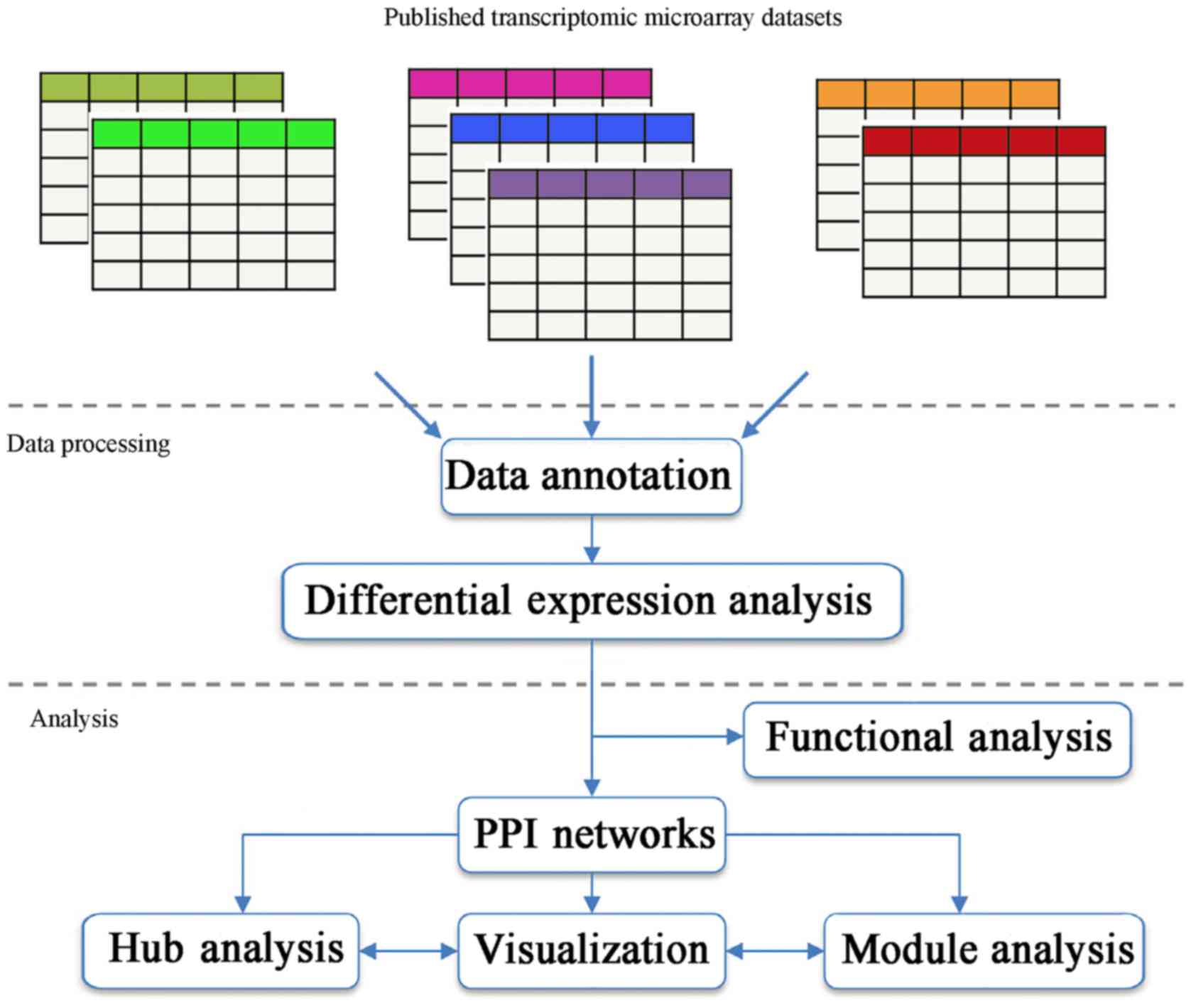
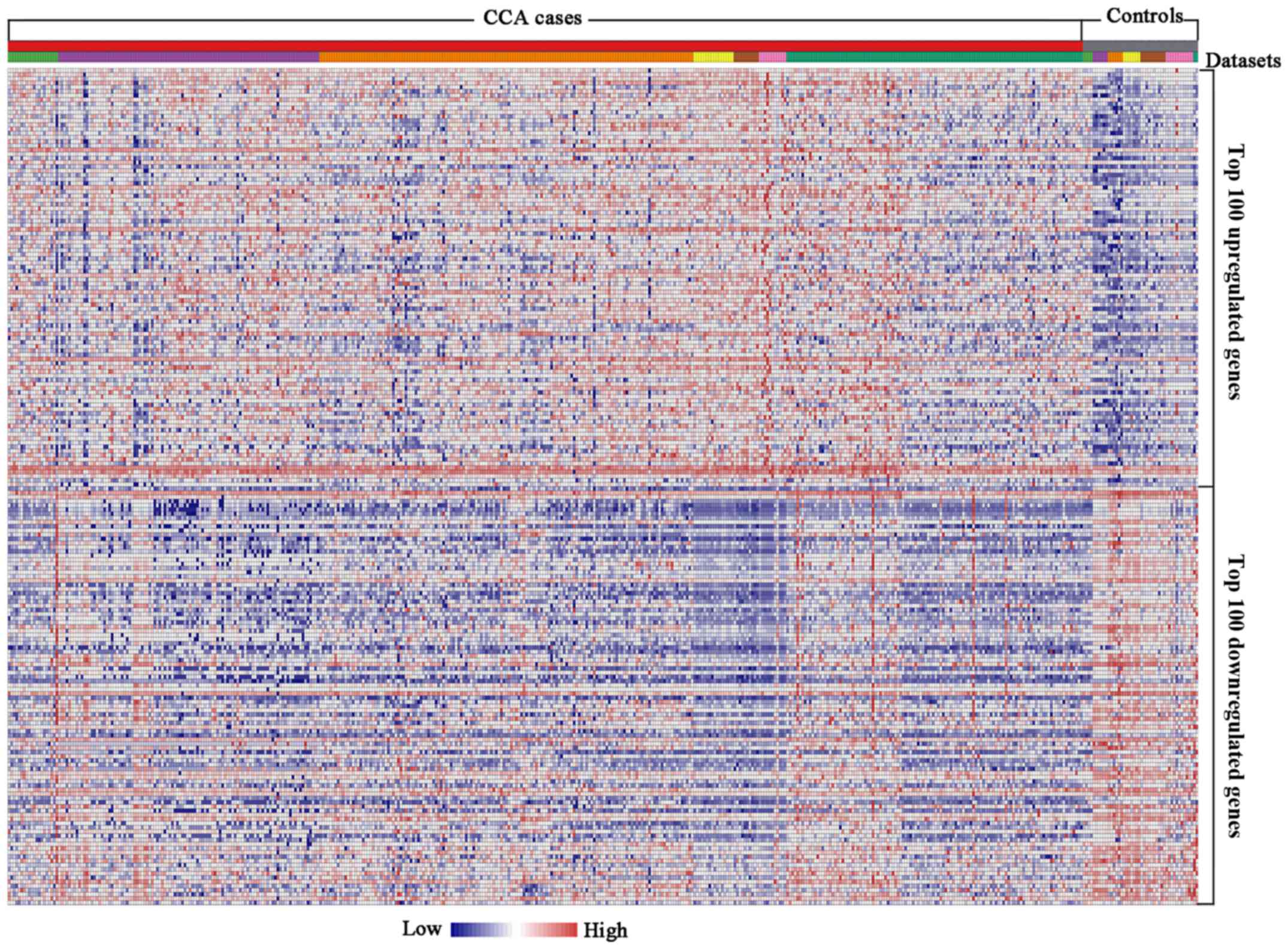
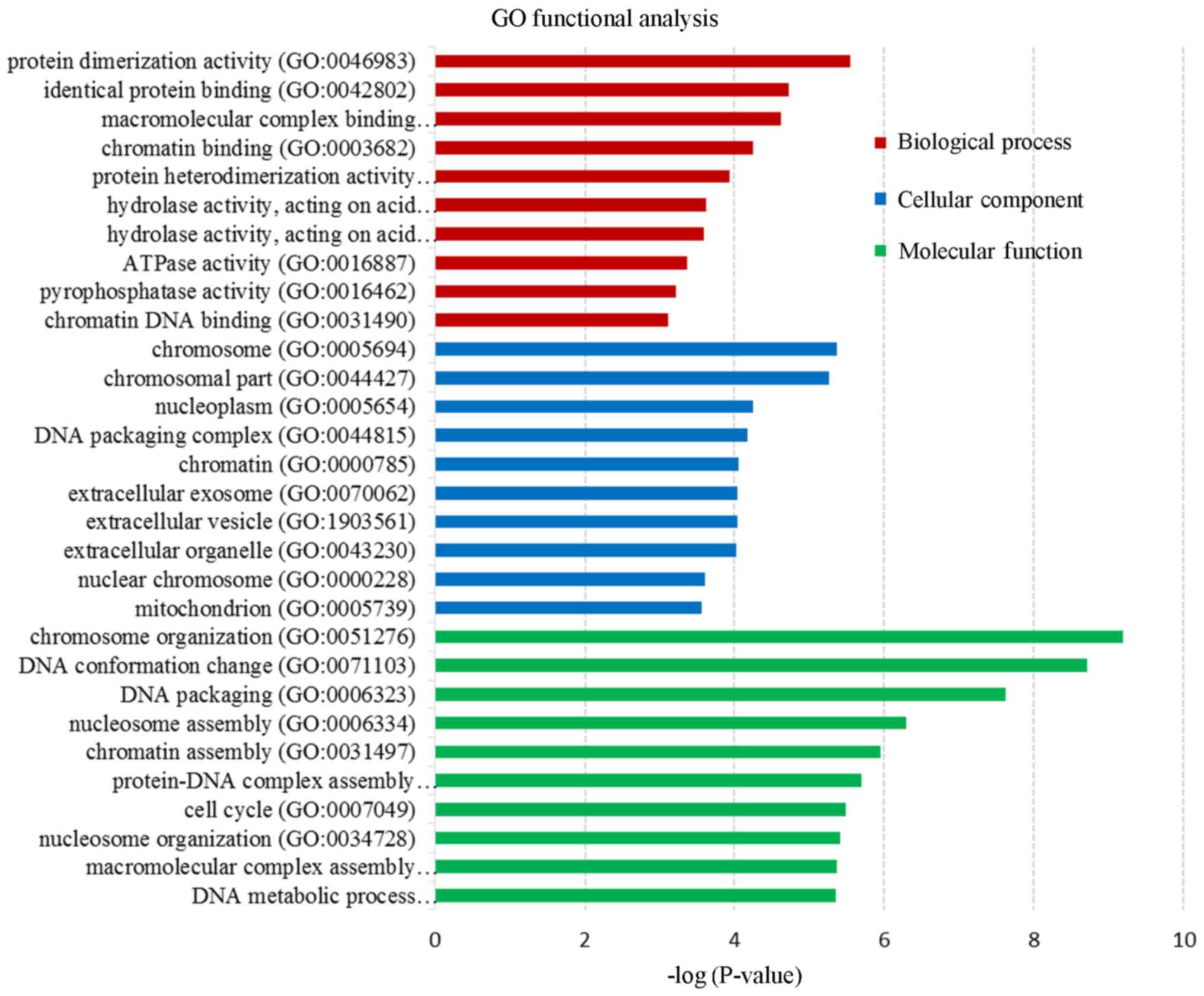
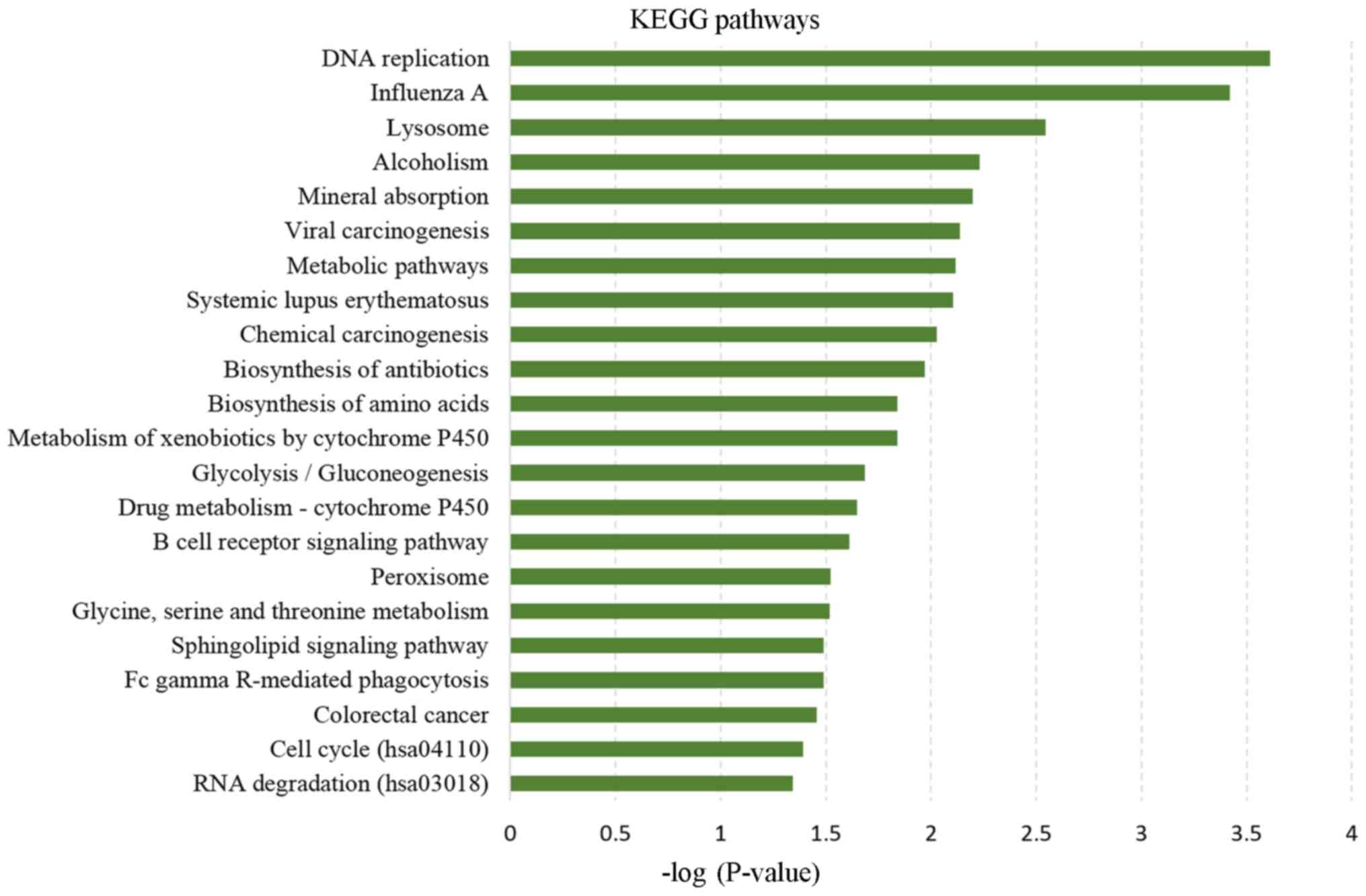
|
1
|
de Groen PC, Gores GJ, LaRusso NF,
Gunderson LL and Nagorney DM: Biliary tract cancers. N Engl J Med.
341:1368–1378. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tyson GL and El-Serag HB: Risk factors for
cholangiocarcinoma. Hepatology. 54:173–184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rizvi S and Gores GJ: Pathogenesis,
diagnosis, and management of cholangiocarcinoma. Gastroenterology.
145:1215–1229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saha SK, Zhu AX, Fuchs CS and Brooks GA:
Forty-year trends in cholangiocarcinoma incidence in the U.S.:
Intrahepatic disease on the rise. Oncologist. 21:594–599. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oliveira IS, Kilcoyne A, Everett JM,
Mino-Kenudson M, Harisinghani MG and Ganesan K: Cholangiocarcinoma:
Classification, diagnosis, staging, imaging features, and
management. Abdom Radiol (NY). 42:1637–1649. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Khuntikeo N, Chamadol N, Yongvanit P,
Loilome W, Namwat N, Sithithaworn P, Andrews RH, Petney TN,
Promthet S, Thinkhamrop K, et al: Cohort profile:
Cholangiocarcinoma screening and care program (CASCAP). BMC Cancer.
15:4592015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jusakul A, Cutcutache I, Yong CH, Lim JQ,
Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et
al: Whole-genome and epigenomic landscapes of etiologically
distinct subtypes of cholangiocarcinoma. Cancer Discov.
7:1116–1135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kayhanian H, Smyth EC and Braconi C:
Emerging molecular targets and therapy for cholangiocarcinoma.
World J Gastrointest Oncol. 9:268–280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sia D, Hoshida Y, Villanueva A, Roayaie S,
Ferrer J, Tabak B, Peix J, Sole M, Tovar V, Alsinet C, et al:
Integrative molecular analysis of intrahepatic cholangiocarcinoma
reveals 2 classes that have different outcomes. Gastroenterology.
144:829–840. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seok JY, Na DC, Woo HG, Roncalli M, Kwon
SM, Yoo JE, Ahn EY, Kim GI, Choi JS, Kim YB and Park YN: A fibrous
stromal component in hepatocellular carcinoma reveals a
cholangiocarcinoma-like gene expression trait and
epithelial-mesenchymal transition. Hepatology. 55:1776–1786. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murakami Y, Kubo S, Tamori A, Itami S,
Kawamura E, Iwaisako K, Ikeda K, Kawada N, Ochiya T and Taguchi YH:
Comprehensive analysis of transcriptome and metabolome analysis in
intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Sci
Rep. 5:162942015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Andersen JB, Spee B, Blechacz BR, Avital
I, Komuta M, Barbour A, Conner EA, Gillen MC, Roskams T, Roberts
LR, et al: Genomic and genetic characterization of
cholangiocarcinoma identifies therapeutic targets for tyrosine
kinase inhibitors. Gastroenterology. 142:1021–1031.e15. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oishi N, Kumar MR, Roessler S, Ji J,
Forgues M, Budhu A, Zhao X, Andersen JB, Ye QH, Jia HL, et al:
Transcriptomic profiling reveals hepatic stem-like gene signatures
and interplay of miR-200c and epithelial-mesenchymal transition in
intrahepatic cholangiocarcinoma. Hepatology. 56:1792–1803. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seol MA, Chu IS, Lee MJ, Yu GR, Cui XD,
Cho BH, Ahn EK, Leem SH, Kim IH and Kim DG: Genome-wide expression
patterns associated with oncogenesis and sarcomatous
transdifferentation of cholangiocarcinoma. BMC Cancer. 11:782011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sulpice L, Desille M, Turlin B, Fautrel A,
Boudjema K, Clément B and Coulouarn C: Gene expression profiling of
the tumor microenvironment in human intrahepatic
cholangiocarcinoma. Genom Data. 7:229–232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xia J, Gill EE and Hancock RE:
NetworkAnalyst for statistical, visual and network-based
meta-analysis of gene expression data. Nat Protoc. 10:823–844.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xia J, Benner MJ and Hancock RE:
NetworkAnalyst-integrative approaches for protein-protein
interaction network analysis and visual exploration. Nucleic Acids
Res. 42(Web Server Issue): W167–W174. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Breuer K, Foroushani AK, Laird MR, Chen C,
Sribnaia A, Lo R, Winsor GL, Hancock RE, Brinkman FS and Lynn DJ:
InnateDB: Systems biology of innate immunity and beyond-recent
updates and continuing curation. Nucleic Acids Res. 41(Database
Issue): D1228–D1233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Orchard S, Kerrien S, Abbani S, Aranda B,
Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G,
et al: Protein interaction data curation: The International
Molecular Exchange (IMEx) consortium. Nat Methods. 9:345–350. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hermjakob H, Montecchi-Palazzi L,
Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B,
Roepstorff P, Valencia A, et al: IntAct: An open source molecular
interaction database. Nucleic Acids Res. 32(Database Issue):
D452–D455. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Licata L, Briganti L, Peluso D, Perfetto
L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP,
Santonico E, et al: MINT, the molecular interaction database: 2012
update. Nucleic Acids Res. 40(Database Issue): D857–D861. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Salwinski L, Miller CS, Smith AJ, Pettit
FK, Bowie JU and Eisenberg D: The database of interacting proteins:
2004 update. Nucleic Acids Res. 32(Database Issue): D449–D451.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Isserlin R, El-Badrawi RA and Bader GD:
The biomolecular interaction network database in PSI-MI 2.5.
Database (Oxford). 2011:baq0372011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chatr-Aryamontri A, Breitkreutz BJ,
Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A,
Kolas N, O'Donnell L, et al: The BioGRID interaction database: 2015
update. Nucleic Acids Res. 43(Database Issue): D470–D478. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Blechacz B, Komuta M, Roskams T and Gores
GJ: Clinical diagnosis and staging of cholangiocarcinoma. Nat Rev
Gastroenterol Hepatol. 8:512–522. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hass HG, Nehls O, Jobst J, Frilling A,
Vogel U and Kaiser S: Identification of osteopontin as the most
consistently over-expressed gene in intrahepatic
cholangiocarcinoma: Detection by oligonucleotide microarray and
real-time PCR analysis. World J Gastroenterol. 14:2501–2510. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sulpice L, Rayar M, Desille M, Turlin B,
Fautrel A, Boucher E, Llamas-Gutierrez F, Meunier B, Boudjema K,
Clément B and Coulouarn C: Molecular profiling of stroma identifies
osteopontin as an independent predictor of poor prognosis in
intrahepatic cholangiocarcinoma. Hepatology. 58:1992–2000. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tongtawee T, Kaewpitoon SJ, Loyd R,
Chanvitan S, Leelawat K, Praditpol N, Jujinda S and Kaewpitoon N:
High expression of matrix metalloproteinase-11 indicates poor
prognosis in human cholangiocarcinoma. Asian Pac J Cancer Prev.
16:3697–3701. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang QX, Cui JY, Ma H, Jia XM, Huang FL
and Jiang LX: Screening of potential biomarkers for
cholangiocarcinoma by integrated analysis of microarray data sets.
Cancer Gene Ther. 23:48–53. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sawanyawisuth K, Wongkham C, Araki N, Zhao
Q, Riggins GJ and Wongkham S: Serial analysis of gene expression
reveals promising therapeutic targets for liver fluke-associated
cholangiocarcinoma. Asian Pac J Cancer Prev. 13 Suppl:S89–S93.
2012.
|
|
33
|
Batmunkh E, Tátrai P, Szabó E, Lódi C,
Holczbauer A, Páska C, Kupcsulik P, Kiss A, Schaff Z and Kovalszky
I: Comparison of the expression of agrin, a basement membrane
heparan sulfate proteoglycan, in cholangiocarcinoma and
hepatocellular carcinoma. Hum Pathol. 38:1508–1515. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang H, Jiang S, Zhang Y, Pan K, Xia J and
Chen M: High expression of thymosin beta 10 predicts poor prognosis
for hepatocellular carcinoma after hepatectomy. World J Surg Oncol.
12:2262014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Ren D, Guo L, Wang L, Wu S, Lin
C, Ye L, Zhu J, Li J, Song L, et al: Thymosin beta 10 is a key
regulator of tumorigenesis and metastasis and a novel serum marker
in breast cancer. Breast Cancer Res. 19:152017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mattu S, Fornari F, Quagliata L, Perra A,
Angioni MM, Petrelli A, Menegon S, Morandi A, Chiarugi P,
Ledda-Columbano GM, et al: The metabolic gene HAO2 is downregulated
in hepatocellular carcinoma and predicts metastasis and poor
survival. J Hepatol. 64:891–898. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fan K, Li N, Qi J, Yin P, Zhao C, Wang L,
Li Z and Zha X: Wnt/β-catenin signaling induces the transcription
of cystathionine-γ-lyase, a stimulator of tumor in colon cancer.
Cell Signal. 26:2801–2808. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
You J, Shi X, Liang H, Ye J, Wang L, Han
H, Fang H, Kang W and Wang T: Cystathionine-γ-lyase promotes
process of breast cancer in association with STAT3 signaling
pathway. Oncotarget. 8:65677–65686. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nakagawa S, Okabe H, Sakamoto Y, Hayashi
H, Hashimoto D, Yokoyama N, Sakamoto K, Kuroki H, Mima K, Nitta H,
et al: Enhancer of zeste homolog 2 (EZH2) promotes progression of
cholangiocarcinoma cells by regulating cell cycle and apoptosis.
Ann Surg Oncol. 20 Suppl 3:S667–S675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giussani M, Merlino G, Cappelletti V,
Tagliabue E and Daidone MG: Tumor-extracellular matrix
interactions: Identification of tools associated with breast cancer
progression. Semin Cancer Biol. 35:3–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kuo DS, Labelle-Dumais C and Gould DB:
COL4A1 and COL4A2 mutations and disease: Insights into pathogenic
mechanisms and potential therapeutic targets. Hum Mol Genet.
21:R97–R110. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Morine Y, Shimada M, Iwahashi S,
Utsunomiya T, Imura S, Ikemoto T, Mori H, Hanaoka J and Miyake H:
Role of histone deacetylase expression in intrahepatic
cholangiocarcinoma. Surgery. 151:412–419. 2012. View Article : Google Scholar : PubMed/NCBI
|