Introduction
Lung cancer is one of the most frequently diagnosed
cancers with high mortality worldwide, with >1,500,000 new cases
diagnosed annually (1,2). Based on the cancer statistics data
from 2015, lung and bronchial cancers are expected to have the
highest mortality among all cancers in the USA, with the estimated
mortality rate of 158,040 (3).
Lung squamous cell carcinoma (LSQCC) is the second most common type
of lung cancer, with an annual mortality rate of ~400,000 worldwide
(4). Chemotherapy and radiotherapy
are the most common treatments for LSQCC; however, patient
responses to these therapies are limited (5). Therefore, there is a requirement for
additional agents to be developed that may enhance the response to
these treatments.
Cyclooxygenase (COX)-2 serves an important role in
the tumorigenesis of various types of cancer, and COX-2 inhibitors
may effectively prevent tumor progression (6). Celecoxib is a selective COX-2
inhibitor; at the early stages of non-small-cell lung cancer
(NSCLC), celecoxib was reported to increase the anti-cancer
properties of preoperative chemotherapies, such as paclitaxel and
carboplatin (7). In addition,
celecoxib treatment upregulated the expression of death receptor 5
(DR5), decreased cell survival and induced apoptosis in NSCLCs
(8). Increased expression of COX-2
has been reported in LSQCC, and its inhibitor celecoxib is
predicted as the beneficial target for chemotherapy (9). Although these previous studies have
implied that celecoxib enhances the response sensitivity of
chemotherapy in LSQCC, the specific molecular mechanisms of this
inhibitor are still unknown. Several previous studies have
indicated that celecoxib treatment may result in cell cycle arrest
by downregulating the expression of p21 and p27, which may account
for the reduction of cyclin-dependent kinase activity (10,11).
In addition, celecoxib may contribute to the inhibition of
angiogenesis by suppressing the expression of angiogenic factors in
tumor cells (12). However,
additional studies are required to comprehensively determine the
mechanism of celecoxib on chemoprevention.
Long noncoding RNAs (lncRNAs) are a recently
described RNA transcript species that is different from mRNAs and
microRNAs. They are not transcriptional ‘noise’, but are important
genes that function in numerous biological processes (13,14).
Currently, several lncRNAs have been identified with significant
functions in lung cancer; for example, the upregulation of
PVT1 was reported to promote oncogenesis in NSCLCs (15), and prostate cancer-associated
transcript 6 was predicted as an oncogenic lncRNA in the growth and
invasion of lung cancer cells (16). In addition, a recent study
demonstrated that the novel lncRNA onco-lncRNA 230 was able
to induce invasion and apoptosis in LSQCC, and it was suggested as
a possible new diagnostic marker for the disease (17). These imply that regulation of
lncRNAs serves a key role in LSQCC. However, lncRNA expression
under celecoxib treatment has not been reported. A previous study
demonstrated that celecoxib treatment (50 µM) induced significant
overexpression of Bcl-2, Bcl-extra large and survivin
following 24 h treatment, whereas no significant alterations in
expression were identified in the activation of caspase-3,
caspase-8 or caspase-9 (18).
Another study revealed that the expression of multidrug
resistance-associated-4, a member of the ATP-binding cassette
transporters, was significantly upregulated in human LSQCC SK-MES-1
cells following treatment with 5 and 50 µmol/l of celecoxib for 24,
48 and 72 h (19). These data
suggested that celecoxib treatment may induce a series of
variations in the metabolism of SK-MES-1 cells; however, no lncRNAs
have been identified. Therefore, the present study used
RNA-sequencing (RNA-seq), which facilitates transcript analysis in
various cancers (20), to identify
differentially expressed genes (DEGs) and differentially expressed
lncRNAs (DE-LNRs) between SK-MES-1 cells cultured with or without
celecoxib treatment. In addition, potential correlations were
calculated, followed by pathway exploration of the lncRNAs. This
study aimed to provide novel insight into celecoxib chemoprevention
and to identify potential targeting markers for COX-2 induced
LSQCC.
Materials and methods
Cell culture and drug treatment
The human LSQCC cell line SK-MES-1 was purchased
from the Cell Bank of Type Culture Collection Chinese Academy of
Sciences (Shanghai, China). Cells were cultured in the RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (Gibco; Thermo Fisher Scientific,
Inc.), at 37°C and 5% CO2.
Cells at the logarithmic growth phase (at a
confluency of 70–75%) were divided into two groups: i) Two
identical SK-MES-1 cell samples were treated with 10 µM celecoxib
in 1% DMSO medium (celecoxib-treated group); and ii) two identical
SK-MES-1 cell samples were treated with equal amounts of DMSO
(Control group). Both groups were cultured for 48 h at 37°C.
RNA extraction and RNA-seq
A total of 5×107 cells were utilized to
isolate RNA using the RNeasy kit (catalog no. 74106; Qiagen
Sciences, Inc., Gaithersburg, MD, USA) according to the
manufacturer's protocol. RNA purity was analyzed with a NanoDrop
2000 spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.), and the RNA was reverse transcribed into cDNA
for library preparation using NEBNext Ultra RNA Library Prep kit
for Illumina (catalog no. E7530L; New England BioLabs, Inc.,
Ipswich, MA, USA), following the manufacturer's protocol. Briefly,
RNA (5 µg) from each sample was sheared into small fragments (200
nucleotides) prior to cDNA synthesis using fragment buffer.
Subsequently, the cDNA was blunt-ended and phosphorylated. A single
3′ adenosine moiety and Illumina adapters were added on the
repaired ends, followed by 15 cycles of polymerase chain reaction
(PCR) according to the protocol of the kit. preamplification were
performed using the NEB Phusion DNA polymerase (New England
BioLabs, Inc.). RNA-seq was performed on the Illumina Hiseq 2000
Sequencing System (Illumina, Inc., San Diego, CA, USA) using the
2×50 paired-end sequencing method.
Pretreatment of RNA-seq data
Quality control (QC) of raw sequencing reads was
performed using the next generation sequencing (NGS) QC Toolkit, as
previously described (21).
Briefly, the adaptor sequences in the reads were removed, and the
low-quality reads with the base quality score <20 were filtered
out. High quality sequences were defined having bases with a
quality score >20 that accounted for >90% of its length.
Subsequently, the clean reads were aligned against the University
of California Santa Cruz Homo sapiens reference genome (hg19
assembly, http://www.genome.ucsc.edu/index.html) using TopHat2
software (v2.0.9; http://ccb.jhu.edu/software/tophat) with the default
parameters (22).
Identification of DEGs
Cufflinks (v2.2.1; http://cufflinks.cbcb.umd.edu/index.html) software was
used to calculate the fragments per kilobase of exon per million
fragments mapped (FPKM), from which the gene expression values were
obtained (23). The linear models
for microarray analysis (limma; v3.10.3) package in R (http://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used to select DEGs between the two groups (24), with the thresholds of
q-value<0.05 and |log2(FC)|>0.58; where FC is fold
change.
Selection of DE-LNRs
LNCipedia 3.0 (http://www.lncipedia.org), an online storage of lncRNA
annotation (25), was used to
acquire information on lncRNAs. Subsequently, Cufflinks was used to
screen the DE-LNRs. Similar to the selection of DEGs, the cut-off
values were q<0.05 and |log2(FC)|> 0.58.
Enrichment analysis of the DEGs
To explore potential functions and pathways that the
DEGs may participate in, function enrichment and pathway enrichment
were implemented based on the Gene Ontology (GO; http://www.geneontology.org) (26) database and the Kyoto Encyclopedia
of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html) database,
respectively, and the Database for Annotation, Visualization and
Integration Discovery (DAVID; v6.8; http://david.abcc.Ncifcrf.gov) (27). Selection criteria for a significant
GO or KEGG pathway category were P<0.05 with ≥2 genes enriched
in a category.
Protein-protein interaction (PPI)
network construction
The Search Tool for the Retrieval of Interacting
Genes (STRING; v10.0, http://string-db.org) database was searched to
discover potential interactions of proteins encoded by the
identified DEGs (28). With the
selection criterion of a combined score >0.7, a PPI network was
established, which was drawn using Cytoscape (v3.2.0; http://cytoscape.org) software (29).
Co-expression analysis of lncRNAs and
mRNAs
For the identified DE-LNRs and DEGs, their
correlation coefficient (CC) was calculated by Pearson correlation.
The co-expressed DE-LNRs and DEGs were selected under the condition
of |CC|>0.98. Subsequently, enrichment analysis of the
co-expressed DEGs was performed to predict biological functions of
the DE-LNRs.
Validation of identified DEGs and
lncRNA
To further confirm the identification of DEGs and
DE-LNRs, expression levels of fibronectin 1 (FN1), vascular
endothelial growth factor A (VEGFA) and lncAP000769.1-2:10 were
determined using reverse transcription-quantitative PCR (RT-qPCR)
in SK-MES-1 cells treated with 10 µM celecoxib or DMSO for 48 h.
Total RNA was isolated from cells at a confluency of 70–75% using
TRIzol agent (Takara Biotechnology Co., Ltd., Dalian, China), and
reverse transcribed to cDNA using the PrimeScript RT Reagent kit
(Takara Biotechnology Co., Ltd.), according to the manufacturer's
protocol. qPCR was performed using a SYBR Green kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) on a ViiA7 PCR
instrument (Applied Biosystems; Thermo Fisher Scientific, Inc.)
with the following thermocycling conditions: 1 cycle of 50°C for 3
min and 95°C for 3 min; followed by 40 cycles of 95°C for 10 sec
and 60°C for 30 sec. GAPDH was used as the internal control during
expression analysis, and gene expression was calculated using the
2−∆∆Cq method (30).
Primer sequences are provided in Table
I.
 | Table I.Primer sequences of genes and lncRNA
determined using reverse transcription-quantitative polymerase
chain reaction. |
Table I.
Primer sequences of genes and lncRNA
determined using reverse transcription-quantitative polymerase
chain reaction.
| Gene | Primer sequence
(5′→3′) |
|---|
| VEGFA | F:
CTGTCTAATGCCCTGGAGCC |
|
| R:
ACGCGAGTCTGTGTTTTTGC |
| FN1 | F:
TTGCTCCTGCACATGCTTTG |
|
| R:
CATGAAGCACTCAATTGGGCA |
| Lnc-AP000769. | F:
GGGGAAGTAGTCTCGGGTAT |
| 1-2:10 | R:
GTCGTTATGAAGGCAATGTG |
| GAPDH | F:
TGACAACTTTGGTATCGTGGAAGG |
|
| R:
AGGCAGGGATGATGTTCTGGAGAG |
Statistical analysis
DEGs were screened using the limma package in R with
the thresholds of q<0.05 and |log2(FC)|>0.58;
DE-LNRs were screened using Cufflinks software with the same
thresholds. Continuous variables are presented as the mean ±
standard deviation, and difference between groups was calculated
using Student's t-test. P<0.05 was considered to indicate a
statistically significant difference.
Results
DEGs and DE-LNRs identification
According to the aforementioned criteria, a set of
261 upregulated and 56 downregulated DEGs were identified in
celecoxib-treated group, compared with the untreated Control cells.
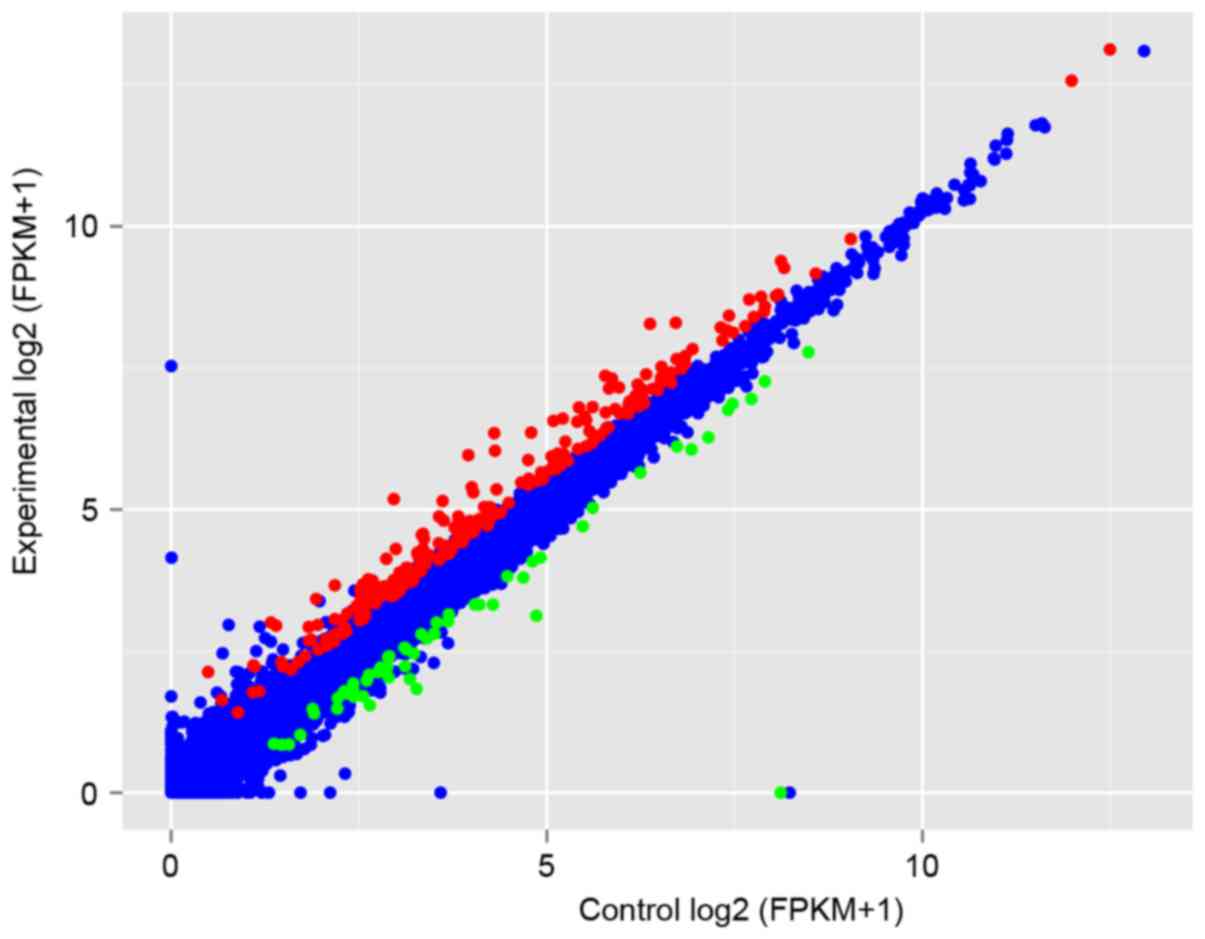
Gene expressions in each group are presented in a scatter plot of
FPKM (Fig. 1). Based on the
predefined selection criterion, 17 lncRNAs were upregulated and 8
lncRNAs were downregulated in celecoxib-treated group compared with
the untreated Control cells (Table
II).
 | Table II.Differentially expressed lncRNAs in
the celecoxib-treated group and Control group. |
Table II.
Differentially expressed lncRNAs in
the celecoxib-treated group and Control group.
| A, Upregulated |
|---|
|
|---|
| lncRNA | Value_1 | Value_2 | Log2 (FC) | q-value |
|---|
|
lnc-C14orf166B-3:4 | 183.118 | 400.277 | 112.823 |
5.35×10−4 |
| lnc-CNN3-3:1 | 112.248 | 179.237 | 0.675 |
5.06×10−8 |
| lnc-CTSL1-2:2 | 327.718 | 549.936 | 0.747 |
2.63×10−2 |
| lnc-CXCL3-1:1 | 128.057 | 34.035 | 141.023 |
8.89×10−13 |
| lnc-ENTPD6-2:1 | 101.243 | 188.522 | 0.897 |
1.7×10−2 |
| lnc-ERN1-1:1 | 750.668 | 164.504 | 113.187 |
8.84×10−8 |
| lnc-HES1-10:1 | 258.042 | 694.213 | 142.777 |
5.96×10−4 |
| lnc-HFE2-2:1 | 343.839 | 851.858 | 130.888 | 0 |
|
lnc-KIAA1257-3:1 | 132.568 | 21.211 | 0.678 |
6.09×10−4 |
| lnc-KSR1-1:1 | 108.716 | 165.887 | 0.610 |
1.57×10−2 |
| lnc-MOGAT2-5:1 | 930.449 | 140.722 | 0.597 |
2.81×10−2 |
| lnc-MT2A-1:2 | 1990.13 | 3034.1 | 0.608 |
2.03×10−8 |
| lnc-RAB44-3:1 | 517.827 | 879.917 | 0.765 |
1.22×10−8 |
| lnc-RBM3-1:1 | 726.858 | 139.598 | 0.942 |
1.57×10−2 |
|
lnc-RP11-231C14.2.1–3:1 | 164.477 | 551.775 | 174.619 |
1.89×10−2 |
| lnc-RSPH9-4:1 | 658.082 | 105.15 | 0.676 |
1.33×10−8 |
| lnc-TRIB3-1:2 | 366.972 | 569.299 | 0.634 |
7.30×10−3 |
|
| B,
Downregulated |
|
|
lnc-AP000769.1-2:10 | 895.237 | 589.174 | −0.604 |
6.98×10−5 |
| lnc-BOLA3-2:2 | 136.839 | 0 |
−1.80×10−8 |
4.31×10−2 |
| lnc-E2F2-1:1 | 200.807 | 118.647 | −0.759 |
1.50×10−6 |
| lnc-FOXG1-7:1 | 39.749 | 0 |
−1.80×10−8 |
4.16×10−13 |
| lnc-GNLY-4:2 | 90.639 | 306.009 | −156.656 |
3.67×10−3 |
|
lnc-KIAA0226-2:1 | 219.713 | 0.367 | −258.257 |
1.62×10−4 |
| lnc-LTBP3-2:5 | 11.504 | 730.114 | −0.656 |
5.95×10−3 |
| lnc-TOR1A-2:1 | 23.017 | 131.966 | −0.803 |
1.40×10−2 |
Pathway enrichment analysis of
DEGs
KEGG pathway analysis indicated that the upregulated
DEGs were significantly enriched in 12 pathway categories,
including Aminoacyl-tRNA biosynthesis, protein processing in
endoplasmic reticulum (ER), protein export, amino sugar and
nucleotide sugar metabolism and mammalian target of rapamycin
(mTOR) signaling pathway. The downregulated DEGs were enriched in
17 pathways, such as ‘transforming growth factor (TGF)-β signaling
pathway’, ‘extracellular matrix (ECM)-receptor interaction’,
‘cytokine-cytokine receptor interaction’ and ‘p53 signaling
pathway’ (Table III).
 | Table III.Top 10 identified KEGG enrichment
pathways of identified DEGs. |
Table III.
Top 10 identified KEGG enrichment
pathways of identified DEGs.
| A, Upregulated
DEGs |
|---|
| KEGG ID | Pathway name | Counta | Genes | P-value |
|---|
| 970 | Aminoacyl-tRNA
biosynthesis | 10 | AARS, CARS,
EPRS, GARS, IARS |
4.49×10−7 |
| 4141 | Protein processing
in endoplasmic reticulum | 15 | ATF4, DDIT3,
DNAJB2, ERN1, HERPUD1 |
1.05×10−6 |
| 260 | Glycine, serine and
threonine metabolism | 6 | CBS, CTH, PHGDH,
PSAT1, PSPH, SHMT2 |
3.78×10−5 |
| 3060 | Protein export | 4 | HSPA5, SEC11C,
SEC63, SRPRB |
1.09×10−3 |
| 520 | Amino sugar and
nucleotide sugar metabolism | 5 | GFPT1, GFPT2,
GMPPB, HKDC1, NAGK |
2.79×10−3 |
| 250 | Alanine, aspartate
and glutamate metabolism | 4 | ASNS, GFPT1,
GFPT2, GPT2 |
3.84×10−3 |
| 4150 | mTOR signaling
pathway | 5 | DDIT4, EIF4EBP1,
RPS6KA2, ULK1, VEGFA |
3.97×10−3 |
| 860 | Porphyrin and
chlorophyll metabolism | 4 | EPRS, FTH1,
HMOX1, UROS |
1.11×10−2 |
| 5020 | Prion diseases | 3 | EGR1, HSPA5,
IL1A |
3.39×10−2 |
| 450 | Selenocompound
metabolism | 2 | CTH,
MARS |
4.62×10−2 |
|
| B, Downregulated
DEGs |
|
| KEGG ID | Pathway name | Counta | Genes | P-value |
|
| 4350 | TGF-β signaling
pathway | 4 | ID1, ID3, TGFβ2,
THBS1 |
1.94×10−4 |
| 5323 | Rheumatoid
arthritis | 4 | CCL2, CSF1,
CXCL6, TGFβ2 |
2.65×10−4 |
| 5144 | Malaria | 3 | CCL2, TGFβ2,
THBS1 |
7.36×10−4 |
| 5200 | Pathways in
cancer | 6 | AXIN2, E2F2,
EGLN3, FN1, MITF, TGFβ2 |
7.47×10−4 |
| 4512 | ECM-receptor
interaction | 3 | COL1A1, FN1,
THBS1 |
3.23×10−3 |
| 5146 | Amoebiasis | 3 | COL1A1, FN1,
TGFβ2 |
6.01×10−3 |
| 5219 | Bladder cancer | 2 | E2F2,
THBS1 |
9.63×10−3 |
| 4380 | Osteoclast
differentiation | 3 | CSF1, MITF,
TGFβ2 |
1.01×10−2 |
| 4060 | Cytokine-cytokine
receptor interaction | 4 | CCL2, CSF1,
CXCL6, TGFβ2 |
1.32×10−2 |
| 4115 | p53 signaling
pathway | 2 | RRM2,
THBS1 |
2.41×10−2 |
PPI network of DEGs
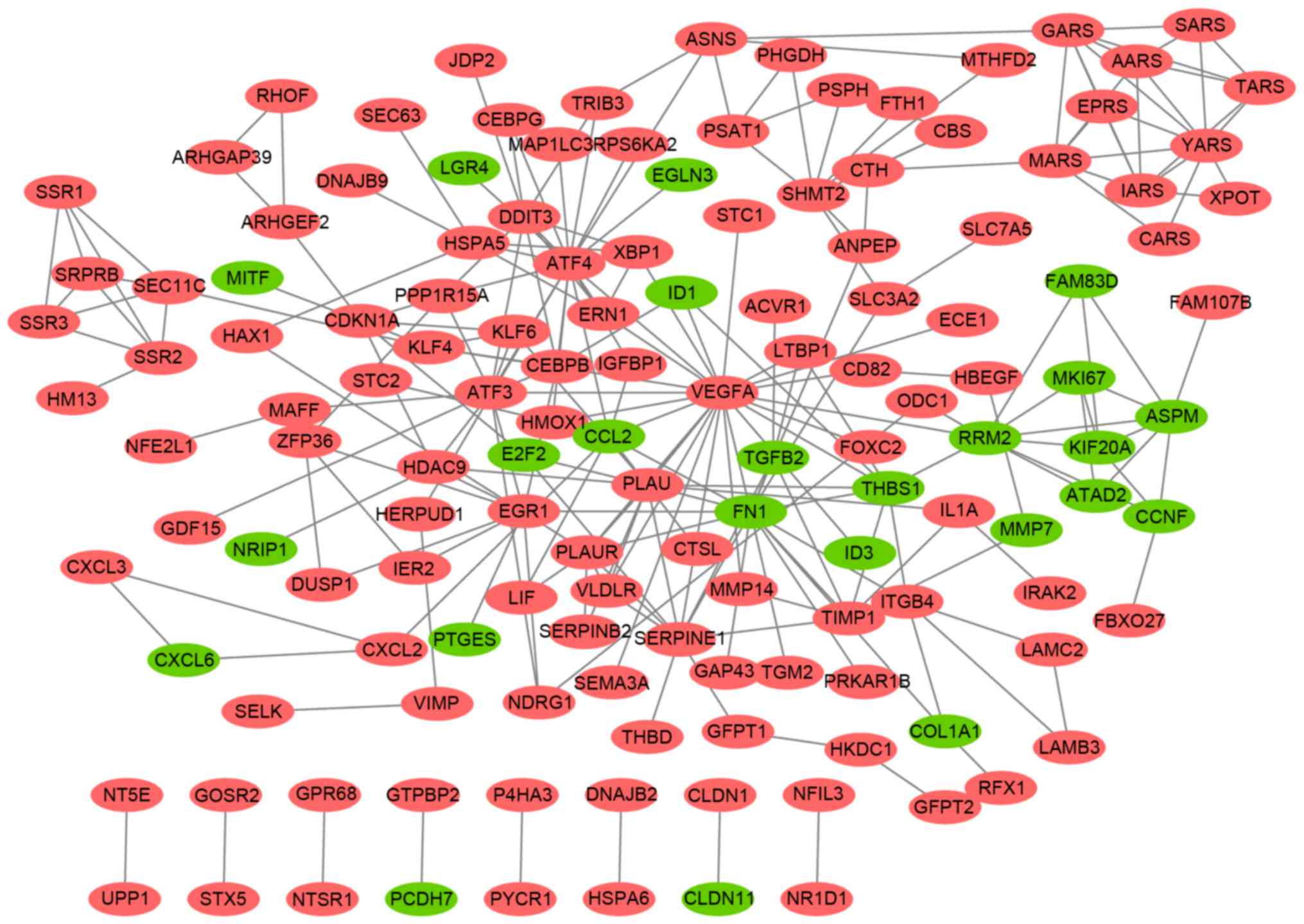
In the established PPI network (Fig. 2), several genes were highlighted
that had high degrees; that is, a high number of connections
between one gene and the others, such as VEGFA (degree=23),
activating transcription factor (ATF)-4 (degree=19), FN1
(degree=16), urokinase-type plasminogen activator PLAU (degree=13),
ATF3 (degree=11) and serpin E1 (SERPINE1; degree=10).
Co-expressed DE-LNRs and DEGs and
their enriched functions
Using the criterion of |CC|>0.98, the
co-expressed DE-LNRs and DEGs were screened out. As presented in
Table IV, SERPINE1 was
co-expressed with lnc-CTSL1-2:2 (CC=0.998577) and lnc-CXCL3-1:1
(CC=0.986928); VEGFA was linked with lnc-HES1-10:1 (CC=0.98906) and
lnc-AP000769.1-2:10 (CC=−0.99227); lnc-HFE2-2:1 was co-expressed
with ATF4 (CC=0.996159) and FN1 (CC= −0.98714); ATF3 was
co-expressed with lnc-KIAA1257-3:1 (CC=0.990212), lnc-KSR1-1:1
(CC=0.99655) and lnc-RP11-231C14.2.1-3:1 (CC= 0.993415);
lnc-BOLA3-2:2 was linked with PLAU (CC= −0.99288); and
lnc-KIAA0226-2:1 was co-expressed with FN1 (CC=0.981655).
 | Table IV.The most highly correlated
co-expressed DE-LNRs and DEGs. |
Table IV.
The most highly correlated
co-expressed DE-LNRs and DEGs.
| DE-LNR | DEG | CC |
|---|
|
lnc-C14orf166B-3:4 | VEGFA |
0.999351 |
|
| CLDN11 | −0.98538 |
| lnc-CNN3-3:1 | ACOT8 |
0.994477 |
|
| ATAD2 | −0.99951 |
| lnc-CTSL1-2:2 | SERPINE1 |
0.998577 |
|
| AMER1 | −0.99246 |
| lnc-CXCL3-1:1 | SERPINE1 |
0.986928 |
|
| AXIN2 | −0.9893 |
| lnc-ENTPD6-2:1 | ACVR1 |
0.995922 |
|
| ASPN | −0.98602 |
| lnc-ERN1-1:1 | ABTB2 |
0.996183 |
|
| AXIN2 | −0.98832 |
| lnc-HES1-10:1 | VEGFA |
0.98906 |
|
| AMER1 | −0.99383 |
| lnc-HFE2-2:1 | ATF4 |
0.996159 |
|
| FN1 | −0.98714 |
|
lnc-KIAA1257-3:1 | ATF3 |
0.990212 |
|
| LRFN1 | −0.98734 |
| lnc-KSR1-1:1 | ATF3 |
0.99655 |
|
| CCDC80 | −0.9997 |
| lnc-MOGAT2-5:1 | ACVR1 |
0.986953 |
|
| ASPN | −0.9935 |
| lnc-MT2A-1:2 | AASR |
0.991019 |
|
| ATAD2 | −0.99032 |
| lnc-RAB44-3:1 | ACOT8 |
0.997679 |
|
| ATAD2 | −0.99846 |
| lnc-RBM3-1:1 | ACOT8 |
0.990015 |
|
| ATAD2 | −0.99475 |
|
lnc-RP11-231C14.2.1–3:1 | ATF3 |
0.993415 |
|
| CCDC80 | −0.98512 |
| lnc-RSPH9-4:1 | ACOT8 |
0.990293 |
|
| CCNF | −0.988 |
| lnc-TRIB3-1:2 | ACOT8 |
|
|
| ATAD2 | −0.98366 |
|
lnc-AP000769.1-2:10 | ASPN |
0.987202 |
|
| VEGFA | −0.99227 |
| lnc-BOLA3-2:2 | CCNF |
0.985247 |
|
| PLAU | −0.99288 |
| lnc-E2F2-1:1 | AMER1 |
0.987329 |
|
| AARS | −0.98601 |
| lnc-FOXG1-7:1 | AMER1 |
0.988729 |
|
| ANTXR2 | −0.9897 |
| lnc-GNLY-4:2 | COL1A1 |
0.989359 |
|
| CDH13 | −0.99856 |
|
lnc-KIAA0226-2:1 | FN1 |
0.981655 |
|
| AARS | −0.99647 |
| lnc-LTBP3-2:5 | ASPN |
0.993248 |
|
| BTG1 | −0.993 |
| lnc-TOR1A-2:1 | ATOH8 |
0.98542 |
|
| CDH13 | −0.99533 |
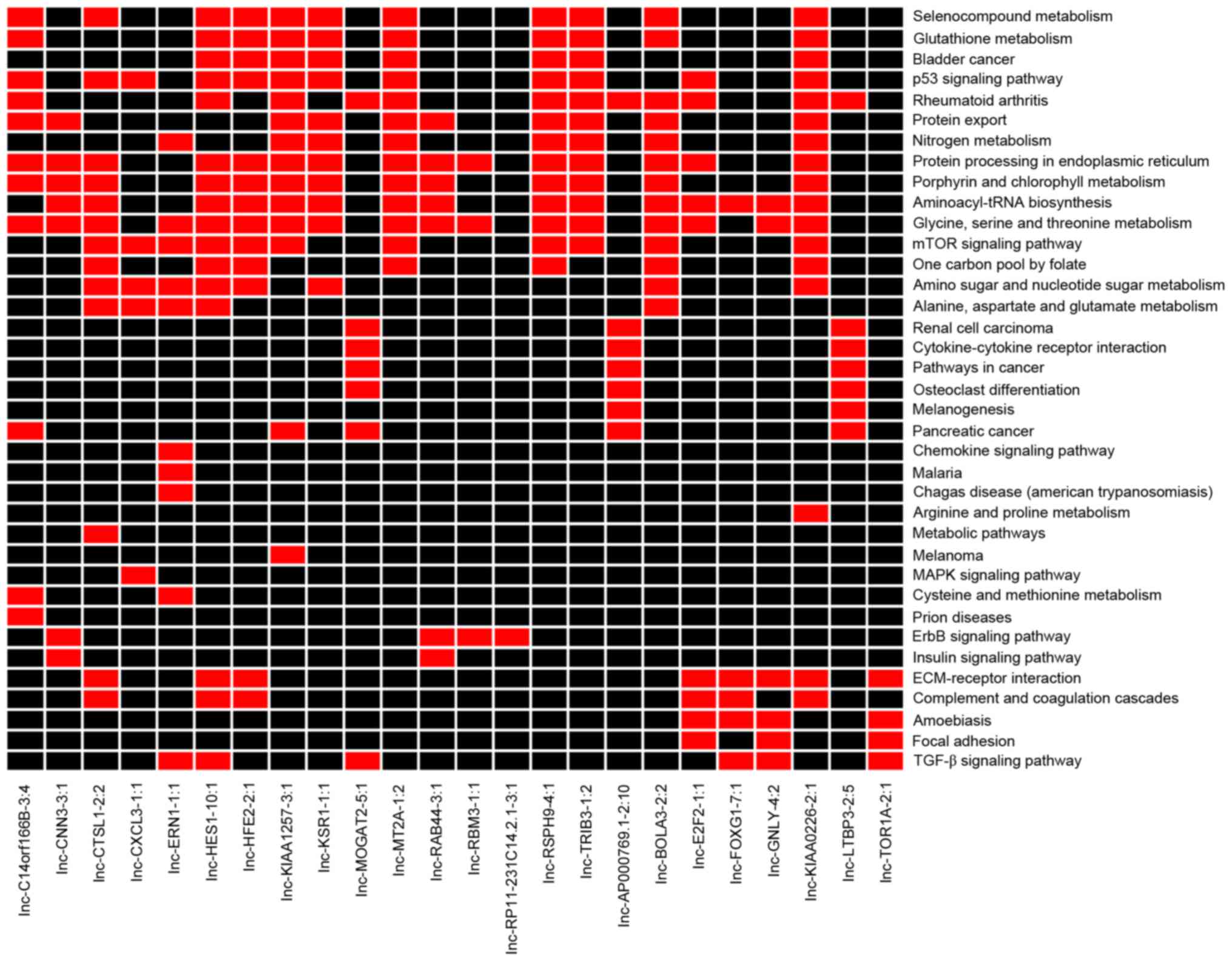
According to the enrichment analysis results of the
co-expressed DEGs, the co-expressed DE-LNRs were mainly enriched in
the ‘p53 signaling pathway’ (for example, lnc-HES1-10:1,
lnc-HFE2-2:1, lnc-KIAA1257-3:1 and lnc-KSR1-1:1; Fig. 3) and the ‘mTOR signaling pathway’
(for example, lnc-CTSL1-2:2, lnc-CXCL3-1:1, lnc-ERN1-1:1,
lnc-HES1-10:1 and lnc-HFE2-2:1; Fig.
3).
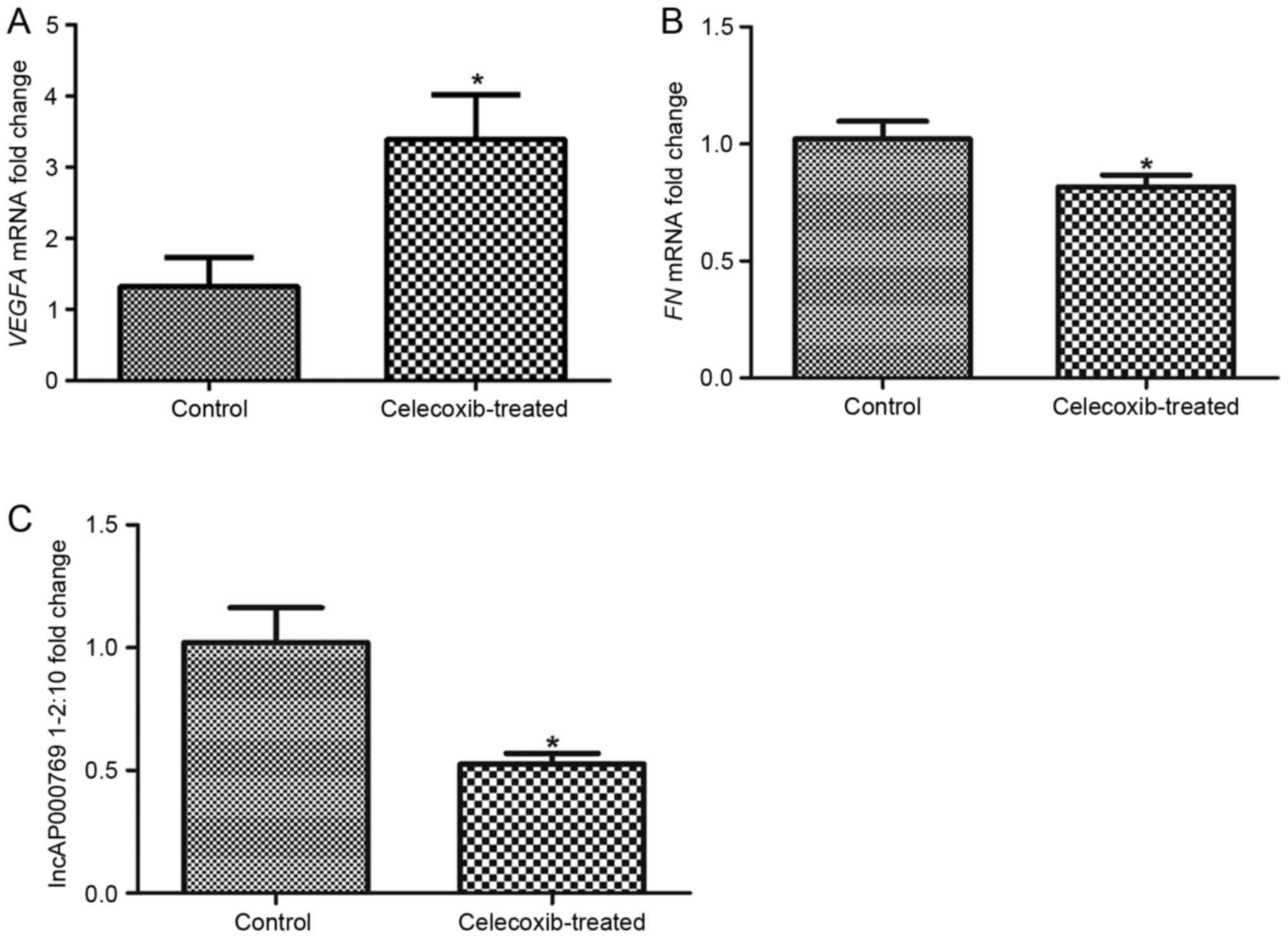
Validations of DEGs and lncRNA
To verify the identification of possible DEGs and
DE-LNRs, the expression levels of VEGFA, FN1 and
lnc-AP000769.1-2:10 were analyzed by RT-qPCR. The results revealed
that the expression level of VEGFA was significantly increased in
celecoxib-treated SK-MES-1 cells compared with untreated Control
cells (Fig. 4A). However, the
expression levels of FN1 and lnc-AP000769.1-2:10 were significantly
decreased in the celecoxib-treated group compared with the Control
group (Fig. 4B and C,
respectively). These results were consistent with the
aforementioned bioinformatics analytical results.
Discussion
The present study identified a number of genes and
lncRNAs that exhibited differential expression levels in
celecoxib-treated human LSQCC SK-MES-1 cells, compared with
untreated cells, including the genes VEGFA, ATF4 and
FN1, and the lncRNAs lnc-AP000769.1-2:10 and lnc-HFE2-2:1.
Notably, many of the identified DEGs and DE-LNRs were significantly
enriched in pathways like ‘protein processing in endoplasmic
reticulum’, ‘mTOR signaling pathway’ and ‘ECM-receptor
interaction’.
VEGFA is an essential growth factor for
stimulating angiogenesis, which often accompanies tumoral growth
(31). A previous study reported
that by upregulating the expression of FLJ10540, VEGFA activates
the phosphatidylinositol 3-kinase/AKT signaling pathway,
subsequently promoting cell invasion and migration in lung cancer
(32). mTOR is located downstream
of AKT signaling and functions in the control of angiogenesis and
cell proliferation during tumor progression (33). VEGFA was also reported to
activate the downstream mTOR signaling pathway to promote cancer
growth (34). Notably, several
therapeutic drugs for lung cancer have been reported to function
through this pathway. 2-(18F)-fluoro-2-deoxy-d-glucose
(18F-FDG) is a radiolabelled sugar molecule that is
commonly used to monitor the therapeutic effects of chemotherapy
for many malignant tumors, and the accumulation of
18F-FDG is regulated by the activation of mTOR signaling
in NSCLC (35).
Cis-diamminedichloroplatinum (II) (CDDP; also known as
cisplatin) is an effective drug for the treatment of many cancers
(36). However, resistance to this
drug limited its potential use in lung cancer treatment. It was
previously reported that overexpression of AKT activates the mTOR
signaling pathway, which induces CDDP resistance in lung cancer
cells (37). Therefore, inhibitors
of the AKT/mTOR signaling pathway may provide a promising
therapeutic target. In lung cancer, targeting of mTOR signaling was
suggested as an effective method in developing therapeutic drugs
(38). Curcumin, a natural extract
of turmeric, is considered as an antitumoral agent, which was
reported to enhance the anti-cancer ability of chemotherapy in
LSQCC by regulating multiple pathways such as VEGF signaling
(39). Notably, VEGFA was
also indicated to be enriched in the mTOR signaling pathway
(36). In the present study,
VEGFA was identified as an upregulated DEG in
celecoxib-treated LSQCC cells, and was demonstrated to be
significantly enriched in the mTOR signaling pathway. These data
suggested that VEGFA may be a sensitive gene in response to
celecoxib, and the increased expression may inhibit the activation
of mTOR signaling, which may improve the anti-tumor effects of
celecoxib on LSQCC cells.
In addition, lncRNAs may also serve crucial roles in
the amplification of anti-tumor effects. Nuclear paraspeckle
assembly transcript 1 (Neat1; ENST00000501122.2) is a factor
required for the assembly of paraspeckle compartments in the cell
(40). Neat1-containing
paraspeckles were reported to be responsible for the regulation of
chemosensitivity and may be induced by p53 (41). The biofunction of Neat1 is similar
to lnc-AP000769.1-2:3. However, no information about
lnc-AP000769.1-2:10 has yet been reported. In the present study,
lnc-AP000769.1-2:10 was closely correlated with VEGFA, which
was enriched in the mTOR signaling pathway following celecoxib
treatment. In addition, the expression of lnc-AP000769.1-2:10 was
significantly decreased in celecoxib treated SK-MES-1 cells.
Therefore, the present study hypothesized that this lncRNA may
regulate VEGFA gene expression in the mTOR signaling
pathway, which may facilitate to the enhancement of anti-tumor
effect of celecoxib for LSQCC treatment.
ATF4 was reported to be associated with
cisplatin sensitivity in lung cancer cell lines (42). Celecoxib has been demonstrated to
induce the expression of DR5 (43). In addition, C/EBP homologous
protein (CHOP) was revealed to serve a crucial role in
celecoxib-induced DR5 expression and may also be upregulated by
celecoxib (44). Inhibition of
ATF4 expression by small interfering RNAs was able to
abolish CHOP induction, which indicated the involvement of ATF4 in
celecoxib-induced apoptosis (45).
The ER is an essential site for protein processing. In many types
cancer, the ER serves an important role in the structural
maintenance of proteins in pivotal signaling pathways (46). Control of these proteins may offer
promising target therapies. In the present study, ATF4 was
indicated as enriched in the protein processing in ER pathway,
which suggested that it may influence the sensitivity of celecoxib
in LSQCC through the regulation of protein processing.
The FN1 protein may be involved in several cellular
activities, such as cell adhesion and migration (47,48).
In lung cancer, knockdown of FN1 was previously reported to
increase the chemosensitivity of cisplatin and promote apoptosis in
tumor cells (49). Notably, in the
present study, FN1 expression was downregulated in the
celecoxib-treated LSQCC cells, which suggested that the reduced
expression of this gene may also enhance the sensitivity of tumor
cells in response to celecoxib. In addition, FN1 has been
implicated in pathways such as the ECM-receptor interaction pathway
in many types cancer (50,51). Consistent with these results,
FN1 was significantly enriched in the ECM-receptor
interaction pathway in the present study, which indicated that
FN1 may exert its function through its involvement in this
pathway. Results from the present study also predicted that both
FN1 and ATF4 were targets of lnc-HFE2-2:1, which
suggested that the two genes may be regulated by this lncRNA in
LSQCC cells following celecoxib treatment. However, there is still
little information available about this lncRNA. Based on the
present results, lnc-HFE2-2:1 may be a novel target to predict
sensitivity of celecoxib for the treatment of LSQCC, through the
regulation of FN1 and ATF4 expressions.
There were some limitations to the present study.
First, although some genes and lncRNA had been validated in this
study, the regulatory relationships between lncRNAs and DEGs have
yet to be confirmed with in vitro experiments. Second, as
LSQCC may develop from a number of lung cell dysfunctions, SK-MES-1
cells may not reflect the wider results of celecoxib. Therefore,
different types of LSQCC cell lines should be used in future
studies, and the intersection of DEGs and lncRNAs may be focused.
Finally, an appropriate animal model should be used to confirm the
identified DEGs and DE-LNRs, including investigations on the
predicted signaling pathways.
In conclusion, genes (such as VEGFA, ATF4 and
FN1), and lncRNAs (such as lnc-AP000769.1-2:10 and
lnc-HFE2-2:1) may be crucial molecules to enhance the anti-cancer
effects of celecoxib treatment on LSQCC, and may be used as
predictors for chemosensitivity of celecoxib. However, additional
validation experiments are required in further studies.
Glossary
Abbreviations
Abbreviations:
|
18F-FDG
|
2-(18F)-fluoro-2-deoxy-d-glucose
|
|
ATF4
|
activating transcription factor 4
|
|
CC
|
correlation coefficient
|
|
CDDP
|
cis-diamminedichloroplatinum (II)
|
|
CHOP
|
C/EBP homologous protein
|
|
COX
|
cyclooxygenase
|
|
DEG
|
differentially expressed gene
|
|
DR5
|
death receptor 5
|
|
ER
|
endoplasmic reticulum
|
|
FC
|
fold change
|
|
FN1
|
fibronectin 1
|
|
GO
|
gene ontology
|
|
lncRNA
|
long noncoding RNA
|
|
LSQCC
|
lung squamous cell carcinoma
|
|
mTOR
|
mammalian target of rapamycin
|
|
NSCLC
|
non-small-cell lung cancer
|
|
PCR
|
polymerase chain reaction
|
|
PPI
|
protein-protein interaction
|
|
RNA-seq
|
RNA-sequencing
|
|
VEGFA
|
vascular endothelial growth factor
A
|
References
|
1
|
Henley SJ, Richards TB, Underwood JM,
Eheman CR, Plescia M and McAfee TA: Centers for Disease Control and
Prevention (CDC): Lung cancer incidence trends among men and
women-United States, 2005–2009. MMWR Morb Mortal Wkly Rep. 63:1–5.
2014.PubMed/NCBI
|
|
2
|
de Groot P and Munden RF: Lung cancer
epidemiology, risk factors, and prevention. Radiol Clin North Am.
50:863–876. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–298. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cancer Genome Atlas Research Network:
Comprehensive genomic characterization of squamous cell lung
cancers. Nature. 489:519–525. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bendale Y, Bendale V, Birari-Gawande P,
Kadam A and Gund P: Tumor regression with ayurvedic rasayana
therapy in squamous cell carcinoma of lungs. Rasamruta. 7:1–5.
2015.
|
|
6
|
Mascaux C, Martin B, Verdebout JM, Ninane
V and Sculier JP: COX-2 expression during early lung squamous cell
carcinoma oncogenesis. Eur Respir J. 26:198–203. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Altorki NK, Keresztes RS, Port JL, Libby
DM, Korst RJ, Flieder DB, Ferrara CA, Yankelevitz DF, Subbaramaiah
K, Pasmantier MW and Dannenberg AJ: Celecoxib, a selective
cyclo-oxygenase-2 inhibitor, enhances the response to preoperative
paclitaxel and carboplatin in early-stage non-small-cell lung
cancer. J Clin Oncol. 21:2645–2650. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu X, Yue P, Zhou Z, Khuri FR and Sun SY:
Death receptor regulation and celecoxib-induced apoptosis in human
lung cancer cells. J Natl Cancer Inst. 96:1769–1780. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gold KA, Kim ES, Lee JJ, Wistuba II,
Farhangfar CJ and Hong WK: The BATTLE to personalize lung cancer
prevention through reverse migration. Cancer Prev Res (Phila).
4:962–972. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fu SL, Wu YL, Zhang YP, Qiao MM and Chen
Y: Anti-cancer effects of COX-2 inhibitors and their correlation
with angiogenesis and invasion in gastric cancer. World J
Gastroenterol. 10:1971–1974. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mineo TC, Ambrogi V, Cufari ME and Pompeo
E: May cyclooxygenase-2 (COX-2), p21 and p27 expression affect
prognosis and therapeutic strategy of patients with malignant
pleural mesothelioma? Eur J Cardiothorac Surg. 38:245–252. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Khan Z, Khan N, Tiwari RP, Sah NK, Prasad
GB and Bisen PS: Biology of Cox-2: An application in cancer
therapeutics. Curr Drug Targets. 12:1082–1093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumorigenesis in non-small cell lung cancer. Int J Clin Exp Pathol.
7:6929–6935. 2014.PubMed/NCBI
|
|
16
|
Wan L, Zhang L, Fan K, Cheng ZX, Sun QC
and Wang JJ: Knockdown of long noncoding RNA PCAT6 inhibits
proliferation and invasion in lung cancer cells. Oncol Res.
24:161–170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang CY, Silva-Fisher JM, Dang HX, White
NM and Maher CA: Abstract 971: A novel long noncoding RNA,
onco-lncRNA 230, induces apoptosis and invasion in lung squamous
cell carcinoma. Cancer Res. 76 14 Suppl:S9712016. View Article : Google Scholar
|
|
18
|
Gradilone A, Silvestri I, Scarpa S,
Morrone S, Gandini O, Pulcinelli FM, Gianni W, Frati L, Aglianò AM
and Gazzaniga P: Failure of apoptosis and activation on NFkappaB by
celecoxib and aspirin in lung cancer cell lines. Oncol Rep.
17:823–828. 2007.PubMed/NCBI
|
|
19
|
Gradilone A, Pulcinelli FM, Lotti LV,
Martino S, Mattiello T, Frati L, Aglianò AM and Gazzaniga P:
Celecoxib induces MRP-4 in lung cancer cells: Therapeutic
implications. J Clin Oncol. 25:4318–4320. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin L, Abo R, Dolcen D, Paquette R, Laing
A, de Waal L, Thorner A, Ducar M, Ziaugra L, Wollison B, et al:
Abstract 1115: Targeted RNA sequencing improves transcript analysis
in cancer samples. Cancer Res. 75 15 Suppl:S11152015. View Article : Google Scholar
|
|
21
|
Patel RK and Jain M: NGS QC Toolkit: A
toolkit for quality control of next generation sequencing data.
PLoS One. 7:e306192012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14:R362013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Volders PJ, Verheggen K, Menschaert G,
Vandepoele K, Martens L, Vandesompele J and Mestdagh P: An update
on LNCipedia: A database for annotated human lncRNA sequences.
Nucleic Acids Res. 43:(Database Issue). D174–D180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gene Ontology Consortium: Gene ontology
consortium: Going forward. Nucleic Acids Res. 43:(Database Issue).
D1049–D1056. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dennis G, Sherman BT, Hosack DA, Yang J,
Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization and Integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:(Database Issue). D561–DD68. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using rea-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2011.
View Article : Google Scholar
|
|
31
|
Claesson-Welsh L and Welsh M: VEGFA and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen CH, Lai JM, Chou TY, Chen CY, Su LJ,
Lee YC, Cheng TS, Hong YR, Chou CK, Whang-Peng J, et al: VEGFA
upregulates FLJ10540 and modulates migration and invasion of lung
cancer via PI3K/AKT pathway. PLoS One. 4:e50522009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Inoki K, Corradetti MN and Guan KL:
Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet.
37:19–24. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen B, Zhang C, Dong P, Guo Y and Mu N:
Molecular regulation of cervical cancer growth and invasion by
VEGFa. Tumor Biol. 35:11587–11593. 2014. View Article : Google Scholar
|
|
35
|
Kaira K, Serizawa M, Koh Y, Takahashi T,
Yamaguchi A, Hanaoka H, Oriuchi N, Endo M, Ohde Y, Nakajima T and
Yamamoto N: Biological significance of 18F-FDG uptake on PET in
patients with non-small-cell lung cancer. Lung Cancer. 83:197–204.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Petryk AA, Giustini AJ, Gottesman RE,
Kaufman PA and Hoopes PJ: Magnetic nanoparticle hyperthermia
enancement of cisplatin chemotherapy cancer treatment. Int J
Hyperthermia. 29:845–851. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu LZ, Zhou XD, Qian G, Shi X, Fang J and
Jiang BH: AKT1 amplification regulates cisplatin resistance in
human lung cancer cells through the mammalian target of
rapamycin/p70S6K1 pathway. Cancer Res. 67:6325–6332. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ekman S, Wynes MW and Hirsch FR: The mTOR
pathway in lung cancer and implications for therapy and biomarker
analysis. J Thorac Oncol. 7:947–953. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao W, Wang Y, Wang Y, Gao N, Han Z and
Yu H: Potential anti-cancer effect of curcumin in human lung
squamous cell carcinoma. Thoracic Cancer. 6:508–516. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Imamura K, Imamachi N, Akizuki G, Kumakura
M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et
al: Long noncoding RNA NEAT1-dependent SFPQ relocation from
promotor region to paraspeckle mediates IL8 expression upon immune
stimuli. Mol Cell. 53:393–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Adriaens C, Standaert L, Barra J, Latil M,
Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W,
et al: p53 induces formation of NEAT1 lncRNA-containing
paraspeckles that modulate replication stress response and
chemosensitivity. Nat Med. 22:861–868. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tanabe M, Izumi H, Ise T, Higuchi S,
Yamori T, Yasumoto K and Kohno K: Activating transcription factor 4
increases the cisplatin resistance of human cancer cell lines.
Cancer Res. 63:8592–8595. 2003.PubMed/NCBI
|
|
43
|
He Q, Luo X, Jin W, Huang Y, Reddy MV,
Reddy EP and Sheikh MS: Celecoxib and a novel COX-2 inhibitor
ON09310 upregulate death receptor 5 expression via GADD153/CHOP.
Oncogene. 27:2656–2660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim SJ, Ha GH, Bae JH, Kim GR, Son CH,
Park YS, Yang K, Oh SO, Kim SH and Kang CD: COX-2 and endoplasmic
reticulum stree-independent induction of ULBP-1 and enhancement of
sensitivity to NK cell-mediated cytotocity by celecoxib in colon
cancer cells. Exp Cell Res. 330:451–459. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Oh YT, Liu X, Yue P, Kang S, Chen J,
Taunton J, Khuri FR and Sun SY: ERK/ribosomal S6 kinase (RSK)
signaling positively regulates death receptor 5 expression through
co-activation of CHOP and Elk1. J Biol Chem. 285:41310–41319. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dong HS, Kim MK, Kim HS, Chung HH and Yong
SS: Unfolded protein response to autophagy as a promising druggable
target for anticancer therapy. Ann N Y Acad Sci. 1271:20–32. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Veluscek G, Li Y, Yang SH and Sharrocks
AD: Jun-mediated changes in cell adhesion contribute to mouse
embryonic stem cell exit from ground state pluripotency. Stem
Cells. 34:1213–1224. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lou X, Xu H, Jin C, Tian W, Yu W, Dong D,
Cheng L, Huang B, Jiang H and Lin B: SOX2 targets fibronectin 1 to
promote cell migration and invasion in ovarian cancer: New
molecular leads for therapeutic intervention. OMICS. 17:520–508.
2013. View Article : Google Scholar
|
|
49
|
Gao W, Liu Y, Qin R, Liu D and Feng Q:
Silence of fibronectin 1 increases cisplatin sensitivity of
non-small cell lung cancer cell line. Biochem Biophys Res Commun.
476:35–41. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fang L, Gao X, Jing W, Chao G, Li X, Li X,
Gong X and Zeng X: Transcriptome sequencing to identify
transcription factor regulatory network and alternative splicing in
endothelial cells under VEGF stimulation. J Mol Neurosci.
58:170–177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang Y and Li Y: Analysis of molecular
pathways in pancreatic ductal adenocarcinomas with a bioinformatics
approach. Asian Pac J Cancer Prev. 16:2561–2567. 2015. View Article : Google Scholar : PubMed/NCBI
|