Introduction
Spinal cord injury (SCI) entails severe physical and
social consequences for patients and their families, and may be of
a traumatic or non-traumatic etiology (1,2).
Traumatic SCI, which is primarily caused by traffic accidents,
occurs when an external impact acutely damages the spinal cord, and
may be temporally divided into four stages: i) Acute at <48 h
following injury; ii) subacute at 48 h-14 days following injury;
iii) intermediate at 14 days-6 months following injury; and iv)
chronic at >6 months following injury (1). Although numerous studies have focused
on neuroprotective and neuroregenerative therapies, no major
breakthroughs have been achieved (3–5).
Therefore, it is necessary to elucidate the underlying mechanism of
SCI.
Pathophysiologically, the initial mechanical injury
damages neurons and initiates a complex secondary injury cascade,
leading to progressive cell death, ischemia and inflammation
(6). It has been demonstrated that
the transcriptome may reflect the pathophysiological state of the
cell (7). In a number of recent
bioinformatics studies, transcriptome analysis at different time
points post-SCI was performed and various molecular events were
characterized (8–12). The immune response,
inflammatory-associated functions, vasculature development and
neurological functions were demonstrated to serve roles in the
development of SCI (6–12). However, certain molecular
alterations that occur in a temporal and spatial manner remain to
be elucidated, particularly those that occur during the secondary
injury process.
In the present study, transcriptome data under
accession no. GSE5296 was used to identify SCI-specific molecular
programs. Temporally, three different time points were evaluated,
including 24 and 72 h, and 7 days post-injury for pathway and
functional enrichment analysis. Furthermore, a protein-protein
interaction (PPI) network was constructed. Spatially,
differentially expressed genes (DEGs) in different locations,
including the trauma site (M), and immediately adjacent rostral (R)
and caudal (C) regions were determined at the aforementioned time
points. The present study revealed molecular mechanisms that may be
associated with SCI and provided an insight into potential
therapeutic targets for treatment of SCI.
Materials and methods
Transcriptome data
The transcriptome data under accession number
GSE5296 based on the GPL1261 platform (Affymetrix Mouse Genome 430
2.0 Array; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) was obtained from the National Center for Biotechnology
Information Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo). In the original
dataset (GSE5296), C57BL6 mice were subjected to moderate contusion
injury at the T8 spinal segment. Sections of the spinal cord (4 mm
in length) were analyzed from the site of the trauma and at the
immediately adjacent R and C regions, at 0.5, 4, 24 and 72 h, and 7
and 28 days following injury. A total of 96 chips were available in
this dataset data, including 18 SCI samples for each M, R and C
region (n=3/time-point), 12 sham-injury samples for each M, R and C
region (n=2/time-point), and two samples in each region obtained
from naive mice (data not shown).
Data preprocessing
The Robust Multichip Average algorithm in the Oligo
package, version 1.42.0 (http://www.bioconductor.org) was used to preprocess
the raw transcriptome data included in the GSE5296 dataset
(13). Data were subjected to
background correction, normalization, probe summary and
log2 transformation. If there were several probes
annotated to same gene symbol, the average value was used to
represent the expression level of this gene. There were 45,037
probes in the raw data and 21,812 genes remained following data
processing.
Identification and analysis of
DEGs
In the present study, data were divided into the
following paired groups: i) Post-SCI group vs. sham group in the M
region at different time-points (0.5, 4, 24 and 72 h, and 7 and 28
days); ii) post-SCI group vs. post-sham group in the R region at
different time-points (24 and 72 h, and 7 days); and iii) post-SCI
group vs. sham group in the C region at different time-points (24
and 72 h, and 7 days). Fold change |log2FC| and P-values
from a Student's t-test were used to identify the DEGs. An average
fold-change >2.0 and P<0.05 were used as cutoff criteria.
Subsequently, Venny 2.1 (bioinfogp.cnb.csic.es/tools/venny/index.html) was
used to compare lists of DEGs and to construct Venn diagrams.
Pathway and functional enrichment
analysis
To identify pathways and biological processes
enriched by DEGs, the Database for Annotation, Visualization and
Integrated Discovery (DAVID 6.8; http://david.abcc.ncifcrf.gov) was used to perform
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene
Ontology (GO) functional analysis (14). GO terms were identified under
categories of biological process. The threshold was set to
P<0.05.
PPI network construction
A PPI network was constructed using the Search Tool
for the Retrieval of Interacting Genes/Proteins (String 10.5;
www.string-db.org). The network was visualized
using the Cytoscape software platform (Cytoscape 3.6) based on
functional analysis information, including fold-change in
gene/protein expression, PPIs and GO/KEGG pathway enrichment
(15). A default confidence cutoff
of 400 was used in the present study. Experimentally determined
interactions were presented as solid lines between genes/proteins,
and the dashed lines represent database predicted interactions
(16).
Trend charts of neuronal function- and
inflammatory response-associated genes
In the present study, neuronal function and synaptic
transmission-associated genes were defined as cholinergic receptor
nicotinic α7 subunit (Chrna7), synapsin II (Syn2),
potassium voltage-gated channel subfamily C member 1
(Kcnc1), ATPase plasma membrane Ca2+ transporting
2 (Atp2b2), unc-13 homolog A (Unc13a), regulating
synaptic membrane exocytosis 1 (Rims1), calcium/calmodulin
dependent protein kinase IIγ (Camk2g), calcium/calmodulin
dependent protein kinase IIα (Camk2a), thyrotropin releasing
hormone receptor (Trhr) and glutamate metabotropic receptor
1 (Grm1) (3,4,9).
Furthermore, based on literature review, inflammation-associated
genes were identified, including interleukin (IL)1β, IL6,
IL7, IL4, IL10, CD44 molecule (Indian blood group)
(Cd44), cytochrome b-245 β-chain (Cybb),
intercellular adhesion molecule 1 (Icam1), cytochrome b-245
α-chain (Cyba), HCK proto-oncogene, Src family tyrosine
kinase (Hck), caspase 1 (Casp1), transforming growth
factor β1 (Tgfb1), Rac family small GTPase 2 (Rac2),
integrin subunit β2 (Itgb2) and C-X-C motif chemokine
receptor 4 (Cxcr4) (3,4,9). The
fold-change of expression of each gene (post-SCI data vs. sham data
in each region) at three different time-points (24 and 72 h, and 7
days) was determined.
Results
Data preprocessing and DEG
screening
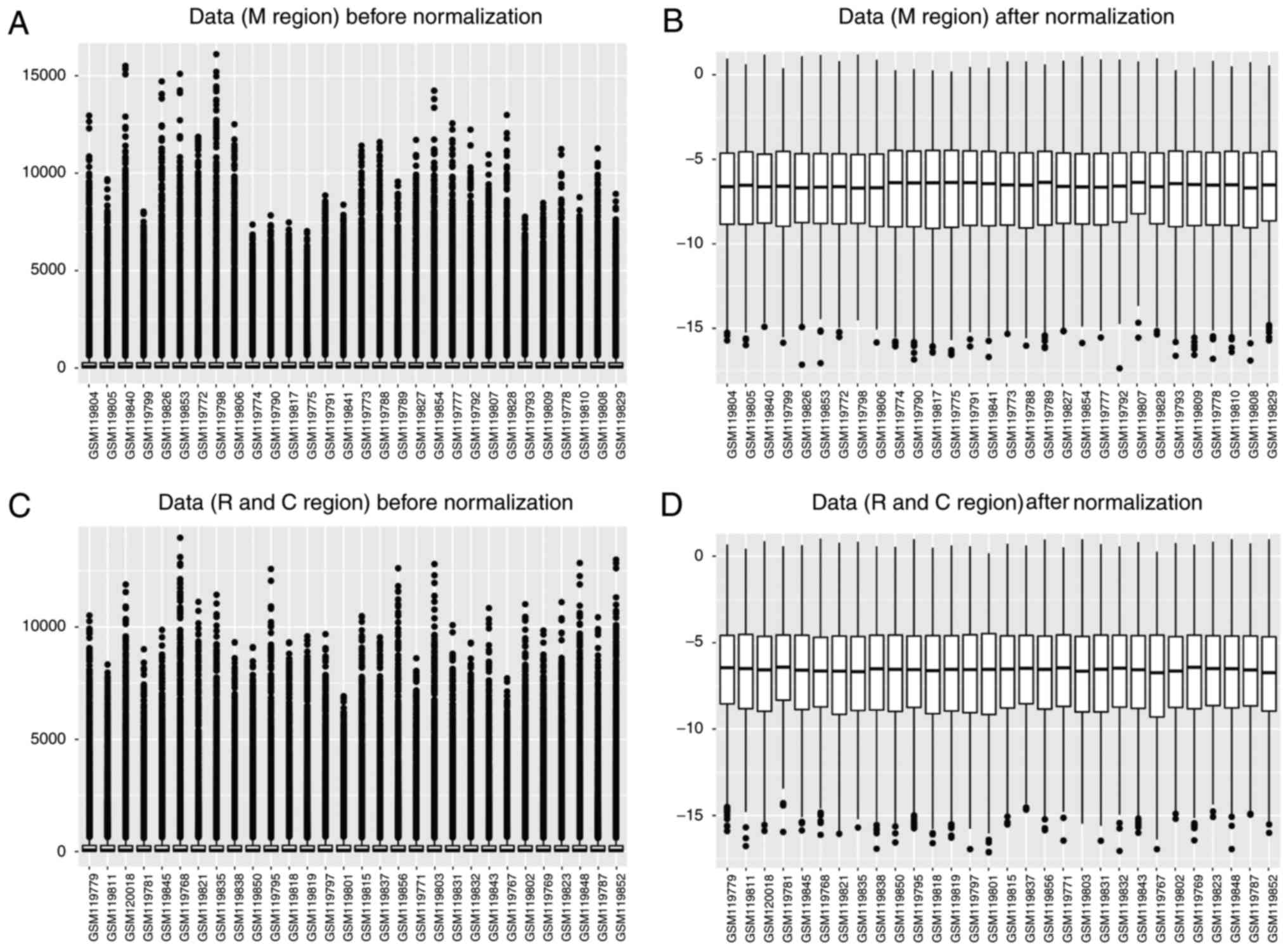
Box plots presenting post-SCI and sham surgery data
in the different regions at different time points (M region: 0.5,
4, 24 and 72 h, and 7 and 28 days; R and C regions: 24 and 72 h,
and 7 days) prior to and following data normalization are presented
in Fig. 1. The results
demonstrated that the gene expression values in each sample were
similar following normalization. Following data pre-processing,
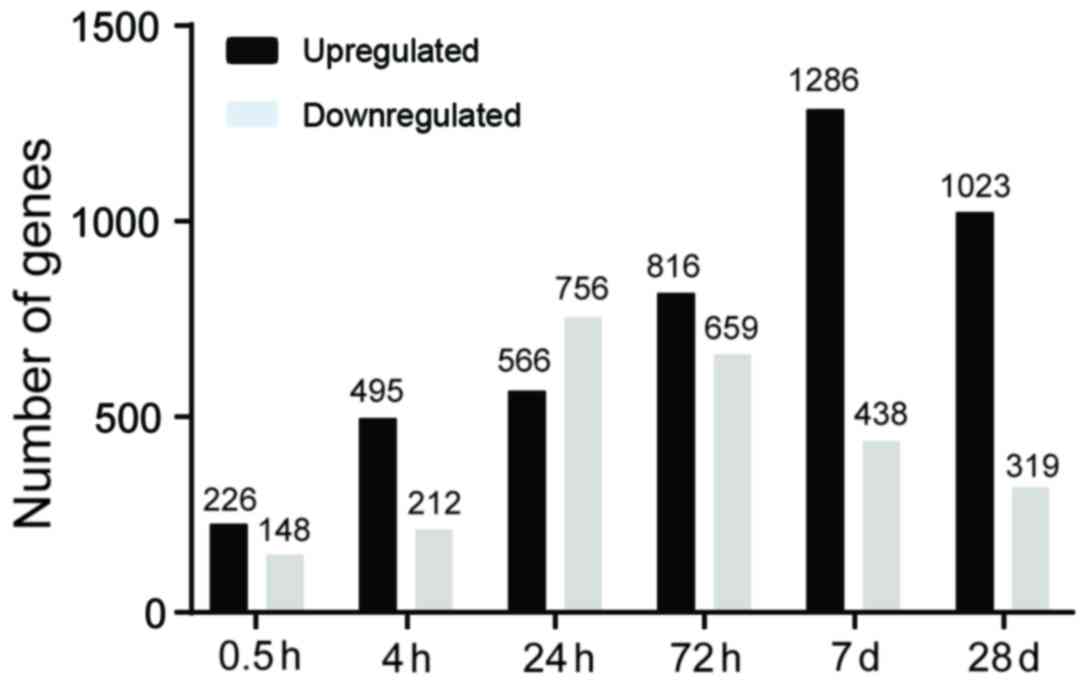
DEGs between the post-SCI groups and sham groups in the M region at
six time points were analyzed. There were 226, 495, 566, 816, 1,286
and 1,023 upregulated DEGs at 0.5, 4, 24, and 72 h and 7 and 28
days, respectively. Additionally, a total of 148, 212, 756, 659,
438 and 319 downregulated DEGs were identified at the six time
points, respectively (Fig. 2).
There was an increased number of upregulated DEGs compared with
downregulated DEGs at the five time points (0.5, 4 and 72 h, and 7
and 28 days) and the number of upregulated DEGs reached a peak on
day 7. By contrast, the number of downregulated DEGs peaked at 24 h
and subsequently decreased over time. The above results indicated
that gene expression alterations occurred primarily at 24 and 72 h,
and 7 days following injury.
KEGG pathway and GO enrichment
analysis of up- and downregulated DEGs
A number of studies have investigated alterations in
gene expression between 0.5 and 6 h following SCI in mice (17,18).
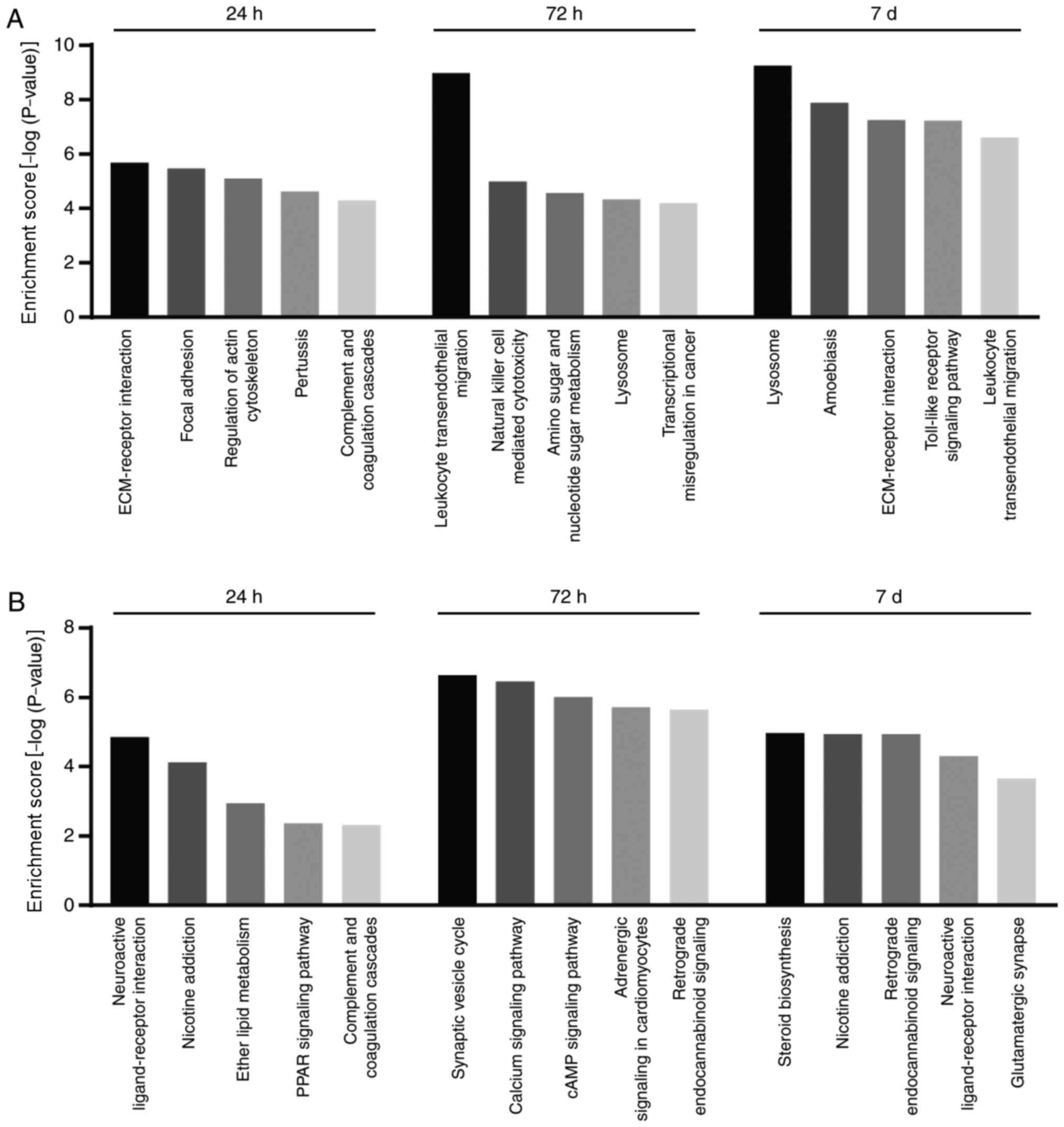
The present study focused on DEGs at 24 and 72 h, and 7 days
following injury. The top five enriched KEGG pathways of up- and
downregulated DEGs at each time point (24 and 72 h, and 7 days) are
presented in Fig. 3A and B,
respectively. At 24 h, upregulated DEGs were enriched in pathways
including ‘extracellular matrix-receptor interaction’
(P=2.08×10−6), ‘focal adhesion’ (P=3.33×10−6)
and ‘regulation of the actin cytoskeleton’
(P=7.98×10−6). At 72 h and 7 days, upregulated DEGs were
primarily involved in inflammation- and immunity-associated
pathways. ‘Leukocyte transendothelial migration’
(P=1.05×10−9) and ‘natural killer cell-mediated
cytotoxicity’ (P=1.02×10−5) were enriched at 72 h.
‘Lysosome’ (P=5.57×10−10), ‘Toll-like receptor signaling
pathway’ (P=5.93×10−8) and ‘leukocyte transendothelial
migration’ (P=2.48×10−7) were enriched at 7 days
following injury. Notably, downregulated DEGs were enriched in the
neuronal function and synaptic transmission-associated pathways,
including ‘neuroactive ligand-receptor interactions’
(P=1.43×10−5) at 24 h, ‘synaptic vesicle cycle’
(P=2.32×10−7) and ‘calcium signaling pathway’
(P=3.45×10−7) at 72 h, and ‘neuroactive ligand-receptor
interaction’ (P=4.95×10−5) and ‘glutamatergic synapse’
(P=2.23×10−4) at 7 days following injury. Other
downregulated DEGs were most significantly enriched in ‘steroid
biosynthesis’ (P=1.08×10−5).
The GO terms (biological process) of up- and
downregulated DEGs are summarized in Table I. The results demonstrated that
upregulated DEGs were most enriched in ‘response to stress’
(P=2.30×10−27) at 24 h. Additionally, ‘immune system
process’ was the most enriched function at 72 h and 7 days
(P=3.59×10−27 and P=7.77×10−68,
respectively). By contrast, downregulated DEGs were most enriched
in ‘single-organism cellular process’ (P=9.30×10−8) at
24 h following injury. Furthermore, downregulated DEGs were
enriched in neuronal function- and synaptic transmission-associated
biological process terms at 72 h and 7 days following injury. At 72
h ‘synaptic signaling’ (P=1.21×10−30) and ‘chemical
synaptic transmission’ (P=7.68×10−25) were most
enriched. At 7 days following injury, ‘synaptic signaling’
(P=5.83×10−9) and ‘anterograde trans-synaptic signaling’
(P=2.51×10−8) were most enriched. ‘Sterol biosynthetic
process’ (P=4.96×10−10) was the most significantly
enriched biological process at 7 days following injury.
Collectively, in the present study, upregulated DEGs were
predominantly associated with immune and inflammatory functions,
while downregulated DEGs were involved in neuronal function,
synaptic transmission and steroid biosynthesis.
 | Table I.GO terms enriched by differentially
expressed genes at three time-zpoints following spinal cord
injury. |
Table I.
GO terms enriched by differentially
expressed genes at three time-zpoints following spinal cord
injury.
| A, Injury vs. sham
(24 h) |
|---|
|
|---|
| Category | Term | Biological
process | No. genes | P-value |
|---|
|
|---|
| Upregulated |
|
|
|
|
|
| GO:0006950 | Response to
stress | 144 |
2.30×10−27 |
|
| GO:0070887 | Cellular response
to chemical stimulus | 107 |
9.95×10−22 |
|
| GO:0050896 | Response to
stimulus | 233 |
3.47×10−21 |
|
| GO:0010033 | Response to organic
substance | 110 |
9.68×10−21 |
|
| GO:0006952 | Defense
response | 73 |
3.99×10−20 |
| Downregulated |
|
|
|
|
|
| GO:0044763 | Single-organism
cellular process | 329 |
9.30×10−8 |
|
| GO:0044699 | Single-organism
process | 363 |
1.70×10−7 |
|
| GO:0048512 | Circadian
behavior | 9 |
3.21×10−7 |
|
| GO:0007622 | Rhythmic
behavior | 9 |
6.19×10−7 |
|
| GO:0007275 | Multicellular
organism development | 154 |
1.11×10−6 |
|
| B, Injury vs.
sham (72 h) |
|
|
Category | Term | Biological
process | No.
genes | P-value |
|
| Upregulated |
|
|
|
|
|
| GO:0002376 | Immune system
process | 148 |
3.59×10−27 |
|
| GO:0006952 | Defense
response | 90 |
4.21×10−18 |
|
| GO:0006955 | Immune
response | 87 |
5.70×10−18 |
|
| GO:0006954 | Inflammatory
response | 52 |
9.35×10−18 |
|
| GO:0002684 | Positive regulation
of immune system process | 64 |
6.12×10−16 |
| Downregulated |
|
|
|
|
|
| GO:0099536 | Synaptic
signaling | 56 |
1.21×10−30 |
|
| GO:0098916 | Anterograde
trans-synaptic signaling | 54 |
3.26×10−29 |
|
| GO:0099537 | Trans-synaptic
signaling | 54 |
3.65×10−29 |
|
| GO:0007268 | Chemical synaptic
transmission | 57 |
7.68×10−25 |
|
| GO:0007267 | Cell-cell
signaling | 74 |
2.96×10−20 |
|
| C, Injury vs.
sham (day 7) |
|
|
Category | Term | Biological
process | No.
genes | P-value |
|
| Upregulated |
|
|
|
|
|
| GO:0002376 | Immune system
process | 286 |
7.77×10−68 |
|
| GO:0006952 | Defense
response | 188 |
7.30×10−52 |
|
| GO:0006950 | Response to
stress | 319 |
6.25×10−45 |
|
| GO:0006955 | Immune
response | 166 |
2.74×10−41 |
|
| GO:0002682 | Regulation of
immune system process | 164 |
1.22×10−39 |
| Downregulated |
|
|
|
|
|
| GO:0016126 | Sterol biosynthetic
process | 9 |
4.96×10−10 |
|
| GO:0099536 | Synaptic
signaling | 19 |
5.83×10−9 |
|
| GO:1902653 | Secondary alcohol
biosynthetic process | 7 |
2.09×10−8 |
|
| GO:0098916 | Anterograde
trans-synaptic signaling | 18 |
2.51×10−8 |
|
| GO:0099537 | Trans-synaptic
signaling | 18 |
2.59×10−8 |
|
PPI network construction and
functional module analysis
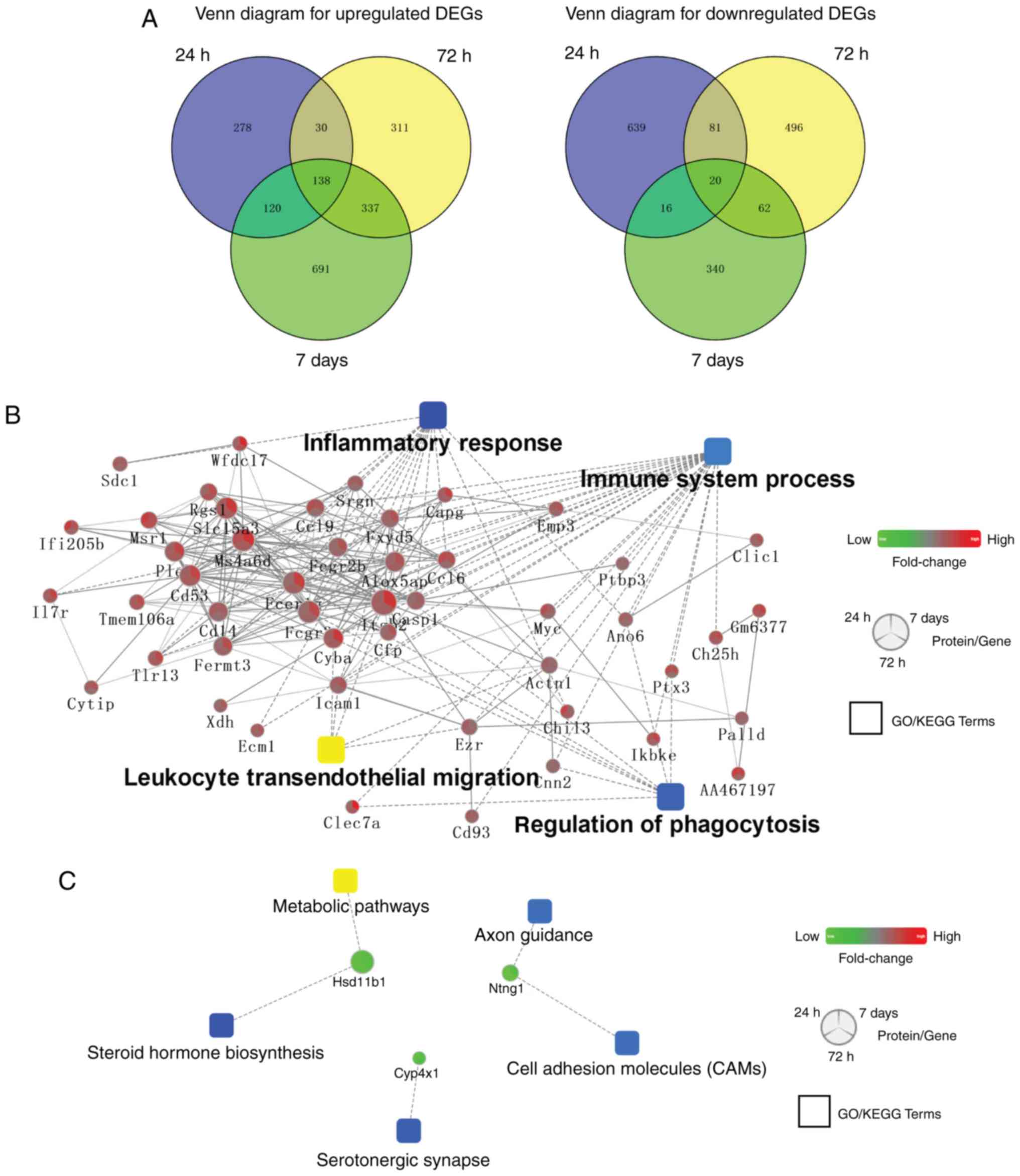
Venn diagram analysis of DEGs at 24 and 72 h, and 7
days is presented in Fig. 4A.
There were 138 common upregulated and 20 overlapping downregulated
DEGs at these three time points. Subsequently, a more comprehensive
bioinformatics analysis was performed using Cytoscape software, a
tool for predicting PPI networks. The results revealed that immune
and inflammatory functions were enriched in co-upregulated DEGs
(Fig. 4B). Additionally, neuronal
functions and ‘steroid hormone biosynthesis’ were enriched in the
co-downregulated DEGs (Fig.
4C).
Trends in the expression levels of
neuronal function- and inflammatory response-associated genes
The results of pathway, functional enrichment and
PPI network analyses demonstrated that inflammatory- and
neuronal-associated functions serve roles in post-SCI mice in the M
region at three different time points (24 and 72 h, and 7 days).
Alterations in these functions were further investigated via
temporal and spatial analysis of the expression of numerous genes.
Neuronal function- and synaptic transmission-associated genes,
including Chrna7, Atp2b2, Rims1, Camk2g, and Trhr,
were downregulated at 72 h compared with the expression at 24 h
post-SCI in the M region (Fig.
5A). Furthermore, neuronal function associated genes,
Chrna7, Syn2 and Unc13a, were significantly upregulated on
day 7 compared with the expression at 72 h post-SCI. Similar
alterations were observed in R and C regions at three different
time points post-SCI, although the overall fold-changes with time
were minimal (Fig. 5A). Genes
involved in inflammatory processes exhibited different and complex
alterations (Fig. 5B). The
expression levels of inflammatory-associated genes (IL1b and
IL6) decreased significantly at 72 h following injury in the
M region compared with levels at the 24 h time interval. However,
similar alterations were not observed in the R and C regions. By
contrast, the expression levels of other genes associated with
inflammatory processes (Cd44, Cybb, Cyba, Hck, Casp1, Itgb2
and Cxcr4) significantly increased at 7 days post-SCI in the
M region compared with expression levels at the 72 h time interval.
Similar trends were observed in the C region, although not in the R
region (Fig. 5C). Therefore, the
results of the present study suggest that alterations in expression
of genes associated with inflammatory and neuronal functions were
primarily observed in the M region at different time points
post-SCI and these two events occurred in a temporally and
spatially specific manner respectively, as reflected in the trend
charts.
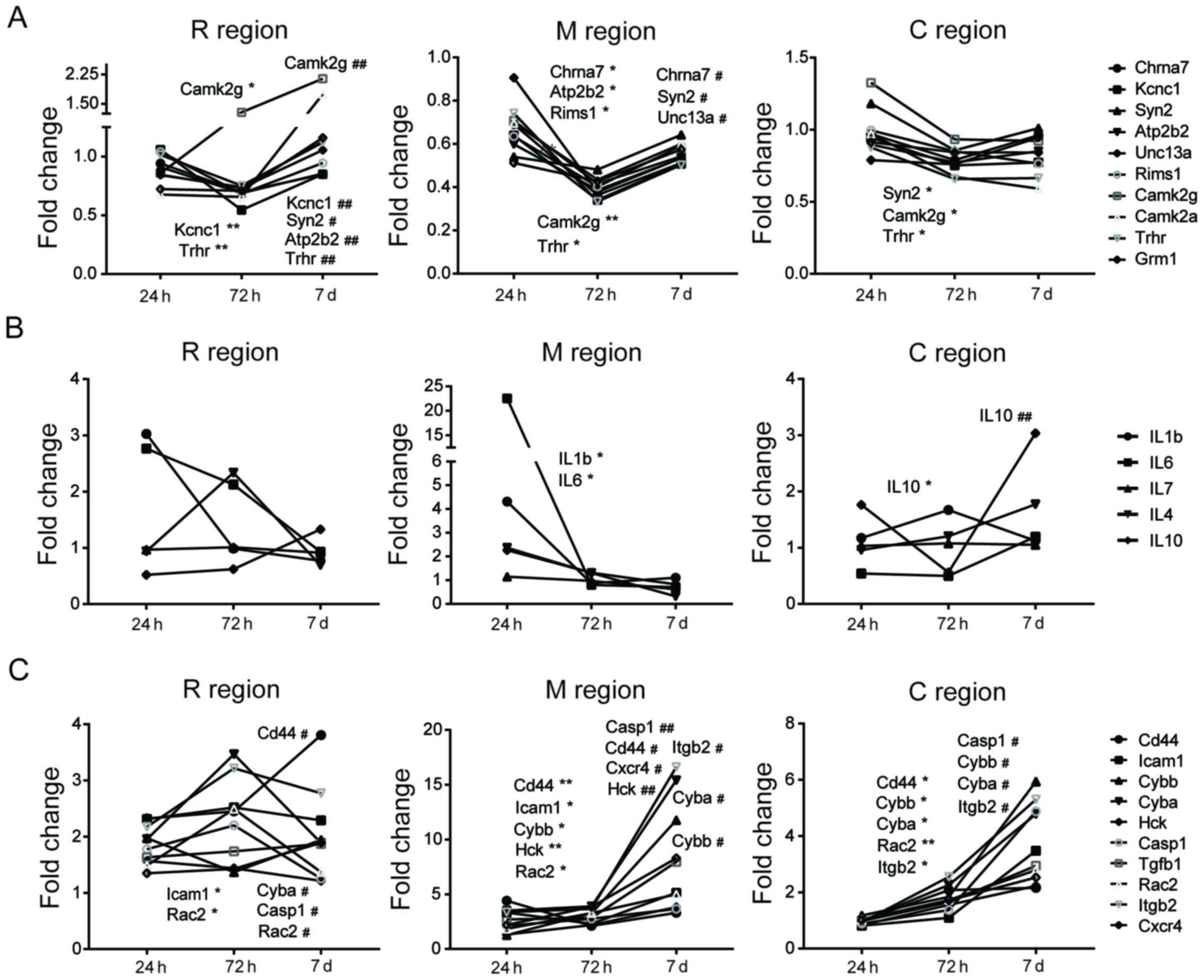 | Figure 5.Trend charts of gene expression in R,
M and C regions. The trend charts of expression of genes associated
with (A) neuronal function, including Chrna7, Syn2, Kcnc1, Atp2b2,
Unc13a, Rims1, Camk2g, Camk2a, Trhr and Grm1 and inflammatory
response, including (B) IL1b, IL6, IL7, IL4 and IL10, and (C) CD44,
Icam1, Cybb, Cyba, Hck, Casp1, Tgfb1, Rac2, Itgb2 and Cxcr4.
Significant differences between expression levels at different time
points were identified (*P<0.05 and **P<0.01, 24 h vs. 72 h;
#P<0.05 and ##P<0.01, 72 h and 7 days).
R, rostral; C, caudal; M, trauma site. IL, interleukin; Cd44, CD44
molecule (Indian blood group); Cybb, cytochrome b-245 β-chain;
Icam1, intercellular adhesion molecule 1; Cyba, cytochrome b-245
α-chain; Hck, HCK proto-oncogene, Src family tyrosine kinase;
Casp1, caspase 1; Tgfb1, transforming growth factor β1; Rac2, Rac
family small GTPase 2; Itgb2, integrin subunit β2; Cxcr4, C-X-C
motif chemokine receptor 4; Chrna7, cholinergic receptor nicotinic
α7 subunit; Syn2, synapsin II; Kcnc1, potassium voltage-gated
channel subfamily C member 1; Atp2b2, ATPase plasma membrane
Ca2+ transporting 2; Unc13a, unc-13 homolog A; Rims1,
regulating synaptic membrane exocytosis 1; Camk2g,
calcium/calmodulin dependent protein kinase II γ; Camk2a,
calcium/calmodulin dependent protein kinase IIα; Trhr, thyrotropin
releasing hormone receptor; Grm1, glutamate metabotropic receptor
1. |
Discussion
Experimental modeling of SCI in animals has been
widely used to investigate the complex secondary injury cascade
(1,19). The GSE5296 database consists of
transcriptome data obtained from spinal cord sections from injury
sites, and immediately adjacent R and C regions at 0.5, 4, 24 and
72 h, and 7 and 28 days post-injury. In the present study,
bioinformatics analysis was used to determine molecular events and
pathological states in SCI. Based on analysis of gene expression at
different time points, DEGs were determined, and KEGG pathway and
GO enrichment analyses were performed. Additionally, a PPI network
and gene expression trend charts were constructed to further
analyze the molecular processes underlying SCI. The analyses
performed in the present study may contribute to a better
understanding of SCI.
In the present study, at the M site, there were 374,
707, 1,322, 1,475, 1,724 and 1,342 DEGs identified at 0.5, 4, 24
and 72 h, and 7 and 28 days following injury, respectively. The
results of the present study indicated that the majority of
alterations in molecular events occurred at 7 days following injury
and decreased over time. In addition, Wang et al (8) demonstrated that the number of DEGs
decreased in a time-dependent manner (1,942, 396, 188 and 193 DEGs
were identified at 3, 7, 14 days and 1 month, respectively).
Although the R and C regions exhibited decreased fold changes in
gene expression compared with the M region, further studies of
these two areas may improve the understanding of the overall
process of SCI.
Numerous molecular events were detected by analyzing
up- and downregulated DEGs. As demonstrated in a previous study,
downregulated DEGs were primarily enriched in neuronal functions
(9). The spectra of expression
changes in neuronal function- and synaptic transmission-associated
genes (Chrna7, Syn2, Kcnc1, Atp2b2, Unc13a, Rims1, Camk2g,
Camk2a, Trhr and Grm1) from 24 h to 7 days post-SCI
reflects the regulation between the degeneration and survival of
injured tissues. Insight into time-dependent alterations in
structural and functional neuronal biomarkers may be useful for
developing protective or regenerative therapeutic
interventions.
As a regulator of degeneration and regeneration of
the spinal cord, inflammation is a hallmark of the secondary SCI
process (20). In the present
study, KEGG pathway and GO enrichment analyses, and the PPI
network, revealed that the upregulated DEGs were primarily
associated with ‘immune system’ process and the inflammatory
response, particularly at 72 h and 7 days post-SCI. As previously
demonstrated, there is an association between the severity of SCI
and the intensity of the acute inflammatory response, which
includes proinflammatory cytokines and immune cells (20,21).
A significant increase in the expression of proinflammatory
(IL-1b, IL-6 and IL-7) and anti-inflammatory
cytokines (IL-4 and IL-10) in the present study
reflects both the regulation between degeneration and survival of
injured tissues. A protective strategy is to target the process of
inflammation and limit the infiltration of immune cells into the
injury site (22–24). Notably, another group of
inflammation-associated genes, including Cd44, Cybb, Icam1,
Cyba, Hck, Casp1, Tgfb1, Rac2, Itgb2 and Cxcr4 exhibited
a different temporal pattern compared with proinflammatory genes
(IL-1β, IL-6 and IL-7), indicating a complex
inflammatory immune microenvironment at the damaged site that
requires further analysis. These findings the importance of
monitoring inflammation over time following SCI (20).
In the present study, downregulated DEGs at day 7
were primarily enriched in the ‘steroid biosynthesis’ process.
Steroids may be functionally divided into cholesterol,
corticosteroids, sex steroids, neuroactive steroids and vitamin D
(25,26). A number of studies have
investigated the association between steroid metabolism and SCI
(27–34). Estrogen may attenuate inflammation
and promote neural survival and regeneration following SCI
(27,28). Statins, known as
cholesterol-controlling drugs, may significantly enhance neuronal
and oligodendrocyte survival, in addition to decreasing the levels
of proinflammatory cytokines (29). Previous studies additionally
suggested that individuals with SCI are at an increased risk of
vitamin D deficiency (30–32). Furthermore, neuroactive steroids
are naturally occurring steroids that impact behavior, alter the
excitability of neurons and interact with specific neurotransmitter
receptors (33,34). Therefore, targeting steroid
biosynthesis as a therapeutic approach for neuroprotective and
neuroregenerative purposes merits further investigation.
However, one limitation of the present study was
that the raw data did not include gene expression data from samples
at 3 or 6 months following injury, limiting the information
regarding molecular processes during the progression of SCI. KEGG
pathway and GO enrichment analyses of data for the very acute phase
(0.5 and 4 h post-SCI) or at 1 month post-SCI were not included in
the present study. Additionally, future comprehensive analysis of
transcriptome data from the adjacent R and C regions at each time
point post-SCI may reveal the complex alterations that occur during
the pathophysiological process.
In conclusion, the present study revealed that
inflammatory response, immune processes, neuronal-associated
functions and ‘steroid biosynthesis’ serve roles in the progression
of SCI. Furthermore, the M region exhibited increased fold-changes
in the expression of genes associated with inflammatory responses
and neuronal function compared with the R and C regions at
different time-points post-SCI. However, in vivo and in
vitro studies are required to determine the specific roles of
these molecular events in SCI.
Acknowledgements
We would like to thank Dr Yu-qing Jiang (Department
of Orthopedics, the Affiliated Hospital of Nanjing Medical
University, Changzhou No. 2 People's Hospital) for assistance with
the present study.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 8177235,
81520108018 and 81472080), the Jiangsu Committee of Science and
Technology-Social Development Plan (grant no. BE2017755) and the
Nanjing Committee of Science and Technology (grant no.
201505005).
Availability of data and materials
All data generated or analysed during this study are
included in this published article.
Authors' contributions
GYY designed the present study. SJZ, WZ, JC and YJL
performed the data analysis and statistical analysis. SJZ and WZ
wrote and revised the manuscript. GYY supervised the present study.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ahuja CS, Wilson JR, Nori S, Kotter MRN,
Druschel C, Curt A and Fehlings MG: Traumatic spinal cord injury.
Nat Rev Dis Primers. 3:170182017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jia X, Kowalski RG, Sciubba DM and
Geocadin RG: Critical care of traumatic spinal cord injury. J
Intensive Care Med. 28:12–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siddiqui AM, Khazaei M and Fehlings MG:
Translating mechanisms of neuroprotection, regeneration and repair
to treatment of spinal cord injury. Prog Brain Res. 218:15–54.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ahuja CS and Fehlings M: Concise review:
Bridging the gap: Novel neuroregenerative and neuroprotective
strategies in spinal cord injury. Stem Cells Transl Med. 5:914–924.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim YH, Ha KY and Kim SI: Spinal cord
injury and related clinical trials. Clin Orthop Surg. 9:1–9. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bareyre FM and Schwab ME: Inflammation,
degeneration and regeneration in the injured spinal cord: Insights
from DNA microarrays. Trends Neurosci. 26:555–563. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ydens E, Palmers I, Hendrix S and Somers
V: The next generation of biomarker research in spinal cord injury.
Mol Neurobiol. 54:1482–1499. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang W, Liu R, Xu Z, Niu X, Mao Z, Meng Q
and Cao X: Further insight into molecular mechanism underlying
thoracic spinal cord injury using bioinformatics methods. Mol Med
Rep. 12:7851–7858. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen G, Fang X and Yu M: Regulation of
gene expression in rats with spinal cord injury based on microarray
data. Mol Med Rep. 12:2465–2472. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wen T, Hou J, Wang F, Zhang Y, Zhang T and
Sun T: Comparative analysis of molecular mechanism of spinal cord
injury with time based on bioinformatics data. Spinal Cord.
54:431–438. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu Z, Shen Q, Zhu L and Wei X:
Identification of pivotal genes and pathways for spinal cord injury
via bioinformatics analysis. Mol Med Rep. 16:3929–3937. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duan H, Ge W, Zhang A, Xi Y, Chen Z, Luo
D, Cheng Y, Fan KS, Horvath S, Sofroniew MV, et al: Transcriptome
analyses reveal molecular mechanisms underlying functional recovery
after spinal cord injury. Proc Natl Acad Sci USA. 112:13360–13365.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Carvalho BS and Irizarry RA: A framework
for oligonucleotide microarray preprocessing. Bioinformatics.
26:2363–2367. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun N, Sun W, Li S, Yang J, Yang L, Quan
G, Gao X, Wang Z, Cheng X, Li Z, et al: Proteomics analysis of
cellular proteins co-immunoprecipitated with nucleoprotein of
influenza a virus (H7N9). Int J Mol Sci. 16:25982–25998. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Di Giovanni S, Knoblach SM, Brandoli C,
Aden SA, Hoffman EP and Faden AI: Gene profiling in spinal cord
injury shows role of cell cycle in neuronal death. Ann Neurol.
53:454–468. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carmel JB, Galante A, Soteropoulos P,
Tolias P, Recce M, Young W and Hart RP: Gene expression profiling
of acute spinal cord injury reveals spreading inflammatory signals
and neuron loss. Physiol Genomics. 7:201–213. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kjell J and Olson L: Rat models of spinal
cord injury: From pathology to potential therapies. Dis Model Mech.
9:1125–1137. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saghazadeh A and Rezaei N: The role of
timing in the treatment of spinal cord injury. Biomed Pharmacother.
92:128–139. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kinoshita T, Nakamura T, Umemoto Y, Kojima
D, Moriki T, Mitsui T, Goto M, Ishida Y and Tajima F: Increase in
interleukin-6 immediately after wheelchair basketball games in
persons with spinal cord injury: Preliminary report. Spinal Cord.
51:508–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu JZ, Huang JH, Xiao ZM, Li JH, Li XM and
Lu HB: Tetramethylpyrazine accelerates the function recovery of
traumatic spinal cord in rat model by attenuating inflammation. J
Neurol Sci. 324:94–99. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
David BT, Ratnayake A, Amarante MA, Reddy
NP, Dong W, Sampath S, Heary RF and Elkabes S: A toll-like receptor
9 antagonist reduces pain hypersensitivity and the inflammatory
response in spinal cord injury. Neurobiol Dis. 54:194–205. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jin W, Wang J, Zhu T, Yuan B, Ni H, Jiang
J, Wang H and Liang W: Anti-inflammatory effects of curcumin in
experimental spinal cord injury in rats. Inflamm Res. 63:381–387.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schroepfer GJ Jr: Sterol biosynthesis. Ann
Rev Biochem. 50:585–621. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ghayee HK and Auchus RJ: Basic concepts
and recent developments in human steroid hormone biosynthesis. Rev
Endocr Metab Disord. 8:289–300. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Elkabes S and Nicot AB: Sex steroids and
neuroprotection in spinal cord injury: A review of preclinical
investigations. Exp Neurol. 259:28–37. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brotfain E, Gruenbaum SE, Boyko M, Kutz R,
Zlotnik A and Klein M: Neuroprotection by estrogen and progesterone
in traumatic brain injury and spinal cord injury. Curr
Neuropharmacol. 14:641–653. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Eftekharpour E, Nagakannan P, Iqbal MA and
Chen QM: Mevalonate cascade and small Rho GTPase in spinal cord
injury. Curr Mol Pharmacol. 10:141–151. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Flueck JL and Perret C: Vitamin D
deficiency in individuals with a spinal cord injury: A literature
review. Spinal Cord. 55:428–434. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bauman WA, Morrison NG and Spungen AM:
Vitamin D replacement therapy in persons with spinal cord injury. J
Spinal Cord Med. 28:203–207. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Oleson CV, Patel PH and Wuermser LA:
Influence of season, ethnicity and chronicity on vitamin D
deficiency in traumatic spinal cord injury. J Spinal Cord Med.
33:202–213. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Giatti S, Garcia-Segura LM and Melcangi
RC: New steps forward in the neuroactive steroid field. J Steroid
Biochem Mol Biol. 153:127–134. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tuem KB and Atey TM: Neuroactive steroids:
Receptor interactions and responses. Front Neurol. 8:4422017.
View Article : Google Scholar : PubMed/NCBI
|