Introduction
Long non-coding RNAs (lncRNAs) are untranslated
transcripts longer than 200 nucleotides that structurally resemble
mRNAs but do not encode proteins (1). They are composed of several typical
mRNA structural characteristics, including a polyA tail,
5′-capping, and a promoter structure (2). Previous studies have revealed that
lncRNAs have important effects on the regulation of gene expression
at the epigenetic, transcriptional, and post-transcriptional level
(3). In addition, it has been
revealed that lncRNAs display important roles in a number of
physiological and pathological processes, including the
pathogenesis of human cancers (1,2).
Aberrant expression of lncRNAs is correlated with tumorigenesis
through several distinct modes of action (4). It is hypothesized that the regulation
of lncRNAs is a crucial part of tumorigenesis, however, the details
of lncRNAs regulation and their mechanisms of action in specific
cancers remain unclear (4).
Recent studies have demonstrated that microRNAs,
small noncoding RNAs comprising of 20–22 nucleotides, function as
oncogenes or as tumor suppressors; they inhibit cell proliferation
by binding to mRNA sequences and preventing their translation
(5–9). Given the structural similarities with
mRNAs, miRNAs could also potentially target lncRNAs, suggesting a
novel mode of regulatory interactions between noncoding RNA
families (10). In addition, the
reciprocal regulation of lncRNAs and miRNAs has been correlated
with tumor invasion and metastasis (2). For example, both miR-31 and its host
gene lncRNA LOC554202 were downregulated in triple-negative breast
cancer lines (11). The loss of
miR-31 expression was demonstrated to be mediated by the
hypermethylation of its promoter-associated CpG islands (11). Mitochondrial dynamic related
lncRNAs (MDRL) inhibit mitochondrial fission and apoptosis by
directly binding to miR-361 and downregulating its expression,
which in turn relieves miR-361-mediated inhibition of miR-484
processing (12). The lncRNA
urothelial carcinoma-associated 1 (UCA1) functions by directly
binding to miR-216b and downregulating miR-216b expression. In
addition, UCA1 downregulates fibroblast growth factor receptor 1
(FGFR1) expression to reverse the inhibitory effect of miR-216b on
the growth and metastasis of human hepatocellular carcinoma cells
(13). Several studies have
demonstrated that miRNA-148a inhibits cell proliferation and
promotes cell apoptosis in pancreatic (14), colorectal (15), bladder (16), ovarian (17), gastric (18), and hepatocellular carcinoma
(19).
Prostate cancer is the most common malignancy
afflicting men in the United States and the second leading cause of
cancer mortality (20). Prostate
cancer gene expression marker 1 (PCGEM1), a prostate-specific gene,
is a novel class of androgen-regulated lncRNAs (2). Previous studies have revealed that
elevated expression of PCGEM1 is associated with high-risk prostate
cancer (21,22). PCGEM1 was expressed exclusively or
in higher levels in primary prostate tumor specimens than in
matched normal tissues. In addition, PCGEM1 expression was detected
exclusively in the androgen receptor-positive cell line LNCaP among
various prostate cancer cell lines analyzed (20).
The myocyte enhancer factor 2 (MEF2) profoundly
influences cell differentiation, proliferation and metastasis
(23,24). MEF2 directly binds to muscle
A-kinase anchoring protein (mAKAP) which leads to inhibition of
MEF2 activation during the early stages of muscle cell
differentiation (23). In
addition, class I myosin-epididymal binding protein 1 (E12)
heterodimers interact with MEF2, resulting in the activation of
myogenesis. However, homodimers of E12 do not interact with MEF2
due to lack of the conserved alanine and threonine residues in the
basic domain. The interaction between the myogenic basic
helix-loop-helix and MEF2 is uncoupled from transcriptional
activation (25). A gene
expression analysis study of the prostate cancer cell line LNCaP
demonstrated that MEF2 is differentially expressed following
exposure to androgen (26). MEF2
transcription factors binding site is present in the promoter of
co-expressed genes (26).
To date, the effect of MEF2 on PCGEM1 regulation
remains unclear. Furthermore, functional analysis of lncRNAs PCGEM1
potential interactions with miRNAs is warranted by previous
studies. Therefore, in the present study, the regulatory
interaction of MEF2 with PCGEM1, and of PCGEM1 with miR-148a were
explored. The results indicated novel insights in the function of
PCGEM1 on prostate cancer cells.
Materials and methods
Experimental sample collection
60 random cases of prostate cancer and adjacent
tumor-free prostate cancer tissue specimens were collected between
April 2016 and November 2016 at Ji'nan Central Hospital Affiliated
to Shandong University (Ji'nan, China). All cases were confirmed by
pathological diagnosis, and the surgery during which specimens were
obtained was the first surgical treatment in each case. No
chemotherapy, radiotherapy or other treatments for prostate cancer
were performed prior to surgery. All tissues were placed
immediately in liquid nitrogen and stored in the central laboratory
of Ji'nan Central Hospital. All experiments were approved by the
Medical Ethics Committee of Ji'nan Central Hospital and written
informed consent documents were signed by all patients. The large
samples of prostate cancer tissue and tumor surrounding tissue were
processed, and RNAs were extracted for sequencing analysis. Using
an Illumina HiSeq sequencing platform (Illumina, Inc., San Diego,
CA, USA), the sequencing data were obtained, the quality of the
original sequencing data was evaluated, and processed to obtain the
clean reads. The fragments per kilobase of transcript per million
mapped reads (FPKM) method was used for quantitative analysis, and
the prostate cancer tissue samples were used for analysis of
differential gene expression (27).
Cell culture and treatments
LNCaP, DU145, and PC-3 prostate cancer cell lines
were obtained from the American Type Culture Collection (Manassas,
VA, USA). PrEC normal human prostate epithelial cells were obtained
from Clonetics (Lonza, Basel, Switzerland) and cultured as
recommended by the supplier. All cell lines were cultured in RPMI
1640 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% (v/v) fetal bovine serum (FBS; HyClone; GE
Healthcare Life Sciences, Logan, UT, USA), and were maintained at
37°C in a humidified atmosphere of 5% CO2. PC-3 cells
(1.5×104 cells/well) were seeded into 96-well plates.
The slow-growing LNCaP cells were seeded at a density of
2.0×104 cells/well into 96-well plates. Cells were
cultured to attach to the wells for 24 h.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
The tissue samples were ground to powder by a frozen
tissue pulverizer. The cells were harvested following wash with PBS
twice and 12,000 × g centrifugation for 15 min at 4°C. The
harvested cells were resuspended in a solution containing 4 M
guanidine isothiocyanate, 100 mM β-mercaptoethanol, 25 mM sodium
citrate pH 7.0 and 0.5% sarcosyl. Total RNA extraction was
performed as previously described (26). The quality of total RNA was
analyzed by 1% agarose gel electrophoresis and ethidium bromide
staining. qPCR was performed according to the manufacturer's
protocol of the PCR kit (cat. no. 0960211; Beijing Kuangbo
Biotechnology Co., Ltd., Beijing, China). qPCR reactions were
performed using a preheated 7500 RT-PCR System (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with SYBR-Green detection. Reaction
conditions were as follows: Stage 1, 95°C for 30 sec; stage 2, 40
cycles of 95°C for 5 sec and 60°C for 34 sec; and stage 3
(dissolution curve), 95°C for 15 sec, 60°C for 1 min and 95°C for
15 sec. The primers for miR-148a were designed by Applied
Biosystems (Thermo Fisher Scientific, Inc.). Other primers were as
follows: PCGEM1, 5′-tccacccaatacacaggat-3′ (forward),
5′-aattgggagctgatgaggac-3′ (reverse); U6,
5′-agcttcggcagcacatatactaaaattggaat-3′ (forward),
5′-tcttcacgaatttgcgtgtcatccttga-3′ (reverse); β-actin,
5′-aaactggaacggtgaaggtg-3′ (forward) and 5′-agagaagtggggtggctttt-3′
(reverse). The relative level of miR-148a was normalized to U6, and
the relative amount of PCGEM1 was normalized to β-actin. The data
were analyzed using the 2−ΔΔCq method (28).
Western blot analysis
Cells were lysed by radio immunoprecipitation assay
buffer containing protease inhibitor cocktail (Beyotime Institute
of Biotechnology, Haimen, China). The total protein concentration
was determined using a bicinchoninic acid protein assay kit
(Shanghai Haoyang Biological Technology Co., Ltd., Shanghai,
China), and were subjected to 10% SDS-PAGE and transferred to
polyvinylidene difluoride membranes with Western Blocking Buffer
(100 µg; Shanghai Yansheng Industrial Co., Ltd, Shanghai, China).
Proteins were electrophoretically resolved on 8–10% Tris-Glycine
gels and transferred onto a nitrocellulose membrane. After blocking
the non-specific binding sites with 5% skimmed milk for 1 h at
20°C, the membrane was incubated with the primary antibody (1:800
dilution; cat. no. ADI-950-100-0001; Enzo Life Sciences, Inc.,
Farmingdale, NY, USA) at 4°C overnight. The membranes were then
incubated with a horseradish peroxidase-conjugated secondary
antibody (1:800 dilution; rabbit secondary antibodies, 6 vials;
cat. no. NB910-95603; Novus Biologicals, LLC, Littleton, CO, USA)
for 1 h at 20°C. Immunoreactive proteins were detected with the ECL
Plus western blotting Detection System (GE Healthcare Life
Sciences, Little Chalfont, UK) and exposed to X-ray film. The
samples were analyzed using enhanced chemiluminescence (cat. no.
320002; Best Biotechnology Co., Ltd., Shanghai, China) and
quantified using an image analyzer (LabWorks LLC, Lehi, UT, USA).
The density of the bands on the membrane was quantified using
Quantity One software (version 4.62; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). β-actin (1:2,000 dilution; cat. no. A01010;
Abbkine Scientific Co., Ltd., Wuhan, China) was used as a
control.
Cell transfection and luciferase
reporter assay
Human MEF2-directed and PCGEM1-directed small
interfering RNA (siRNA) (cat. no. sc-29528) and a non-specific
control siRNA (cat. no. sc-29533) were obtained from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). PC-3 cells were seeded one
day prior to transfection. siRNA (100 nm) was transfected into the
cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
medium was replaced by complete RPMI 1640 medium 3 h
post-transfection and cultured for another 48 h. Following
transfection, cells were harvested after 12,000 × g centrifugation
for 15 min at 4°C and analyzed for mRNA and protein expression,
using the aforementioned RT-qPCR and western blotting protocols.
For transfection of the pcDNA3.1 expression vector, PC3 cells were
cultured in a mixture of Dulbecco's Modified Eagle Medium: Nutrient
Mixture F-12 (DMEM/F12, 1:1) (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany), supplemented with 10% FBS (HyClone; GE Healthcare Life
Sciences) and 25 µg/ml gentamicin (Gibco; Thermo Fisher Scientific,
Inc.). PC3 cells were transfected with pcDNA3.1-MEF2 plasmid or
pcDNA3.1-PCGEM1 plasmid (Shanghai Kyrgyzstan Biological Technology
Co., Ltd., Shanghai, China) and control cells were transfected with
pcDNA3.1 empty vector. Stable transfectants were selected by adding
400 µg/ml of G418 (Geneticin) in the medium. Individual colonies
were picked and maintained in RPMI-1640 media enriched with 5% FBS,
penicillin-streptomycin and 200 µg/ml of G418 (29). Luciferase activity was measured
using the dual luciferase reporter assay system kit (Promega
Corporation, Madison, WI, USA), according to the manufacturer's
instructions, on a Tecan M200 luminescence reader (Tecan Group,
Ltd. Zurich, Switzerland).
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was carried out as previously described
(30). Native protein-DNA
complexes were cross-linked by 1% formaldehyde treatment for 10
min. Equal aliquots of isolated chromatin were used for
immunoprecipitation with specific antibodies (cat. no. FHP004-100,
FHP003-100 and FHP003-050; Beijing Zheng Bo Biological Technology
Co., Ltd., Beijing, China). DNA associated with specific
immunoprecipitates or with mouse immunoglobulin G as a negative
control was isolated and used as a template for PCR amplifying the
PCGEM1 promoter sequence containing the MEF2 binding site (30).
Apoptosis assay
PC3 cells were seeded for 24 h in 24-well plates and
transfected with non-specific control siRNA or PCGEM1 siRNA, in the
presence (5 nmol/l) or absence of miRNA-148a inhibitor (cat. no.
M101; Shanghai Tuoran Biotechnology Co., Ltd., Shanghai, China), in
serum free RPMI-1640 for 5 h. Following transfection, each well was
supplemented with 500 µl of appropriate growth medium containing
20% FBS. PC3 cells (5×105) were harvested after
incubating for another 48 h, via 12,000 × g centrifugation for 10
min and washed with PBS at room temperature. Cells were then
stained with Annexin V-fluorescein isothiocyanate (10 µM;
SouthernBiotech, Birmingham, AL, USA) and 50 µg/ml propidium iodide
(PI) for 1 h at 25°C and analyzed by flow cytometry. All the
experiments were performed at least three times.
Statistical analysis
Values are presented as the means + standard
deviation. Statistical significance analysis between two groups was
carried out by the student's t-test using GraphPad Prism software
version 4.0 (GraphPad Software, Inc., La Jolla, CA, USA).
Significance among multiple groups was analyzed by one-way analysis
of variance followed by Tukey's test, using DPS software (31). Significance analysis for the
clinical samples was performed by Wilcoxon rank sum test. P<0.05
was considered to indicate a statistically significant
difference.
Results
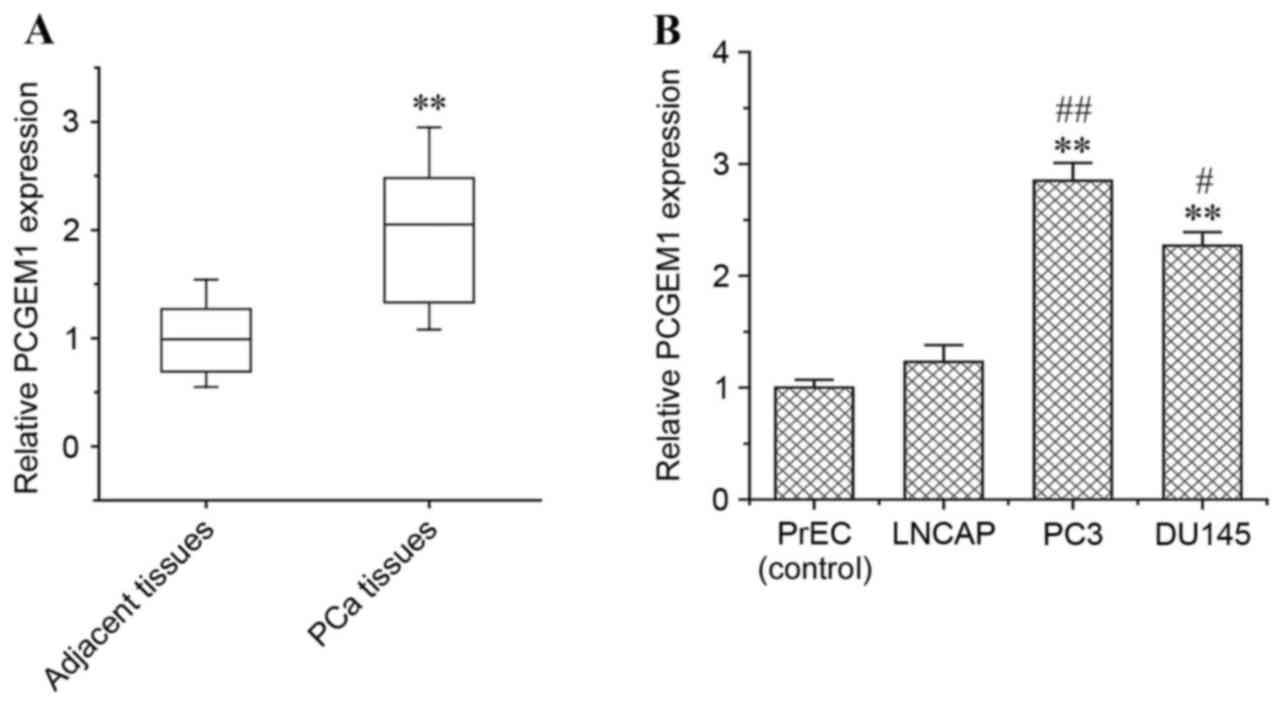
PCGEM1 is overexpressed in prostate
tumor tissues and cell lines
To determine the difference in PCGEM1 expression in
prostate cancer vs normal prostate, semi-quantitative RT-PCR
analysis was performed in prostate cancer tissue samples and
adjacent non-cancerous tissue samples (Fig. 1A). PCGEM1 mRNA expression was
significantly higher in prostate carcinoma tissues than in the
adjacent non-cancerous tissues (P<0.01; Fig. 1A), which is consistent with a
previous report demonstrating that PCGEM1, a prostate specific
gene, is overexpressed in prostate cancer (20). In addition, PCGEM1 mRNA expression
was investigated in a normal prostate epithelial cell line (PrEC),
in the LNCaP hormone-sensitive prostate cancer cell line, and in
the PC3 and DU145 hormone-refractory prostate cancer cell lines.
PCGEM1 mRNA expression was significantly higher in PC3 and DU145
cells compared with LNCaP cells and with the normal PrEC cells
(Fig. 1B). In addition, PC3 cells
expressed the highest levels of PCGEM1 mRNA among the cell lines
tested (Fig. 1B). Thus, the PC3
cell line was chosen for further examinations of the function of
PCGEM1 in prostate cancer.
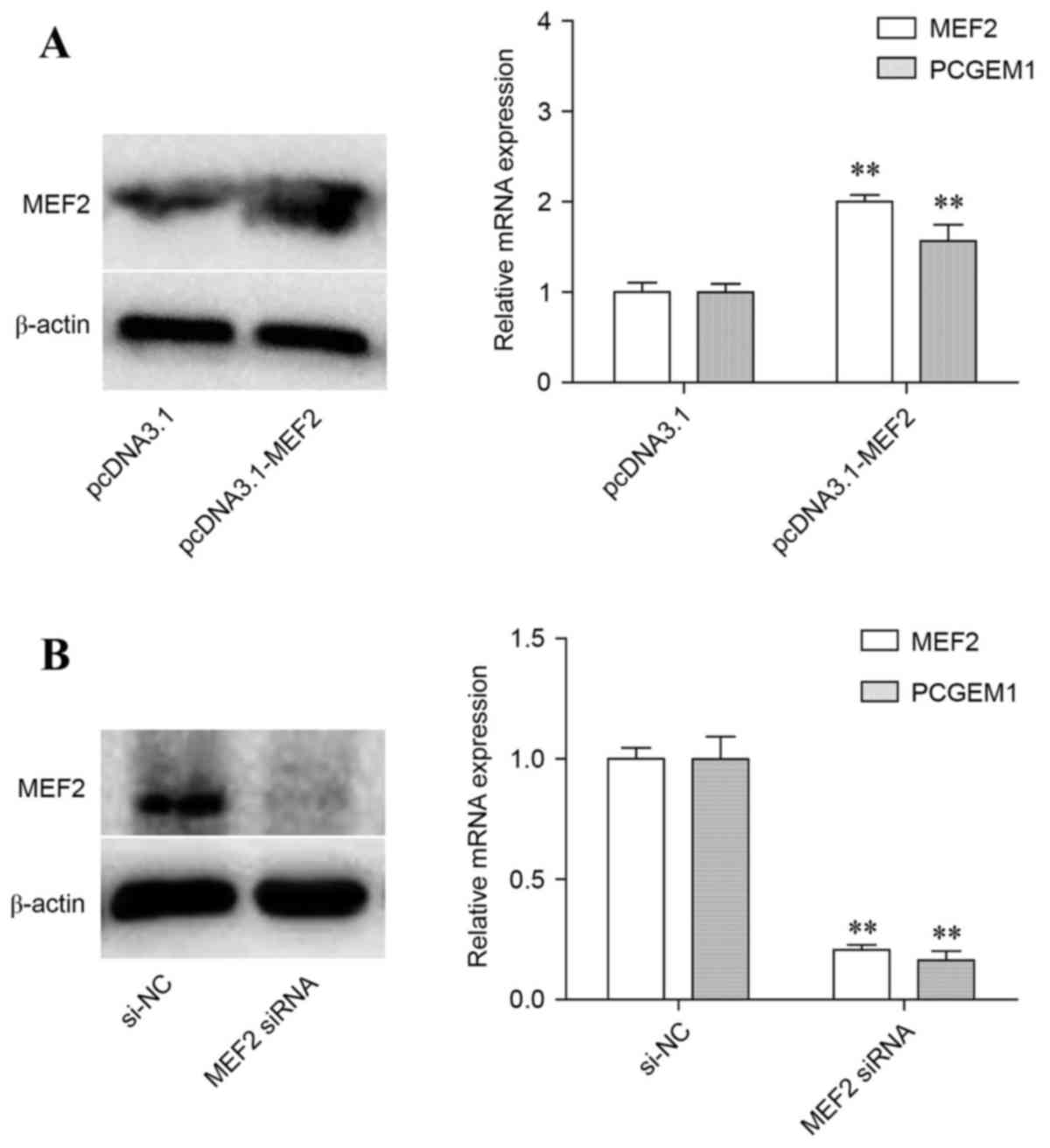
MEF2 effect on PCGEM1 expression
MEF2 expression was evaluated at the mRNA and
protein level in PC3 cells following transfection with
pcDNA3.1-MEF2 overexpression vector or with MEF2 siRNA, in order to
confirm successful overexpression or silencing respectively. PC3
cells transfected with empty pcDNA3.1 vector or non-targeting siRNA
were used as the respective controls. As expected, MEF2 mRNA and
protein levels were significantly increased by pcDNA3.1-MEF2
transfection compared with control (Fig. 2A). Similarly, MEF2 mRNA and protein
expression was markedly decreased following MEF2 siRNA
transfection, compared with control (Fig. 2B). The mRNA expression levels of
PCGEM1 were then analyzed. The results demonstrated that PCGEM1
mRNA expression was significantly increased by MEF2 overexpression
(Fig. 2A), but significantly
decreased by MEF2 silencing (Fig.
2B), suggesting that MEF2 regulated expression of PCGEM1.
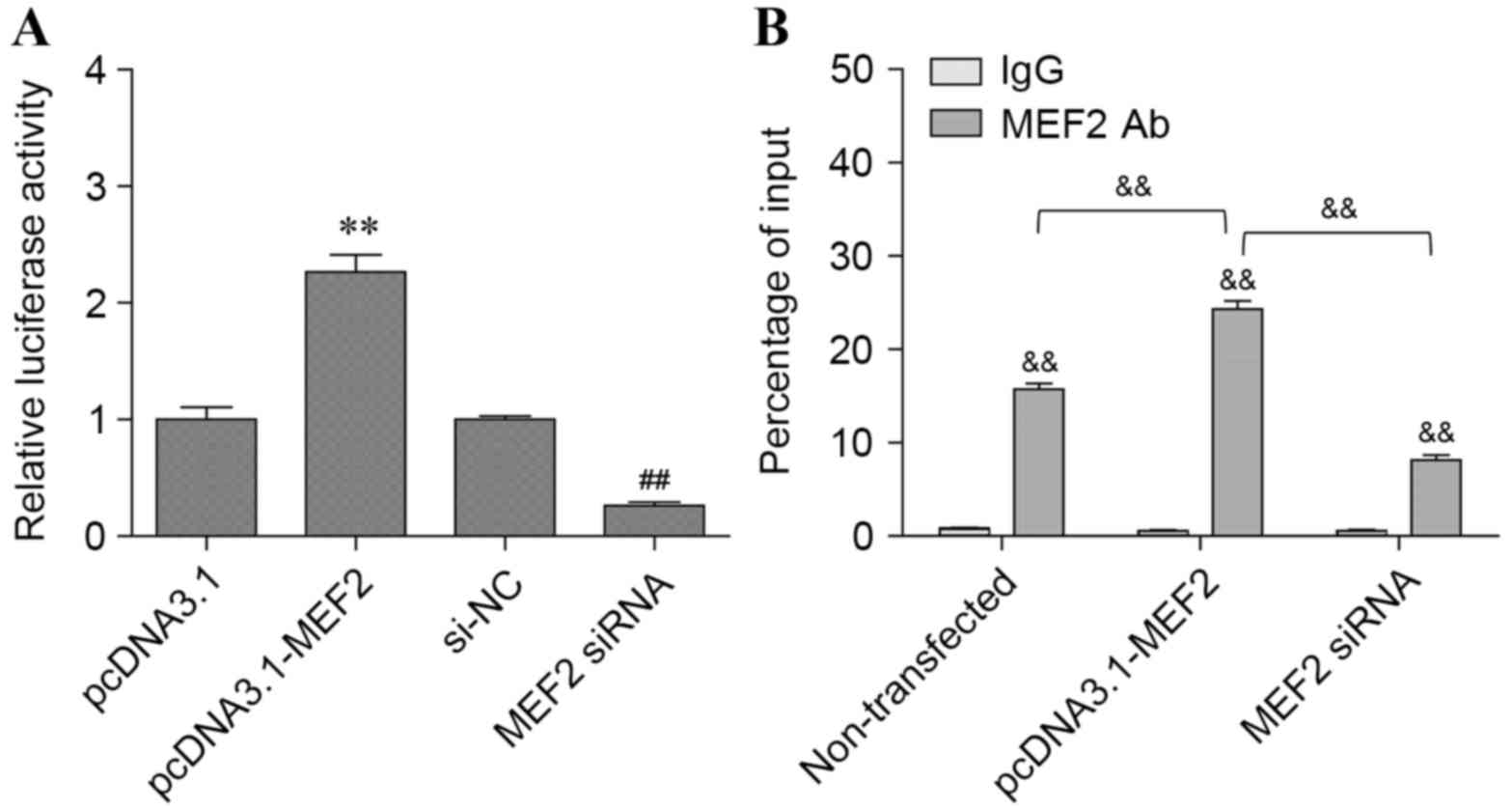
MEF2 effect on PCGEM1 promoter
activity
Previous studies have indicated that MEF2 is
differentially expressed by androgen exposure and that MEF2 binding
sites are present in the promoters of co-expressed genes (26). Therefore, it was hypothesized that
MEF2 could regulate PCGEM1 expression by directly interacting with
its promoter and driving gene transcription. To test this
hypothesis, pcDNA3.1 and pcDNA3.1-MEF2 vectors were transfected
into PC3 cells to induce MEF2 overexpression, and transcription
efficiency by the PCGEM1 promoter was determined by luciferase
assay (Fig. 3A). The results
demonstrated that MEF2 overexpression increased PCGEM1 promoter
activity ~2.0–2.5-fold compared with control (P<0.01; Fig. 3A). By contrast, promoter activity
was significantly reduced in PC3 cells transfected with MEF2 siRNA
compared with control (P<0.01; Fig.
3A).
To validate the existence of MEF2 binding sites on
the PCGEM1 promoter, ChIP analysis was used to evaluate its
enrichment. As expected, PCGEM1 promoter sequences were highly
enriched in PC3 cells immunoprecipitated with the MEF2 specific
antibody compared with IgG control (P<0.01; Fig. 3B). To determine whether PCGEM1
enrichment was dependent on MEF2, ChIP analysis was repeated
following MEF2 overexpression or silencing (Fig. 3B). The results indicated that
enrichment of PCGEM1 promoter in pcDNA3.1-MEF2 transfected cells
was increased by ~2-fold compared with control cells (P<0.01;
Fig. 3B). In addition, PCGEM1
enrichment was significantly decreased in MEF2 siRNA transfected
cells compared with control (P<0.01; Fig. 3B). In conclusion, the present
results indicated that MEF2 activated the lncRNA PCGEM1 expression
via targeting its promoter.
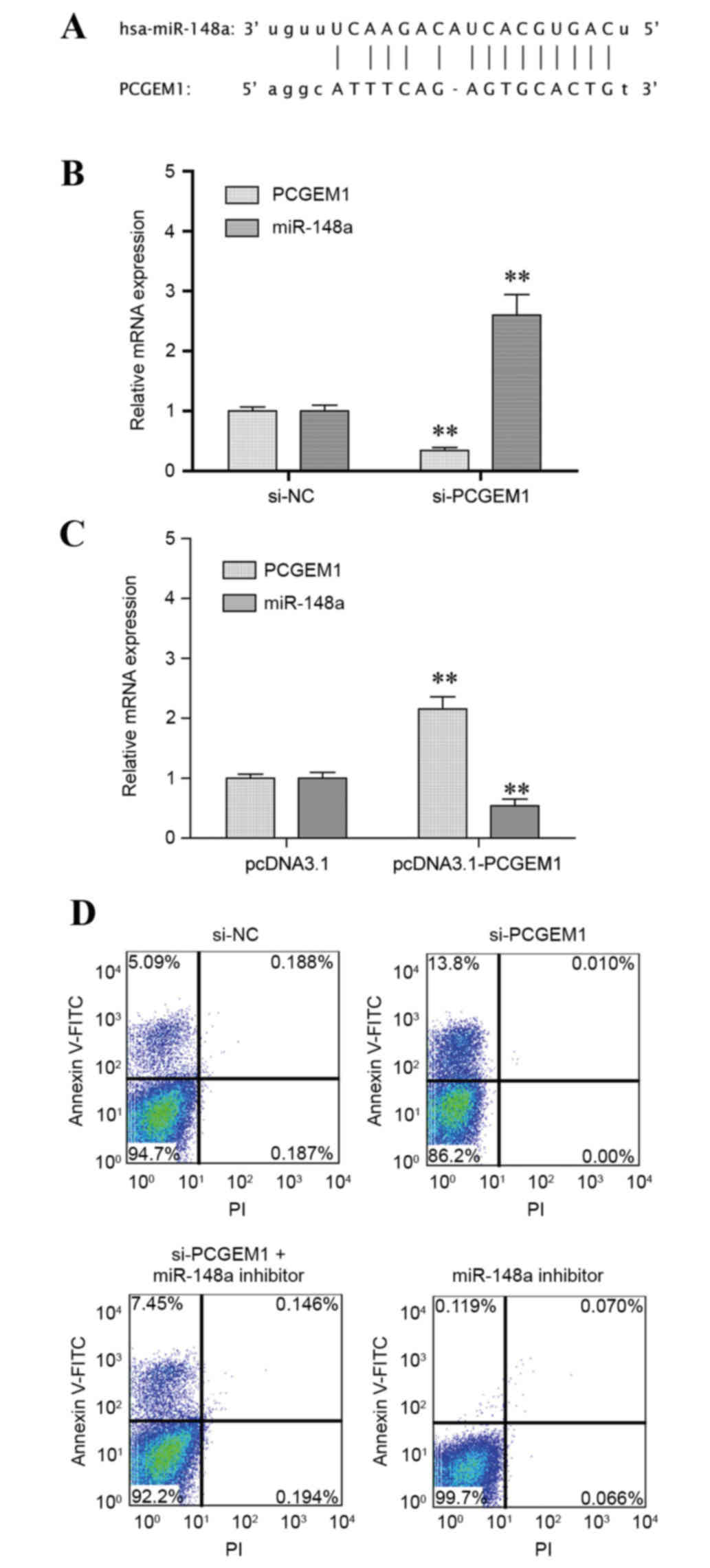
Identification of miR-148a as a target
of PCGEM1
To elucidate the molecular mechanism underlying the
role of PCGEM1 in prostate cancer cells, the effect of PCGEM1 in
regulation of miR-148a expression was analyzed. Based on the
web-based RegRNA analysis for prediction of functional RNA motifs
(32), a putative PCGEM1 binding
site was identified in the 5′ untranslated region (UTR) of miR-148a
(Fig. 4A). To confirm the
regulation of miR-148a by PCGEM1, PC3 cells were transfected with
PCGEM1 siRNA and miR-148a expression was analyzed. Expression of
miR-148a was significantly increased following PCGEM1 silencing
compared with control (P<0.05; Fig.
4B). By contrast, when PCGEM1 was overexpressed, miR-148a
expression was significantly downregulated compared with control
(P<0.05; Fig. 4C). The results
indicated a negative regulation between PCGEM1 and miR-148a.
Finally, the effect of PCGEM1 and miR-148a on PC3 cell apoptosis
was evaluated by flow cytometry (Fig.
4D). The results demonstrated that the number of early-stage
apoptotic cells was increased in the PCGEM1 siRNA-transfected cells
compared with control (13.8% vs. 5.09%, respectively; Fig. 4D). However, when cells were
additionally treated with a miR-148a inhibitor, the cell apoptosis
rate was reduced compared with PCGEM1 siRNA transfection alone
treatment (7.45% vs. 13.8%, respectively; Fig. 4D).
Discussion
The present study demonstrated that MEF2-induced
activation of PCGEM1 altered the apoptosis rate in PC3 cancer cells
by downregulating miR-148a. PCGEM1 has been demonstrated to serve a
role in castration-resistant proliferation of cancer cells
(33). It has been reported that
expression of the lncRNA PCGEM1 is significantly higher in prostate
cancer tissues of African-American patients (33). In the present study, it was
demonstrated that this prostate cancer-associated noncoding RNA
gene was also highly expressed in prostate cancer tissues of Asian
patients. PCGEM1 expression was significantly elevated in tumor
tissues compared with non-cancerous tissues, which was in
accordance with a previous report (20).
Although the number of reports related to lncRNAs
has sharply risen in recent years, their role in enhancing cell
growth and promoting cell proliferation still remains to be
elucidated (34–36). The putative PCGEM1 promoter
contains a 5′ flanking region. The interaction between
ligand-induced enhancer and promoter is impaired by depletion of
PCGEM1 (37).
Based on previous bioinformatics analyses, MEF2
transcription factor binding sites are present in the promoters of
co-expressed genes (38). In
addition, various members of the MEF2 family of transcription
factors have been detected in diverse cell types and display an
important regulatory role in cell development and differentiation
(39). Proteins of the MEF2 family
are calcium-dependent regulators of cell division, differentiation
and death (40). The regulatory
function of MEF2 in accelerating myeloid leukemia has also been
confirmed. However, its role in multiple human cancers remains
largely unknown (41). In the
present study, the interaction between MEF2 and PCGEM1 was assessed
and MEF2 was demonstrated to positively regulate PCGEM1 expression
by targeting the PCGEM1 promoter. MEF2 directly bound to the
promoter of PCGEM1 and enhanced its activity. Taken together, MEF2
regulated the expression of PCGEM1 at the mRNA level by activating
transcription. Further studies will be required to fully explore
their reciprocal regulation and the underlying mechanisms.
Of note, miR-148a has been identified as a tumor
suppressor in human cancer cell lines (40). Its expression was significantly
reduced in the PC3 and DU145 hormone-refractory prostate cancer
cell lines compared with the PrEC normal prostate epithelial cell
line and the LNCaP hormone-sensitive prostate cancer cell line
(42). Using the prediction
analysis RegRNA software (43) and
an online prostate cancer genomic database (cbio.mskcc.org/cancergenomics/prostate/data) (44,45),
a complementary sequence of miR-148a was identified against the
5′-UTR of PCGEM1. In order to understand the regulation of miR-148a
expression by PCGEM1, their interaction was further examined
following PCGEM1 overexpression or silencing. RT-qPCR results
revealed that PCGEM1 silencing in PC3 cells significantly elevated
miR-148a expression. By contrast, PCGEM1 overexpression in PC3
cells resulted in miR-148a downregulation, indicating that miR-148a
expression was regulated by a PCGEM1-dependent mechanism. Apoptosis
of PC3 cancer cells was also evaluated by flow cytometry. PCGEM1
silencing increased the number of PC3 apoptotic cells, while
simultaneous treatment with a miR-148a inhibitor reduced cell
apoptosis. Thus, downregulation of miR-148a mediated by the lncRNA
PCGEM1 may be a potential strategy promoting cell proliferation in
prostate cancer cells.
In conclusion, the present study demonstrated a
reciprocal regulation between MEF2 and PCGEM1. MEF2 enhanced the
activity of the PCGEM1 promoter and upregulated PCGEM1 expression.
Furthermore, the tumor-promoting lncRNA PCGEM1 promoted
downregulation of the tumor suppressor miR-148a, resulting in
reduced cell apoptosis in PC3 prostate cancer cells. In conclusion,
it was demonstrated that lncRNA PCGEM1 and miR-148a may be novel
biomarkers and targets for the early prevention and treatment of
prostate cancer.
References
|
1
|
Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li
YJ, Yan B and Jiang Q: Pathogenic role of lncRNA-MALAT1 in
endothelial cell dysfunction in diabetes mellitus. Cell Death Dis.
5:e15062014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He JH, Zhang JZ, Han ZP, Wang L, Lv YB and
Li YG: Reciprocal regulation of PCGEM1 and miR-145 promote
proliferation of LNCaP prostate cancer cells. J Exp Clin Cancer
Res. 33:722014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang
XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al: Human colorectal
cancer-specific CCAT1-L lncRNA regulates long-range chromatin
interactions at the MYC locus. Cell Res. 24:513–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jin M, Zhang T, Liu C, Badeaux MA, Liu B,
Liu R, Jeter C, Chen X, Vlassov AV and Tang DG: miRNA-128
suppresses prostate cancer by inhibiting BMI-1 to inhibit
tumor-initiating cells. Cancer Res. 74:4183–4195. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rathod SS, Rani SB, Khan M, Muzumdar D and
Shiras A: Tumor suppressive miRNA-34a suppresses cell proliferation
and tumor growth of glioma stem cells by targeting Akt and Wnt
signaling pathways. FEBS Open Bio. 4:485–495. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishihara T, Seki N, Inoguchi S, Yoshino H,
Tatarano S, Yamada Y, Itesako T, Goto Y, Nishikawa R, Nakagawa M
and Enokida H: Expression of the tumor suppressive miRNA-23b/27b
cluster is a good prognostic marker in clear cell renal cell
carcinoma. J Urol. 192:1822–1830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen Z, Tang ZY, He Y, Liu LF, Li DJ and
Chen X: miRNA-205 is a candidate tumor suppressor that targets ZEB2
in renal cell carcinoma. Oncol Res Treat. 37:658–664. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Babae N, Bourajjaj M, Liu Y, Van Beijnum
JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH,
Van Haastert RJ, et al: Systemic miRNA-7 delivery inhibits tumor
angiogenesis and growth in murine xenograft glioblastoma.
Oncotarget. 5:6687–6700. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jalali S, Bhartiya D, Lalwani MK,
Sivasubbu S and Scaria V: Systematic transcriptome wide analysis of
lncRNA-miRNA interactions. PLoS One. 8:e538232013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang K, Sun T, Li N, Wang Y, Wang JX, Zhou
LY, Long B, Liu CY, Liu F and Li PF: MDRL lncRNA regulates the
processing of miR-484 primary transcript by targeting miR-361. PLoS
Genet. 10:e10044672014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang F, Ying HQ, He BS, Pan YQ, Deng QW,
Sun HL, Chen J, Liu X and Wang SK: Upregulated lncRNA-UCA1
contributes to progression of hepatocellular carcinoma through
inhibition of miR-216b and activation of FGFR1/ERK signaling
pathway. Oncotarget. 6:7899–7917. 2015.PubMed/NCBI
|
|
14
|
Zhang R, Li M, Zang W, Chen X, Wang Y, Li
P, Du Y, Zhao G and Li L: MiR-148a regulates the growth and
apoptosis in pancreatic cancer by targeting CCKBR and Bcl-2. Tumour
Biol. 35:837–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang H, Li Y, Huang Q, Ren X, Hu H, Sheng
H and Lai M: MiR-148a promotes apoptosis by targeting Bcl-2 in
colorectal cancer. Cell Death Differ. 18:1702–1710. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lombard AP, Mooso BA, Libertini SJ, Lim
RM, Nakagawa RM, Vidallo KD, Costanzo NC, Ghosh PM and Mudryj M:
miR-148a dependent apoptosis of bladder cancer cells is mediated in
part by the epigenetic modifier DNMT1. Mol Carcinog. 55:757–767.
2016. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao S, Wen Z, Liu S, Liu Y, Li X, Ge Y
and Li S: MicroRNA-148a inhibits the proliferation and promotes the
paclitaxel-induced apoptosis of ovarian cancer cells by targeting
PDIA3. Mol Med Rep. 12:3923–3939. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhu A, Xia J, Zuo J, Jin S, Zhou H, Yao L,
Huang H and Han Z: MicroRNA-148a is silenced by hypermethylation
and interacts with DNA methyltransferase 1 in gastric cancer. Med
Oncol. 29:2701–2709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heo MJ, Kim YM, Koo JH, Yang YM, An J, Lee
SK, Lee SJ, Kim KM, Park JW and Kim SG: microRNA-148a dysregulation
discriminates poor prognosis of hepatocellular carcinoma in
association with USP4 overexpression. Oncotarget. 5:2792–2806.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Srikantan V, Zou Z, Petrovics G, Xu L,
Augustus M, Davis L, Livezey JR, Connell T, Sesterhenn IA, Yoshino
K, et al: PCGEM1, a prostate-specific gene, is overexpressed in
prostate cancer. Proc Natl Acad Sci USA. 97:12216–12221. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Petrovics G, Zhang W, Makarem M, Street
JP, Connelly R, Sun L, Sesterhenn IA, Srikantan V, Moul JW and
Srivastava S: Elevated expression of PCGEM1, a prostate-specific
gene with cell growth-promoting function, is associated with
high-risk prostate cancer patients. Oncogene. 23:605–611. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fu X, Ravindranath L, Tran N, Petrovics G
and Srivastava S: Regulation of apoptosis by a prostate-specific
and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell
Biol. 25:135–141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vargas MA, Tirnauer JS, Glidden N,
Kapiloff MS and Dodge-Kafka KL: Myocyte enhancer factor 2 (MEF2)
tethering to muscle selective A-kinase anchoring protein (mAKAP) is
necessary for myogenic differentiation. Cell Signal. 24:1496–1503.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pallavi SK, Ho DM, Hicks C, Miele L and
Artavanis-Tsakonas S: Notch and Mef2 synergize to promote
proliferation and metastasis through JNK signal activation in
Drosophila. EMBO J. 31:2895–2907. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Black BL, Molkentin JD and Olson EN:
Multiple roles for the MyoD basic region in transmission of
transcriptional activation signals and interaction with MEF2. Mol
Cell Biol. 18:69–77. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Coutinho-Camillo CM, Salaorni S, Sarkis AS
and Nagai MA: Differentially expressed genes in the prostate cancer
cell line LNCaP after exposure to androgen and anti-androgen.
Cancer Genet Cytogenet. 166:130–138. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Manteniotis S, Lehmann R, Flegel C, Vogel
F, Hofreuter A, Schreiner BS, Altmüller J, Becker C, Schöbel N,
Hatt H and Gisselmann G: Comprehensive RNA-Seq expression analysis
of sensory ganglia with a focus on ion channels and GPCRs in
Trigeminal ganglia. PLoS One. 8:e795232013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanigan MH, Gallagher BC, Townsend DM and
Gabarra V: Gamma-glutamyl transpeptidase accelerates tumor growth
and increases the resistance of tumors to cisplatin in vivo.
Carcinogenesis. 20:553–559. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Daems C, Martin LJ, Brousseau C and
Tremblay JJ: MEF2 is restricted to the male gonad and regulates
expression of the orphan nuclear receptor NR4A1. Mol Endocrinol.
28:886–898. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang QY and Feng MG: DPS Data Processing
System for Practical Statistics. Science Press; Beijing, China:
2002
|
|
32
|
Chang TH, Huang HY, Hsu JB, Weng SL, Horng
JT and Huang HD: An enhanced computational platform for
investigating the roles of regulatory RNA and for identifying
functional RNA motifs. BMC Bioinformatics. 14 Suppl 2:S42013.
|
|
33
|
Park JY, Lee JE, Park JB, Yoo H, Lee SH
and Kim JH: Roles of Long Non-Coding RNAs on Tumorigenesis and
Glioma Development. Brain Tumor Res Treat. 2:1–6. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jiang H, Wang Y, Ai M, Wang H, Duan Z,
Wang H, Zhao L, Yu J, Ding Y and Wang S: Long noncoding RNA CRNDE
stabilized by hnRNPUL2 accelerates cell proliferation and migration
in colorectal carcinoma via activating Ras/MAPK signaling pathways.
Cell Death Dis. 8:e28622017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang B, Luo T, Zhang M, Lu Z, Xue X and
Fang G: The novel long noncoding RNA RP11-357H14.17 acts as an
oncogene by promoting cell proliferation and invasion in
diffuse-type gastric cancer. Onco Targets Ther. 10:2635–2643. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen S, Shao C, Xu M, Ji J, Xie Y, Lei Y
and Wang X: Macrophage infiltration promotes invasiveness of breast
cancer cells via activating long non-coding RNA UCA1. Int J Clin
Exp Pathol. 8:9052–9061. 2015.PubMed/NCBI
|
|
37
|
Ho TT, Huang J, Zhou N, Zhang Z, Koirala
P, Zhou X, Wu F, Ding X and Mo YY: Regulation of PCGEM1 by p54/nrb
in prostate cancer. Sci Rep. 6:345292016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sacilotto N, Chouliaras KM, Nikitenko LL,
Lu YW, Fritzsche M, Wallace MD, Nornes S, García-Moreno F, Payne S,
Bridges E, et al: MEF2 transcription factors are key regulators of
sprouting angiogenesis. Genes Dev. 30:2297–2309. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schroeter A, Walzik S, Blechschmidt S,
Haufe V, Benndorf K and Zimmer T: Structure and function of splice
variants of the cardiac voltage-gated sodium channel Na(v)1.5. J
Mol Cell Cardiol. 49:16–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
McKinsey TA, Zhang CL and Olson EN: MEF2:
A calcium-dependent regulator of cell division, differentiation and
death. Trends Biochem Sci. 27:40–47. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Laddha SV, Nayak S, Paul D, Reddy R,
Sharma C, Jha P, Hariharan M, Agrawal A, Chowdhury S, Sarkar C and
Mukhopadhyay A: Genome-wide analysis reveals downregulation of
miR-379/miR-656 cluster in human cancers. Biol Direct. 8:102013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fujita Y, Kojima K, Ohhashi R, Hamada N,
Nozawa Y, Kitamoto A, Sato A, Kondo S, Kojima T, Deguchi T and Ito
M: MiR-148a attenuates paclitaxel resistance of hormone-refractory,
drug-resistant prostate cancer PC3 cells by regulating MSK1
expression. J Biol Chem. 285:19076–19084. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chang TH, Huang HY, Hsu JB, Weng SL, Horng
JT and Huang HD: An enhanced computational platform for
investigating the roles of regulatory RNA and for identifying
functional RNA motifs. BMC bioinformatics. 14 Suppl 2:S42013.
|
|
44
|
Robinson JT, Thorvaldsdóttir H, Winckler
W, Guttman M, Lander ES, Getz G and Mesirov JP: Integrative
genomics viewer. Nat Biotechnol. 29:24–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Thorvaldsdóttir H, Robinson JT and Mesirov
JP: Integrative Genomics Viewer (IGV): High-performance genomics
data visualization and exploration. Brief Bioinform. 14:178–192.
2013. View Article : Google Scholar : PubMed/NCBI
|