|
1
|
McDougall C, Hurd K and Barnabe C:
Systematic review of rheumatic disease epidemiology in the
indigenous populations of Canada, the United States, Australia, and
New Zealand. Semin Arthritis Rheum. 46:675–686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bijlsma JW, Berenbaum F and Lafeber FP:
Osteoarthritis: An updata with relevance for clinical practice.
Lancet. 377:2115–2126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sulzbacher I: Osteoarthritis: Histology
and pathogenesis. Wien Med Wochenschr. 163:212–219. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hashimoto S, Ochs RL, Komiya S and Lotz M:
Linkage of chondrocyte apoptosis and cartilage degradation in human
osteoarthritis. Arthritis Rheum. 41:1632–1638. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schroeppel JP, Crist JD, Anderson HC and
Wang J: Molecular regulation of articular chondrocyte function and
its significance in osteoarthritis. Histol Histopathol. 26:377–394.
2011.PubMed/NCBI
|
|
6
|
Kleine SA and Budsberg SC: Synovial
membrane receptors as therapeutic targets: A review of receptor
localization, structure, and function. J Orthop Res. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Hunter DJ, Jin X and Ding C: The
importance of synovial inflammation in osteoarthritis: Current
evidence from imageing assessments and clinical trial.
Osteoarthritis Cartilage. 26:165–167. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhattaram P and Chandrasekharan U: The
joint synovium: A critical determinant of articular cartilage fate
in inflammatory joint diseases. Semin Cell Dev Biol. 62:86–93.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scanzello CR and Goldring SR: The role of
synovitis in osteoarthritis pathogenesis. Bone. 51:249–257. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goldenberg DL and Cohen AS: Synovial
membrane histopathology in the differential diagnosis of rheumatoid
arthritis, gout, pseudogout, systemic lupus erythematosus,
infectious arthritis and degenerative joint disease. Medicine
(Baltimore). 57:239–252. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sellam J and Berenbaum F: The role of
synovitis in pathophysiology and clinical symptoms of
osteoarthritis. Nat Rev Rheumatol. 6:625–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Myers SL, Brandt KD, Ehlich JW, Braunstein
EM, Shelbourne KD, Heck DA and Kalasinski LA: Synovial inflammation
in patients with early osteoarthritis of the knee. J Rheumatol.
17:1662–1669. 1990.PubMed/NCBI
|
|
13
|
Ayral X, Pickering EH, Woodworth TG,
Mackillop N and Dougados M: Synovitis: A potential predictive
factor of structural progression of medial tibiofemoral knee
osteoarthritis-results of a 1 year longitudinal arthroscopic study
in 422 patients. Osteoarthritis Cartilage. 13:361–367. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fernandez-Madrid F, Karvonen RL, Teitge
RA, Miller PR, An T and Negendank WG: Synovial thickening detected
by MR imaging in osteoarthritis of the knee confirmed by biopsy as
synovitis. Magn Reson Imaging. 13:177–183. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Felson DT, Niu J, Neoqi T, Goggins J,
Nevitt MC, Roemer F, Torner J, Lewis CE and Guermazi A: MOST
Investigators Group. Synovitis and the risk of knee osteoarthritis:
The MOST Study. Osteoarthritis Cartilage. 24:458–464. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen D, Shen J, Zhao W, Wang T, Han L,
Hamilton JL and Lm HJ: Osteoarthritis: Toward a comprehensive
understanding of pathological mechanism. Bone Res. 5:160442017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li M, Zhi L, Zhang Z, BIan W and Qiu Y:
Identification of potential target genes associated with the
pathogenesis of osteoarthritis using microarray based analysis. Mol
Med Rep. 16:2799–2806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kong R, Gao J, Si Y and Zhao D:
Combination of circulating miR-19b-3p, miR-122-5p and miR-486-5p
expressions correlates with risk and disease severity of knee
osteoarthritis. Am J Transl Res. 9:2852–2864. 2017.PubMed/NCBI
|
|
19
|
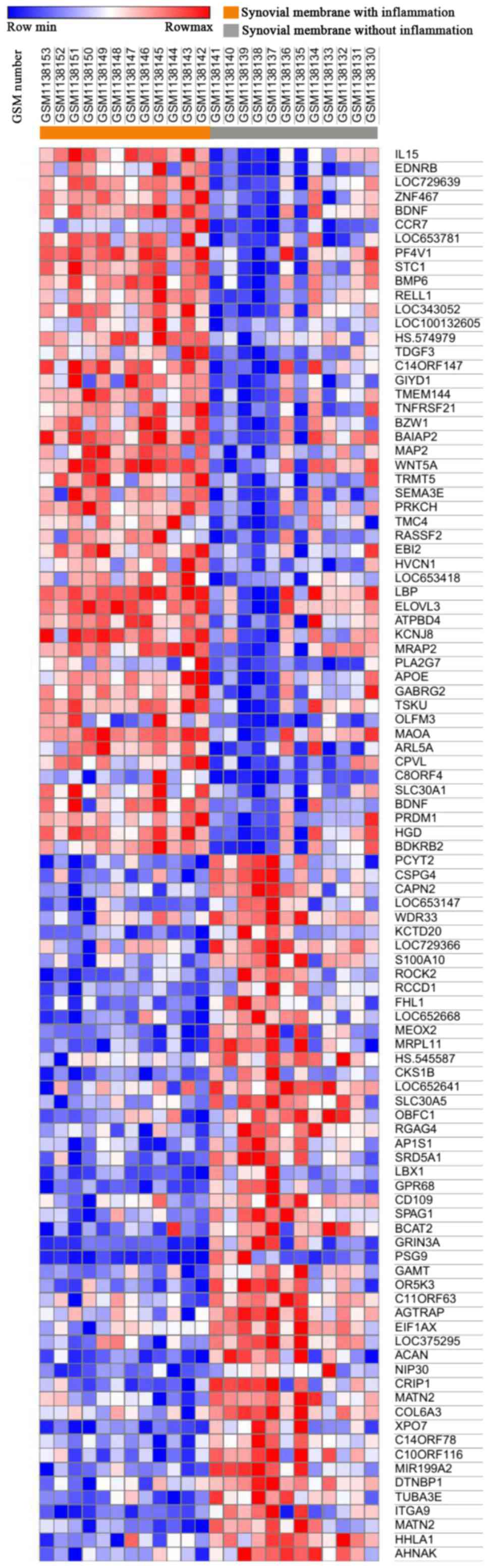
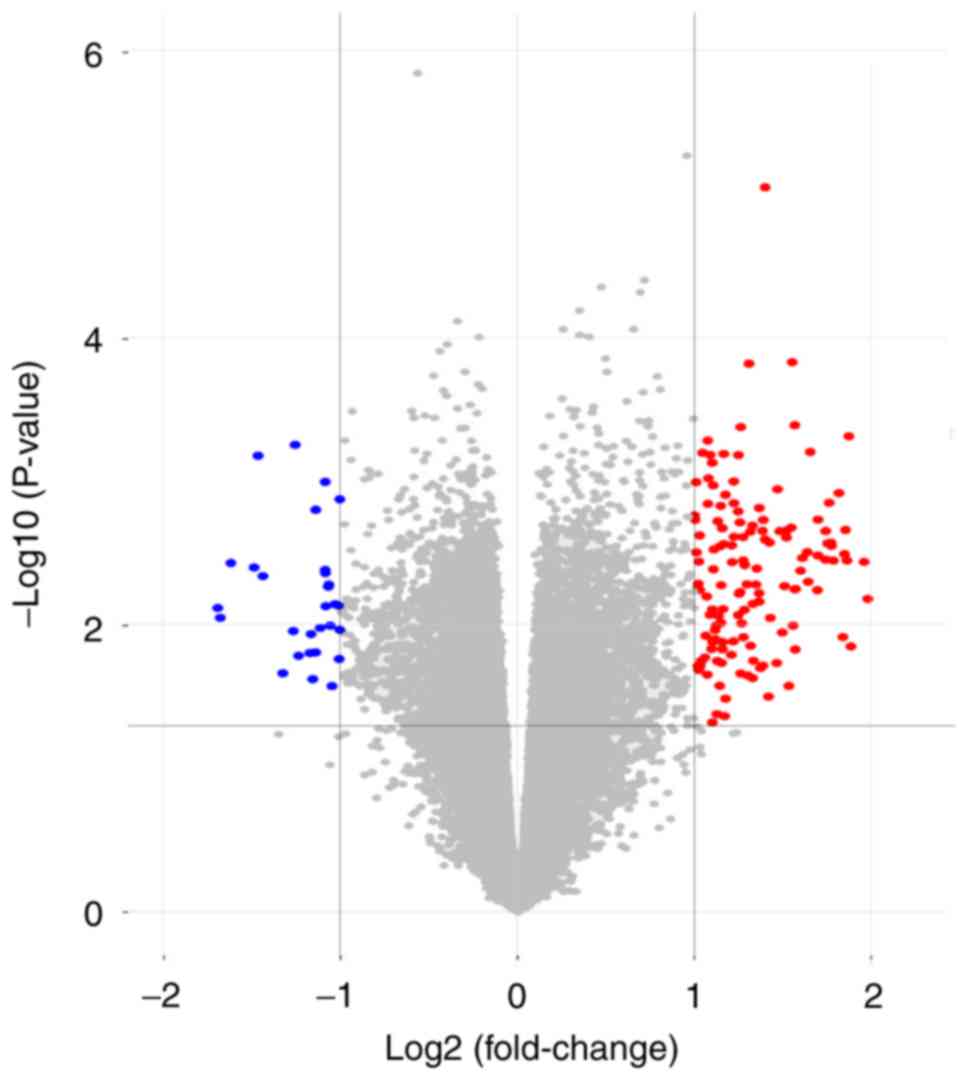
Lambert C, Dubuc JE, Montell E, Verges J,
Munaut C, Noel A and Henrotin Y: Gene expression pattern of cell
from inflamed and normal areas of osteoarthritis synovial membrane.
Arthritis Rheumatol. 66:960–968. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
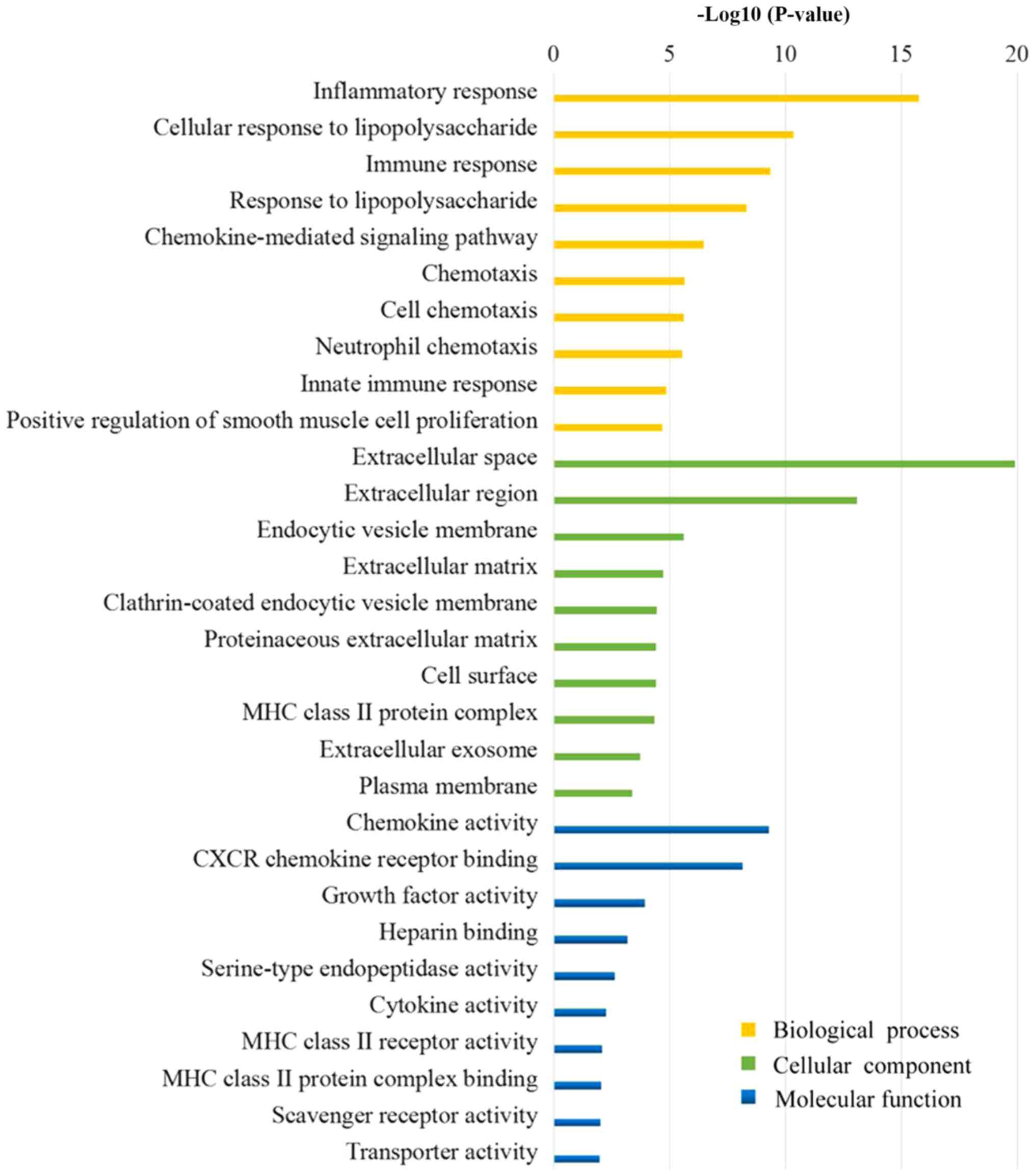
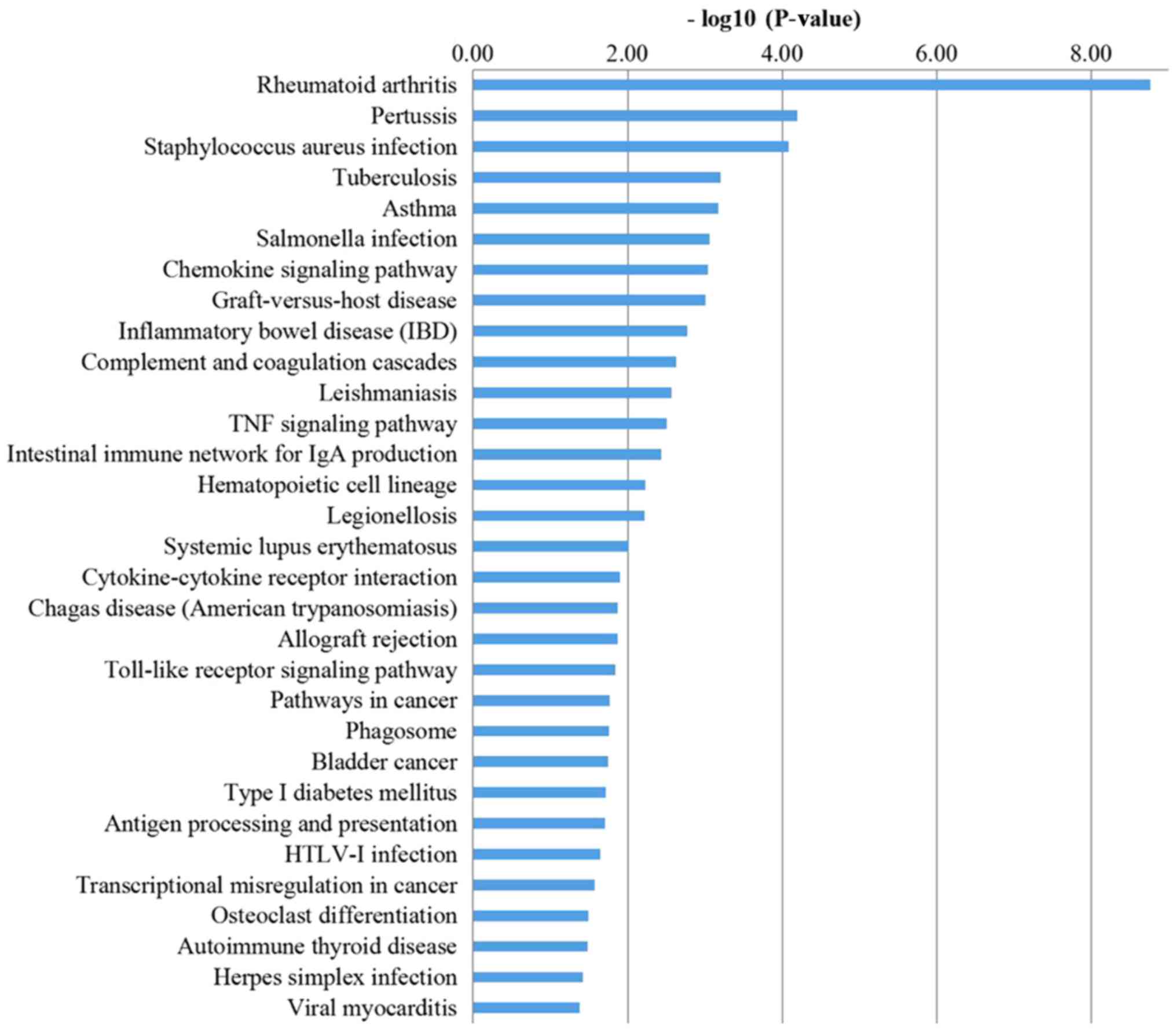
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:(Database Issue). D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li YH, Yu CY, Li XX, Zhang P, Tang J, Yang
Q, Fu T, Zhang X, Cui X, Tu G, et al: Therapeutic target database
update 2018: Enriched resource for facilitating bench-to-clinic
research of targeted therapeutics. Nucleic Acids Res.
46:D1121–D1127. 2018.PubMed/NCBI
|
|
24
|
Johnson VL and Hunter DJ: The epidemiology
of osteoarthritis. Best Pract Res Clin Rheumatol. 28:5–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benito MJ, Veale DJ, FitzGerald O, van den
Berg WB and Bresnihan B: Synovial tissue inflammation in early and
late osteoarthritis. Ann Rheum Dis. 64:1263–1267. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lopes EBP, Filiberti A, Husain SA and
Humphrey MB: Immune contributions to osteoarthritis. Curr
Osteoporos Rep. 15:593–600. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kalaitzoglou E, Griffin TM and Humphrey
MB: Innate immune responses and osteoarthritis. Curr Rheumatol Rep.
19:452017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Silawal S, Triebel J, Bertsch T and
Schulze-Tanzil G: Osteoarthritis and the complement cascade. Clin
Med Insights Arthritis Musculoskelet Disord.
11:11795441177514302018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Syx D, Tran PB, Miller RE and Malfait AM:
Peripheral mechanisms contributing to osteoarthritis pain. Curr
Rheumatol Rep. 20:92018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nguyen LT, Sharma AR, Chakraborty C,
Saibaba B, Ahn ME and Lee SS: Review of prospects of biological
fluid biomarkers in osteoarthritis. Int J Mol Sci. 18:pii: E601.
2017. View Article : Google Scholar
|
|
31
|
So AK, Varisco PA, Kemkes-Matthes B,
Herkenne-Morard C, Chobaz-Peclat V, Gerster JC and Busso N:
Arthritis is linked to local and systemic activation of coagulation
and fibrinolysis pathways. J Thromb Haemost. 1:2510–2515. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brocker C, Thompson D, Matsumoto A, Nebert
DW and Vasiliou V: Evolutionary divergence and functions of the
human interlrukin (IL) gene family. Hum Genomics. 5:30–55. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang F, Zhou S, Wang C, Huang Y, Li H,
Wang Y, Zhu Z, Tang J and Yan M: Epigenetic modifications of
interleukin-6 in synovial fibroblasts from osteoarthritis patients.
Sci Rep. 7:435922017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nair A, Gan J, Bush-Joseph C, Verma N,
Tetreault MW, Saha K, Margulis A, Fogg L and Scanzello CR: Synovial
chemokine expression and relationship with knee symptoms in
patients with meniscal tears. Osteoarthritis Cartilage.
23:1158–1164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rohani MG and Parks WC: Matrix remodeling
by MMps during wound repair. Matrix Biol. 44–46:113–121. 2015.
View Article : Google Scholar
|
|
36
|
Kanyama M, Kuboki T, Kojima S, Fujisawa T,
Hattori T, Takigawa M and Yamashita A: Matrix metalloproteinases
and tissue inhibitors of metalloproteinases in synovial fluids of
patients with temporomandibular joint osteoarthritis. J Orofac
Pain. 14:20–30. 2000.PubMed/NCBI
|
|
37
|
Yang CC, Lin CY, Wang HS and Lyu SR:
Matrix metalloproteases and tissue inhibitors to knee
osteoarthritis progression. PLoS One. 8:e796622013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meyers MJ, Pelc M, Kamtekar S, Day J, Poda
GI, Hall MK, Michener ML, Reitz BA, Mathis KJ, Pierce BS, et al:
Structure-based drug design enables conversion of a DFG-in bingding
CSF-1R kinase inhibitor to a DFG-out binding mode. Bioorg Med Chem
Lett. 20:1543–1547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Campbell IL, Rich MJ, Bioschof RJ and
Hamilton JA: The colony-stimulating factors and collagen-induced
arthritis: Exacerbation of disease by M-CSF and G-CSF and
requirement for endogenous M-CSF. J Leukoc Biol. 68:144–150.
2000.PubMed/NCBI
|
|
40
|
Chung L: A brief introduction to the
transduction of neural activity into fos signal. Dev Reprod.
19:61–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kinne RW, Boehm S, Iftner T, Aigner T,
Vornehm S, Weseloh G, Bravo R, Emmrich F and Kroczek RA: Synovial
fibroblast-like cells strongly express jun-B and C-fos
proto-oncogenes in rheumatoid- and osteoarthritis. Scand J
Rheumatol Suppl. 101:121–125. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Annunziata M, Granata R and Ghiqo E: The
IGF system. Acta Diabetol. 48:1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schneiderman R, Rosenberg N, Hiss J, Lee
P, Liu F, Hintz RL and Maroudas A: Concentration and size
distribution of insulin-like growth factor-1 in human normal and
osteoarthritis synovial fluid and cartilage. Arch Biochem Biophys.
324:173–188. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tomasello E and Vivier E:
KARAP/DAP12/TYROBP: Three names and a multiplicity of biological
functions. Eur J Immunol. 35:1670–1677. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Crotti YN, Dharmapatni AA, Alias E,
Zannettino AC, Smith MD and Haynes DR: The immunoreceptor
tyrosine-based activation motif (IYAM)-related factors are
increased in synovial tissue and vasculature of rheumatoid
arthritic joints. Arthritis Res Ther. 14:R2452012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Daghestani HN, Pieper CF and Kraus VB:
Solube macrophage biomarkers indicate inflammatory phenotypes in
patients with knee osteoarthritis. Arthritis Rheumatol. 67:956–965.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nair A, Kanda V, Bush-Joseph C, Verma N,
Chubinskaya S, Mikecz K, Glant TT, Malfair AM, Crow MK, Spear GT,
et al: Synovial fluid from patients with early osteoarthritis
modulates fibroblast-like synoviocyte responses to toll-like
recepyor 4 and toll-like receptor 2 ligands via soluble CD14.
Arthritis Rheum. 64:2268–2277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fabriek BO, Dijkstra CD and van den Berg
TK: The macrophage scavenger receptor CD163. Immunobiology.
210:153–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim JR, Yoo JJ and Kim HA: Therapeutics in
osteoarthritis based on an understanding of its molecular
pathogenesis. Int J Mol Sci. 19:E6742018. View Article : Google Scholar : PubMed/NCBI
|