Introduction
Osteoarthritis (OA) is one of the most common types
of chronic arthritis in elderly people, and has been identified to
affect ≥6–32% people worldwide (1). There are multiple risk factors for OA
development, including abnormal mechanical stress, aging, obesity
and genetic factors (2).
Pathological features of OA primarily include articular cartilage
degeneration, synovial inflammation and atypical bone formation
(3). In the past, research has
primarily focused on cartilage tissue and chondrocytes in the study
of OA mechanisms and treatment (4,5).
With the rapid development of medicine, synovial tissue and
subchondral bone have additionally been investigated. Previous
studies suggested that synovial tissue serves an important role in
OA (6,7). As part of the joint structure, the
synovial membrane may produce and regulate synovial fluid, maintain
joint activity, and be adversely affected in joint diseases
(8). Synovial lesions may
additionally be identified in a number of joint diseases, which may
serve a role in promoting the occurrence and progression of the
disease (9).
Synovitis is an aseptic inflammation that occurs at
the synovial membrane and is observed in rheumatoid arthritis, OA,
lupus OA and gout (10). Synovitis
leads to abnormal synovial fluid production and absorption,
resulting in joint effusion, causing pain, swelling and other
reactions (11). Previous studies
observed synovitis in early OA (12), and that the severity of synovitis
may be associated with different stages of OA (13). Fernandez-Madrid et al
(14) suggested that OA synovitis
is caused by the degeneration of cartilage stimulation. However,
Felson et al (15)
suggested that synovitis occurs not only in the early stages of OA;
however, even prior to imaging. Additionally, the occurrence of
synovitis may further promote cartilage degeneration, which would
in turn exacerbate synovitis (11). Synovitis serves an important role
in the symptoms, progression and development of OA, and is a
concern for the treatment of OA.
With the development of modern biomedicine,
increasing evidence suggested that the occurrence and development
of OA may be mediated by a number of genes and signaling pathways
(16). In order to develop clearer
diagnostic criteria and more effective treatment options, it is
essential to fully understand the molecular mechanism of OA. With
the aim of fully understanding the gene expression alterations in
OA, previous studies used DNA microarray technology to analyze gene
expression profiles (17,18). The results demonstrated that
molecules encoded by differentially expressed genes (DEGs) located
in different cell structures and with different molecular functions
(MF) were associated with different biological processes (BP)
during their involvement in the disease process. The availability
of bioinformatics analysis based on high-throughput technology
enabled the investigation of the alterations in mRNA expression and
the interaction between differential genes in OA, to provide novel
insights for further in-depth OA studies.
The Gene Expression Omnibus (GEO) is a database and
online resource for the gene expression of any species. The present
study obtained genetic microarray dataset no. GSE46750 from GEO.
The samples in GSE46750 were divided into two groups: Synovial
cells with and without inflammation in OA. The two groups were
compared and analyzed to identify the DEGs. Functional enrichment
analysis, protein-protein interaction (PPI) networks and module
analysis were conducted on the DEGs. Subsequently, text-mining of
OA treatment drugs and their target genes were performed to
initially validate the results. The results of the present study
may enable us to recognize the effects of synovial membrane
inflammation in the development of OA, and to provide certain
possible OA target molecules for subsequent validation.
Materials and methods
Gene chip data
GSE46750 gene expression data (19) was obtained from the GEO database
(http://www.ncbi.nlm.nih.gov/geo/), which
was expressed on the GPL10558 platform [(Illumina HumanHT-12 V 4.0)
Bead chip; Illumina, Inc., San Diego, CA, USA]. The GSE46750
dataset samples, which were synovial cells, were derived from 12
patients with OA, specifically from those with synovial membrane
with inflammation (n=12) and synovial membrane without inflammation
(n=12).
Identifying DEGs
The original micro array data was analyzed through
heat mapping using Morpheus (https://software.broadinstitute.org/morpheus/) to
visually observe gene expression. The chip data were divided into
an inflammatory synovial membrane group and a non-inflammatory
synovial membrane group for analysis. GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc=GSE46750)
was used to identify the DEGs in OA synovial membrane. The criteria
for a DEG was |log2 (fold change)|≥1 and P<0.05.
Gene Ontology (GO) enrichment and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis
GO enrichment analysis enables the annotation of
cell components (CC), MF and BP of DEGs. KEGG (http://www.genome.jp/) is a database that includes a
KEGG path database for examining the path of the gene cluster and
associated functions. The present study used Database for
Annotation, Visualization and Integrated Discovery (DAVID 6.8;
http://david.ncifcrf.gov/) (20) to perform GO enrichment analysis and
KEGG pathway analysis. P<0.05 was considered to indicate a
statistically significant difference.
Construction of the PPI network and
module analysis
The Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) database (http://www.string-db.org/) may be searched for
associations between known and predicted proteins, and is commonly
used to predict PPI information in molecular biology (21). The DEGs were mapped to STRING, and
those with interaction scores >0.4 were selected for further
examination. Cytoscape 3.5.0 (http://www.cytoscape.org/) was used to visualize the
results from the PPI network. Module analysis on the PPI network
results was performed using the molecular complex detection (MCODE)
(22) clustering algorithm that
comes with Cytoscape. Module analysis may be used to identify more
connected gene groups. In addition, the module analysis results
were further analyzed for function and pathway enrichment, and
P<0.05 was considered to indicate a statistically significant
difference.
Text-mining
Drugs commonly used for the treatment of OA were
searched in The Therapeutic Target Database (https://db.idrblab.org/ttd/) (23) with the key word ‘osteoarthritis’.
The molecular targets of the detected drugs were identified in The
Drug Gene Interaction database (DGIdb; http://dgidb.genome.wustl.edu/). These known molecules
that may serve a therapeutic role in OA were compared with the
present results.
Results
DEGs
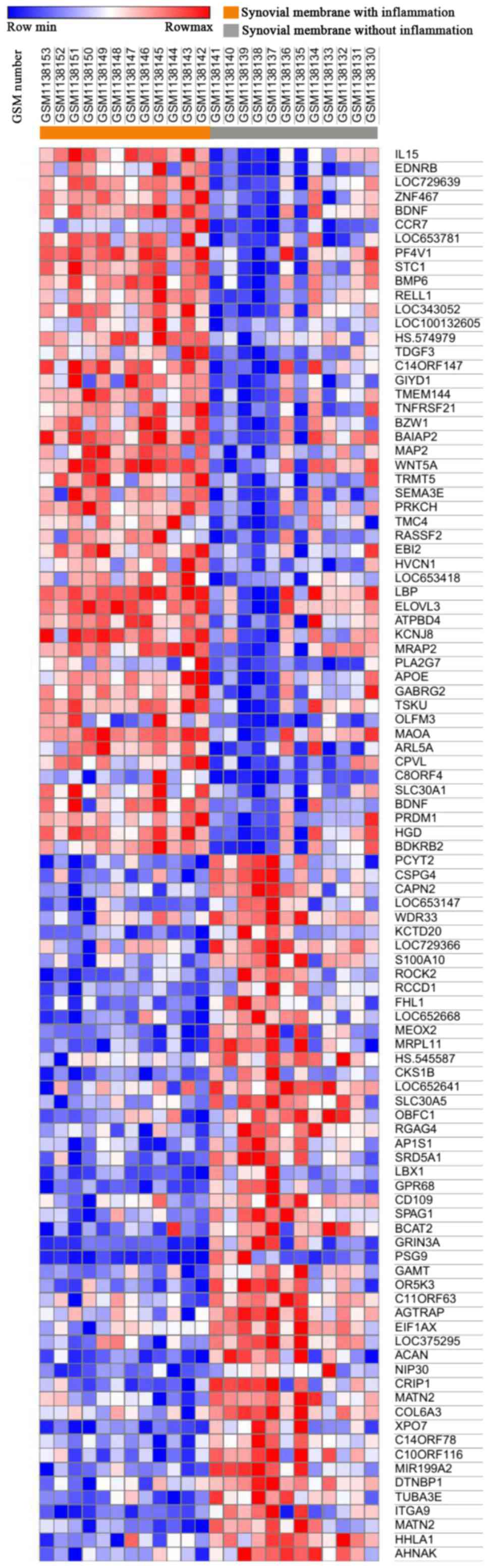
Morpheus was used to observe the overall gene
expression of all samples (Fig.
1), and differences in gene expression between inflammatory
synovial cells (n=12) and non-inflammatory synovial cells (n=12)
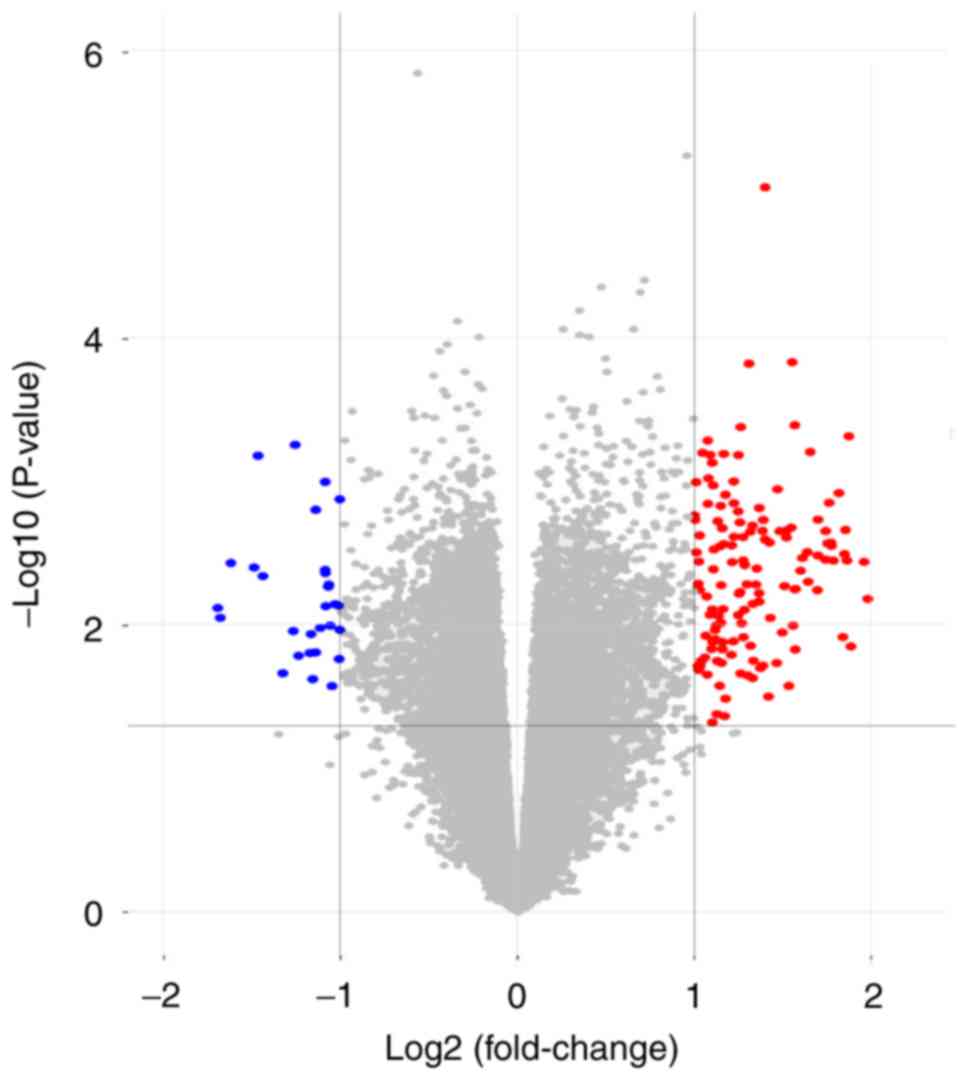
were identified. A total of 174 DEGs were identified by GEO2R
analysis, of which 145 were upregulated and 29 were downregulated
(Fig. 2).
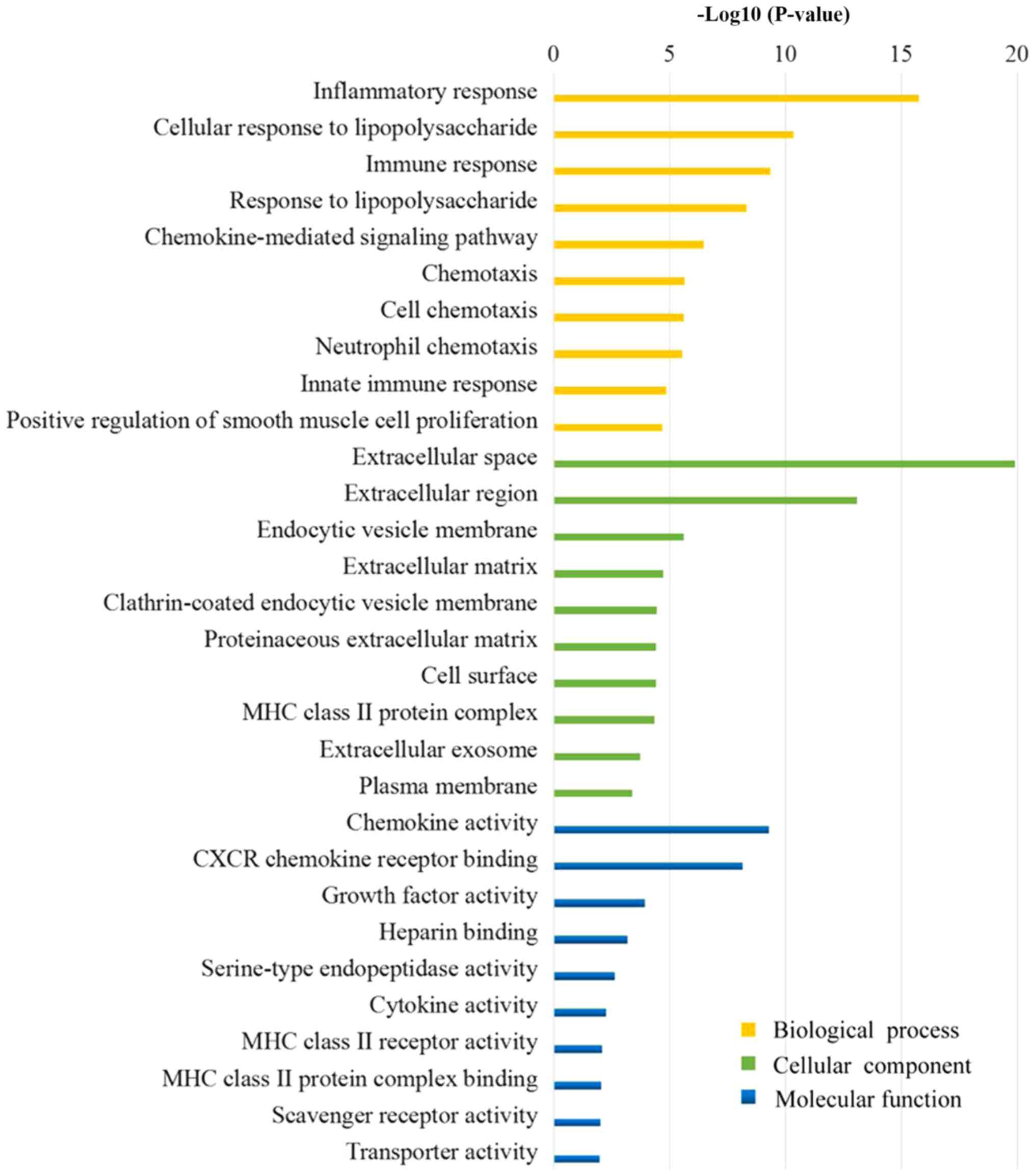
GO term enrichment analysis
Fig. 3 presents the
results of the GO analysis. The majority of the DEGs were
determined to be located in the ‘extracellular region’, the ‘plasma
membrane’ and the ‘vesicle’. The DEGs were demonstrated to exert
molecular functions by regulating ‘chemokine activity’, ‘chemokine
receptor binding’, ‘growth factor activity’ and other ‘cytokine
activity’. Furthermore, the DEGs may regulate the OA membrane
inflammatory response by involvement in the regulation of
‘inflammatory response’, ‘immune response’, ‘response to
lipopolysaccharide’ and ‘chemotaxis’.
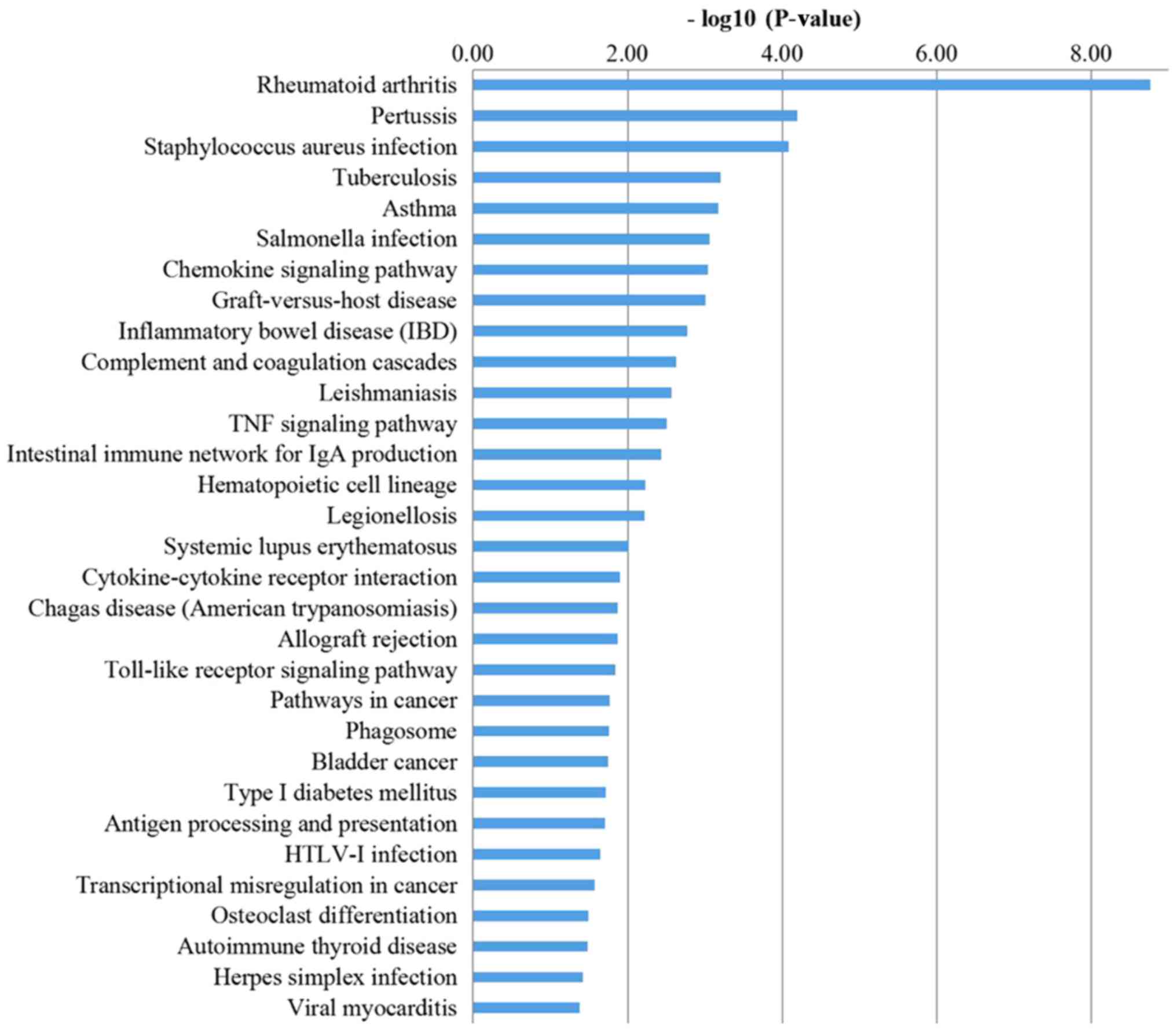
KEGG pathway analysis
The KEGG analysis results (Fig. 4) suggested that DEGs, through
pathways, including ‘chemokine signaling pathway’, ‘complement and
coagulation cascades’, ‘TNF signaling pathway’, ‘intestinal immune
network for IgA production’, ‘cytokine-cytokine receptor
interaction’, ‘Toll-like receptor signaling pathway’ and ‘antigen
processing and presentation’, serve a role in regulating the
synovial inflammation of OA.
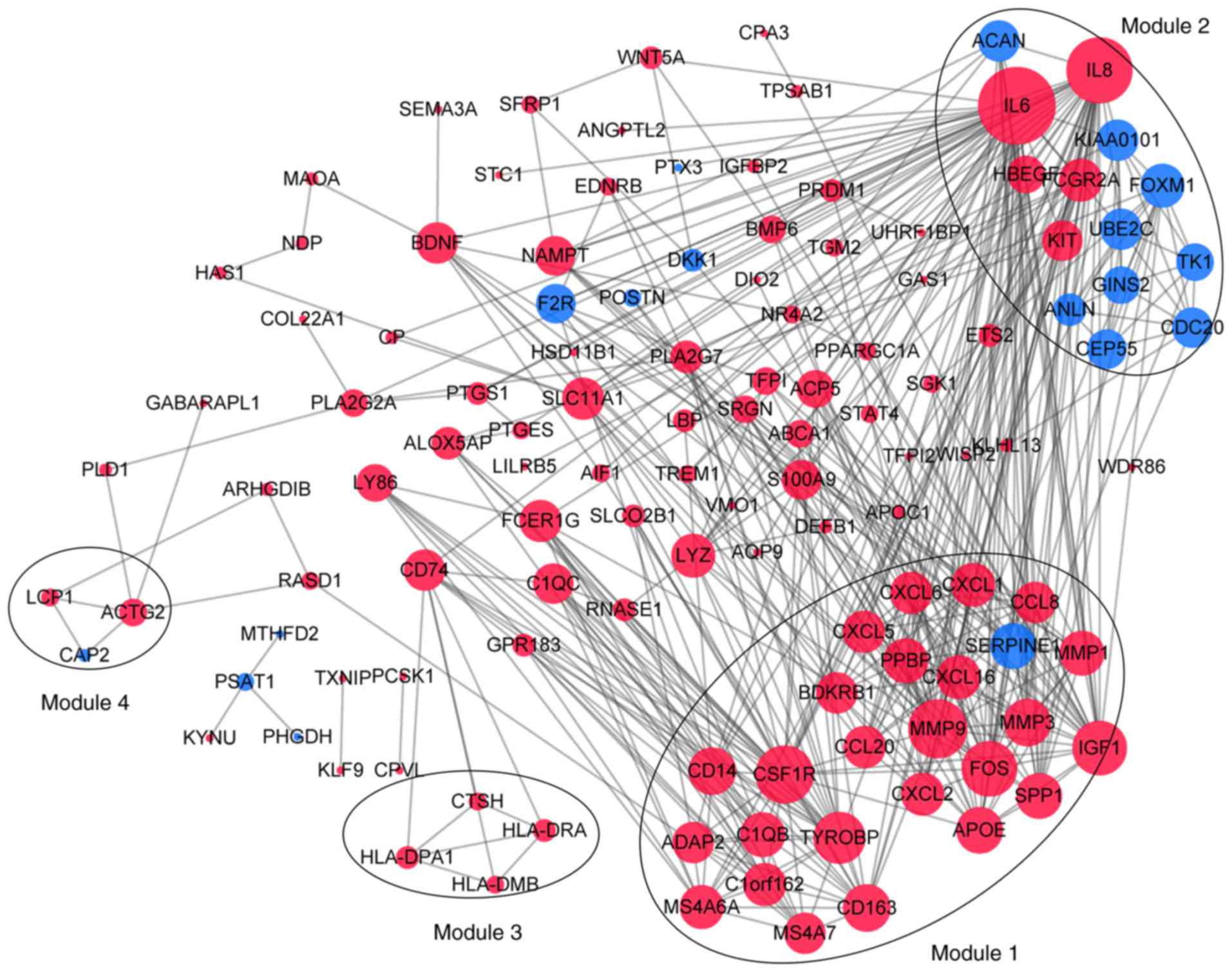
Analysis of hub genes and modules
Following the analysis based on the PPI networks,
122 nodes (DEGs) and 444 edges (interactions between DEGs) were
identified in Cytoscape (Fig. 5).
The genes with higher scores were the hub genes, as the genes of
higher degree may be associated with OA. The top 10 hub genes were
interleukin (IL)6, IL8, matrix metallopeptidase (MMP)9, colony
stimulating factor 1 receptor (CSF1R), FOS proto-oncogene, AP1
transcription factor subunit (FOS), insulin-like growth factor 1
(IGF1), TYRO protein tyrosine kinase binding protein (TYROBP),
MMP3, cluster of differentiation (CD) 14 and CD163. A total of four
modules were selected through MCODE analysis (Fig. 5). Enrichment analysis demonstrated
that modules 1 and 2 may be associated with ‘cytokine-cytokine
receptor interactions’, ‘NOD-like receptor signaling pathways’ and
‘toll-like receptor signaling pathways’ (Table I).
 | Table I.Enriched pathways of modules 1–3. |
Table I.
Enriched pathways of modules 1–3.
| A, Module 1 |
|---|
|
|---|
| Pathway | FDR | Genes |
|---|
| Cytokine-cytokine
receptor interaction |
2.80×10−9 | CCL20, CCL8, CSF1R,
CXCL1, CXCL16, CXCL2, CXCL5, CXCL6, PPBP |
| Rheumatoid
arthritis |
2.80×10−9 | CCL20, CXCL1,
CXCL5, CXCL6, FOS, MMP1, MMP3 |
| Chemokine signaling
pathway |
3.57×10−9 | CCL20, CCL8, CXCL1,
CXCL16, CXCL2, CXCL5, CXCL6, PPBP |
| TNF signaling
pathway |
3.57×10−9 | CCL20, CXCL1,
CXCL2, CXCL5, FOS, MMP3, MMP9 |
| Pertussis |
1.35×10−6 | C1QB, CD14, CXCL5,
CXCL6, FOS |
| Transcriptional
misregulation in cancer |
8.55×10−5 | CD14, CSF1R, IGF1,
MMP3, MMP9 |
| Salmonella
infection | 0.000131 | CD14, CXCL1, CXCL2,
FOS |
| Legionellosis | 0.00145 | CD14, CXCL1,
CXCL2 |
| Pathways in
cancer | 0.00145 | CSF1R, FOS, IGF1,
MMP1, MMP9 |
| Complement and
coagulation cascades | 0.00248 | BDKRB1, C1QB,
SERPINE1 |
| Chagas disease
(American trypanosomiasis) | 0.00628 | C1QB, FOS,
SERPINE1 |
| Toll-like receptor
signaling pathway | 0.00669 | CD14, FOS,
SPP1 |
| Osteoclast
differentiation | 0.011 | CSF1R, FOS,
TYROBP |
| Bladder cancer | 0.0206 | MMP1, MMP9 |
| NOD-like receptor
signaling pathway | 0.0439 | CXCL1, CXCL2 |
|
| B, Module
2 |
|
| Pathway | FDR | Genes |
|
| Cytokine-cytokine
receptor interaction | 0.0431 | IL6, IL8, KIT |
| NOD-like receptor
signaling pathway | 0.0431 | IL6, IL8 |
| Hematopoietic cell
lineage | 0.0431 | IL6, KIT |
| Epithelial cell
signaling in Helicobacter pylori infection | 0.0431 | HBEGF, IL8 |
| Salmonella
infection | 0.0431 | IL6, IL8 |
| Pertussis | 0.0431 | IL6, IL8 |
| Legionellosis | 0.0431 | IL6, IL8 |
| Malaria | 0.0431 | IL6, IL8 |
| Pathways in
cancer | 0.0431 | IL6, IL8, KIT |
| Rheumatoid
arthritis | 0.0431 | IL6, IL8 |
| Toll-like receptor
signaling pathway | 0.0494 | IL6, IL8 |
| Chagas disease
(American trypanosomiasis) | 0.0494 | IL6, IL8 |
| Amoebiasis | 0.0494 | IL6, IL8 |
|
| C, Module
3 |
|
| Pathway | FDR | Genes |
|
| Intestinal immune
network for IgA production | 0.00149 | HLA-DPA1,
HLA-DRA |
| Type I diabetes
mellitus | 0.00149 | HLA-DPA1,
HLA-DRA |
| Staphylococcus
aureus infection | 0.00149 | HLA-DPA1,
HLA-DRA |
| Asthma | 0.00149 | HLA-DPA1,
HLA-DRA |
| Autoimmune thyroid
disease | 0.00149 | HLA-DPA1,
HLA-DRA |
| Allograft
rejection | 0.00149 | HLA-DPA1,
HLA-DRA |
| Graft-versus-host
disease | 0.00149 | HLA-DPA1,
HLA-DRA |
| Viral
myocarditis | 0.00152 | HLA-DPA1,
HLA-DRA |
| Inflammatory bowel
disease | 0.00172 | HLA-DPA1,
HLA-DRA |
| Antigen processing
and presentation | 0.00175 | HLA-DPA1,
HLA-DRA |
| Leishmaniasis | 0.00175 | HLA-DPA1,
HLA-DRA |
| Rheumatoid
arthritis | 0.00244 | HLA-DPA1,
HLA-DRA |
| Systemic lupus
erythematosus | 0.00275 | HLA-DPA1,
HLA-DRA |
| Toxoplasmosis | 0.00363 | HLA-DPA1,
HLA-DRA |
| Cell adhesion
molecules | 0.00499 | HLA-DPA1,
HLA-DRA |
| Phagosome | 0.0051 | HLA-DPA1,
HLA-DRA |
| Tuberculosis | 0.00645 | HLA-DPA1,
HLA-DRA |
| Influenza A | 0.00645 | HLA-DPA1,
HLA-DRA |
| Herpes simplex
infection | 0.00645 | HLA-DPA1,
HLA-DRA |
| Epstein-Barr virus
infection | 0.00722 | HLA-DPA1,
HLA-DRA |
| HTLV-I
infection | 0.012 | HLA-DPA1,
HLA-DRA |
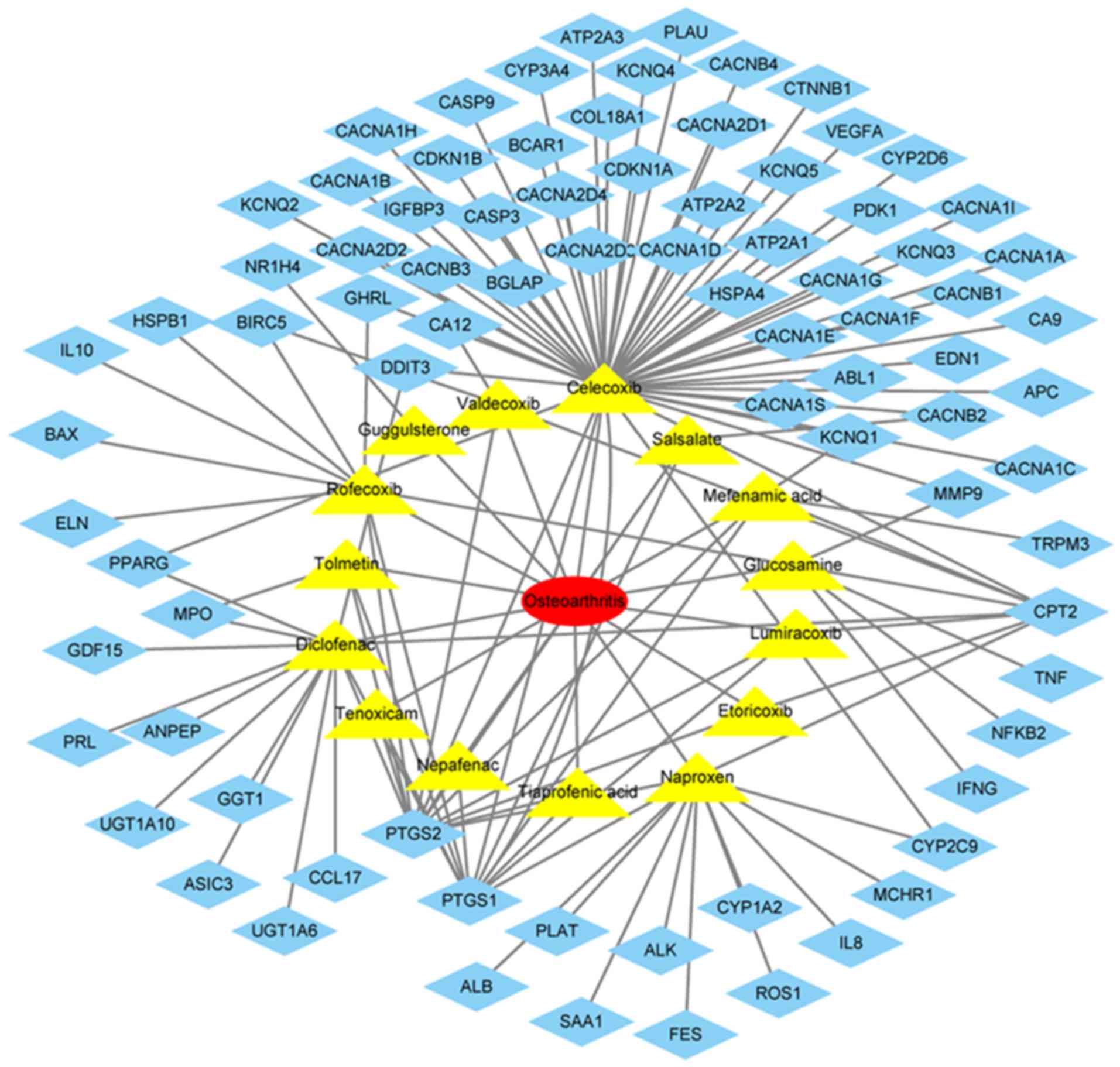
Common therapeutic drugs and their
targets
The drugs commonly used in the treatment of OA
included lumiracoxib, rofecoxib, guggulsterone, nepafenac,
glucosamine, diclofenac, valdecoxib, naproxen, tiaprofenic acid,
celecoxib, tolmetin, etoricoxib, tenoxicam, salsalate and mefenamic
acid (Fig. 6). The target genes of
these drugs were identified in DGIdb, which included IL8, MMP9,
IL10, BCL2 associated X, apoptosis regulator, cyclin dependent
kinase inhibitor 1B (Fig. 6).
Discussion
At present, factors, including heredity, age and
mechanical alterations, are involved in the complex pathogenesis of
OA (24). The pathogenesis remains
unclear, and thus, the treatment of OA primarily relieves symptoms
and no specific treatment has been identified. Microarray and
sequencing technology that provide expression levels of thousands
of genes in humans, have been widely used to predict the potential
therapeutic targets for diseases. A thorough understanding of the
molecular mechanisms of OA may provide insight for diagnosis and
treatment. In the present study, 174 DEGs were identified between
synovial membrane with and without inflammation in OA, among which,
145 genes were upregulated and 29 were downregulated. Synovial
inflammation has been identified as one of the pathological
manifestations of the development and progression of OA (25). These DEGs were involved in the
regulation of synovial inflammation. In the present study, an
enrichment analysis was conducted to further understand the
regulatory roles of DEGs in OA.
The top 10 CC terms of the DEGs demonstrated that
they are primarily located in the ‘extracellular region’, ‘plasma
membrane’, ‘vesicles membrane’. The roles of DEGs in MF are to
activate ‘cytokine activity’, promote ‘chemokine receptor binding’,
and regulate ‘receptor binding’ and ‘phospholipid binding’. The top
10 BP terms involved in DEGs are primarily ‘inflammatory response’,
‘immune response’ and ‘cell-cell signaling’. The KEGG pathways
involved in synovial inflammation development of OA by DEGs include
‘chemokine signaling pathway’, ‘complement and coagulation
cascades’, ‘TNF signaling pathway’, ‘cytokine-cytokine receptor
interaction’, ‘allograft rejection’, ‘Toll-like receptor signaling
pathway’ and ‘antigen processing and presentation’. Previous
studies suggested that immune responses, including complement
activation, cytokines and immune cell populations are involved in
the occurrence and development of OA (26–28).
Additionally, previous studies suggested that cytokines are
involved in the pathological process of OA, and cause inflammatory
reactions and pain (29,30). A previous study demonstrated that
the occurrence of coagulation cascades in the synovium was observed
in joint diseases, including OA, and the associated molecules were
more highly expressed in inflammatory arthritis (31).
PPI networks containing the DEGs were additionally
constructed, and the top 10 hub genes were IL6, IL8, MMP9, CSF1R,
FOS, IGF1, TYROBP, MMP3, CD14 and CD163. IL6 and IL8 primarily
regulate immune responses and inflammatory responses (32). Previous studies demonstrated that
IL6 and IL8 expression levels were increased in OA synovial tissues
(33,34). The role of MMP9 is primarily to
degrade and remodel the extracellular matrix, and the role of MMP3
is to degrade extracellular matrix proteins (35). The results of the present study
were consistent with the results of previous studies, which
demonstrated high expression of MMP9 and MMP3 in OA synovial
tissues (36,37). CSF1R encodes a receptor protein for
colony stimulating factor 1, which mediates the majority of the
biological effects of this cytokine (38). A previous study revealed
macrophage-CSF and macrophage-CSF and granulocyte-CSF are involved
in the inflammatory response in a rheumatoid arthritis mouse model
(39). FOS is a nuclear protein
transcription factor that regulates the growth, division,
differentiation, proliferation and apoptosis of cells (40). Compared with normal human synovial
tissues, immunohistochemistry demonstrated strong staining of c-FOS
in OA synovial tissues (41). IGF1
encodes an active protein peptide substance that promotes cell
growth and is essential for sugar, lipid, protein metabolism and
inorganic salt metabolism (42). A
previous study demonstrated that the IGF1 content in OA synovial
fluid was twice that in normal fluid and that all forms of IGF1
were highly concentrated in the cartilage of OA, which may affect
the progression of the disease by regulating the anabolic
metabolism of cartilage macromolecules (43). TYROBP is involved in immune and
inflammatory reactions (44). A
previous study demonstrated that TYROBP was more highly expressed
in OA synovial tissues compared with healthy synovial tissues
(45). A previous clinical study
demonstrated that the expression levels of CD14 and CD163 in the
serum and joint fluid were increased in patients with OA, and were
directly proportional to the activation of macrophages, joint
space, osteophytes and the severity of pain (46). CD14, as a co-receptor, was involved
in the signaling pathway of lipopolysaccharide binding and may
trigger inflammatory activation of synovial cells by stimulating
toll-like receptors (47). CD163
is a marker for cells from the monocyte/macrophage lineage and is
also hemoglobin scavenger receptor, with pro- and anti-inflammatory
effects (48). Comparing the
results of the present study with the text-mining results, IL8 and
MMP9 were identified as useful therapeutic targets for OA.
Module analysis demonstrated that the development of
OA synovitis is associated with ‘cytokine-cytokine receptor
interactions’, ‘NOD-like receptor signaling pathway’, ‘Toll-like
receptor signaling pathway’. Previous studies have demonstrated
that the occurrence and development of OA were associated with a
number of cytokines, leading to synovial inflammation, cartilage
damage and osteophyte production (30,49).
The present study demonstrated that OA synovial inflammation is
associated with ‘NOD-like receptor signaling pathway’, ‘Toll-like
receptor signaling pathway’ and ‘TNF signaling pathway’, and OA
progression is additionally associated with ‘osteoclast
differentiation’.
In the present study, bioinformatics analysis was
performed to investigate synovial inflammation. The DEGs
identification and in-depth analysis may help to identify useful
targets and pathways in OA pathogenesis. These results may provide
insight for the study of OA therapeutic targets. A limitation of
the present study was that the screened genes and pathways were not
experimentally validated, which may become a focus of future
studies.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81573801), the Natural
Science Foundation of Fujian Province (grant no. 2017J06018) and
the 2017 Science and Technology project of Traditional Chinese
Medicine of Fujian Province (grant no. 2017FJZYJC204).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL and JL conceived and designed the study. GW, ZZ
and YH collected the expression data and screened for the
differentially expressed genes. JC, CF and JY analyzed and
interpreted the data. JL wrote the manuscript. GW and XL reviewed
and edited the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McDougall C, Hurd K and Barnabe C:
Systematic review of rheumatic disease epidemiology in the
indigenous populations of Canada, the United States, Australia, and
New Zealand. Semin Arthritis Rheum. 46:675–686. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bijlsma JW, Berenbaum F and Lafeber FP:
Osteoarthritis: An updata with relevance for clinical practice.
Lancet. 377:2115–2126. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sulzbacher I: Osteoarthritis: Histology
and pathogenesis. Wien Med Wochenschr. 163:212–219. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hashimoto S, Ochs RL, Komiya S and Lotz M:
Linkage of chondrocyte apoptosis and cartilage degradation in human
osteoarthritis. Arthritis Rheum. 41:1632–1638. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schroeppel JP, Crist JD, Anderson HC and
Wang J: Molecular regulation of articular chondrocyte function and
its significance in osteoarthritis. Histol Histopathol. 26:377–394.
2011.PubMed/NCBI
|
|
6
|
Kleine SA and Budsberg SC: Synovial
membrane receptors as therapeutic targets: A review of receptor
localization, structure, and function. J Orthop Res. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang X, Hunter DJ, Jin X and Ding C: The
importance of synovial inflammation in osteoarthritis: Current
evidence from imageing assessments and clinical trial.
Osteoarthritis Cartilage. 26:165–167. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bhattaram P and Chandrasekharan U: The
joint synovium: A critical determinant of articular cartilage fate
in inflammatory joint diseases. Semin Cell Dev Biol. 62:86–93.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Scanzello CR and Goldring SR: The role of
synovitis in osteoarthritis pathogenesis. Bone. 51:249–257. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Goldenberg DL and Cohen AS: Synovial
membrane histopathology in the differential diagnosis of rheumatoid
arthritis, gout, pseudogout, systemic lupus erythematosus,
infectious arthritis and degenerative joint disease. Medicine
(Baltimore). 57:239–252. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sellam J and Berenbaum F: The role of
synovitis in pathophysiology and clinical symptoms of
osteoarthritis. Nat Rev Rheumatol. 6:625–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Myers SL, Brandt KD, Ehlich JW, Braunstein
EM, Shelbourne KD, Heck DA and Kalasinski LA: Synovial inflammation
in patients with early osteoarthritis of the knee. J Rheumatol.
17:1662–1669. 1990.PubMed/NCBI
|
|
13
|
Ayral X, Pickering EH, Woodworth TG,
Mackillop N and Dougados M: Synovitis: A potential predictive
factor of structural progression of medial tibiofemoral knee
osteoarthritis-results of a 1 year longitudinal arthroscopic study
in 422 patients. Osteoarthritis Cartilage. 13:361–367. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fernandez-Madrid F, Karvonen RL, Teitge
RA, Miller PR, An T and Negendank WG: Synovial thickening detected
by MR imaging in osteoarthritis of the knee confirmed by biopsy as
synovitis. Magn Reson Imaging. 13:177–183. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Felson DT, Niu J, Neoqi T, Goggins J,
Nevitt MC, Roemer F, Torner J, Lewis CE and Guermazi A: MOST
Investigators Group. Synovitis and the risk of knee osteoarthritis:
The MOST Study. Osteoarthritis Cartilage. 24:458–464. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen D, Shen J, Zhao W, Wang T, Han L,
Hamilton JL and Lm HJ: Osteoarthritis: Toward a comprehensive
understanding of pathological mechanism. Bone Res. 5:160442017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li M, Zhi L, Zhang Z, BIan W and Qiu Y:
Identification of potential target genes associated with the
pathogenesis of osteoarthritis using microarray based analysis. Mol
Med Rep. 16:2799–2806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kong R, Gao J, Si Y and Zhao D:
Combination of circulating miR-19b-3p, miR-122-5p and miR-486-5p
expressions correlates with risk and disease severity of knee
osteoarthritis. Am J Transl Res. 9:2852–2864. 2017.PubMed/NCBI
|
|
19
|
Lambert C, Dubuc JE, Montell E, Verges J,
Munaut C, Noel A and Henrotin Y: Gene expression pattern of cell
from inflamed and normal areas of osteoarthritis synovial membrane.
Arthritis Rheumatol. 66:960–968. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:(Database Issue). D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li YH, Yu CY, Li XX, Zhang P, Tang J, Yang
Q, Fu T, Zhang X, Cui X, Tu G, et al: Therapeutic target database
update 2018: Enriched resource for facilitating bench-to-clinic
research of targeted therapeutics. Nucleic Acids Res.
46:D1121–D1127. 2018.PubMed/NCBI
|
|
24
|
Johnson VL and Hunter DJ: The epidemiology
of osteoarthritis. Best Pract Res Clin Rheumatol. 28:5–15. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Benito MJ, Veale DJ, FitzGerald O, van den
Berg WB and Bresnihan B: Synovial tissue inflammation in early and
late osteoarthritis. Ann Rheum Dis. 64:1263–1267. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lopes EBP, Filiberti A, Husain SA and
Humphrey MB: Immune contributions to osteoarthritis. Curr
Osteoporos Rep. 15:593–600. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kalaitzoglou E, Griffin TM and Humphrey
MB: Innate immune responses and osteoarthritis. Curr Rheumatol Rep.
19:452017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Silawal S, Triebel J, Bertsch T and
Schulze-Tanzil G: Osteoarthritis and the complement cascade. Clin
Med Insights Arthritis Musculoskelet Disord.
11:11795441177514302018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Syx D, Tran PB, Miller RE and Malfait AM:
Peripheral mechanisms contributing to osteoarthritis pain. Curr
Rheumatol Rep. 20:92018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nguyen LT, Sharma AR, Chakraborty C,
Saibaba B, Ahn ME and Lee SS: Review of prospects of biological
fluid biomarkers in osteoarthritis. Int J Mol Sci. 18:pii: E601.
2017. View Article : Google Scholar
|
|
31
|
So AK, Varisco PA, Kemkes-Matthes B,
Herkenne-Morard C, Chobaz-Peclat V, Gerster JC and Busso N:
Arthritis is linked to local and systemic activation of coagulation
and fibrinolysis pathways. J Thromb Haemost. 1:2510–2515. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Brocker C, Thompson D, Matsumoto A, Nebert
DW and Vasiliou V: Evolutionary divergence and functions of the
human interlrukin (IL) gene family. Hum Genomics. 5:30–55. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang F, Zhou S, Wang C, Huang Y, Li H,
Wang Y, Zhu Z, Tang J and Yan M: Epigenetic modifications of
interleukin-6 in synovial fibroblasts from osteoarthritis patients.
Sci Rep. 7:435922017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nair A, Gan J, Bush-Joseph C, Verma N,
Tetreault MW, Saha K, Margulis A, Fogg L and Scanzello CR: Synovial
chemokine expression and relationship with knee symptoms in
patients with meniscal tears. Osteoarthritis Cartilage.
23:1158–1164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rohani MG and Parks WC: Matrix remodeling
by MMps during wound repair. Matrix Biol. 44–46:113–121. 2015.
View Article : Google Scholar
|
|
36
|
Kanyama M, Kuboki T, Kojima S, Fujisawa T,
Hattori T, Takigawa M and Yamashita A: Matrix metalloproteinases
and tissue inhibitors of metalloproteinases in synovial fluids of
patients with temporomandibular joint osteoarthritis. J Orofac
Pain. 14:20–30. 2000.PubMed/NCBI
|
|
37
|
Yang CC, Lin CY, Wang HS and Lyu SR:
Matrix metalloproteases and tissue inhibitors to knee
osteoarthritis progression. PLoS One. 8:e796622013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Meyers MJ, Pelc M, Kamtekar S, Day J, Poda
GI, Hall MK, Michener ML, Reitz BA, Mathis KJ, Pierce BS, et al:
Structure-based drug design enables conversion of a DFG-in bingding
CSF-1R kinase inhibitor to a DFG-out binding mode. Bioorg Med Chem
Lett. 20:1543–1547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Campbell IL, Rich MJ, Bioschof RJ and
Hamilton JA: The colony-stimulating factors and collagen-induced
arthritis: Exacerbation of disease by M-CSF and G-CSF and
requirement for endogenous M-CSF. J Leukoc Biol. 68:144–150.
2000.PubMed/NCBI
|
|
40
|
Chung L: A brief introduction to the
transduction of neural activity into fos signal. Dev Reprod.
19:61–67. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kinne RW, Boehm S, Iftner T, Aigner T,
Vornehm S, Weseloh G, Bravo R, Emmrich F and Kroczek RA: Synovial
fibroblast-like cells strongly express jun-B and C-fos
proto-oncogenes in rheumatoid- and osteoarthritis. Scand J
Rheumatol Suppl. 101:121–125. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Annunziata M, Granata R and Ghiqo E: The
IGF system. Acta Diabetol. 48:1–9. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Schneiderman R, Rosenberg N, Hiss J, Lee
P, Liu F, Hintz RL and Maroudas A: Concentration and size
distribution of insulin-like growth factor-1 in human normal and
osteoarthritis synovial fluid and cartilage. Arch Biochem Biophys.
324:173–188. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tomasello E and Vivier E:
KARAP/DAP12/TYROBP: Three names and a multiplicity of biological
functions. Eur J Immunol. 35:1670–1677. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Crotti YN, Dharmapatni AA, Alias E,
Zannettino AC, Smith MD and Haynes DR: The immunoreceptor
tyrosine-based activation motif (IYAM)-related factors are
increased in synovial tissue and vasculature of rheumatoid
arthritic joints. Arthritis Res Ther. 14:R2452012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Daghestani HN, Pieper CF and Kraus VB:
Solube macrophage biomarkers indicate inflammatory phenotypes in
patients with knee osteoarthritis. Arthritis Rheumatol. 67:956–965.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Nair A, Kanda V, Bush-Joseph C, Verma N,
Chubinskaya S, Mikecz K, Glant TT, Malfair AM, Crow MK, Spear GT,
et al: Synovial fluid from patients with early osteoarthritis
modulates fibroblast-like synoviocyte responses to toll-like
recepyor 4 and toll-like receptor 2 ligands via soluble CD14.
Arthritis Rheum. 64:2268–2277. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fabriek BO, Dijkstra CD and van den Berg
TK: The macrophage scavenger receptor CD163. Immunobiology.
210:153–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim JR, Yoo JJ and Kim HA: Therapeutics in
osteoarthritis based on an understanding of its molecular
pathogenesis. Int J Mol Sci. 19:E6742018. View Article : Google Scholar : PubMed/NCBI
|