Introduction
Skin functions as an important natural barrier
between an organism and its external environment, thus it has a
unique biological structure and specific immune functions (1). Skin is composed of two distinct
regions, the epidermis and dermis. The predominant cell type of the
epidermis is keratinocytes (KCs), whereas the dermis contains cells
of the immune system, including dendritic cells (DCs), T helper
cells, γδT cells, natural killer T cells, macrophages and
fibroblasts (2). A skin immune
system imbalance can cause several immune-mediated skin diseases,
such as psoriasis (1). Therefore,
in-depth analysis of the skin immune system would be of great
clinical significance for the treatment of these diseases.
Psoriasis is a common chronic inflammatory skin
disease triggered by a dysregulated immune response, which
typically manifests as plaques with adherent silvery scales and has
a great impact on the physical and mental health of patients
(3,4). This disease is characterized by
excessive growth and aberrant differentiation of KCs. In psoriasis,
the pathological changes in KCs include hyperkeratosis,
parakeratosis, a loss of the normal granular layer and acanthosis
with elongation of the epidermal rete ridges (5). Thus, as the skin specific cells, KCs
are the main cell type affected by psoriasis.
The immunopathogenesis of psoriasis outlines the
important role KCs serve in the induction and amplification of
psoriatic inflammation. KCs respond to different signals and
release the cathelicidin antimicrobial peptide (LL-37), which
activates plasmacytoid DCs (pDCs) via Toll-like receptors (5–7).
Myeloid DCs are activated by interferon (IFN)-α/β from the pDCs,
stimulating psoriatic T cells that produce interleukin (IL)-17,
IFN-γ and IL-22. KCs also produce chemokines and antimicrobial
peptides, which attract myeloid DCs and T cells that produce IL-17.
The cytokines released by these cells further stimulate the KCs,
and the immune circuit is further amplified by feedback cytokines
produced by the KCs (1,5,8).
Thus, KCs have a central role in the immune circuits of psoriatic
skin.
Several immunosuppressants are currently available
for psoriasis treatment; their targets include cytokines, signaling
molecules and receptors in the immune system (7). However, as with any drug designed to
suppress the general immune system, the main concern is the risk of
serious side effects. Efalizumab, a monoclonal antibody targeting
cluster of differentiation (CD)11a, has inhibitory effects against
broad T cell subsets, resulting in systemic immune suppression,
which can lead to serious infection, cancer and other severe
complications (9,10). As a result, it was withdrawn 6
years after approval (FDA Issues Boxed Warning for Efalizumab,
2008) (11). The clinical
development of briakinumab, an antibody targeting IL-23/P40, was
halted after a series of major cardiovascular events occurred
during clinical trials (9). In
clinical application, other psoriasis drugs, including infliximab,
[a tumor necrosis factor (TNF)-α blocker], corticosteroids,
antibiotics and vitamins, can also cause different degrees of
toxicity (12). In addition,
IL-17-targeting antibodies and small molecule drugs targeting Janus
kinases also have limitations in the treatment of psoriasis. The
most fundamental reason is that these drugs do not have specific
targets or cannot regulate the cutaneous immunity (13,14).
Therefore, identifying novel psoriasis-specific molecules will help
treatment.
In the present study, novel psoriatic skin-specific
genes were screened from the transcriptome of psoriasis-like KCs.
An integrative approach, combining psoriatic transcriptome data,
psoriasis-associated genes information, genetic loci linked to
psoriasis and human tissue expression pattern, was used to screen
novel psoriasis-associated genes that were highly expressed in the
skin/epidermis. The present study provides a novel way of
identifying novel skin-specific genes for cutaneous diseases.
Materials and methods
Human skin samples
The present study was performed in accordance with
the principles of the Helsinki Declaration and approved by the
Ethics Committee of the West China Hospital, Sichuan University
(Chengdu, Sichuan, China). Written informed consent was obtained
from all study participants prior to the study. All patients were
diagnosed based on the clinically apparent symptoms (fairly easily
diagnosed as characteristic red colored plaques with well-defined
borders and silvery-white dry scale), and histopathological
criteria (abnormal proliferation and differentiation of the
epidermis, hyperkeratosis and parakeratosis of keratinocytes)
(5). All patients were assessed
according to the Psoriasis Area and Severity Index (15). Skin samples ~0.5–1.0 cm were
collected from eight patients with psoriasis (four females and four
males; age, 21–63 years) at the West China Hospital, Sichuan
University between March 2016 and November 2017. The lesional and
non-lesional psoriatic skin (~0.5–1.0 cm) were taken from each
patient, one was obtained from lesional skin of patients and the
other from non-lesional skin of the same patients. The fresh skin
samples were snap-frozen in liquid nitrogen and stored at −80°C.
All participants had not been treated with systemic therapy
including investigational agents for at least 4 weeks prior to the
study entry. Patients with a history of other autoimmune diseases,
immunologic deficiency diseases or tumors were excluded.
Cell culture
The human HaCaT KCs were obtained from the China
Center for Type Culture Collection (Wuhan, China; CCTCC no. 0106).
HaCaT cells were cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% (v/v) fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), 100 U/ml penicillin G
and 0.1 mg/ml streptomycin sulfate (Thermo Fisher Scientific,
Inc.). The conditions of cell culture were 37°C and 5%
CO2. All cells were demonstrated to be free from
mycoplasma contamination.
Induction of psoriatic keratinocytes
model in vitro
The five proinflammatory cytokines (termed M5),
including IL-17A, oncostatin-M, TNF-α, IL-22, and IL-1a (ProSpec
Bio, East Brunswick, NJ, USA) were used to induce psoriasis-like
KCs inflammation that recapitulates the features of psoriasis
(16), as described in a previous
study (17). Briefly, KCs were
cultured to 80% confluency and then starved for 24 h in DMEM
without serum prior to stimulation. The cells were then stimulated
with 10 ng/ml recombinant IL-17A, 10 ng/ml recombinant
oncostatin-M, 10 ng/ml recombinant TNF-α, 10 ng/ml recombinant
IL-22 and 10 ng/ml recombinant IL-1α in combination or untreated
for 24 h prior to the whole transcriptome gene expression
analysis.
Microarray expression profiling
Gene array analysis was performed using Human Expr
12×135K Arr Del (Roche-NimbleGen; Roche Diagnostics, Basel,
Switzerland) by KangChen Bio-tech, Inc. (Shanghai, China). In
brief, total mRNA was isolated from KCs at 24 h post-stimulation
with M5 using PureYield™ RNA Midiprep System (Promega Corporation,
Madison, WI, USA). Total RNA was quantified by the NanoDrop
ND-1000A, and RNA integrity and gDNA contamination was assessed by
standard denaturing 1% agarose gel electrophoresis. Total mRNA of
each sample was used for labeling and array hybridization were
performed according to the manufacturers' protocols: Reverse
transcription using SuperScript Double-Stranded cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.); ds-cDNA labeling with NimbleGen
one-color DNA labeling kit (Roche Diagnostics, Mannheim, Germany);
array hybridization using the NimbleGen Hybridization System
followed by washing with the NimbleGen wash buffer kit (Roche
Diagnostics); array scanning using the Axon GenePix 4000B
microarray scanner (Molecular Devices LLC, Sunnyvale, CA, USA). Raw
signal intensities were extracted and normalized using the Robust
Multichip Average (RMA) method by NimbleScan v2.5 software (Roche
NimbleGen Inc., Madison, WI), and low intensity (<100.0) genes
were filtered. Further data analysis was performed using Agilent
GeneSpring GX 11.5.1 software (Agilent Technologies, Inc., Santa
Clara, CA, USA). Two biological replicates were used for each
sample, expression values were normalized based on the mean
expression value for each probe set, differently expressed probe
sets were identified based on Student's t-test for paired samples'
normalized expression values using the following cutoff: Absolute
fold change (FC) >3 and a P<0.01, false discovery rate
<0.05. In addition, 3,577 differentially expressed genes (DEGs)
were obtained from psoriatic lesional and normal skin in the study
by Li et al (18), and
1,446 DEGs in psoriatic lesional epidermis compared with
non-lesional psoriatic epidermis in the study by Mitsui et
al (19). To compare the
present keratinocytes microarray data with previously published
reports data, these two transcriptome data sets and the
transcriptome dataset in the psoriasis-like KCs model were further
analyzed for enriched Gene Ontology (GO) terms using Gorilla
(cbl-gorilla.cs.technion.ac.il/). Biological terms that have many
genes in common can be grouped into a module of associated terms
and genes, with a significance threshold of 0.001.
Gene expression datasets
GEO DataSets (GSE40263) were obtain of peripheral
blood mononuclear cells (PBMCs) of psoriasis (n=5) and healthy
controls (n=5) from National Center for Biotechnology Information
(NCBI; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE40263).
The ID_REF of EREG, NIPAL4, SERPINB7 and WFDC12 were
retrieved in website of Affymetrix, Inc (https://www.affymetrix.com/analysis/netaffx/index.affx).
Based on the gene of ID_REF, RMA signal intensity, which is a form
of quantile normalization applicable to gene expression
(microarray) experiments, was searched in the data table of
psoriasis and healthy controls. The PBMCs expression levels of
these four genes in the psoriasis and healthy controls were
analyzed.
Sources of genetic loci linked to
psoriasis
Genetic loci associated with psoriasis were obtained
from a knowledgebase of Human Genes and Genetic Disorders [Online
Mendelian Inheritance in Man (OMIM); search, ‘psoriasis’; entries
with ‘gene map locus’; retrieve, ‘gene map’; www.omim.org/search?index=entry&search=psoriasis&filter=gm_exists%3Atrue&sort=chromosome_number+asc%2C+chromosome_sort+asc&start=1&limit=100&retrieve=geneMap;
accessed February 2018], and previous research reports prior to
February 2018; these included 1p36, 1q21.3, 2p16, 5q15, 5q31.1,
5q33.3, 5q33.3, 8p23.2, 9q34.13, 13q12.11, 13q13.3, 14q13.2,
16p11.2, 17q11.2, 14q32.13, 18q21.2, 18q21.33, 18q22.1, 19p13,
19q13.41 and 20q13 (20–29).
Psoriasis-associated genes in
GeneCards database
Using the GeneCards database (www.genecards.org/Search/Keyword?queryString=PSORIASIS;
accessed February 2018), psoriasis-associated genes were obtained
and the approximate degree of correlation was inferred from the
score. A score ≥1 was considered to be a psoriasis-associated
gene.
Expressed sequence tag (EST)
sources
National Center for Biotechnology Information
Unigene (www.ncbi.nlm.nih.gov/unigene/) was used to search the
indicated homo candidate genes, their EST profiles were entered and
the approximate gene expression of various tissues was inferred
from the transcripts per million (TPM). Indicated genes with
relatively high specific expression in skin tissue were selected by
screening the indicated gene EST profiles inferred from TPM
value.
Ethics statement
Wild-type C57BL/6 female mice 8–12 weeks old,
weight, 17–18 g, n=180, were purchased from Vital River Laboratory
Animal Technology Co., Ltd. (Beijing, China). The animals were
housed under the following controlled conditions: 12 h light-dark
cycle at a steady temperature of 25±1°C, with free access to water
and food. The animal protocols were approved by the Committee on
the Ethics of Animal Experiments of Sichuan University. The
experimental procedures were conducted according to the ethical
Guidelines For The Care And Use Of Laboratory Animals of the
National Institutes of Health (https://grants.nih.gov/grants/olaw/guide-for-the-care-and-use-of-laboratory-animals.pdf)
and the International Association for the Study of Pain (IASP).
Every effort was made to decrease the number of animals used and to
reduce animal suffering.
Tissue dissection
Following the sacrifice of the mice, various tissues
(large intestine, lung, liver, testis, ovary, brain, spleen, and
kidney, small intestine and heart) were collected as previous
described (30). The backs of mice
were shaved, the skin was wiped with alcohol prior to its removal,
then subcutaneous fat was removed and was cut into small pieces
convenient for digestion and separation, and the samples (~0.3 cm)
were then incubated in dispase II (2.5 U/ml; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) at 4°C overnight followed by immersion in
DMEM containing 50% (v/v) FBS to inactivate the dispase II. The
epidermis and dermis were then separated at the epidermal-dermal
interface under magnification with a dissecting microscope. Only
pieces that consisted entirely of epidermis or dermis were
used.
Imiquimod (IMQ)-induced psoriasis-like
skin inflammation
The psoriasis animal model used in the present study
was the IMQ-induced psoriasis-like skin inflammation. The IMQ mouse
model of psoriasis-like skin inflammation was induced as previously
described (31). Briefly, the day
before induction, the backs of the mice were shaved. Subsequently,
the backs of the mice were treated with Aldara cream (Sichuan
MingXin Pharmaceutical Co., LTD., Sichuan, China) containing 5% IMQ
(55 mg) once daily for 1–6 days.
Lipopolysaccharide (LPS)-induced
systemic inflammatory response syndrome (SIRS)
SIRS was induced by intraperitoneal injection of
dose of 2.5 mg/kg of LPS from Escherichia coli 0111:B4
(Sigma-Aldrich; Merck KGaA). Groups of animals were sacrificed 3 h
after a single injection of LPS (n=5) or normal saline (control
group, n=5), three independent experiments were performed. Various
tissues (large intestine, lung, liver, testis, ovary, brain,
spleen, and kidney, small intestine, heart and skin) were collected
as previous described (30). The
systemic expression of the proinflammatory cytokine TNF-α in
various tissues (large intestine, lung, liver, testis, ovary,
brain, spleen, kidney, small intestine and heart) were induced by
LPS and the systemic inflammatory response was activated.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Cells or mouse tissues (large intestine, lung,
liver, testis, ovary, brain, spleen, and kidney, small intestine,
heart and skin) were obtained and TRIzol (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to extract the total RNA, according to
the manufacturer's protocol. Gel electrophoresis was performed to
detect the integrity of the total RNA extracted. Total RNA (2 µg)
was reverse transcribed into cDNA, PrimeScript RT reagent kit with
gDNA Eraser (Takara Bio, Inc., Otsu, Japan) was used for RT to
produce cDNA at 42°C for 50 min and at 85°C for 5 min, according to
the manufacturer's protocol. cDNA (20 ng) was subjected to qPCR
analysis with TB Green™ Premix Ex Taq™ II (Tli RNaseH Plus; Takara
Bio, Inc.) according to the manufacturer's protocol. PCR was run
under the following conditions: An initial denaturation at 95°C for
30 sec, 35 cycles of 95°C for 5 sec, annealing and extension at
60°C for 30 sec, and final extension at 72°C for 5 min. β-actin was
used as the internal control. All primers were obtained from
Chengdu Qing Ke Zi Xi Biotechnology Co. (Chengdu, China). Human
primers included C-X-C motif chemokine ligand 1 (CXCL1
forward, 5′-GCCAGTGCTTGCAGACCCT-3′ and reverse,
5′-GGCTATGACTTCGGTTTGGG-3′), CXCL2 (forward,
5′-CAAACCGAAGTCATAGCCAC-3′ and reverse,
5′-TCTGGTCAGTTGGATTTGCC-3′), CXCL8 (forward,
5′-TCTGTCTGGACCCCAAGGAA-3′ and reverse,
5′-GCATCTGGCAACCCTACAACA-3′), C-C motif chemokine ligand 20
(CCL20 forward, 5′-TGACTGCTGTCTTGGATACACAGA-3′ and reverse,
5′-TGATAGCATTGATGTCACAGCCT-3′), CCL27 (forward,
5′-AGCACTGCCTGCTGTACTCA-3′ and reverse,
5′-TCTTGGTGCTCAAACCACTG-3′), S100 calcium binding protein
A7 (S100A7 forward, 5′-CCTTAGTGCCTGTGACAA-3′ and
reverse, 5′-CTGCTTGTGGTAGTCTGT-3′), S100A8 (forward,
5′-AGTGTCCTCAGTATATCA-3′ and reverse, 5′-CATCTTTATCACCAGAATG-3′),
S100A9 (forward, 5′-CAACACCTTCCACCAATAC-3′ and reverse,
5′-TCATTCTTATTCTCCTTCTTGAG-3′), S100A12 (forward,
5′-CAATACTCAGTTCGGAAGG-3′ and reverse,
5′-CTTTGATATTCTTGATGGTGTTT-3′), LL-37 (forward,
5′-GATAACAAGAGATTTGCCCTGCTG-3′ and reverse,
5′-TTTCTCAGAGCCCAGAAGCCTG-3′), β-defensin 2 [BD2
(forward, 5′-TTCTCGTTCCTCTTCATA-3′ and reverse,
5′-ATATGGCTCCACTCTTAA-3′)], serpin family B member 7
(SERPINB7 forward, 5′-TTGGTGAAGGTGGCATAA-3′ and reverse,
5′-CAGAGCACTTGGGAGATT-3′), β-actin (forward,
5′-CCACGAAACTACCTTCAACTCC-3′ and reverse,
5′-GTGATCTCCTTCTGCATCCTGT-3′). Mouse primer sequences including
epiregulin (EREG forward, 5′-ACCGCCTTAGTTCAGATG-3′ and
reverse, 5′-ATGTCCACCAGGTAGATG-3′), NIPA like domain
containing 4 (NIPAL4 forward, 5′-GCACCCTGTCTGGCTTCGT-3′ and
reverse, 5′-AGTTTAATGACTGTGGGCTCTGG-3′), phospholipase A2
group IVE (PLA2G4E forward, 5′-GATGGTGACAGACTCCTT-3′
and reverse, 5′-GCAGCAAAGCCTAAAGTTA-3′), SERPINB7 (forward,
5′-AATAATCAGCCAGGACTTC-3′ and reverse,
5′-CACACTCAATGTAGTTCTTATG-3′), solute carrier family 1 member 6
(SLC1A6 forward, 5′-GGCATCATCATCTGGTATG-3′ and reverse,
5′-GGTGACGAGGAAGTAGATA-3′), WAP four-disulfide core domain 12
(WFDC12 forward, 5′-GACAACAGTGAAGAACAGAT-3′ and reverse,
5′-GGAGTCCAAGATCAAGGT-3′), β-actin (forward,
5′-CCTCTATGCCAACACAGTGC-3′ and reverse,
5′-ACATCTGCTGGAAGGTGGAC-3′). Relative mRNA expression changes were
calculated using the 2−ΔΔCq method (32).
Statistical analysis
The data are expressed as the mean ± standard
deviation. All statistical analyses were performed using GraphPad
Prism 7.0 software (GraphPad Prism Inc., La Jolla, CA, USA).
Comparison between two groups was performed by unpaired Student's
t-test. Comparison among multiple groups was performed by one-way
analysis of variance followed by a Tukey's post-hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Identification of differential
expression genes in psoriatic KCs
To induce a psoriasis-like KCs model in
vitro, KCs were stimulated with M5 combination (containing
IL-1a, IL-17A, IL-22, oncostatin-M and TNF-α). The mRNA levels of
the chemokines (CXCL1, CXCL2, CXCL8, CCL20 and CCL27) and
antimicrobial peptides (S100A7, S100A8, S100A9, S100A12, LL-37 and
BD2) were significantly increased by M5 stimulation of KCs
(Fig. 1A). The result suggested
that an in vitro model of psoriasis-like KCs was established
(16). Subsequently, using a human
gene expression microarray, a transcriptomic profile of the
psoriasis-like KCs was generated. Different colors represent gene
expression levels. Scatter plots provided a profile of
psoriasis-like KC mRNAs that were upregulated, downregulated or
unaffected compared with the control (Fig. 1B). These data identified 2,957 DEGs
with a FC>3 (P<0.01) as a cutoff, of which 1,735 were
upregulated and 1,222 were downregulated (Fig. 1B).
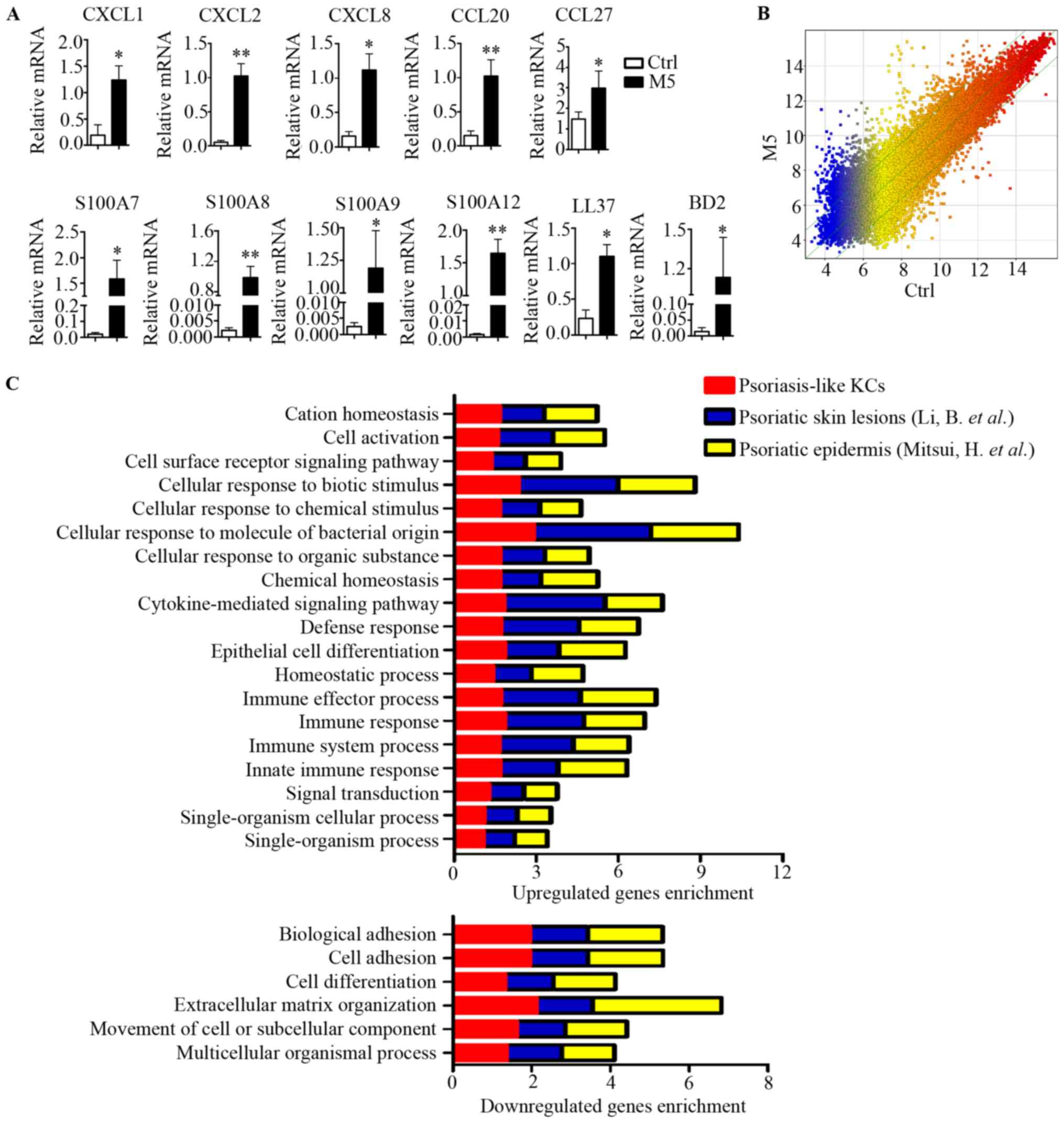 | Figure 1.Comparison of transcriptomes in
psoriasis-like KCs and patients with psoriasis. (A) KCs were
stimulated with M5 for 24 h, reverse transcription-quantitative
polymerase chain reaction analysis was performed for each indicated
gene. (B) Whole transcriptome gene expression analysis visualizing
the variation in the expression between M5 and control groups.
Different colors represent gene expression levels. The values on
the X and Y axes in the scatter plot are the averaged log2-scaled
signal values of the groups. The green lines showed a fold change
of 3.0. The levels of the mRNAs above the top green line and below
the bottom green line were >3.0-fold different between the two
groups. (C) DEGs of psoriasis-like KCs, reported DEGs of psoriatic
skin lesions by Li et al (18), and reported DEGs of psoriatic
epidermis by Mitsui et al (19), were separately sorted according to
biological process. Enrichment analysis was performed using Gorilla
and P<0.001 was the enrichment significance threshold. Bars
represent different biological processes. Data are presented as the
mean ± standard deviation, n=3, *P<0.05, **P<0.01 vs. Ctrl
group. DEGs, differentially expressed genes; CXCL, C-X-C motif
chemokine ligand; CCL, C-C motif chemokine ligand; Ctrl, control;
BD, β-defensin; KCs, keratinocytes. |
Comparative analysis of DEGs in
psoriatic KCs, human psoriatic skin and human psoriatic
epidermis
In previous studies, the DEGs between psoriatic
lesional and normal skin samples reported by Li et al
(18), and DEGs between lesional
and non-lesional epidermis samples reported by Mitsui et al
(19), were analyzed further. In
the present study, these two data sets were analyzed and compared
with the transcriptome dataset in the current psoriasis-like KCs
model. GO enrichment analysis was performed on these three
transcriptome data sets. The result produced a similar distribution
pattern of biological processes and similar genes enrichment among
the three datasets (Fig. 1C),
indicating that the changes in the biological process observed in
psoriasis-like KCs were also presents in patients with
psoriasis.
The most significantly enriched biological processes
among the upregulated genes included ‘cellular response to biotic
stimulus’, ‘cellular response to molecule of bacterial stimulus’,
‘cytokine-mediated signaling pathway’, ‘defense response’,
‘epithelial cell differentiation’, ‘immune effector process’,
‘immune response’, ‘immune system process’ and ‘innate immune
response’ (Fig. 1C). By contrast,
the most significantly enriched biological processes among the
downregulated genes included ‘biological adhesion’, ‘cell adhesion’
and ‘extracellular matrix organization’ (Fig. 1C). All these biological processes
have been implicated in psoriasis. These results suggested that the
psoriasis-like KCs model is an appropriate model for with similar
changes observed as in clinical samples.
Screening of key novel
psoriasis-associated genes from differential expression genes that
are identified in psoriatic keratinocytes
The human gene expression microarray identified
2,957 DEGs in the induced psoriasis-like KCs model in vitro
(Fig. 2, step 1). DEGs in the
induced psoriasis-like KCs model (n=2,957, psoriasis-like vs.
normal KCs) were integrated with the DEGs identified in previous
studies [n=3,577, psoriatic lesions vs. normal skin] (18); and n=1,446, psoriatic lesional
epidermis vs. non-lesional psoriatic epidermis (19)]. A core set of 506 overlapping genes
(329+100+77) were identified, which were not only differently
expressed in psoriasis-like KCs, but also in psoriatic lesions or
lesional epidermis of patients with psoriasis (Fig. 2, step 2). Thus, 506 psoriasis
disease-associated genes were obtained.
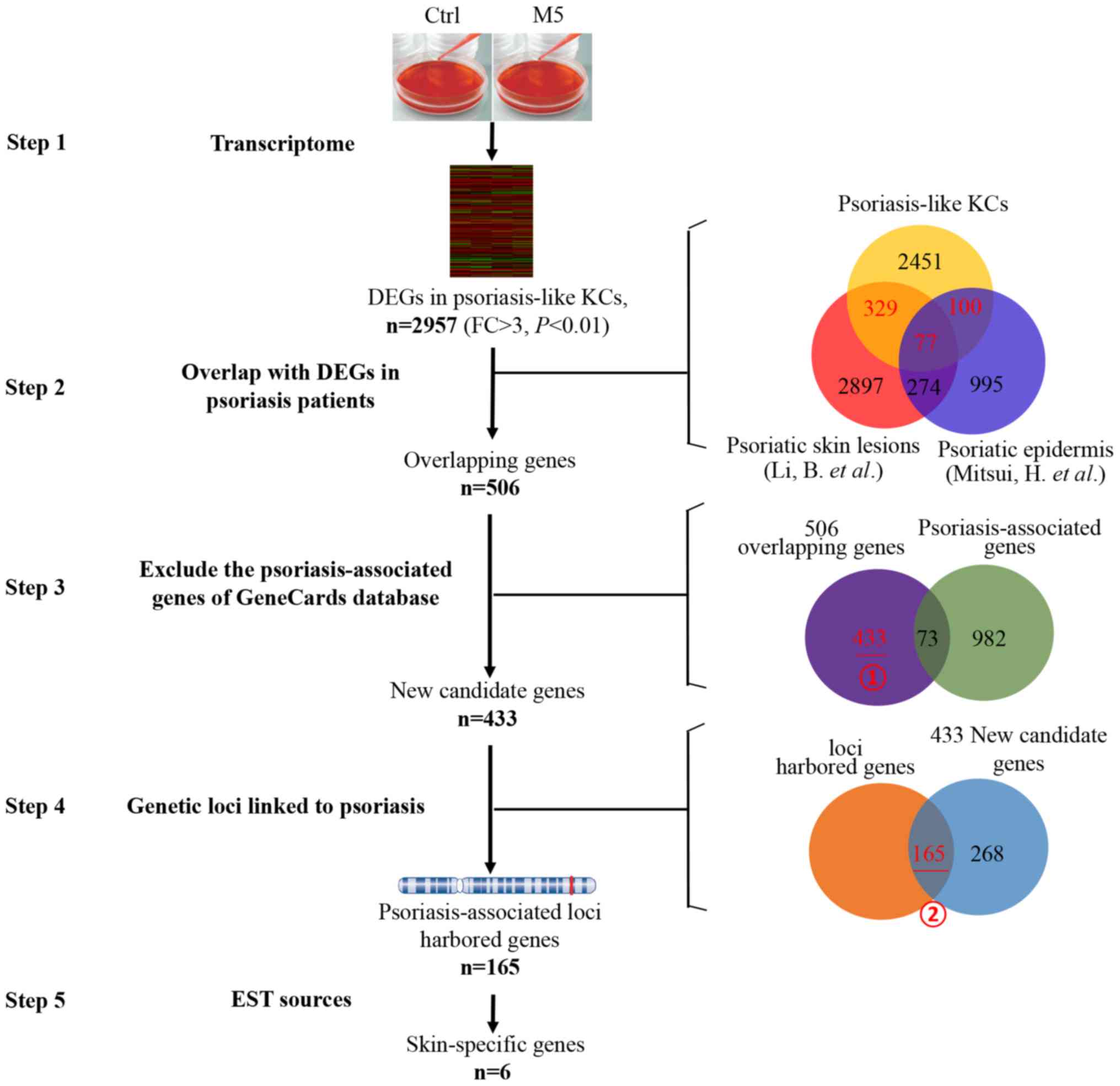 | Figure 2.Screening of key skin-specific
candidate genes from transcriptome of psoriasis-like KCs. Step 1:
2,957 DEGs were identified in psoriasis-like KCs. Step 2: Using the
overlap between the 2,957 DEGs in psoriasis-like KCs and the
reported DEGs in psoriasis patients [DEGs from Li et al
(18) and by Mitsui et al
(19)], and 506 overlapping genes
were identified. Red number represents selected overlapping genes.
Step 3: Further screening was performed to overlap between the 506
genes and the psoriasis-associated genes from GeneCards database
excluding 73 psoriasis-associated genes. Step 4: In an additional
screening was performed by genetic locus retrieval, and 165 genes
were located near summary genetic loci linked to psoriasis. Step 5:
These 165 genes were identified by analyses expression patterns in
multiple human tissues inferred from EST sources. Finally, six
genes were identified as skin-specific candidate genes. Ctrl,
control; DEGs, differentially expressed genes; KCs, keratinocytes;
FC, fold change; EST, expressed sequence tag. |
In order to screen for novel psoriasis-associated
genes, an additional screening was performed to overlap between the
506 and the known psoriasis-associated genes from the GeneCards
database. In the 506 psoriasis disease-associated genes, 73
psoriasis-associated genes that had been well described in previous
studies according to GeneCards database were excluded (Fig. 2, step 3), and the remaining 433
genes (Fig. 2, step 3, marking i)
were selected as novel psoriasis disease-associated genes.
If the candidate genes were present in the psoriasis
susceptibility region, which genetic loci were also linked to
psoriasis as described in the ‘Materials and methods’ section,
these genes were likely to serve an important role in psoriasis.
More than 100 genetic loci linked to psoriasis were obtained from
OMIM (www.omim.org/search?index=entry&search=psoriasis&filter=gm_exists%3Atr
ue&
sort=chromosome_number+asc%2C+chromosome_sort+asc&start=1&limit=100
&retrieve=geneMap) and from previous studies (20–29).
These 433 novel psoriasis-associated genes were further screened by
genetic locus retrieval, and it was revealed that 165 genes were
located near the summary genetic loci linked to psoriasis (Fig. 2, step 4, marking ii), and these
were identified as key novel psoriasis-associated genes.
Screening of skin-specific candidate
genes associated with psoriasis
To screen skin-specific genes from the 433 novel
psoriasis-associated genes, an additional screening was performed
to assess expression patterns of 165 genes in normal human tissue
by referring to the EST resources. Based on the TPM value of
indicted gene in various tissues, six genes (EREG, NIPAL4,
PLA2G4E, SERPINB7, SLC1A6, WFDC12) that were relatively
tissue-specific and highly expressed in human skin were selected
(Fig. 2, step 5). The tissue
expression pattern of these six genes was demonstrated by their EST
profile and the heat map analysis of log2-FC of TPM (Fig. 3).
 | Figure 3.EST profiles of screened
skin-specific candidate genes. Heat map analysis of the
differential expression of six skin-specific candidate genes in
normal human tissues. Black, no expression; red, high expression.
Color gradient, log2-fold-changes of transcriptions per million
from EST sources. Red indicates the genetic loci linked to
psoriasis. EST, expressed sequence tag; EREG, epiregulin; NIPAL4,
NIPA like domain containing 4; PLA2G4E, phospholipase A2 group IVE;
SERPINB7, serpin family B member 7; SLC1A6, solute carrier family 1
member 6; WFDC12, WAP four-disulfide core domain 12. |
As shown in Fig. 2,
an integrated approach was used, combining psoriasis transcriptome
data derived from the GeneCards database, psoriasis-associated
locus and EST resources. Through the above screening process, six
significantly differentially expressed genes (EREG, NIPAL4,
PLA2G4E, SERPINB7, SLC1A6 and WFDC12) in psoriatic KCs and
lesional skin/epidermis of patients with psoriasis were identified.
As novel psoriasis-associated candidate genes, these six genes were
located near the psoriasis-associated locus, and they were
relatively tissue-specific and highly expressed in human skin
tissue.
Identification of candidate genes
specifically and highly expressed in mouse skin epidermis
The tissue expression profile of the six novel
candidate genes in the mouse was confirmed by RT-qPCR, and the
results showed that four genes (EREG, NIPAL4,
SERPINB7 and WFDC12) were highly expressed in the
skin, but their expression levels were lower in other major organs
and tissues, including the large intestine, lung, liver, testis,
ovary, brain, spleen, and kidney, small intestine and heart
(Fig. 4A). This result was
consistent with their EST profiles (Fig. 3). However, PLA2G4E and
SLC1A6 exhibited no such skin specificity (Fig. 4A).
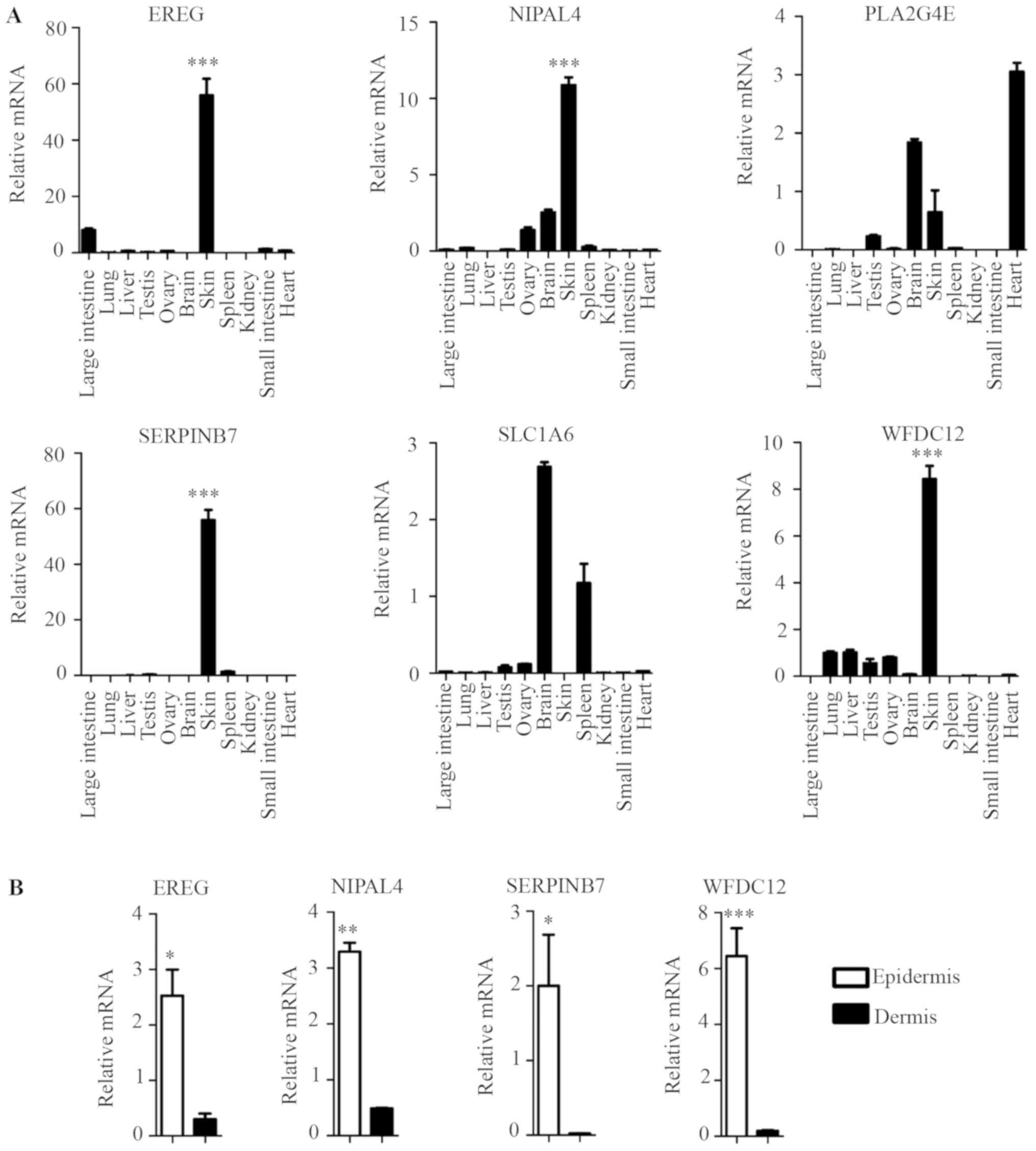 | Figure 4.Identification of key skin-specific
candidate genes. (A) RT-qPCR analysis was performed for each
indicated gene from C57BL/6 WT mice different tissues. Data
presented the mean ± standard deviation of three independent
experiments, n=5 mice, ***P<0.001 skin vs. the other major
organs and tissues (large intestine, lung, liver, testis, ovary,
brain, spleen, and kidney, small intestine and heart). (B) RT-qPCR
analysis was performed for each indicated gene from dissected mice
epidermis and dermis. Data presented the mean ± standard deviation
of three independent experiments, n=5 mice, *P<0.05,
**P<0.01, ***P<0.001 vs. dermis. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; EREG,
epiregulin; NIPAL4, NIPA like domain containing 4; PLA2G4E,
phospholipase A2 group IVE; SERPINB7, serpin family B member 7;
SLC1A6, solute carrier family 1 member 6; WFDC12, WAP
four-disulfide core domain 12. |
The epidermis is composed primarily of KCs that are
key skin-specific immune cells. The dermis is primarily composed of
other immunocyte cell types, such as fibroblasts, dendritic cells
(DCs), T helper cells, γδT cells (1,2).
EREG, NIPAL4, SERPINB7 and WFDC12 exhibited higher
expression levels in mouse epidermis than in dermis (Fig. 4B). This result suggested that these
genes exhibited higher expression levels in the key skin-specific
immune cells (KCs) than in other dermis immunocyte cell types,
indicating their skin specificity.
Identification of candidate genes that
are specifically increased in psoriasis-like skin epidermis
In order to define whether the candidate genes were
differentially regulated in psoriasis-like skin, mRNA from
IMQ-treated or untreated dorsal skin (day 0) was isolated and
analyzed by RT-qPCR. Upon treatment with IMQ, the four candidate
genes (EREG, NIPAL4, SERPINB7 and WFDC12) were
strongly induced on day 4 (EREG, NIPAL4, SERPINB7) and day 6
(WFDC12) (Fig. 5A). This result
suggested that these four candidate molecules may be involved in
local immune responses in psoriatic skin tissue.
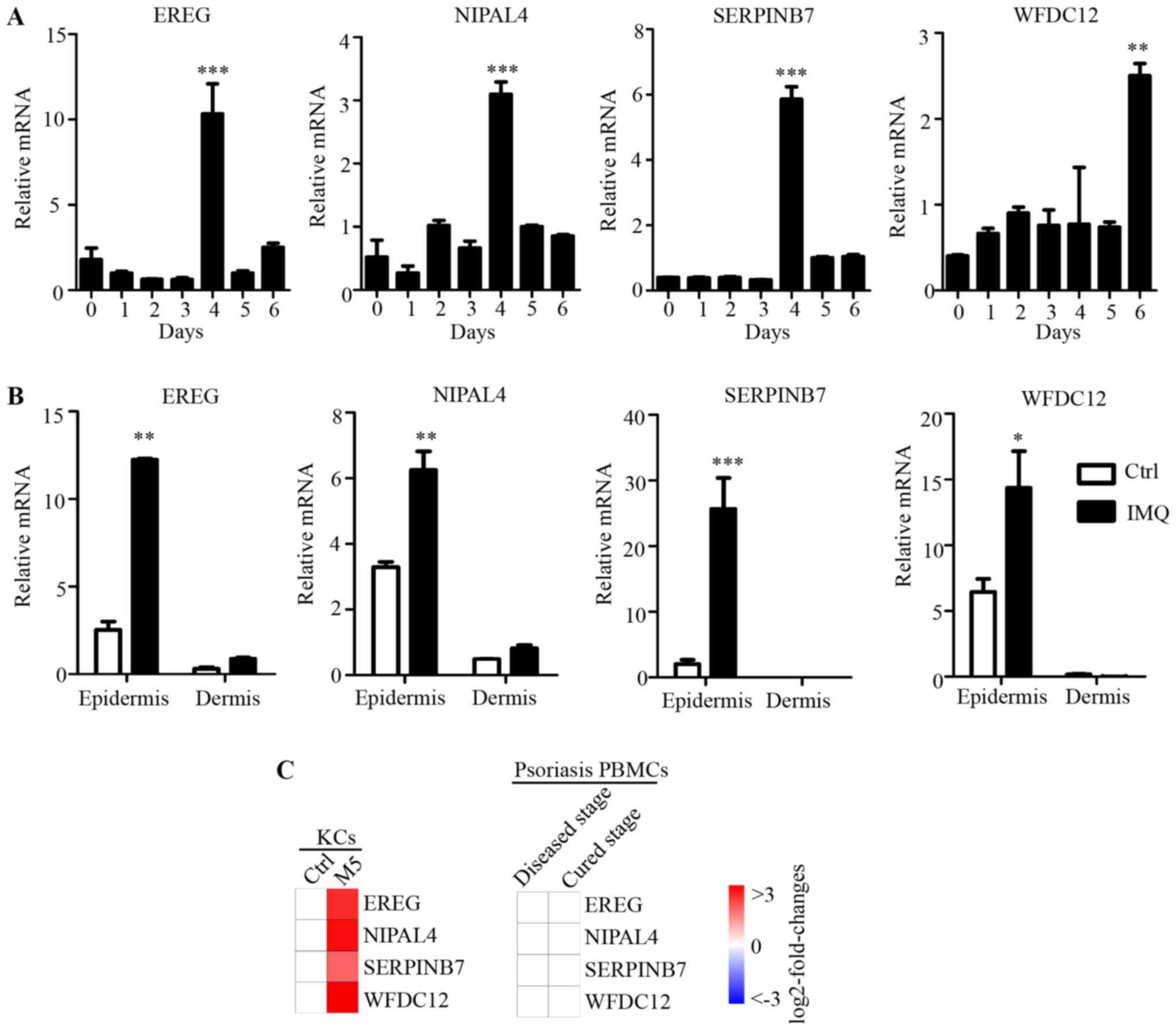 | Figure 5.Expression of key candidate genes
specifically increased in psoriatic epidermis. (A) The dorsal skin
of C57BL/6 (n=5) mice was treated daily with IMQ (55 mg) for 6
days, RT-qPCR analysis was performed for each indicated gene at day
0, 1, 2, 3, 4, 5 and 6. Data are presented the mean ± standard
deviation of three independent experiments, ***P<0.001, 4 day
vs. other groups; **P<0.01, 6 day vs. other groups. (B) The
dorsal skin of the C57BL/6 (n=5) mice were treated daily with IMQ
(55 mg) for 6 days, then the dermis and epidermis were obtained by
digested separation. RT-qPCR analysis of EREG, NIPAL4 and
SERPINB7 at 4 days after IMQ treatment, WFDC12 at 6
days after IMQ treatment. Data are presented the mean ± standard
deviation of three independent experiments, *P<0.05,
**P<0.01, ***P<0.001 vs. Ctrl. (C) Heat map of four key
candidate genes in psoriasis-like KCs and psoriasis PBMCs from
published microarray data (GSE40263). White, no change; green,
downregulated; red, upregulated. Color gradient, log2-fold-changes.
RT-qPCR, reverse transcription-quantitative polymerase chain
reaction; EREG, epiregulin; NIPAL4, NIPA like domain containing 4;
SERPINB7, serpin family B member 7; WFDC12, WAP four-disulfide core
domain 12; SLC1A6, solute carrier family 1 member 6; Ctrl, control;
IMQ, imiquimod; KC, keratinocytes; PBMCs, peripheral blood
mononuclear cells. |
Subsequently, in order to evaluate the psoriatic
skin specificity of the four candidate genes, mRNA expression
levels in dissected epidermis and dermis of IMQ-treated dorsal skin
or normal dorsal skin was analyzed by RT-qPCR. In normal skin and
IMQ-treated skin, the four genes were all highly expressed in
epidermis compared with the dermis (Fig. 5B). Furthermore, the mRNA expression
levels of EREG, NIPAL4 and SERPINB7 after 4 days, and
WFDC12 after 6 days of IMQ treatment were dramatically
increased in the epidermis of dorsal skin. However, in dermis,
there was no significant difference in the expression levels of
these four genes between normal and IMQ-treated skin. As
immunocytes are recruited to the dermis in psoriasis, this result
indicated that the expressions levels of these molecules were
differentially regulated in psoriatic epidermis (which is
predominantly KCs), but not in immunocytes that infiltrate the
psoriatic dermis. In addition, as many important immune molecules
may exhibit expression alteration in peripheral blood immunocytes,
published microarray data (GSE40263) of peripheral blood
mononuclear cells (PBMCs) from patients with psoriasis, was used in
the current study. There was no significant difference in the PBMCs
expression levels of these four genes in the psoriasis and healthy
control groups (Fig. 5C). Combined
with differences in expression of these four genes induced by M5 in
the psoriasis-like KC model (Fig.
5C), these results suggested that the expression of EREG,
NIPAL4, SERPINB7 and WFDC12 were specifically regulated
and highly expressed in psoriatic KCs (psoriatic skin-specific
immune cells), but not in the dermis or PBMCs involved in the local
immune response to psoriasis. Thus, these genes also were novel
psoriatic skin-specific genes, and may serve a unique role in the
pathogenesis of psoriasis, but this needs to be investigated
further.
Differential expression pattern of
candidate genes in SIRS
In order to confirm whether the candidate genes were
involved in local immune responses in the skin, the expression
levels of the five genes (TNF-α, EREG, NIPAL4,
SERPINB7 and WFDC12) were determined by RT-qPCR in the
LPS-induced SIRS and normal saline group. There was no difference
in expression of the five genes in skin tissue between the two
groups, but significantly higher levels of TNF-α were detected in
the large intestine, lung, liver, testis, ovary, brain, spleen,
kidney, small intestine, and heart of the SIRS group compared with
the saline-treated control. These results suggested that LPS can
induce inflammatory response of numerous tissues, but not in the
skin in the SIRS model. Furthermore, it was found that the
expression levels of EREG in lung, kidney, small intestine
and heart of the SIRS group were higher compared with those of
normal saline group. The expression levels of NIPAL4 in lung
and small intestine were significantly higher in the SIRS model
compared with the control mice. WFDC12 exhibited higher
expression in lung, liver and heart of the SIRS model compared with
those of the control mice. By contrast, there was no significant
difference in the SERPINB7 expression levels in all tissues
between the two groups. This result suggested that SERPINB7
may not participate in the local immune responses in the various
tissues (large intestine, lung, liver, testis, ovary, brain,
spleen, kidney, small intestine and heart) of the SIRS model
examined (Fig. 6).
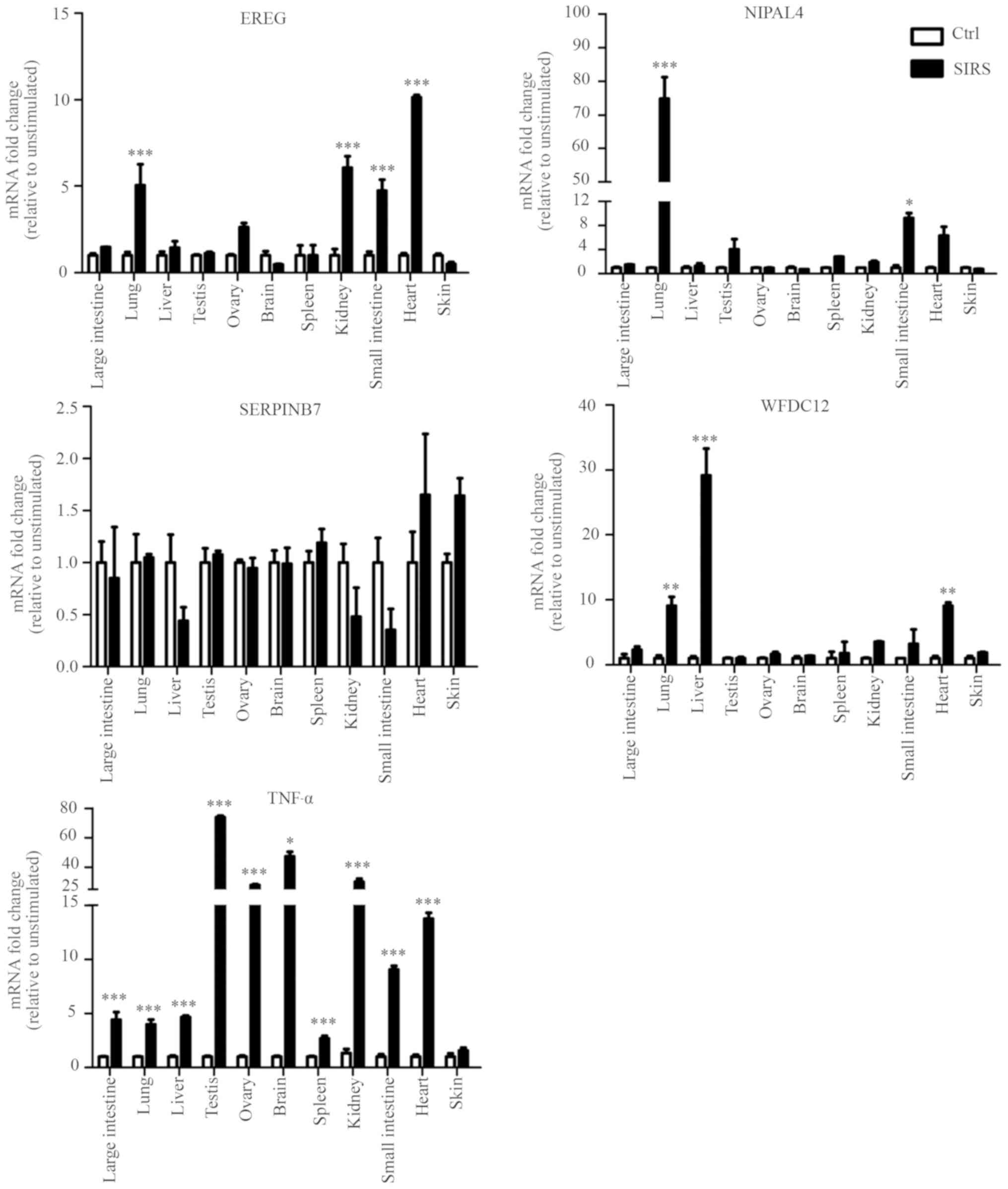 | Figure 6.Differential expression patterns of
candidate genes in SIRS. Tissue expression patterns of
psoriatic-specific candidate genes in lipopolysaccharide-induced
SIRS and normal saline group (n=5, each). Reverse
transcription-quantitative polymerase chain reaction analysis of
the indicated genes in various tissues was performed, and data
presented the mean ± standard deviation of three independent
experiments, *P<0.05, **P<0.01, ***P<0.001 vs. the Ctrl
group. EREG, epiregulin; NIPAL4, NIPA like domain containing 4;
Ctrl, control; SIRS, systemic inflammatory response syndrome;
SERPINB7, serpin family B member 7; WFDC12, WAP four-disulfide core
domain 12; TNFα, tumor necrosis factor-α. |
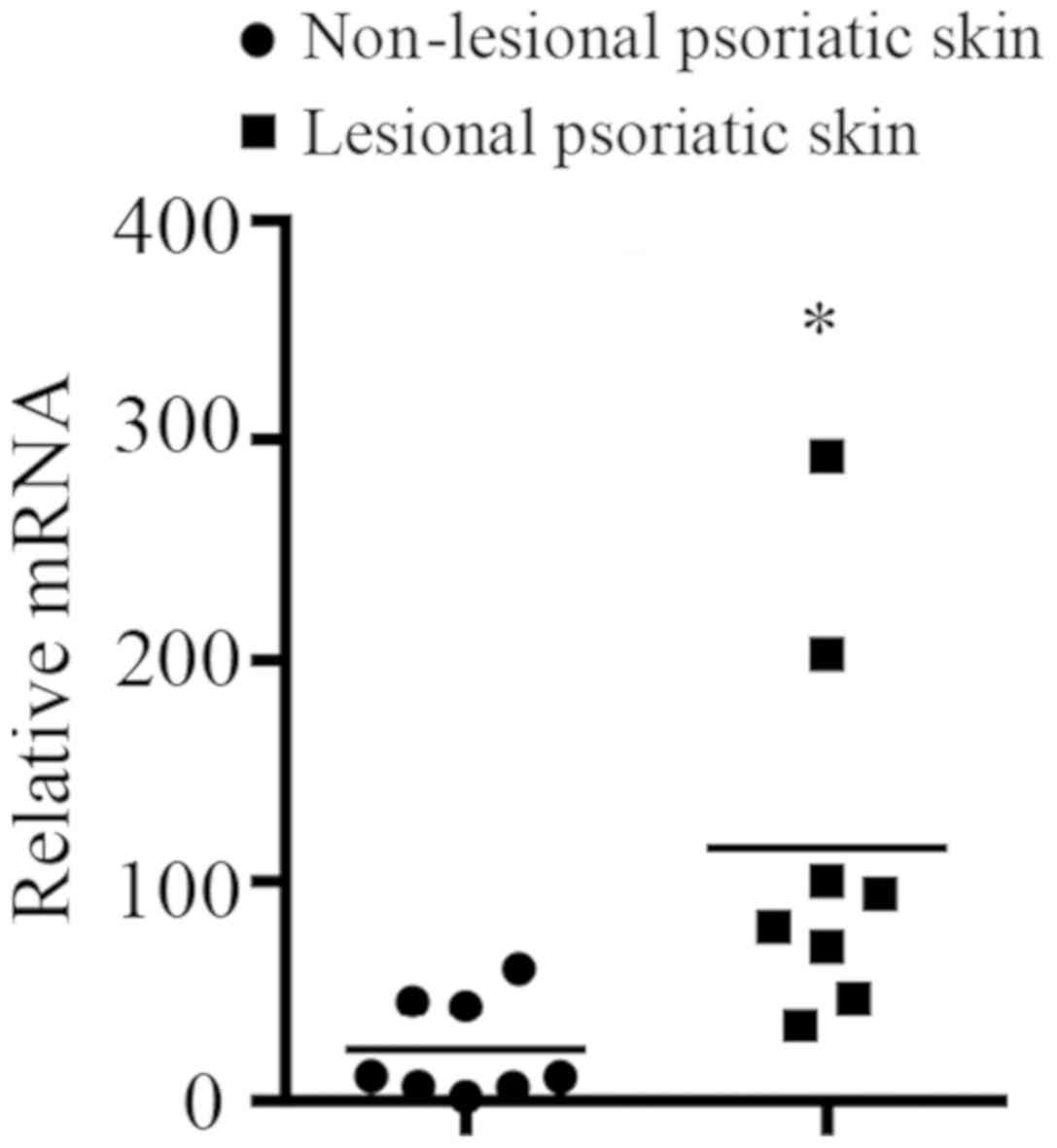
SERPINB7 mRNA expression levels in
lesional and non-lesional psoriatic skin of patients with
psoriasis
mRNA from lesional and non-lesional psoriatic skin
of patients with psoriasis was isolated and was examined by
RT-qPCR. The mRNA expression level of SERPINB7 (selected as
it was involved in the activated immune response of the skin, not
other tissues) in lesional psoriatic skin of patients with
psoriasis was significantly higher than in non-lesional psoriatic
skin of the same patients (Fig.
7).
Discussion
Psoriasis is a common skin disease affecting 2% of
the population worldwide and is characterized by increased
proliferation and abnormal differentiation of KCs (33). There are not many effective
psoriatic skin-specific targeted drugs to control the symptoms. In
the present study, DEGs were detected from KCs (a skin-specific
immune cell type), derived from psoriatic and non-psoriatic human
and mice tissues, from a SIRS model, and skin-specific genes were
identified. The present study introduced a cutaneous
tissue-specific target for skin-related diseases treatment and at
the same time provided a novel method for the exploration of
unknown cutaneous tissue-specific targets for disease
treatment.
Many DEGs identified in previous studies were
localized to defined lesional psoriatic skin, or the epidermis and
dermis of psoriatic lesions (18,19),
whereas expression pattern analysis in a psoriasis-like KCs model
has not been reported frequently. In the present study, the gene
expression profile was produced using a microarray analysis of
psoriasis-like KCs. Although there have been data sets produced
single cytokine-induced gene expression changes in cultured KCs
(34,35), it appears that single cytokine
stimulation has a limited effect on KCs, namely, a limited number
and a limited modulated expression of targeted genes, reflecting
only partial features of psoriasis (16). In previously published
transcriptional profiling experiments, gene sets for KC responses
to cytokines involved in psoriasis were curated, including IL-17,
TNF-α, and IL-22, alone and as a combination (36–39).
In the present study, a psoriasis-like KCs model was established by
treatment with IL-17, IL-1α, IL-22, TNF-α and oncostatin-M, which
produced a strong transcriptional effect on KCs chemokines,
cytokines, and antimicrobial peptide production, and these cells
exhibit a psoriasis-like profile (16,40).
In the present study, by comparing transcriptomes of psoriasis-like
KCs and previously published data sets of lesional psoriatic
skin/epidermis, it was revealed that enriched functions of DEGs had
highly similar patterns in the current study and the previously
published data. Thus, psoriatic skin-specific genes were identified
by analysis of DEGs in psoriasis-like KCs.
Psoriasis is a complex multifactorial disease and
the development of this disease remains largely unexplored. Recent
research identified psoriasis susceptibility loci and genes that
are closely associated with the pathogenesis of psoriasis (5). Genome-wide linkage scans and high
throughput studies have been used to identify genes responsible for
familial psoriasis and several susceptibility loci (41). In the present study, an integrated
approach was used, which may be helpful for exploring the
contributory factors involved in the initiation of a complex
multifactorial disease such as psoriasis.
ESTs derived from different cDNA libraries can be
prepared from different tissues, organs or cell types. It provides
a rapid and efficient approach for deciphering gene expression
levels in different tissues and screening tissue-specific molecules
(42,43). Previous studies have used EST data
to identify tissue-specific genes in the human prostate (44), heart (45), retina (46) and in cancer tissue (47). These studies highlight the
advantages of using this approach. In the present study, integrated
analysis of human ESTs provided a robust platform for
psoriasis-like KCs transcriptome screening. Four genes with
skin-specific expression were identified, indicating that EST
assessment was highly accurate.
Gerber et al (48), using the Body Index of Human Gene
Expression database and comparing the ratio of mean gene expression
in skin with other tissue/cell types, investigated eight genes out
of the top 100 genes preferentially expressed in normal human skin.
The expression profiles of these eight candidate genes (mucin
like 1, WFDC5, SERPINB7, chromosome 5 open reading frame 46,
transmembrane protein 45A, G protein-coupled receptor 115, cadherin
related family member 1 and G protein-coupled receptor
87) were analyzed in five tissues (skin, spleen, kidney, brain,
liver) and four cell types (keratinocytes, fibroblasts, PBMCs,
endothelial cells). The expression levels in cytokine-stimulated
keratinocytes and in biopsies of skin diseases were analyzed
(48). In the present study, novel
candidate genes were investigated in another manner. Initially,
based on gene description and publications from the human gene
database (GeneCards), 73 genes that were closely associated with
psoriasis were excluded. Subsequently, based on the summary of
genetic loci linked to psoriasis, 165 candidate genes were
identified. Finally, as skin specific cells, KCs constitute the
majority of cells in the skin's epidermis; therefore, the
expression of the six candidate genes (EREG, NIPAL4, PLA2GE,
SERPINB7, SLC1A6 and WFDC12) was examined in epidermis
and dermis samples form mice.
The skin is composed of two distinct regions, the
epidermis and dermis, it contains a variety of immune cells
(1,2). The KCs are the predominant cell type
in the epidermis, it is highly specialized epithelial cells
designed to perform a very specific function. The dermis contains
cells of the immune system including T cells and DCs (1,2,4). In
the normal condition, four candidate genes (EREG, NIPAL4,
SERPINB7 and WFDC12) were highly expressed in skin
compared with other tissues. In skin tissue, the expression of
these genes was higher in the epidermis than in the dermis. This
suggested that the four candidate genes were highly expressed in
KCs. KCs exhibit hyperproliferation and abnormal differentiation in
psoriatic epidermis, and a large number of inflammatory cells
infiltrate into the dermal lesions (5,6). The
expression levels of the four candidate genes were increased in
psoriasis-like skin lesion. Furthermore, it was revealed that the
expression levels of these candidate genes were increased in
psoriatic epidermis compared with normal controls; however, the
levels were not different in psoriatic dermis compared with normal
controls. Furthermore, in psoriasis-like KCs, the expression levels
of these genes were increased compared with untreated KCs. This
suggested that the four candidate genes were highly expressed in
psoriasis-like KCs, and they may be involved in the local immune
response of psoriatic KCs, suggesting that they have psoriatic
skin-specific roles.
The pathophysiology of SIRS involves a systemic
immune response that affects pulmonary, gastrointestinal and renal
function. As LPS induced the systemic expression of the
proinflammatory cytokine TNF-α in various tissues (large intestine,
lung, liver, testis, ovary, brain, spleen, kidney, small intestine
and heart), the systemic inflammatory response was activated.
However, TNF-α and the four other detected genes (EREG, NIPAL4,
SERPINB7 and WFDC12) exhibited no difference in
expression the skin between the SIRS and control group. This
suggested that LPS may have not induced an inflammatory response in
skin tissue in the SIRS model. EREG, NIPAL4, and
WFDC12 were upregulated in certain tissues in the SIRS model
compared with the normal control. There was no significant
difference in SERPINB7 expression between the two groups in
the all tissues analyzed. These results suggested that
SERPINB7 was not involved in the activated immune response
of various tissues, except for the skin. SERPINB7, a serpin
peptidase inhibitor, has critical roles in the immune system; it
can increase mesangial cell proliferation and extracellular matrix
(ECM) deposition and markedly suppress cell motility and invasion
(49). SERPINB7 appears to
be involved in maintaining tissue integrity by preserving ECM
homeostasis, and loss of expression may lead to loss of cell
adhesion and tissue integrity (49). SERPINB7 exhibits substantial
expression variation in skin disorders, such as palmoplantar
keratosis (50). Additionally, the
mRNA levels of SERPINB were significantly higher in lesional
psoriatic skin than in non-lesional psoriatic skin of patients with
psoriasis. Therefore, SERPINB7 requires further
investigation to clarify its potential role in the pathogenesis of
psoriasis.
As novel psoriatic skin-specific genes, EREG,
NIPAL4 and WFDC12 may also be valuable candidates for
further exploration. It was previously reported that EREG
and WFDC12 serve the critical immuno-regulatory roles in
skin. EREG encodes epiregulin, which is a secreted peptide
hormone and member of the epidermal growth factor family of
proteins, is overexpressed in psoriatic epidermis (51). Functions associated with this gene
include growth factor activity and epidermal growth factor receptor
binding. Secreted epiregulin induces downregulation of inflammatory
cytokine IL-18 mRNA expression in KCs (52) and can also stimulate proliferation
of KCs. WFDC12 is one of an 18-member family of secreted proteins
reported as protease inhibitors, and is an antimicrobial peptide
also reported to participate in inflammation and host defense
(53). However, the expression of
EREG and WFDC12 in psoriatic skin lesions is
currently unknown. In addition, there are few studies on the role
of NIPAL4 in the skin. NIPAL4, also known as
ichthyin, is composed of several transmembrane domains. It is
associated with keratins and desmosomes in KCs and is involved in
lipid metabolism (54,55). Mutations in this gene have been
associated with autosomal recessive congenital ichthyosis (56).
The present data strongly suggests that these genes
(EREG, NIPAL4, SERPINB7 and WFDC12) were specifically
expressed in psoriatic skin, and they may be involved in the local
immune response of psoriatic skin. Further studies are required to
gain further insights into their regulation and potential role in
skin disorders.
In the present study, a pathological
tissue-specific molecular screening method was established. This
method used psoriatic transcriptome data, psoriasis-associated gene
information, genetic loci associated with psoriasis and
normal/pathological tissue expression patterns. Through the
screening procedure, candidate genes with genetic loci associated
with psoriasis that were specific expressed in normal skin and
exhibited high expression in psoriatic KCs, rather than in other
inflammatory cells or tissues, were identified. The present study
identified the potential key pathogenic tissue-specific molecules
for diseases, and the psoriatic skin-specific genes (EREG,
NIPAL4, SERPINB7 and WFDC12) may represent potential
biomarkers or drug targets for the development of future
diagnostics/therapeutics to treat psoriasis.
Acknowledgements
The authors thanks to our research assistants, Ms
Wenling Wu and Mr Yifan Zhou (Department of Biotherapy, Cancer
Center/Collaborative Innovation Center for Biotherapy, West China
Hospital, Sichuan University) for their technical assistance.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 31271483,
81703132, 81472650, 81673061, 81573050, 31872739, 81602763 and
8160070706), the China Postdoctoral Science Foundation funded
project (grant no. 2018M631087), the National Science and
Technology Major Project (grant nos. 2018ZX09303006-001-006 and
2018ZX09201004-003, 2012ZX10002006-003-001 and
2013ZX09301304001-003), the Sichuan Provincial Outstanding Youth
Fund (grant no. 2015JQ0025).
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW and JL designed the experiments and the present
study. ZW, HPZ, HZ, NH, XW and JZ collected data and did
experiments. ZW, XL, XT, ZH, XZ and WL analyzed the data. ZW, HPZ,
HZ, XW and JZ contributed to critical revisions of the text.
Ethics approval and consent to
participate
The present study was performed in accordance with
the principles of the Helsinki Declaration and approved by the
Ethics Committee of the West China Hospital, Sichuan University.
Written informed consent was obtained from all study participants
prior to the present study. The animal protocols were approved by
the Committee on the Ethics of Animal Experiments of the Sichuan
University (Chengdu, China). The experimental procedures were
conducted according to the ethical guidelines for the care and use
of laboratory animals of the National Institutes of Health and the
IASP.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nestle FO, Di Meglio P, Qin JZ and
Nickoloff BJ: Skin immune sentinels in health and disease. Nat Rev
Immunol. 9:679–691. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heath WR and Carbone FR: The skin-resident
and migratory immune system in steady state and memory: Innate
lymphocytes, dendritic cells and T cells. Nature Immunol.
14:978–985. 2013. View Article : Google Scholar
|
|
3
|
Boehncke WH and Schön MP: Psoriasis.
Lancet. 386:983–994. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nestle FO, Kaplan DH and Barker J:
Psoriasis. N Engl J Med. 361:496–509. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lowes MA, Suárez-Fariñas M and Krueger JG:
Immunology of psoriasis. Annu Rev Immunol. 32:227–255. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bos JD, de Rie MA, Teunissen MB and Piskin
G: Psoriasis: Dysregulation of innate immunity. Br J Dermatol.
152:1098–1107. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Perera GK, Di Meglio P and Nestle FO:
Psoriasis. Annu Rev Pathol. 7:385–422. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim J and Krueger JG: The
immunopathogenesis of psoriasis. Dermatol Clin. 33:13–23. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crow JM: Therapeutics: Silencing
psoriasis. Nature. 492 Suppl:S58–S59. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Williams SC: New biologic drugs get under
the skin of psoriasis. Nat Med. 18:6382012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu X, Fang L, Guo TB, Mei H and Zhang JZ:
Drug targets in the cytokine universe for autoimmune disease.
Trends Immunol. 34:120–128. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iborra M, Beltrán B, Bastida G, Aguas M
and Nos P: Infliximab and adalimumab-induced psoriasis in Crohn's
disease: A paradoxical side effect. J Crohns Colitis. 5:157–161.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matos TR, O'Malley JT, Lowry EL, Hamm D,
Kirsch IR, Robins HS, Kupper TS, Krueger JG and Clark RA:
Clinically resolved psoriatic lesions contain psoriasis-specific
IL-17-producing αβ T cell clones. J Clin Invest. 127:4031–4041.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Leonardi C, Matheson R, Zachariae C,
Cameron G, Li L, Edson-Heredia E, Braun D and Banerjee S:
Anti-interleukin-17 monoclonal antibody ixekizumab in chronic
plaque psoriasis. N Engl J Med. 366:1190–1199. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Langley RG and Ellis CN: Evaluating
psoriasis with psoriasis area and severity index, psoriasis global
assessment, and lattice system physician's global assessment. J Am
Acad Dermatol. 51:563–569. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guilloteau K, Paris I, Pedretti N,
Boniface K, Juchaux F, Huguier V, Guillet G, Bernard FX, Lecron JC
and Morel F: Skin inflammation induced by the synergistic action of
IL-17A, IL-22, oncostatinM, IL-1{alpha}, and TNF-{alpha}
recapitulates some features of psoriasis. J Immunol. 184:5263–5270.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Teng X, Hu Z, Wei X, Wang Z, Guan T, Liu
N, Liu X, Ye N, Deng G, Luo C, et al: IL-37 ameliorates the
inflammatory process in psoriasis by suppressing proinflammatory
cytokine production. J Immunol. 192:1815–1823. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li B, Tsoi LC, Swindell WR, Gudjonsson JE,
Tejasvi T, Johnston A, Ding J, Stuart PE, Xing X, Kochkodan JJ, et
al: Transcriptome analysis of psoriasis in a large case-control
sample: RNA-seq provides insights into disease mechanisms. J Invest
Dermatol. 134:1828–1838. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mitsui H, Suárez-Fariñas M, Belkin DA,
Levenkova N, Fuentes-Duculan J, Coats I, Fujita H and Krueger JG:
Combined use of laser capture microdissection and cDNA microarray
analysis identifies locally expressed disease-related genes in
focal regions of psoriasis vulgaris skin lesions. J Invest
Dermatol. 132:1615–1626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Genetic Analysis of Psoriasis Consortium
& the Wellcome Trust Case Control Consortium 2, ; Strange A,
Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band
G, Bellenguez C, et al: A genome-wide association study identifies
new psoriasis susceptibility loci and an interaction between HLA-C
and ERAP1. Nat Genet. 42:985–990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XJ, Huang W, Yang S, Sun LD, Zhang
FY, Zhu QX, Zhang FR, Zhang C, Du WH, Pu XM, et al: Psoriasis
genome-wide association study identifies susceptibility variants
within LCE gene cluster at 1q21. Nat Genet. 41:205–210. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
de Cid R, Riveira-Munoz E, Zeeuwen PL,
Robarge J, Liao W, Dannhauser EN, Giardina E, Stuart PE, Nair R,
Helms C, et al: Deletion of the late cornified envelope LCE3B and
LCE3C genes as a susceptibility factor for psoriasis. Nat Genet.
41:211–215. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nair RP, Duffin KC, Helms C, Ding J,
Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, et
al: Genome-wide scan reveals association of psoriasis with IL-23
and NF-kappaB pathways. Nat Genet. 41:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun LD, Cheng H, Wang ZX, Zhang AP, Wang
PG, Xu JH, Zhu QX, Zhou HS, Ellinghaus E, Zhang FR, et al:
Association analyses identify six new psoriasis susceptibility loci
in the Chinese population. Nat Genet. 42:1005–1009. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu Y, Helms C, Liao W, Zaba LC, Duan S,
Gardner J, Wise C, Miner A, Malloy MJ, Pullinger CR, et al: A
genome-wide association study of psoriasis and psoriatic arthritis
identifies new disease loci. PLoS Genet. 4:e10000412008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stuart PE, Nair RP, Ellinghaus E, Ding J,
Tejasvi T, Gudjonsson JE, Li Y, Weidinger S, Eberlein B, Gieger C,
et al: Genome-wide association analysis identifies three psoriasis
susceptibility loci. Nat Genet. 42:1000–1004. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gudjonsson JE, Ding J, Johnston A, Tejasvi
T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR and Elder JT:
Assessment of the psoriatic transcriptome in a large sample:
Additional regulated genes and comparisons with in vitro models. J
Invest Dermatol. 130:1829–1840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tsoi LC, Spain SL, Knight J, Ellinghaus E,
Stuart PE, Capon F, Ding J, Li Y, Tejasvi T, Gudjonsson JE, et al:
Identification of 15 new psoriasis susceptibility loci highlights
the role of innate immunity. Nat Genet. 44:1341–1348. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Capon F, Bijlmakers MJ, Wolf N, Quaranta
M, Huffmeier U, Allen M, Timms K, Abkevich V, Gutin A, Smith R, et
al: Identification of ZNF313/RNF114 as a novel psoriasis
susceptibility gene. Hum Mol Genet. 17:1938–1945. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Antal C, Teletin M, Wendling O, Dgheem M,
Auwerx J and Mark M: Tissue collection for systematic phenotyping
in the mouse. Curr Protoc Mol Biol Chapter. 29:Unit 29A.4. 2007.
View Article : Google Scholar
|
|
31
|
El Malki K, Karbach SH, Huppert J, Zayoud
M, Reissig S, Schüler R, Nikolaev A, Karram K, Münzel T, Kuhlmann
CR, et al: An alternative pathway of imiquimod-induced
psoriasis-like skin inflammation in the absence of interleukin-17
receptor a signaling. J Invest Dermatol. 133:441–451. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greb JE, Goldminz AM, Elder JT, Lebwohl
MG, Gladman DD, Wu JJ, Mehta NN, Finlay AY and Gottlieb AB:
Psoriasis. Nat Rev Dis Primers. 2:160822016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Swindell WR, Xing X, Stuart PE, Chen CS,
Aphale A, Nair RP, Voorhees JJ, Elder JT, Johnston A and Gudjonsson
JE: Heterogeneity of inflammatory and cytokine networks in chronic
plaque psoriasis. PLoS One. 7:e345942012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yano S, Banno T, Walsh R and Blumenberg M:
Transcriptional responses of human epidermal keratinocytes to
cytokine interleukin-1. J Cell Physiol. 214:1–13. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wolk K, Kunz S, Witte E, Friedrich M,
Asadullah K and Sabat R: IL-22 increases the innate immunity of
tissues. Immunity. 21:241–254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Banno T, Gazel A and Blumenberg M: Effects
of tumor necrosis factor-alpha (TNF alpha) in epidermal
keratinocytes revealed using global transcriptional profiling. J
Biol Chem. 279:32633–32642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Nograles KE, Zaba LC, Guttman-Yassky E,
Fuentes-Duculan J, Suárez-Fariñas M, Cardinale I, Khatcherian A,
Gonzalez J, Pierson KC, White TR, et al: Th17 cytokines interleukin
(IL)-17 and IL-22 modulate distinct inflammatory and
keratinocyte-response pathways. Br J Dermatol. 159:1092–1102.
2008.PubMed/NCBI
|
|
39
|
Chiricozzi A, Guttman-Yassky E,
Suárez-Fariñas M, Nograles KE, Tian S, Cardinale I, Chimenti S and
Krueger JG: Integrative responses to IL-17 and TNF-α in human
keratinocytes account for key inflammatory pathogenic circuits in
psoriasis. J Invest Dermatol. 131:677–687. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rabeony H, Petit-Paris I, Garnier J,
Barrault C, Pedretti N, Guilloteau K, Jegou JF, Guillet G, Huguier
V, Lecron JC, et al: Inhibition of keratinocyte differentiation by
the synergistic effect of IL-17A, IL-22, IL-1α, TNFα and oncostatin
M. PLoS One. 9:e1019372014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chandra A, Ray A, Senapati S and
Chatterjee R: Genetic and epigenetic basis of psoriasis
pathogenesis. Mol Immunol. 64:313–323. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Beaudoing E and Gautheret D:
Identification of alternate polyadenylation sites and analysis of
their tissue distribution using EST data. Genome Res. 11:1520–1526.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boutrot F, Chantret N and Gautier MF:
Genome-wide analysis of the rice and Arabidopsis non-specific lipid
transfer protein (nsLtp) gene families and identification of wheat
nsLtp genes by EST data mining. BMC Genomics. 9:862008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Skrabanek L and Campagne F: TissueInfo:
High-throughput identification of tissue expression profiles and
specificity. Nucleic Acids Res. 29:E1022001. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Mégy K, Audic S and Claverie JM:
Heart-specific genes revealed by expressed sequence tag (EST)
sampling. Genome Biol. 3:RESEARCH00742002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Malone K, Sohocki MM, Sullivan LS and
Daiger SP: Identifying and mapping novel retinal-expressed ESTs
from humans. Mol Vis. 5:51999.PubMed/NCBI
|
|
47
|
Wu TH, Chu LJ, Wang JC, Chen TW, Tien YJ,
Lin WC and Ng WV: Meta-analytical biomarker search of EST
expression data reveals three differentially expressed candidates.
BMC Genomics. 13 Suppl 7:S122012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gerber PA, Hevezi P, Buhren BA, Martinez
C, Schrumpf H, Gasis M, Grether-Beck S, Krutmann J, Homey B and
Zlotnik A: Systematic identification and characterization of novel
human skin-associated genes encoding membrane and secreted
proteins. PLoS One. 8:e639492013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chou RH, Wen HC, Liang WG, Lin SC, Yuan
HW, Wu CW and Chang WS: Suppression of the invasion and migration
of cancer cells by SERPINB family genes and their derived peptides.
Oncol Rep. 27:238–245. 2012.PubMed/NCBI
|
|
50
|
Hida T, Okura M, Kamiya T and Yamashita T:
Nagashima-type palmoplantar keratosis caused by compound
heterozygous mutations in SERPINB7. Eur J Dermatol. 25:202–203.
2015.PubMed/NCBI
|
|
51
|
Shirakata Y, Kishimoto J, Tokumaru S,
Yamasaki K, Hanakawa Y, Tohyama M, Sayama K and Hashimoto K:
Epiregulin, a member of the EGF family, is over-expressed in
psoriatic epidermis. J Dermatol Sci. 45:69–72. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Shirasawa S, Sugiyama S, Baba I, Inokuchi
J, Sekine S, Ogino K, Kawamura Y, Dohi T, Fujimoto M and Sasazuki
T: Dermatitis due to epiregulin deficiency and a critical role of
epiregulin in immune-related responses of keratinocyte and
macrophage. Proc Natl Acad Sci USA. 101:13921–13926. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Haneda T, Imai Y, Uchiyama R, Jitsukawa O
and Yamanishi K: Activation of molecular signatures for
antimicrobial and innate defense responses in skin with
transglutaminase 1 deficiency. PLoS One. 11:e01596732016.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Dahlqvist J, Westermark GT, Vahlquist A
and Dahl N: Ichthyin/NIPAL4 localizes to keratins and desmosomes in
epidermis and Ichthyin mutations affect epidermal lipid metabolism.
Arch Dermatol Res. 304:377–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Klar J, Schweiger M, Zimmerman R, Zechner
R, Li H, Törmä H, Vahlquist A, Bouadjar B, Dahl N and Fischer J:
Mutations in the fatty acid transport protein 4 gene cause the
ichthyosis prematurity syndrome. Am J Hum Genet. 85:248–253. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Maier D, Mazereeuw-Hautier J, Tilinca M,
Cosgarea R and Jonca N: Novel mutation in NIPAL4 in a Romanian
family with autosomal recessive congenital ichthyosis. Clin Exp
Dermatol. 41:279–282. 2016. View Article : Google Scholar : PubMed/NCBI
|