Introduction
Fibroblast growth factor 15 (FGF15) was initially
identified in the developing nervous system of a mouse (1). Subsequently, the human ortholog,
fibroblast growth factor 19 (FGF19), was identified to be expressed
in the fetal brain (2). Previous
studies suggested that FGF15 functioned in cell division and
patterning within specific regions of the embryonic brain, spinal
cord and sensory organs (1,2).
Other previous studies demonstrated that FGF15/19 exhibited high
expression in the small intestine of adult mice and humans, where
they function as an enterohepatic signal to regulate hepatic
homeostasis (3,4). Further studies demonstrated that
circulating FGF15/19 as metabolic hormones functioned through
fibroblast growth factor receptor 4 (FGFR4) in the liver and
regulated numerous metabolism pathways, including hepatic bile acid
(BA), carbohydrate and protein metabolism (3–7).
FGF15 expression is regulated by farnesoid X
receptor (FXR), a BA receptor (3).
Retinoid X receptors (RXRs) are hypothesized to form a heterodimer
with FXR and serve a role in the transcriptional regulation of
Fgf15 (3). Vitamin D may
additionally activate the transcription of Fgf15 through
vitamin D receptor (8), whereas,
vitamin A upregulates Fgf15 expression through the RXR/FXR
heterodimer (8). Notably, Diet1
was demonstrated to promote FGF15 production at the
post-transcriptional level, and an absence of Diet1 was accompanied
by significantly reduced FGF15 secretion (9). These results demonstrate the present
understanding of Fgf15 regulation. Whether other
transcription factors are involved in the regulation of
Fgf15 expression is unclear.
Dexamethasone (Dex), a synthetic analog of
glucocorticoids (GCs), is a potent agonist for the glucocorticoid
receptor (GR). Due to its function of anti-inflammation, Dex has
been widely used in the treatment of acute inflammatory and
autoimmune diseases (10).
Unfortunately, accumulating evidence demonstrated that the clinical
use of GCs or Dex causes a lot of undesirable side effects
(11,12). For example, GC intake may result in
glucose intolerance and insulin resistance (11). Increased GCs may cause liver
steatosis and hyperlipidemia (12). Treatment with GCs additionally
leads to BA overproduction and gallstone disease (13). As FGF15 serves an important role in
glucose metabolism, insulin resistance and BA synthesis (3,14),
the effects of Dex on Fgf15 expression were hypothesized to
affect the homeostasis of BA and glucose, and may lead to the
adverse effects associated with treatments with GCs in the clinical
setting. The present study aimed to investigate the effect of Dex
on the expression of Fgf15 in the mouse ileum. It was
identified that treatment with Dex significantly downregulated
ileal Fgf15 expression, and this downregulation of
Fgf15 expression by Dex is due to GR-mediated suppression of
FXR transcriptional activity.
Materials and methods
Materials
Cholic acid (CA), pregnenolone-16-α-carbonitrile
(PCN) and Dex were purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany). An anti-GR antibody was purchased from
Epitomics (Abcam, Cambridge, UK) and an anti-GAPDH antibody was
purchased from GenScript (Piscataway, NJ, USA).
Animals and treatments
All animal studies were approved by the
Institutional Animal Care and Use Committee of the Wadsworth Center
(State University of New York, Albany, NY, USA) or the Animal
Ethics Committee of the Fujian Agriculture and Forestry University
(Fuzhou, China). In total, 120 adult male C56BL/6J mice (~25 g; 2–3
months old, 3 mice per group; breeding and feeding in the
laboratory) were used for all experiments. Animals were maintained
at 22°C, 50% relative humidity and a 12-h light/dark cycle in a
specific pathogen-free clean room with food and water ad
libitum. Dex dissolved in corn oil was administered to the mice
at various doses via intraperitoneal (i.p.) injection. For
dose-response experiments, mice were treated with Dex at three
different doses (0.8, 8 and 80 mg/kg) for 24 h. Animals were
sacrificed at the indicated timepoints following treatment with Dex
for tissue collection. Additionally, the weights of the mice and
tissue, and tissue morphology were examined to evaluate the
toxicity of treatment with Dex. PCN (25 mg/kg, dissolved in corn
oil) was administered via oral gavage, and mice were sacrificed at
24 h after treatment with PCN for tissue Collection.
For the co-treatment of CA and Dex, Dex at 80 mg/kg
or vehicle (corn oil) was administered by i.p. injection. Mice
fasted for 2 h prior to administration of CA (200 mg/kg in PBS by
oral gavage). After another 4 h, ileal epithelial cells were
collected for RNA preparation.
Sample collection and western blot
analysis
The small intestine was removed and flushed with ice
cold PBS. The small intestine was divided into three segments
designated the duodenum (proximal 5 cm), jejunum (middle 5 cm) and
ileum (distal 5 cm). The three segments were cut open
longitudinally and the epithelial cells were removed by gentle
scraping. The collected cells were washed once in cold PBS and
subsequently lysed in radioimmunoprecipitation assay buffer (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing Protease
& Phosphatase Inhibitor (Thermo Fisher Scientific, Inc.) and 5
mM EDTA, and homogenized using a polytron homogenizer. The
homogenates were mixed for 10 min at 4°C, and subsequently
centrifuged at 14,000 g and 4°C for 15 min. The supernatant was
collected and the protein concentration was determined by the
bicinchoninic acid method (Pierce; Thermo Fisher Scientific,
Inc.).
Proteins (50 µg) were resolved on 10% NuPAGE
Bis-Tris-gels (Invitrogen; Thermo Fisher Scientific, Inc.) and
subsequently transferred to nitrocellulose membranes. For
immunodetection, subsequent to blocking with 5% fat-free milk at
room temperature for 1 h, pre-cut membranes were incubated with
monoclonal rabbit anti-GR (Epitomics; Abcam; cat. no. EPR4595;
1:4,000) and goat anti-GAPDH (GenScript; cat. no. A00191-40;
1:40,000) antibodies at 4°C overnight. Subsequent to washing, the
membranes were incubated with goat anti-rabbit immunoglobulin G
(IgG) heavy and light chain (H&L) horseradish peroxidase
(HRP)-conjugated antibod (GenScript; cat. no. A00098; 1:8,000) or
donkey anti-goat IgG H&L HRP-conjugated antibody (GenScript;
cat. no. A00178; 1:8,000) according to the host species of the
primary antibody at room temperature for 1 h. Pierce™ Enhanced
Chemiluminescent Western Blotting Substrate (Thermo Fisher
Scientific, Inc.) was used for the detection of HRP on the
immunoblots. Immunoblot quantification was performed using a
Bio-Rad ChemiDoc XRS+ System (Bio-Rad Laboratories, Inc., Hercules,
CA, USA) with Image Lab 3.0 (Bio-Rad Laboratories, Inc.).
Ex vivo culture of ileum tissues
Immediately after harvesting the intestine, 5 cm of
the terminal ileum was flushed with PBS to remove the contents and
was subsequently cut into small segments (1×3 mm). The segments
were cultured at 37°C for 4 h in Dulbecco's modified Eagle's medium
(Thermo Fisher Scientific, Inc.) supplemented with 10%
heat-inactivated fetal bovine serum (Thermo Fisher Scientific,
Inc.), 100 units/ml penicillin and 100 µg/ml streptomycin. Dex (1
µM final concentration) or vehicle (ethanol), and CA (100 µg/ml) or
PBS were added to the culture medium. The epithelial cells were
collected at the end of 4-h culture for RNA preparation.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis
Total RNA was prepared from the enterocytes of
individual mice, with the use of TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), as previously
described (15). RNA concentration
and purity were determined spectrally, and the integrity of the RNA
samples was assessed by ethidium bromide staining following agarose
gel electrophoresis. RT-PCR was performed using 3 µg total RNA and
SYBR Green PCR Master PCR Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.), as previously described (16). The comparative 2−∆∆Cq
method was used for quantifying the relative mRNA expression levels
(17,18). PCR primers for FXR (19), PXR (20), Fgf15 (3), small heterodimer partner (SHP)
(3), glucocorticoid-induced
leucine zipper (GILZ) (21), apical sodium dependent bile acid
transporter (ASBT) (22),
ileum bile acid binding protein (IBABP) (22) and cytochrome P450, family 3,
subfamily a, polypeptide 11 Cyp3a11 (23) were as follows: FXR,
5′-CCAACCTGGGTTTCTACCC-3′ (forward) and 5′-CACACAGCTCATCCCCTTT-3′
(reverse); PXR, 5′-CAAGGCCAATGGCTACCA-3′ (forward) and
5′-CGGGTGATCTCGCAGGTT-3′ (reverse); Fgf15,
5′-GAGGACCAAAACGAACGAAATT-3′ (forward) and
5′-ACGTCCTTGATGGCAATCG-3′ (reverse); SHP,
5′-CGATCCTCTTCAACCCAGATG-3′ (forward) and
5′-AGGGCTCCAAGACTTCACACA-3′ (reverse); GILZ,
5′-CAGCAGCCACTCAAACCAGC-3′ (forward) and;
5′-ACCACATCCCCTCCAAGCAG-3′ (reverse); ASBT,
5′-TGGGTTTCTTCCTGGCTAGACT-3′ (forward) and
5′-TGTTCTGCATTCCAGTTTCCAA-3′ (reverse); IBABP,
5′-AGATCATCACAGAGGTCCAGC-3′ (forward) and
5′-GGTAGCCTTGAACTTCTTGCC-3′ (reverse); Cyp3a11,
5′-GACAAACAAGCAGGGATGG-3′ (forward) and 5′-AATGTGGGGGACAGCAAAG-3′
(reverse); and U36B4, 5′-CGTCCTCGTTGGAGTGACA-3′ (forward) and
5′-CGGTGCGTCAGGGATTG-3′ (reverse). The levels of the target mRNAs
in various total RNA preparations were normalized by the level of
U36B4 mRNA (8).
Statistical analysis
All experiments were repeated at least three times.
Data are presented as the mean ± standard deviation. Statistical
analysis was performed using Student's t-test with SPSS 11.5
software (SPSS, Inc., Chicago, IL, USA). For multiple comparisons,
one-way analysis of variance followed by Fisher's Least Significant
Difference test was used. P<0.05 was considered to indicate a
statistically significant difference.
Results
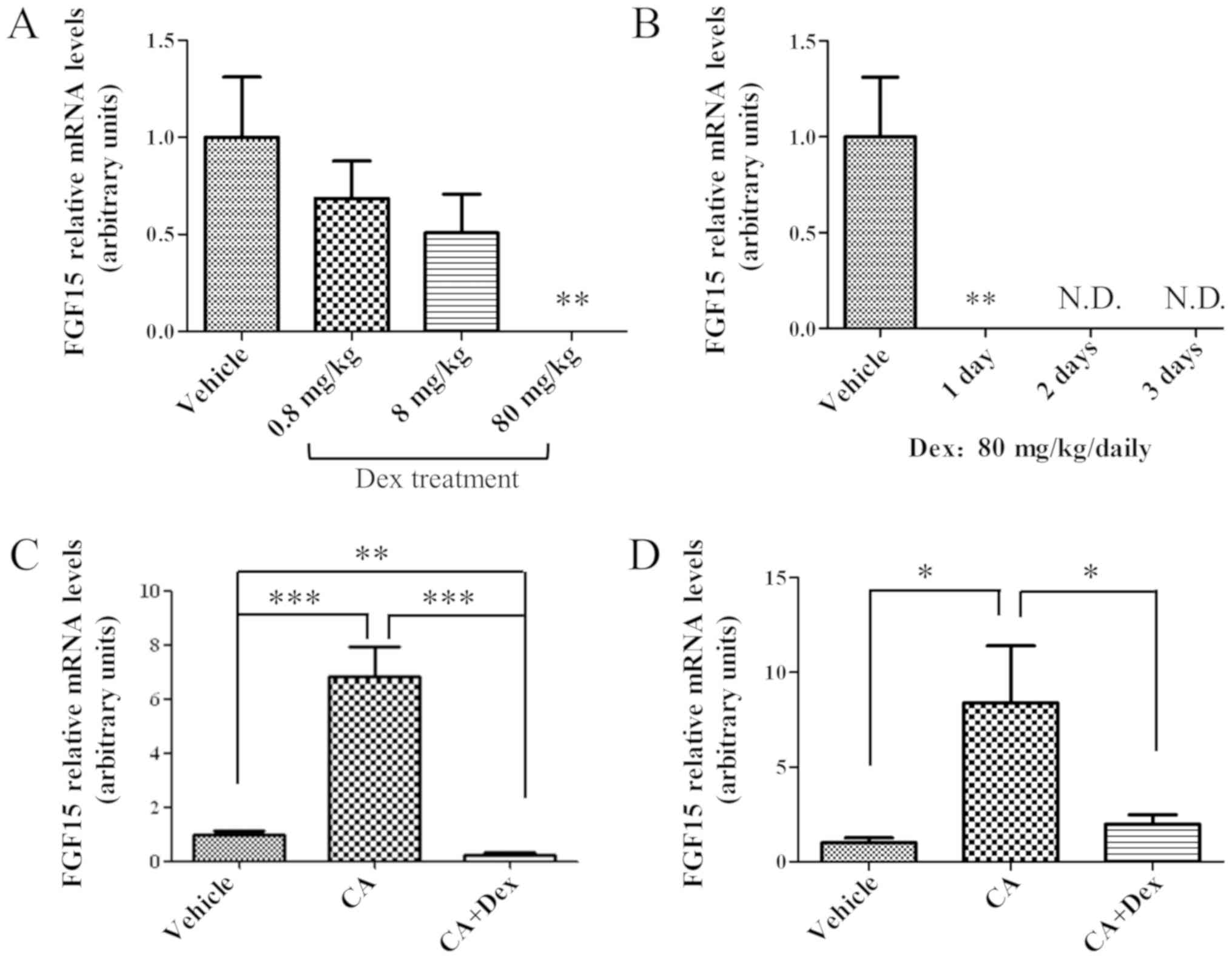
Treatment with Dex significantly
downregulates Fgf15 expression in the mouse ileum
To examine the effect of treatment with Dex on ileal
Fgf15 expression, the mRNA expression levels of Fgf15
were compared prior to and following treatment with Dex. Mice were
treated with Dex at three different doses (0.8, 8 and 80 mg/kg) for
24 h. The ileal Fgf15 expression decreased in a
dose-dependent manner following treatment with Dex (Fig. 1A). At 80 mg/kg Dex, the expression
level of FGF15 mRNA was decreased to an undetectable level two days
later (Fig. 1B). The
downregulation of ileal Fgf15 expression by treatment with
Dex was maintained when the mice were treated with Dex for 3
consecutive days (Fig. 1B). These
results suggested that treatment with Dex was able to suppress
ileal Fgf15 expression at the RNA level. At the same time,
although the highest dosage of Dex used, 80 mg/kg, in the present
study was effective, the potential cytotoxicity of Dex with this
dosage for 3 days requires consideration (data not shown). The
phenomenon of tissue hypertrophy was observed (data not shown),
which may be due to the compensation for the cytotoxicity of
treatment with Dex with high dosage for 3 days.
To investigate whether this downregulation of
Fgf15 expression is mediated by FXR, the critical
transcriptional factor for regulating Fgf15 expression
(3), the effect of treatment with
Dex on the FXR transcriptional activity was examined in
vivo. Mice were pre-treated with Dex or corn oil, and
subsequently FXR was activated with CA. After a 4 h treatment with
CA, as presented in Fig. 1C, the
mRNA expression level of Fgf15 was upregulated. However, the
upregulation of Fgf15 expression by treatment with CA was
significantly inhibited in mice pre-treated with Dex (Fig. 1C; P<0.001), suggesting that Dex
may interfere with FXR activation. Whether this suppression of
Fgf15 expression by Dex directly occurred in ileal
enterocytes, using an ex vivo culture system, was further
investigated. As presented in Fig.
1D, CA induced Fgf15 expression in the ex vivo
culture system, whereas, Dex interfered with the CA-induced FGF15
upregulation in ileal cells.
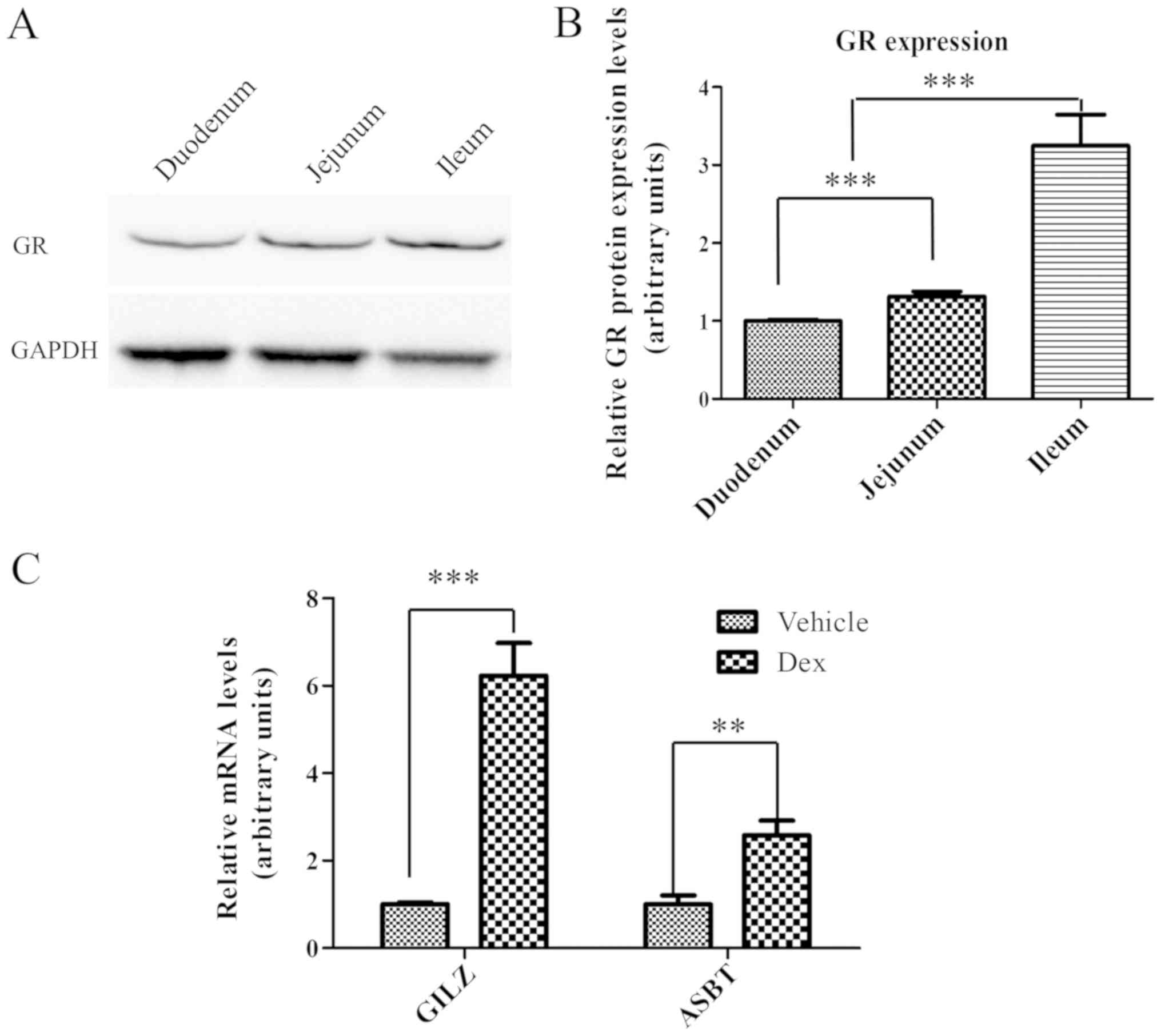
GR is expressed in the intestine and
is able to respond to activation by Dex
GR may be activated by Dex, to trigger the
transactivation or transrepression of GC-responsive genes (24–26).
Therefore, GR protein expression levels in the small intestine were
examined by western blot analysis. As presented in Fig. 2A, GR (~86 kDa) was expressed
throughout the entire small intestine, between the duodenum and the
ileum. Notably, GR expression in the small intestine exhibited a
gradual increase between the duodenum and the ileum. The expression
level of GR in ileum was ~3-times higher compared with the duodenum
and ~2-times higher compared with the jejunum (Fig. 2B), suggesting that GR may have an
important function in the ileum. Subsequently, it was determined
whether ileal GR may be activated by treatment with Dex at a
concentration of 80 mg/kg. It was identified that the mRNA
expression levels of two selected GR target genes, GILZ and ASBT
(27–29), were upregulated by ~6 and
~2.5-times, respectively, in the ileum following treatment with Dex
for 24 h (Fig. 2C). These results
demonstrated that GR was highly expressed in the ileum and may be
activated by Dex, and thus may potentially mediate the repression
of Fgf15 expression by Dex.
Suppression of Fgf15 expression by Dex
is not associated with PXR
As Dex is able to activate GR and PXR, it was
further examined if the effect of Dex on FGF15 expression is
mediated by PXR. As presented in Fig.
3A, the suppression of Fgf15 expression was observed
following treatment with Dex. Since Fgf15 is the downstream
gene for FXR activation, the expression of SHP and
IBABP, another two classical FXR target genes (3,30,31),
was further examined in the ileum following treatment with Dex. As
expected, SHP and IBABP expression were additionally
significantly downregulated following treatment with Dex (Fig. 3A; P<0.01). This downregulation
of Fgf15, SHP and IBABP gene expression reflects the
inhibition of FXR activity; however, not FXR expression, as
there was no significant difference in FXR expression.
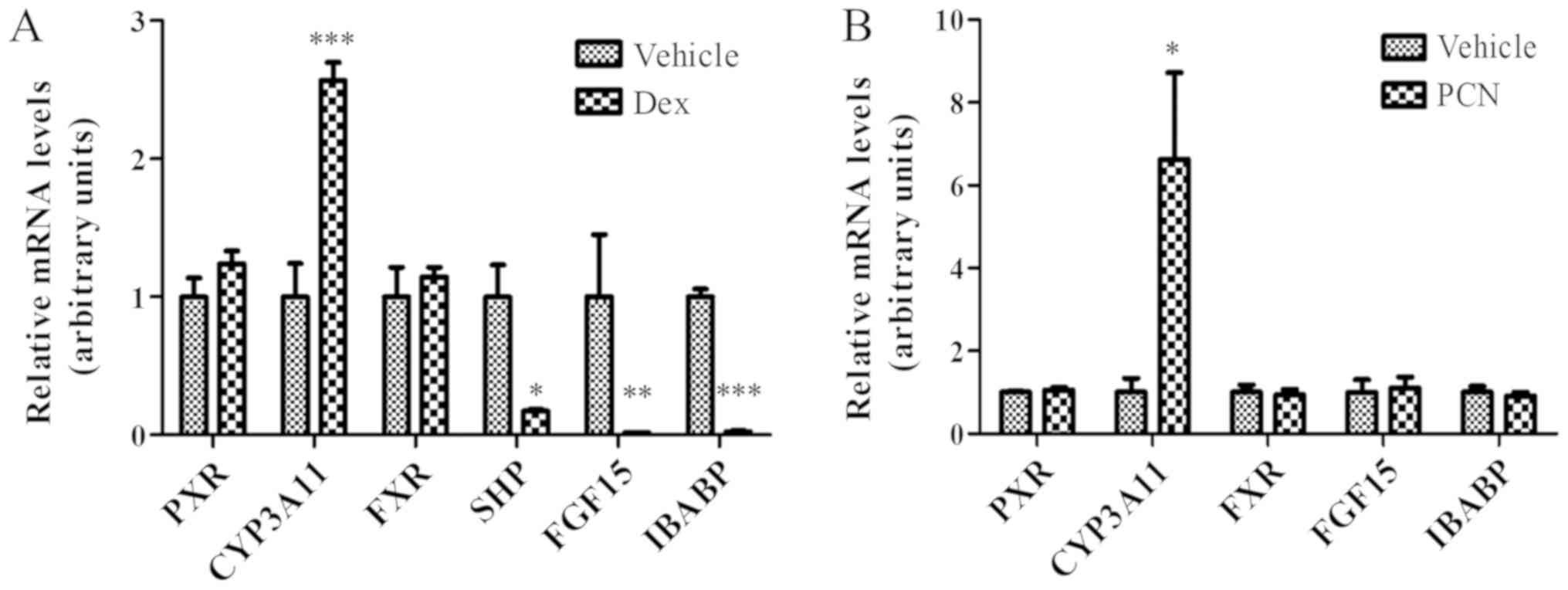 | Figure 3.Suppression of FGF15 expression by
Dex is potentially mediated by GR activation in the ileum. (A) GR
activation by Dex (80 mg/kg) for 24 h induced CYP3A11 expression
and suppressed the expression of FXR target genes (SHP, FGF15 and
IBABP); however, did not affect FXR expression in the ileum. (B)
PXR activation by PCN (25 mg/kg) for 24 h additionally induced
CYP3A11 expression; however, did not affect the expression of FXR
target genes (FGF15 and IBABP). The data were normalized to the
expression levels of mouse U36B4. The data are presented as the
mean ± standard deviation. n=3. *P<0.05, **P<0.01,
***P<0.001 vs. respective vehicle. FGF15, fibroblast growth
factor 15; Dex, dexamethasone; GR, glucocorticoid receptor;
CYP3A11, cytochrome P450, family 3, subfamily a, polypeptide 11;
FXR, farnesoid X receptor; SHP, small heterodimer partner; IBABP,
ileum bile acid binding protein; PXR, pregnane X receptor; PCN,
pregnenolone-16-α-carbonitrile. |
In addition to activating GR, Dex may additionally
activate PXR to induce CYP3A expression (32,33).
As presented in Fig. 3A, the
expression of Cyp3a11, the target gene of PXR, was
significantly increased in the ileum following treatment with Dex;
however, there was no alteration in PXR expression, a result
suggesting that the increased CYP3A expression was due to increased
PXR activation. To confirm whether the suppression of Fgf15
expression by Dex is mediated by PXR activation, PCN, a typical PXR
agonist, was used to treat mice in the same way as Dex. As
presented in Fig. 3B, PCN induced
the expression of Cyp3a11, a PXR target gene; however, it
had no effect on the expression of either FXR or PXR, or FXR target
genes, Fgf15 and IBABP, in the ileum. This result
suggested that PXR activation did not contribute to the suppression
of FGF15 expression by treatment with Dex.
Discussion
The present study demonstrated that treatment with
Dex significantly suppressed the expression of Fgf15 in the
mouse ileum. Although the concentration of serum FGF15 protein was
not determined, the mRNA expression levels of Fgf15 are
routinely used to represent the Fgf15 expression levels
(3,4). Furthermore, as the Fgf15 mRNA
expression levels were below the detectable level for at least 3
days during treatment with Dex, this suggested that the protein
expression level of FGF15 was additionally reduced by treatment
with Dex. As previously demonstrated, FGF15/FGF19 serves an
important role in BA homeostasis, glucose metabolism and hepatic
protein synthesis (3,4,6). For
example, Inagaki et al (3)
identified that FGF15 inhibited BA synthesis by repressing
associated gene expression in the liver (3). Potthoff et al (4) demonstrated that FGF15 inhibited
hepatic gluconeogenesis by blocking the expression of genes
involved in gluconeogenesis (4).
Therefore, Dex intake has the potential to increase the risks of
metabolic disorder by repressing Fgf15 expression. Of note,
GCs or their synthetic analogs (including Dex) have been
demonstrated to cause metabolic diseases, including hyperglycemia
and liver steatosis in specific patients (11–13).
Notably, the effects of Dex on Fgf15 expression were dose
dependent. Therefore, an initial high dosage is occasionally
recommended in the clinical setting; however, the consecutive use
of Dex at high dosage is not recommended. Although the most
effective dose of Dex (80 mg/kg) was studied, is higher compared
with therapeutic doses, which may be as high as ~6 mg/kg in
emergency treatment, according to the US Food and Drug
Administration approved drug dosage information for Dex (34), used in patients, a trend of
inhibition was additionally observed at the lower, therapeutic,
doses. Therefore, the underlying risks require consideration.
The present study further demonstrated that the
downregulation of Fgf15 by Dex is associated with decreased
transcriptional activity of FXR. As previously demonstrated, FXR,
which may be activated by BAs, is considered the principal
transcriptional factor regulating Fgf15 expression (3). Therefore, the decrease in
Fgf15 expression reflected the repression of FXR activity.
The inhibition of FXR activity was further demonstrated by the
result that treatment with Dex additionally suppressed the
expression of SHP and IBABP, another two classical
target genes of FXR in the ileum (3,30,31).
Notably, the suppression of Fgf15 and IBABP by Dex
was restricted to the ileum, as Fgf15 and IBABP are
primarily expressed in the ileum (3,30).
However, the inhibition of FXR activity by Dex may be not
restricted to the ileum based on the preliminary observations of
the present study. Considering that FXR and GR additionally coexist
in other segments of small intestine (3), this hypothesis is reasonable. This
suppression of FXR target gene expression by Dex was not
accompanied by an alteration in the FXR expression level,
which suggested that Dex did not regulate Fgf15 expression
via suppression of FXR expression. However, how Dex interferes with
FXR activation remains unclear.
As Dex is able to activate PXR and GR, it was
additionally determined whether the downregulation of Fgf15
by Dex may be attributed to GR activation alone (33,35).
It was demonstrated that PXR may be activated by higher dosages of
Dex compared with those typically required by classical GR
activation (33,36,37).
In the present study, although PXR additionally responded to Dex in
the ileum, the activation of PXR by PCN had no effect on the
Fgf15 expression, demonstrating that the suppression of
Fgf15 expression was not associated with PXR activation.
Notably, GR, as a classical, accepted receptor of Dex, has the
highest expression in the ileum among the three anatomical parts of
the small intestine. Furthermore, ileal GR responded to treatment
with Dex; the mRNA expression levels of two selected GR target
genes, GILZ and ASBT were significantly upregulated. These results
suggested that GR activation by Dex may be responsible for the
suppression of FXR activation, leading to the downregulation of
Fgf15 expression in ileum. However, the hypothesis that
anti-GC drugs, including RU486, may induce Fgf15 expression
based on the preliminary data of the present study requires further
investigation. This investigation may help to better clarify the
role of GR in regulating Fgf15 expression in the
intestine.
How GR activation by Dex inhibits the
transcriptional activity of FXR remains to be determined. GR is a
classical receptor for GCs, and the activation of GR may repress a
number of pro-inflammatory genes by interacting with nuclear
factor-κB (NF-κB) (10). For
example, GR activation represses TNFα and IL-6 expression by
inhibiting NF-κB transcriptional activity (10). It has additionally been identified
that GR may interact with FXR, by recruiting the C-terminal binding
domain protein and repressing its activity in the liver (38). As GR and FXR are transcriptional
factors that exhibit high expression levels in ileal cells, it is
hypothesized that their crosstalk may serve an important role.
However, further studies are required to provide molecular evidence
and determine the underlying mechanisms.
In summary, the present results suggested that
treatment with Dex may downregulate ileal Fgf15 expression,
and this downregulation is associated with GR-mediated repression
of FXR activity. The present results provide insight for a better
understanding of the mechanism of regulation of Fgf15
expression and suggest the possible occurrence of adverse events
resulting from Fgf15/19 suppression associated with
treatment with Dex in patients.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Institute of General Medical Sciences (grant no. GM082978), The
Chinese National Nature Sciences Foundation (grant no. 31501232),
The Scientific Research Foundation for the Returned Overseas
Chinese Scholars, State Education Ministry, The Fujian Provincial
Nature Science Foundation (grant no. 2015J05052), and The Fujian
Agriculture and Forestry University Science Fund for Distinguished
Young Scholars (grant no. xjq201629; Fuzhou, China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KZ performed the experiments and wrote the
manuscript. QJ, QZ and DZ contributed to the design of the study
and the writing of the manuscript.
Ethics approval and consent to
participate
All animal studies were approved by the
Institutional Animal Care and Use Committee of the Wadsworth Center
(State University of New York, Albany, NY, USA) or the Animal
Ethics Committee of the Fujian Agriculture and Forestry University
(Fuzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
FGF15
|
fibroblast growth factor 15
|
|
Dex
|
dexamethasone
|
|
FXR
|
farnesoid X receptor
|
|
GR
|
glucocorticoid receptor
|
|
CA
|
cholic acid
|
|
PCN
|
pregnenolone-16-α-carbonitrile
|
|
PXR
|
pregnane X receptor
|
|
RXR
|
retinoid X receptors
|
|
GC
|
glucocorticoid
|
References
|
1
|
McWhirter JR, Goulding M, Weiner JA, Chun
J and Murre C: A novel fibroblast growth factor gene expressed in
the developing nervous system is a downstream target of the
chimeric homeodomain oncoprotein E2A-Pbx1. Development.
124:3221–3232. 1997.PubMed/NCBI
|
|
2
|
Nishimura T, Utsunomiya Y, Hoshikawa M,
Ohuchi H and Itoh N: Structure and expression of a novel human FGF,
FGF-19, expressed in the fetal brain. Biochim Biophys Acta.
1444:148–151. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Inagaki T, Choi M, Moschetta A, Peng L,
Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA,
et al: Fibroblast growth factor 15 functions as an enterohepatic
signal to regulate bile acid homeostasis. Cell Metab. 2:217–225.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Potthoff MJ, Boney-Montoya J, Choi M, He
T, Sunny NE, Satapati S, Suino-Powell K, Xu HE, Gerard RD, Finck
BN, et al: FGF15/19 regulates hepatic glucose metabolism by
inhibiting the CREB-PGC-1α pathway. Cell Metab. 13:729–738. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fu L, John LM, Adams SH, Yu XX, Tomlinson
E, Renz M, Williams PM, Soriano R, Corpuz R, Moffat B, et al:
Fibroblast growth factor 19 increases metabolic rate and reverses
dietary and leptin-deficient diabetes. Endocrinology.
145:2594–2603. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kir S, Beddow SA, Samuel VT, Miller P,
Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA and
Mangelsdorf DJ: FGF19 as a postprandial, insulin-independent
activator of hepatic protein and glycogen synthesis. Science.
331:1621–1624. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomlinson E, Fu L, John L, Hultgren B,
Huang X, Renz M, Stephan JP, Tsai SP, Powell-Braxton L, French D
and Stewart TA: Transgenic mice expressing human fibroblast growth
factor-19 display increased metabolic rate and decreased adiposity.
Endocrinology. 143:1741–1747. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schmidt DR, Holmstrom SR, Fon Tacer K,
Bookout AL, Kliewer SA and Mangelsdorf DJ: Regulation of bile acid
synthesis by fat-soluble vitamins A and D. J Biol Chem.
285:14486–14494. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reue K, Lee JM and Vergnes L: Diet1 is a
regulator of fibroblast growth factor 15/19-dependent bile acid
synthesis. Dig Dis. 33:307–313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baschant U, Lane NE and Tuckermann J: The
multiple facets of glucocorticoid action in rheumatoid arthritis.
Nat Rev Rheumatol. 8:645–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andrews RC and Walker BR: Glucocorticoids
and insulin resistance: Old hormones, new targets. Clin Sci (Lond).
96:513–523. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lemke U, Krones-Herzig A, Berriel Diaz M,
Narvekar P, Ziegler A, Vegiopoulos A, Cato AC, Bohl S, Klingmüller
U, Screaton RA, et al: The glucocorticoid receptor controls hepatic
dyslipidemia through Hes1. Cell Metab. 8:212–223. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamanishi Y, Nosaka Y, Kawasaki H,
Hirayama C and Ikawa S: Sterol and bile acid metabolism after
short-term prednisolone treatment in patients with chronic active
hepatitis. Gastroenterol Jpn. 20:246–251. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ge H, Zhang J, Gong Y, Gupte J, Ye J,
Weiszmann J, Samayoa K, Coberly S, Gardner J, Wang H, et al:
Fibroblast growth factor receptor 4 (FGFR4) deficiency improves
insulin resistance and glucose metabolism under diet-induced
obesity conditions. J Biol Chem. 289:30470–30480. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jia K, Li L, Liu Z, Hartog M, Kluetzman K,
Zhang QY and Ding X: Generation and characterization of a novel
CYP2A13-transgenic mouse model. Drug Metab Dispos. 42:1341–1348.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
D'Agostino J, Ding X, Zhang P, Jia K, Fang
C, Zhu Y, Spink DC and Zhang QY: Potential biological functions of
cytochrome P450 reductase-dependent enzymes in small intestine:
Novel link to expression of major histocompatibility complex class
II genes. J Biol Chem. 287:17777–17788. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Schmittgen TD and Livak KJ: Analyszing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cariou B, van Harmelen K, Duran-Sandoval
D, van Dijk TH, Grefhorst A, Abdelkarim M, Caron S, Torpier G,
Fruchart JC, Gonzalez FJ, et al: The farnesoid X receptor modulates
adiposity and peripheral insulin sensitivity in mice. J Biol Chem.
281:11039–11049. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Trousson A, Makoukji J, Petit PX, Bernard
S, Slomianny C, Schumacher M and Massaad C: Cross-talk between
oxysterols and glucocorticoids: Differential regulation of secreted
phopholipase A2 and impact on oligodendrocyte death. PLoS One.
4:e80802009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berrebi D, Bruscoli S, Cohen N, Foussat A,
Migliorati G, Bouchet-Delbos L, Maillot MC, Portier A, Couderc J,
Galanaud P, et al: Synthesis of glucocorticoid-induced leucine
zipper (GILZ) by macrophages: An anti-inflammatory and
immunosuppressive mechanism shared by glucocorticoids and IL-10.
Blood. 101:729–738. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Miyata M, Yamakawa H, Hamatsu M,
Kuribayashi H, Takamatsu Y and Yamazoe Y: Enterobacteria modulate
intestinal bile acid transport and homeostasis through apical
sodium-dependent bile acid transporter (SLC10A2) expression. J
Pharmacol Exp Ther. 336:188–196. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang QY, Dunbar D and Kaminsky LS:
Characterization of mouse small intestinal cytochrome P450
expression. Drug Metab Dispos. 31:1346–1351. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamamoto KR: Steroid receptor regulated
transcription of specific genes and gene networks. Annu Rev Genet.
19:209–252. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamamoto KR, Darimont BD, Wagner RL and
Iñiguez-Lluhí JA: Building transcriptional regulatory complexes:
Signals and surfaces. Cold Spring Harb Symp Quant Biol. 63:587–598.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weikum ER, Knuesel MT, Ortlund EA and
Yamamoto KR: Glucocorticoid receptor control of transcription:
Precision and plasticity via allostery. Nat Rev Mol Cell Biol.
18:159–174. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ayroldi E and Riccardi C:
Glucocorticoid-induced leucine zipper (GILZ): A new important
mediator of glucocorticoid action. FASEB J. 23:3649–3658. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang JC, Derynck MK, Nonaka DF,
Khodabakhsh DB, Haqq C and Yamamoto KR: Chromatin
immunoprecipitation (ChIP) scanning identifies primary
glucocorticoid receptor target genes. Proc Natl Acad Sci USA.
101:15603–15608. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jung D, Fantin AC, Scheurer U, Fried M and
Kullak-Ublick GA: Human ileal bile acid transporter gene ASBT
(SLC10A2) is transactivated by the glucocorticoid receptor. Gut.
53:78–84. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grober J, Zaghini I, Fujii H, Jones SA,
Kliewer SA, Willson TM, Ono T and Besnard P: Identification of a
bile acid-responsive element in the human ileal bile acid-binding
protein gene. Involvement of the farnesoid X
receptor/9-cis-retinoic acid receptor heterodimer. J Biol Chem.
274:29749–29754. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hwang ST, Urizar NL, Moore DD and Henning
SJ: Bile acids regulate the ontogenic expression of ileal bile acid
binding protein in the rat via the farnesoid X receptor.
Gastroenterology. 122:1483–1492. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huss JM and Kasper CB: Two-stage
glucocorticoid induction of CYP3A23 through both the glucocorticoid
and pregnane X receptors. Mol Pharmacol. 58:48–57. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kliewer SA, Moore JT, Wade L, Staudinger
JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM,
Zetterström RH, et al: An orphan nuclear receptor activated by
pregnanes defines a novel steroid signaling pathway. Cell.
92:73–82. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dietzman RH, Ersek RA, Bloch JM and
Lilleheir RC: High-output, low-resistance gram-negative septic
shock in man. Angiology. 20:691–700. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lehmann JM, McKee DD, Watson MA, Willson
TM, Moore JT and Kliewer SA: The human orphan nuclear receptor PXR
is activated by compounds that regulate CYP3A4 gene expression and
cause drug interactions. J Clin Invest. 102:1016–1023. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schuetz EG and Guzelian PS: Induction of
cytochrome P-450 by glucocorticoids in rat liver. II. Evidence that
glucocorticoids regulate induction of cytochrome P-450 by a
nonclassical receptor mechanism. J Biol Chem. 259:2007–2012.
1984.PubMed/NCBI
|
|
37
|
Schuetz EG, Wrighton SA, Barwick JL and
Guzelian PS: Induction of cytochrome P-450 by glucocorticoids in
rat liver. I. Evidence that glucocorticoids and pregnenolone 16
alpha-carbonitrile regulate de novo synthesis of a common form of
cytochrome P-450 in cultures of adult rat hepatocytes and in the
liver in vivo. J Biol Chem. 259:1999–2006. 1984.PubMed/NCBI
|
|
38
|
Lu Y, Zhang Z, Xiong X, Wang X, Li J, Shi
G, Yang J, Zhang X, Zhang H, Hong J, et al: Glucocorticoids promote
hepatic cholestasis in mice by inhibiting the transcriptional
activity of the farnesoid X receptor. Gastroenterology.
143:1630–1640. 2012. View Article : Google Scholar : PubMed/NCBI
|