Introduction
ETS variant 1 (ETV1) [also known as ETS-related 81
(ER81)] is a transcription factor that belongs to a large family of
proteins characterized by a ~85 amino acid-long ETS DNA-binding
domain (1–5). ETV1 expression is developmentally and
tissue-specifically controlled (1,6–9),
suggesting that ETV1 plays an important ontogenetic role.
ETV1-knockout mice display limb ataxia, abnormal flexor-extensor
posturing of their limbs and an anomalous development of sensory
neurons and muscle spindles. Due to these defects, ETV1-knockout
mice die 3–5 weeks after birth (10,11).
The transcriptional activity of ETV1 is regulated by
post-translational modification. Central to this is the activation
of the mitogen-activated protein (MAP) kinase pathway (12). MAP kinases phosphorylate ETV1 on two
serine and two threonine residues and an additional two serine
residues are phosphorylated by MAPKAP kinases that are activated by
MAP kinases. As a consequence of its phosphorylation, ETV1
transcriptional activity becomes greatly enhanced (13–16).
In addition to phosphorylation, ETV1 may be acetylated on two
lysine residues, which is due to p300 (or its paralog CBP) and
PCAF, acetyltransferases that bind to ETV1 and are known
coactivators (17–20). Importantly, MAP kinases appear to
stimulate the acetyltransferase activity of p300 and the resultant
acetylation of ETV1 also increases its transactivation potential
(18). Moreover, ETV1 interacts
with steroid receptor coactivators that are activated by MAP kinase
phosphorylation (21–23). Thus, ETV1 may be an important
downstream effector of a plethora of signals that funnel through
the MAP kinase pathway.
The first indication that ETV1 is involved in
tumorigenesis emerged from the analysis of Ewing tumors, which
arise as a consequence of chromosomal translocations affecting the
EWS gene (24,25). In particular, the N-terminus of EWS,
which represents a potent transactivation domain (26), was found to be translocated onto the
C-terminus of ETV1, including its DNA-binding domain (6). This results in an oncogenic EWS-ETV1
fusion protein that is constitutively activated and not dependent
on MAP kinase stimulation. However, more recently other chromosomal
translocations involving ETV1 were observed in prostate tumors,
leading to the overexpression of ETV1 (27–31).
Mimicking this ETV1 overexpression in transgenic mouse models
induced the development of prostatic intraepithelial neoplasia, the
precursor of prostate carcinomas, indicating that ETV1
overexpression is an underlying cause of prostate cancer initiation
(28,32). Lastly, copy number amplification of
the ETV1 gene was found in ~40% of melanomas (33), suggesting that ETV1 overexpression
also contributes to skin cancer. Consistently, downregulation of
ETV1 in melanoma cells suppressed proliferation and
anchorage-independent growth, whereas the overexpression of ETV1
led to the transformation of immortalized melanocytes. However,
this transforming activity of ETV1 was dependent on the
coexpression of molecules that induce MAP kinase activity, N-Ras or
B-Raf, suggesting that post-translational modification of ETV1 is
required for its transforming activity (33).
ETV1 ultimately affects tumorigenesis by
dysregulating gene transcription. Thus, the identification of ETV1
target genes merits further attention, since this will increase our
understanding of prostate tumorigenesis and point out potential
avenues of therapeutic intervention. Currently, only a few
validated ETV1 target genes are known. This includes human
telomerase reverse transcriptase (34), which is upregulated in the vast
majority of all tumors, where it is responsible for maintaining
telomere length that is crucial for unlimited proliferation
(35,36). Another ETV1 target gene is HER2/Neu
(37), which is overexpressed in
~10% of all prostate tumors. However, HER2/Neu overexpression
increases with the progression of the disease and has been observed
in excess of 50% of all androgen depletion independent prostate
tumors in a number of studies (38–40).
Furthermore, the transforming growth factor-β (TGF-β) signaling
pathway inhibitor Smad7 is an ETV1 target gene (41). TGF-β signaling is a prominent
tumor-suppressing pathway that is intracellularly relayed via the
receptor-regulated R-Smad proteins (42). Smad7 is different from the R-Smads
and antagonizes them by preventing their phosphorylation by TGF-β
type I receptors and by downregulating TGF-β receptors through the
recruitment of ubiquitin ligases. Accordingly, Smad7 overexpression
may be linked to tumor formation and, indeed, has been observed in
several types of cancers (43). In
this study, we investigated whether matrix metalloproteinase-7
(MMP-7) is a bona fide ETV1 target gene in prostate cancer
cells.
Materials and methods
Electrophoretic mobility shift
assays
The following oligonucleotide pairs were hybridized
to generate double-stranded MMP-7 promoter oligonucleotides: -55
ETS, 5′-ATGAGTCACCTATTTCCACATTCGAGGCTG-3′ and
5′-CTCAGCCTCGAATGTGGAAATAGGTGACTC-3′; -144 ETS,
5′-ATAACGATGTAATACTTCCTCGTTTT-3′ and
5′-ACTAAAACGAGGAAGTATTACATCGTT-3′; -168 ETS,
5′-CATTGTGTGCTTCCTGCCAATAACG-3′ and
5′-CATCGTTATTGGCAGGAAGCACACA-3′.
Double-stranded oligonucleotides were labeled with
32P-dATP using Klenow DNA polymerase (44). Binding reactions were performed in
10 μl of 20 mM HEPES pH 7.4, 25 mM NaCl, 12% glycerol, 0.01% Tween
20, 2 mM DTT, 0.1 μg/μl bovine serum albumin and 0.05 μg/μl
poly(dIdC)*(dIdC). Where indicated, bacterially
expressed and purified ETV1 encompassing amino acids 249–477
(13), 0.5 μl anti-ETV1 antibody
(C-20; Santa Cruz Biotechnology, Inc.), unlabeled double-stranded
E74 or mE74 oligonucleotide (45)
were added together with 32P-labeled oligonucleotide to
the reaction mix. Reactions were allowed to proceed for 20 min at
4°C. Resulting DNA-protein complexes were then separated on native
polyacrylamide gels in a cold room and visualized by
autoradiography of the dried gels.
Chromatin immunoprecipitation (ChIP)
assays
Human LNCaP prostate cancer cells were grown in 10%
charcoal-stripped serum with or without 1 nM mibolerone and ChIP
assays were performed as described (46). To amplify a 270-bp fragment of the
human MMP-7 promoter, a nested PCR was performed according to the
following program: 98°C for 2 min; 6 cycles of 98°C for 30 sec,
64°C for 30 sec (−1°C/cycle), 72°C for 25 sec; 20 cycles (first
PCR) or 19 cycles (second PCR) of 98°C for 30 sec, 58°C for 30 sec,
72°C for 25 sec (+1 sec/cycle) and 4 min at 72°C. For the first
PCR, MMP-7pro-for1 (5′-GTCCTGAATGATACCTATGAGAGC-3′; −290 to −267 of
the MMP-7 promoter) and MMP-7pro-rev1
(5′-CCAGAGACAATTGTTCTTGGACC-3′; +38 to +16 of the MMP-7 promoter)
were utilized as primers, and MMP-7pro-for2
(5′-CATGGAGTCAATTTATGCAGCAGAC-3′; −232 to −208 of the MMP-7
promoter) and MMP-7pro-rev1 for the second PCR. For amplification
of a 338-bp fragment of the human MDM2 promoter, the same PCR
program was employed (32 repeats at a 58°C annealing temperature)
with previously described primers (47). Amplified promoter DNA fragments were
visualized by ethidium bromide staining on agarose gels (48).
Luciferase assays
The human MMP-7 promoter (−301 to +52) was amplified
by PCR from genomic DNA and cloned into the luciferase reporter
construct, pGL2-Basic (Promega). Site-directed mutagenesis was
performed to change the ETS core sequence at −55 and/or −168 from
GGAA to CCAA. All constructs were verified by DNA sequencing. Human
embryonic kidney 293T cells were grown in polylysine-coated 12-well
plates (49) and transiently
transfected by the calcium phosphate coprecipitation method
(50). MMP-7 (500 ng) reporter gene
construct, 50 ng CMV-lacZ, 1 μg pBluescript KS+, and
indicated amounts of vector or ETV1 expression construct were
employed. In case of rabbit kidney RK13 cells, 500 ng MMP-7
reporter gene construct, 1.2 μg pBluescript KS+, 30 ng
pEV3S vector or ETV1 expression construct, and 100 ng
HER2/Neu-V664E plasmid (51) were
used. Thirty-six hours after transfection, cells were lysed
(52) and the cleared lysate was
employed to measure luciferase activity as described (53).
Retroviral infection
To downregulate human ETV1, shRNA targeting the
sequence 5′-UUCGATGGAGACAUCAAAC-3′ was cloned into pSIREN-RetroQ
(Clontech). To overexpress ETV1, murine ETV1 cDNA was cloned into
pQCXIP (Clontech). Retrovirus was then produced in 293T cells
according to standard procedures (54) and employed to infect LNCaP cells two
times within 24 h, which were then grown for an additional 72 h
(55). Overexpression or
downregulation of ETV1 was ascertained by standard western blotting
procedures of cell extracts (56)
and utilizing secondary antibodies coupled to horseradish
peroxidase and employment of enhanced chemiluminescence (57). Similarly, retrovirus expressing
MMP-7 shRNA (shRNA#1, 5′-GGGAACAGGCUCAGGACUA-3′; shRNA#4,
5′-CCUACAGGAUCGUAUCAUA-3′) or human MMP-7 cDNA was generated and
utilized.
RT-PCR
Total RNA was extracted from LNCaP cells employing
TRIzol (Invitrogen) and ~50 ng RNA was used for amplification with
the Access Quick RT-PCR kit (Promega) (58). The following PCR program was
utilized: 48°C for 45 min; 96°C for 2 min; 25–35 repeats of 95°C
for 30 sec, 58°C for 45 sec and 68°C for 45 sec; final extension
for 5 min at 68°C. The MMP-7 primers used were
5′-TGTGGAGTGCCAGATGTTGCAG-3′ and 5′-CTAAATGGAGTGGAGGAACAGTGC-3′,
resulting in a 642 bp cDNA fragment. GAPDH mRNA was assayed as
described (59). For determining
MMP-7 mRNA levels in cells expressing MMP-7 shRNA, a two-step
reaction was performed. First, RT-PCR was carried out with primers
MMP-7-b-for (5′-AGATGTGGAG TGCCAGATGT-3′) and MMP-7-a-rev
(5′-CCAATGAATGAA TGAATGGATG-3′) using the PCR program: 48°C for 45
min; 96°C for 2 min; 20 repeats of 95°C for 30 sec, 56°C for 30
sec, and 68°C for 30 sec; final extension for 4 min at 68°C.
Second, PCR was carried out with MMP-7-b-for and MMP-7-b-rev
(5′-TAGACTGCTACCATCCGTCC-3′) primers employing iProof high-fidelity
DNA polymerase (Bio-Rad) with the PCR program: 98°C for 2 min; 30
repeats of 98°C for 30 sec, 56°C for 30 sec, and 72°C for 30 sec;
final extension for 4 min at 72°C, resulting in a 357-bp cDNA
product. Similarly, ETV1 expression was analyzed using hETV1-RT-for
(5′-TCCCTCCATCGCAGT CCATACCAG-3′) and hETV1-RT-rev (5′-GTGGCAGCTAGG
CACTTCTGAGTC-3′) primers in the RT-PCR reaction (15 repeats)
followed by 20 cycles of nested PCR with hETV1-RT-for and
hETV1-RT-rev-new (5′-CATATGCAAAATCTCTGG GTTCCTG-3′) primers,
generating a 291-bp cDNA product. All resultant cDNA fragments were
electrophoresed on 1.5% agarose gels and stained with ethidium
bromide (60).
Proliferation assay
LNCaP cells were infected with the indicated
retrovirus and selected for at least four days with 1 μg/ml
puromycin. Thereafter, cells were seeded into 96-wells (61) and one day later (designated as Day
0) for the first time were analyzed with an MTT
(3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide)
assay (Trevigen) according to the manufacturer’s instructions.
Further measurements were taken 2, 3 or 5 days thereafter.
Migration assay
Puromycin-selected LNCaP cells (2×105)
(as mentioned above) were seeded into a 24-well format cell culture
insert with an 8 μm pore size (353097; Becton-Dickinson) in
serum-free media. Migration towards media containing 10% fetal calf
serum was measured after 24 h. Migrated cells on the bottom of the
membrane were visualized using the Hemacolor staining kit (EMD
Millipore) and then counted.
Results
Binding of the ETS transcription factor
ETV1 to the MMP-7 gene promoter
Previous reports indicate that the MMP-7 gene is
transcriptionally activated by a number of ETS transcription
factors (62,63). Thus, we reasoned that MMP-7 may be a
new ETV1 target gene in prostate cancer cells. To prove this, we
first assessed whether ETV1 directly binds to the MMP-7 gene
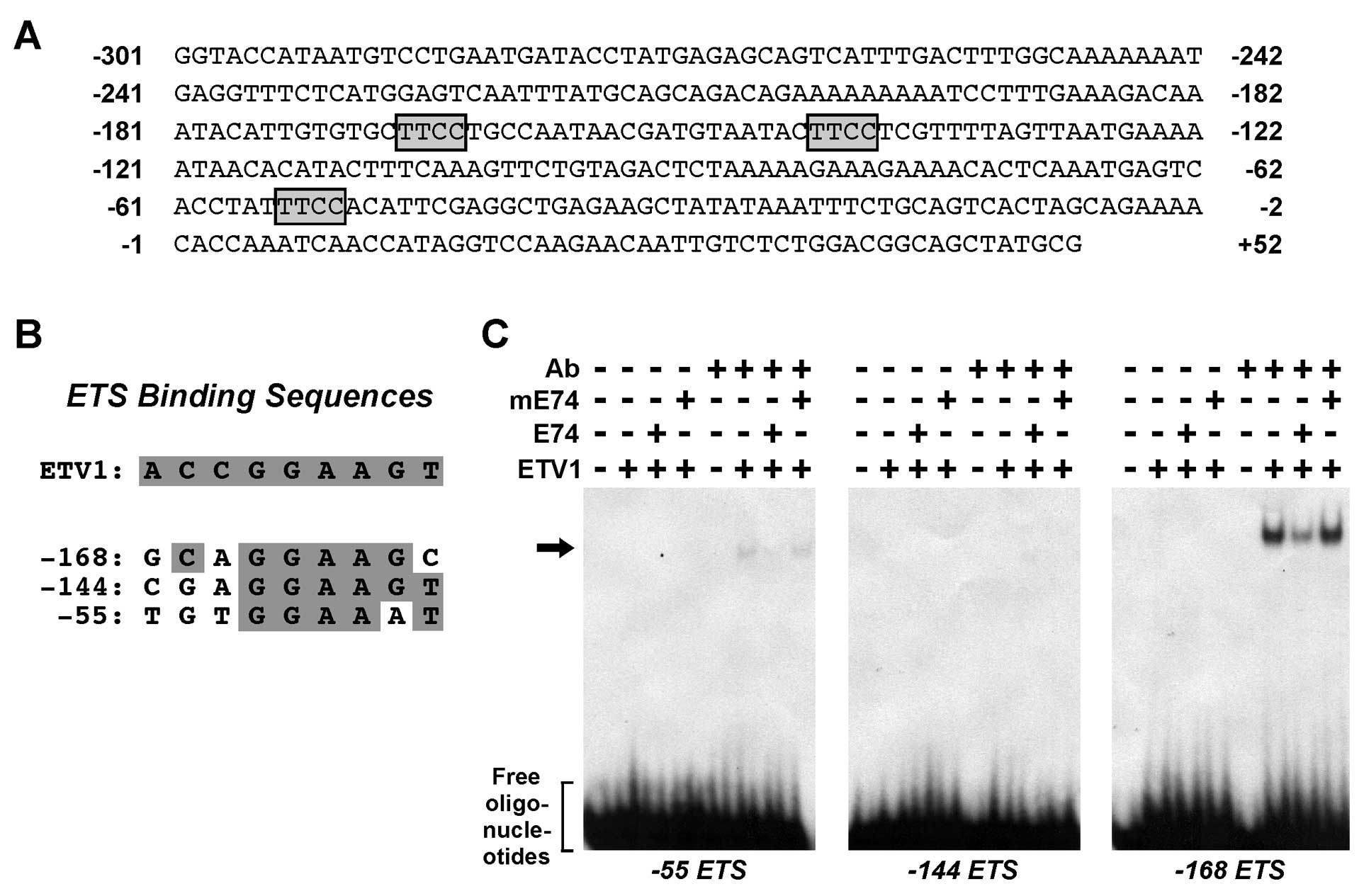
promoter. We analyzed the human MMP-7 promoter for the presence of
potential ETV1-binding sites, which have the consensus sequence
5′-ACCGGAAGT-3′; the GGAA core is the most important determinant of
the DNA-binding affinity (64).
Three such ETS sites are present in the MMP-7 promoter (Fig. 1A), which match to a varying degree
to the ETV1- consensus binding sequence (Fig. 1B).
To study which of these three ETS sites would be
bound by ETV1, we performed electrophoretic mobility shift assays.
When ETV1 was incubated alone with respective radioactively labeled
oligonucleotides, ETV1 binding was not observed (Fig. 1C). However, previous studies showed
that ETV1 DNA binding is inhibited in vitro by its
C-terminus, but an additional antibody recognizing its C-terminus
alleviates this intramolecular inhibition (37,41).
This intramolecular inhibition appears to be especially important
at the ETV1-binding sites that do not match the consensus binding
sequence, as is the case for all three ETS sites within the MMP-7
promoter (Fig. 1B). Thus, we also
included an antibody recognizing the ETV1 C-terminus in the binding
reaction and observed strong binding of ETV1 to the -168 ETS site,
weak binding to the -55 ETS site, while no binding to the -144 ETS
site was observed (Fig. 1C). To
examine binding specificity, we also performed competition
experiments with the E74 site that completely matches the
ETV1-binding site consensus and readily interacts with ETV1 and
other ETS proteins (12,65). Unlabeled E74 oligonucleotide
efficiently reduced ETV1 binding to the radioactively labeled -168
and -55 ETS sites, whereas a mutated E74 oligonucleotide did not
(Fig. 1C). Together, these data
demonstrate that ETV1 is capable of binding to the MMP-7 promoter
at two sites in vitro.
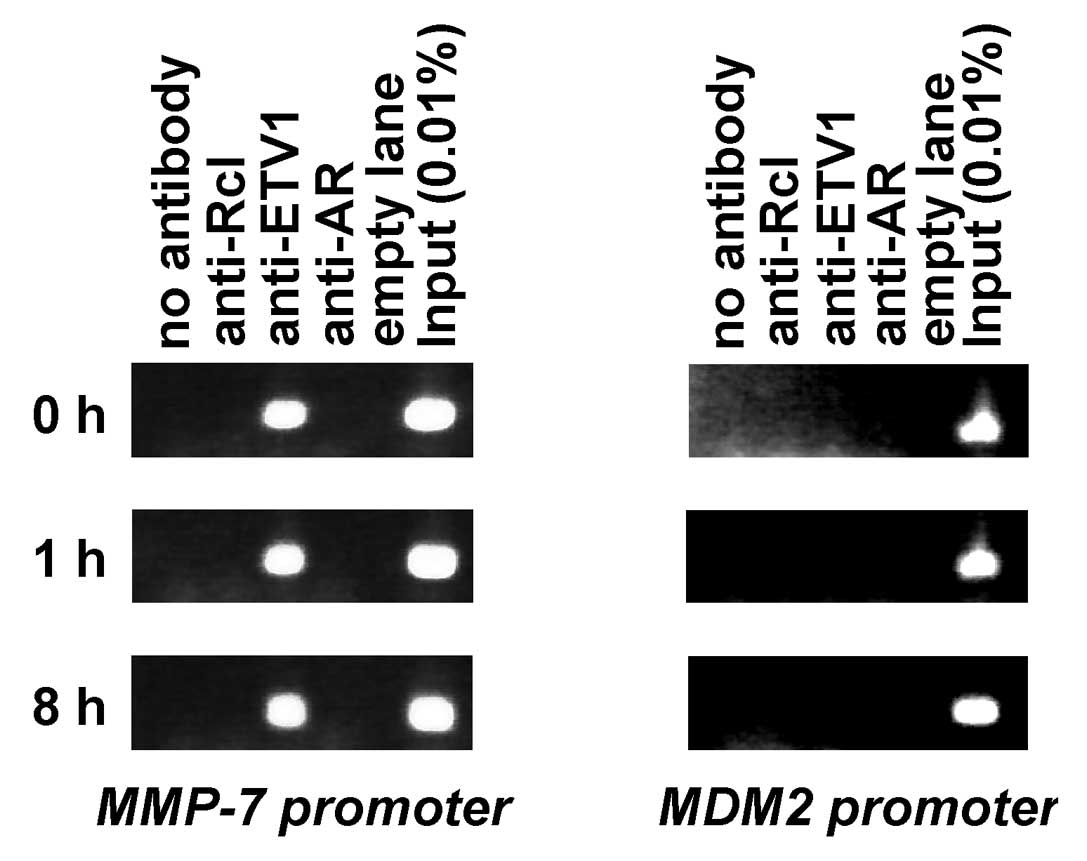
To examine whether ETV1 also binds in vivo to
the MMP-7 promoter, we performed ChIP assays in LNCaP cells. Since
ETV1 binding was previously shown to cooperate with the androgen
receptor (AR) in binding to the prostate-specific antigen enhancer
(32), we also investigated the
influence of androgen stimulation on MMP-7 promoter interaction. We
deprived LNCaP cells of androgen and then induced them for 1 or 8 h
with the synthetic androgen, mibolerone. Utilizing anti-ETV1
antibodies, the MMP-7 promoter was immunoprecipitated before and
after mibolerone stimulation (Fig.
2, left panels), showing that ETV1 may bind to the MMP-7
promoter in cells and that this binding is independent of androgen.
Accordingly, anti-AR antibodies did not immunoprecipitate the MMP-7
promoter, nor did the control anti-Rcl antibodies. Furthermore,
ETV1 did not interact with the MDM2 promoter (Fig. 2, right panels), attesting to the
specificity of our ChIP assay. Thus, ETV1 binds to the MMP-7
promoter in vivo.
Activation of the MMP-7 promoter by
ETV1
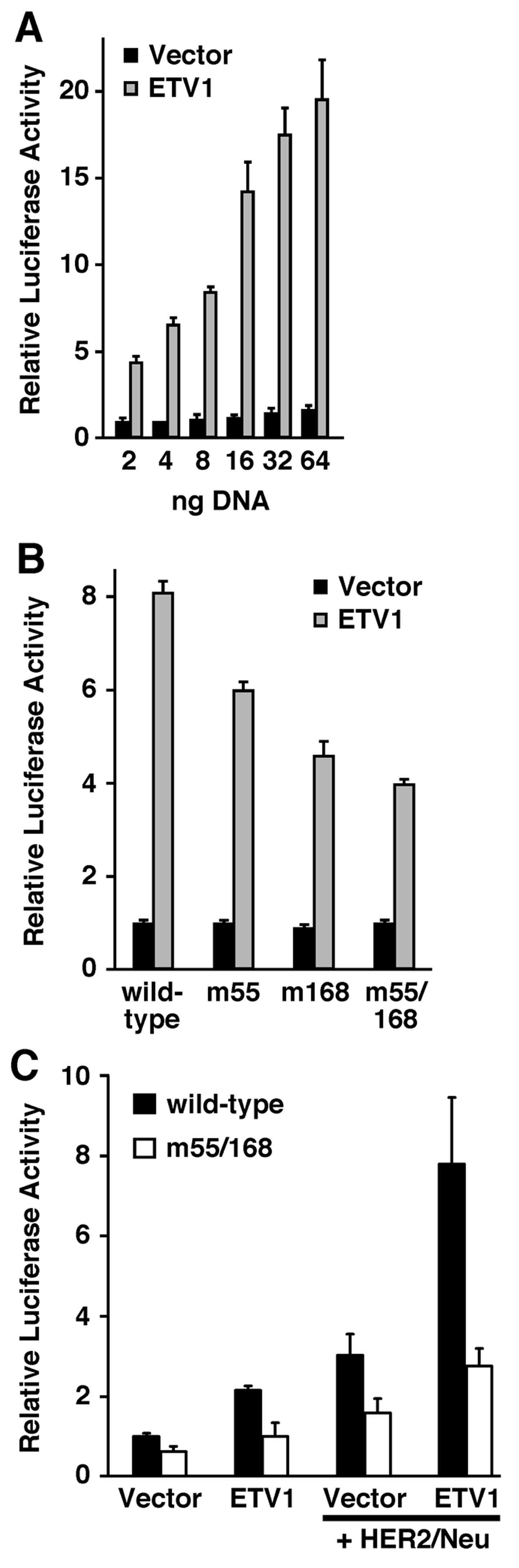
To demonstrate that ETV1 not only binds, but also
stimulates the MMP-7 gene promoter, we cloned respective human DNA
into a luciferase reporter plasmid. This reporter gene was
transfected with increasing amounts of an empty vector or ETV1
expression plasmid into 293T cells. Whereas the empty vector barely
affected the MMP-7 luciferase reporter gene activity, ETV1
stimulated the MMP-7 promoter in a dose-dependent manner up to
~20-fold (Fig. 3A).
Next, we tested how mutation of the ETV1-binding
sites identified above would affect MMP-7 promoter activity. In the
absence of ETV1, mutation of the -55 ETS, -168 ETS site or of both
sites had no impact on MMP-7 reporter gene activity in 293T cells
(Fig. 3B). However, in the presence
of ETV1, mutation of ETS sites at -55 and -168, individually or
combined, reduced MMP-7 promoter activity. As expected, mutation of
the -55 ETS site had a weaker effect compared to the mutation of
the -168 ETS site, while the strongest effect was observed upon
mutation of both ETS sites, reducing ETV1-induced MMP-7 promoter
activity by half (Fig. 3B). The
fact that the MMP-7 promoter remained inducible by ETV1 upon
mutation of both ETV1-binding sites may be due to the indirect
effects of ETV1; such as the induction of another gene encoding a
transcription factor that is also capable of stimulating the MMP-7
promoter.
We also assessed the ability of ETV1 to induce the
MMP-7 promoter in another cell line, RK13. Again, we observed that
ETV1 induced MMP-7 promoter activity, and mutation of the ETS sites
at -55 and -168 severely compromised this activation (Fig. 3C). In contrast to the 293T cells,
mutation of the -55 and -168 ETS sites even suppressed MMP-7
promoter activity in the absence of ETV1, which was probably due to
the fact that RK13 cells harbor endogenous ETV1. We also
demonstrated how the induction of ETV1 transcriptional activity by
HER2/Neu, a receptor tyrosine kinase that stimulates the MAP kinase
pathway and thus the ETV1 transactivation potential (12,13,66),
would affect MMP-7 promoter activity. In the absence of ectopic
ETV1, HER2/Neu stimulated the MMP-7 promoter, possibly due to
endogenous ETV1 in the RK13 cells (Fig.
3C). Moreover, HER2/Neu cooperated with ectopic ETV1 to
stimulate MMP-7 luciferase reporter activity, which was drastically
reduced upon mutation of the -55 and -168 ETS sites (Fig. 3C). Altogether, our promoter studies
strongly suggest that ETV1 activates the MMP-7 promoter through the
-55 and -168 ETS sites.
Endogenous MMP-7 transcription is
activated by ETV1
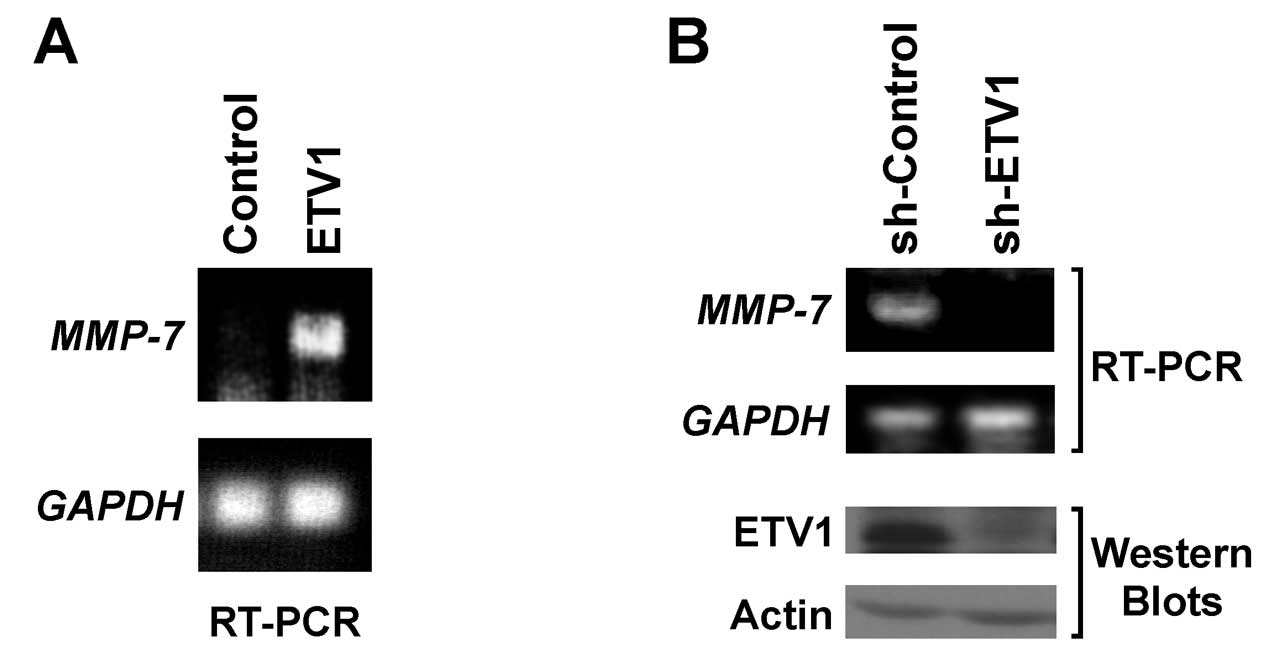
To corroborate that ETV1 also stimulates the
endogenous MMP-7 promoter, we infected human LNCaP prostate cancer
cells with a retrovirus overexpressing ETV1. ETV1 overexpression
led to strongly enhanced MMP-7 mRNA levels in LNCaP cells as
determined by RT-PCR (Fig. 4A). By
utilizing a limited number of PCR amplifications we were unable to
detect MMP-7 mRNA in the absence of ectopic ETV1.
Conversely, we expressed shRNA targeting ETV1 in
LNCaP cells. By utilizing more cycles of amplification than
mentioned above to detect endogenous MMP-7 expression by RT-PCR, we
observed that the downregulation of ETV1 resulted in a robust
decrease in MMP-7 mRNA levels compared to cells expressing a
control shRNA (Fig. 4B, top
panels). Western blotting confirmed that ETV1 protein was reduced
by ETV1 shRNA (Fig. 4B, bottom
panels). Together, these results indicate that MMP-7 transcription
is activated by ETV1 in LNCaP prostate cancer cells.
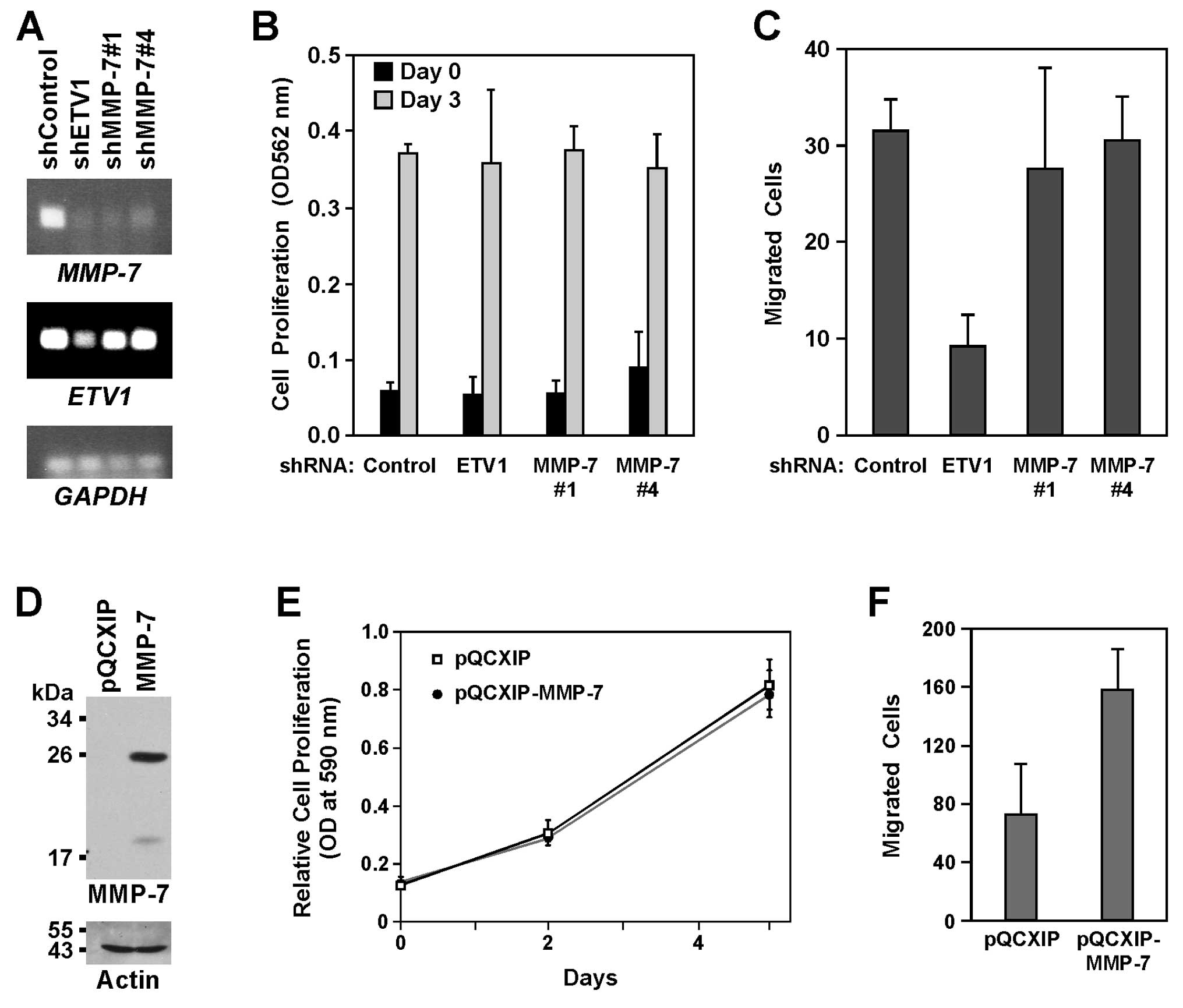
Role of MMP-7 in LNCaP cells
To define the importance of MMP-7 for the biology of
LNCaP cells, we first elected to downregulate MMP-7 with shRNAs.
Similar to ETV1 shRNA, both of the MMP-7 shRNAs employed strongly
reduced MMP-7 mRNA levels in LNCaP cells (Fig. 5A). Downregulation of MMP-7 or ETV1
had no impact on LNCaP cell proliferation (Fig. 5B), and MMP-7 shRNA also did not
affect the ability of LNCaP cells to migrate (Fig. 5C). In contrast, downregulation of
ETV1 reduced LNCaP cell migration (Fig.
5C), which is similar to the reported reduction of LNCaP cell
invasion caused by ETV1 siRNA (28,67).
We next infected LNCaP cells with an
MMP-7-expressing retrovirus. Although we were unable to detect
MMP-7 protein in the control cells, robust MMP-7 expression was
observed upon infection with the MMP-7 retrovirus (Fig. 5D). A change in the cell
proliferation was not noted upon MMP-7 overexpression (Fig. 5E), but LNCaP cell migration was
significantly enhanced (Fig. 5F).
These data suggest that ETV1-mediated MMP-7 upregulation may
particularly contribute to tumor metastasis, which entails cancer
cell migration.
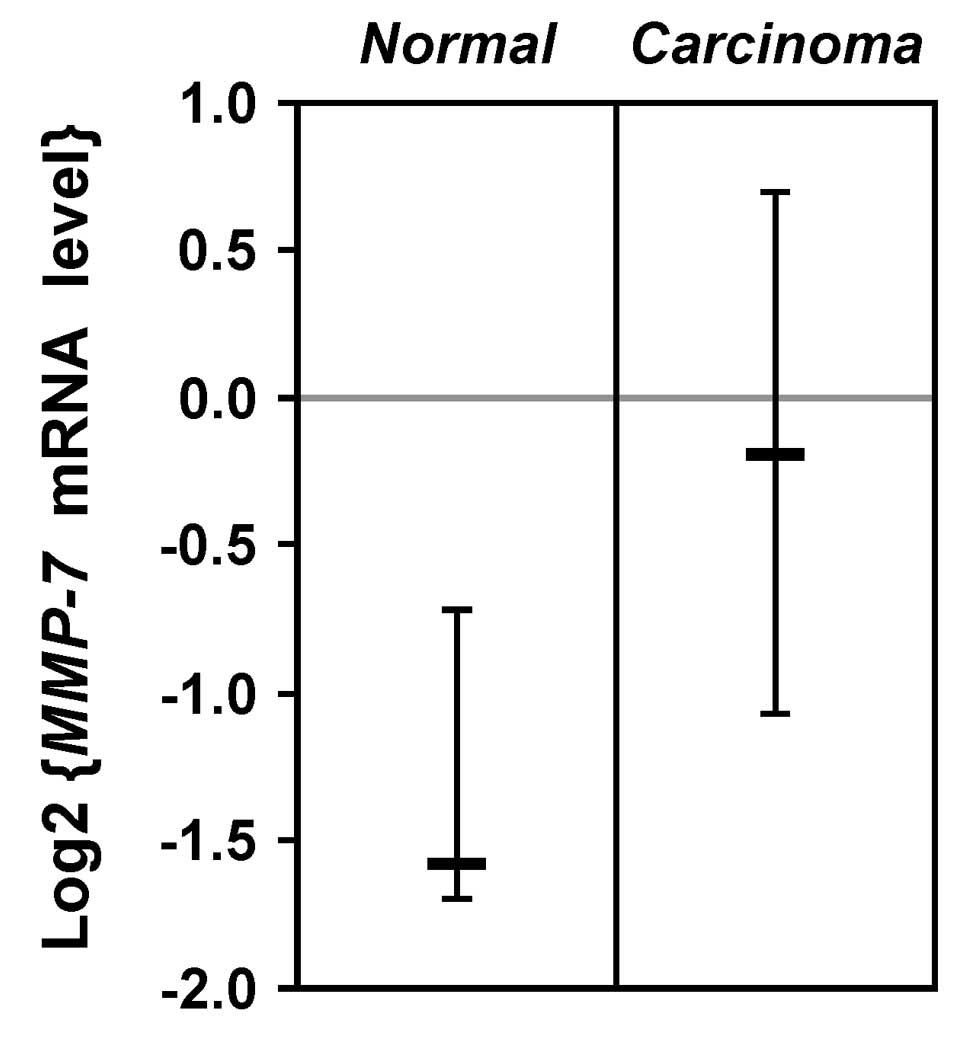
Overexpression of MMP-7 in human prostate
tumors
Recurrent translocation of the ETV1 gene and the
resultant overexpression of the ETV1 protein contributes to
prostate cancer (68). Therefore,
one would predict that MMP-7, as a target gene of ETV1, is
upregulated in prostate tumors. To prove this, we analyzed
published microarray data (69).
The comparison of 23 normal prostate tissues to 65 prostate
carcinomas revealed that MMP-7 mRNA is significantly overexpressed
in prostate tumors (Fig. 6). Thus,
MMP-7 may indeed be a physiologically relevant target gene of ETV1
during prostate cancer formation.
Discussion
In this study, we demonstrated that MMP-7 is a
bona fide target gene of ETV1 in LNCaP prostate cancer cells
based on the following results. First, ETV1 binds to two ETS sites
in the MMP-7 promoter in vitro and also interacts with the
MMP-7 promoter in LNCaP cells in vivo. Second, ETV1
stimulates an MMP-7 luciferase reporter construct and mutation of
two ETV1 binding sites diminished this stimulation. Third, the
overexpression or downregulation of ETV1 in LNCaP cells activated
or repressed, respectively, the endogenous MMP-7 gene
transcription.
Furthermore, our results revealed that MMP-7 gene
transcription is significantly upregulated in human prostate
tumors. These data complement previous studies demonstrating the
presence of MMP-7 mRNA in Northern blot analyses of human prostate
tumor samples (70,71) and analyses in rat models of prostate
cancer showing that MMP-7 is upregulated during carcinogenesis
(72,73). Moreover, it was recently found that
serum levels of MMP-7 are increased in metastatic prostate cancer
patients, but a difference between the control patients and those
with localized disease was not observed (74). Accordingly, high MMP-7 serum levels
were correlated with poor prognosis, strongly indicating that MMP-7
upregulation contributes to the development of aggressive prostate
cancer. This would be similar to other types of cancer, for which
respective recombinant mouse models prove a causal relationship
between MMP-7 and tumorigenesis. For instance, knock out of MMP-7
counteracted colon or pancreatic cancer formation, while MMP-7
overexpression was shown to accelerate breast tumorigenesis induced
by HER2/Neu (75–77). Notably, our results revealed that
HER2/Neu targets ETV1 to stimulate MMP-7 transcription. Thus,
HER2/Neu, which is overexpressed in the majority of prostate tumors
(38–40), may synergize with ETV1
overexpression to induce MMP-7 gene transcription in prostate
cancer.
ETV1 overexpression has been shown to increase the
migration of PNT2C2 and RWPE-1 prostate cells (30,78),
whereas ETV1 downregulation decreased LNCaP cell invasion (28,67).
Similarly, we observed that ETV1 shRNA reduced LNCaP cell
migration, but this was not caused by decreased MMP-7
transcription, since MMP-7 shRNA itself did not have an impact on
LNCaP cell migration. This may be due to the fact that ETV1 not
only affects MMP-7 expression, but potentially the transcription of
several other proteinases involved in cell migration, including the
ETV1 target gene, MMP-1 (13).
Thus, it is conceivable that MMP-7 downregulation alone will not be
sufficient to reduce cell migration. Regardless, the fact that
MMP-7 overexpression increased LNCaP cell migration (in the current
study) and DU-145 prostate cancer cell invasion (79) indicates the physiological relevance
of MMP-7 in prostate cancer. Since MMP-7 antibodies were shown to
inhibit the migration of gastric cancer cells (80) and the overexpression of MMP-7
promoted migration in colorectal cancer cells (81,82),
MMP-7 is not only important for cell migration in prostate cancer,
but also in various other neoplasias.
MMPs such as MMP-7 may also promote cell growth by
fostering the shedding of growth factors or increasing their
bioavailability (83). Furthermore,
MMPs affect apoptosis, angiogenesis and immune surveillance
(84,85). While invasion or angiogenesis is
more important during later cancer stages, growth and survival
effects of MMP-7 may contribute to the development of hyperplasia
and neoplasia at the onset of prostate cell transformation. Thus,
MMP-7 may be one target gene that is critical for the reported
development of prostatic intraepithelial neoplasia upon ETV1
overexpression (28,32).
In conclusion, our data strongly argue that the
upregulation of MMP-7 contributes to the oncogenic phenotype of
ETV1 in the prostate. Since dysregulated ETV1 is also implicated in
skin, breast and gastrointestinal stromal tumors (33,86,87),
our results may not be limited to prostate cancer. As such,
inhibiting MMP-7 enzymatic activity using small-molecule drugs may
be useful in the treatment of various ETV1-overexpressing
tumors.
Acknowledgements
We thank Dr Eric Howard for providing human MMP-7
cDNA. This study was supported by a grant to R.J. from the National
Cancer Institute (R01 CA154745). The content is solely the
responsibility of the authors and does not necessarily represent
the official views of the National Cancer Institute or the National
Institutes of Health. S.A. was supported by the Summer
Undergraduate Research Experience Program at the University of
Oklahoma Health Sciences Center.
References
|
1
|
Brown TA and McKnight SL: Specificities of
protein-protein and protein-DNA interaction of GABP alpha and two
newly defined ets-related proteins. Genes Dev. 6:2502–2512. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Coutte L, Monte D, Imai K, Pouilly L,
Dewitte F, Vidaud M, Adamski J, Baert JL and de Launoit Y:
Characterization of the human and mouse ETV1/ER81 transcription
factor genes: role of the two alternatively spliced isoforms in the
human. Oncogene. 18:6278–6286. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharrocks AD: The ETS-domain transcription
factor family. Nat Rev Mol Cell Biol. 2:827–837. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hollenhorst PC, McIntosh LP and Graves BJ:
Genomic and biochemical insights into the specificity of ETS
transcription factors. Annu Rev Biochem. 80:437–471. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oh S, Shin S and Janknecht R: ETV1, 4 and
5: An oncogenic subfamily of ETS transcription factors. Biochim
Biophys Acta. 1826:1–12. 2012.PubMed/NCBI
|
|
6
|
Jeon IS, Davis JN, Braun BS, Sublett JE,
Roussel MF, Denny CT and Shapiro DN: A variant Ewing’s sarcoma
translocation (7;22) fuses the EWS gene to the ETS gene ETV1.
Oncogene. 10:1229–1234. 1995.
|
|
7
|
Monte D, Coutte L, Baert JL, Angeli I,
Stehelin D and de Launoit Y: Molecular characterization of the
ets-related human transcription factor ER81. Oncogene. 11:771–779.
1995.PubMed/NCBI
|
|
8
|
Chotteau-Lelievre A, Desbiens X, Pelczar
H, Defossez PA and de Launoit Y: Differential expression patterns
of the PEA3 group transcription factors through murine embryonic
development. Oncogene. 15:937–952. 1997. View Article : Google Scholar
|
|
9
|
Chotteau-Lelievre A, Dolle P, Peronne V,
Coutte L, de Launoit Y and Desbiens X: Expression patterns of the
Ets transcription factors from the PEA3 group during early stages
of mouse development. Mech Dev. 108:191–195. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arber S, Ladle DR, Lin JH, Frank E and
Jessell TM: ETS gene Er81 controls the formation of functional
connections between group Ia sensory afferents and motor neurons.
Cell. 101:485–498. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kucera J, Cooney W, Que A, Szeder V,
Stancz-Szeder H and Walro J: Formation of supernumerary muscle
spindles at the expense of Golgi tendon organs in ER81-deficient
mice. Dev Dyn. 223:389–401. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Janknecht R: Analysis of the
ERK-stimulated ETS transcription factor ER81. Mol Cell Biol.
16:1550–1556. 1996.PubMed/NCBI
|
|
13
|
Bosc DG, Goueli BS and Janknecht R:
HER2/Neu-mediated activation of the ETS transcription factor ER81
and its target gene MMP-1. Oncogene. 20:6215–6224. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Janknecht R: Cell type-specific inhibition
of the ETS transcription factor ER81 by mitogen-activated protein
kinase-activated protein kinase 2. J Biol Chem. 276:41856–41861.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu J and Janknecht R: Regulation of the
ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1
and protein kinase A. J Biol Chem. 277:42669–42679. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Janknecht R: Regulation of the ER81
transcription factor and its coactivators by mitogen- and
stress-activated protein kinase 1 (MSK1). Oncogene. 22:746–755.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Papoutsopoulou S and Janknecht R:
Phosphorylation of ETS transcription factor ER81 in a complex with
its coactivators CREB-binding protein and p300. Mol Cell Biol.
20:7300–7310. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goel A and Janknecht R:
Acetylation-mediated transcriptional activation of the ETS protein
ER81 by p300, P/CAF, and HER2/Neu. Mol Cell Biol. 23:6243–6254.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Janknecht R: The versatile functions of
the transcriptional coactivators p300 and CBP and their roles in
disease. Histol Histopathol. 17:657–668. 2002.PubMed/NCBI
|
|
20
|
Lee KK and Workman JL: Histone
acetyltransferase complexes: one size doesn’t fit all. Nat Rev Mol
Cell Biol. 8:284–295. 2007.PubMed/NCBI
|
|
21
|
Goel A and Janknecht R: Concerted
activation of ETS protein ER81 by p160 coactivators, the
acetyltransferase p300 and the receptor tyrosine kinase HER2/Neu. J
Biol Chem. 279:14909–14916. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Font de Mora J and Brown M: AIB1 is a
conduit for kinase-mediated growth factor signaling to the estrogen
receptor. Mol Cell Biol. 20:5041–5047. 2000.PubMed/NCBI
|
|
23
|
Xu J, Wu RC and O’Malley BW: Normal and
cancer-related functions of the p160 steroid receptor co-activator
(SRC) family. Nat Rev Cancer. 9:615–630. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Janknecht R: EWS-ETS oncoproteins: the
linchpins of Ewing tumors. Gene. 363:1–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Toomey EC, Schiffman JD and Lessnick SL:
Recent advances in the molecular pathogenesis of Ewing’s sarcoma.
Oncogene. 29:4504–4516. 2010.
|
|
26
|
Rossow KL and Janknecht R: The Ewing’s
sarcoma gene product functions as a transcriptional activator.
Cancer Res. 61:2690–2695. 2001.
|
|
27
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tomlins SA, Laxman B, Dhanasekaran SM,
Helgeson BE, Cao X, Morris DS, Menon A, Jing X, Cao Q, Han B, et
al: Distinct classes of chromosomal rearrangements create oncogenic
ETS gene fusions in prostate cancer. Nature. 448:595–599. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Attard G, Clark J, Ambroisine L, Mills IG,
Fisher G, Flohr P, Reid A, Edwards S, Kovacs G, Berney D, et al:
Heterogeneity and clinical significance of ETV1 translocations in
human prostate cancer. Br J Cancer. 99:314–320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hermans KG, van der Korput HA, van Marion
R, van de Wijngaart DJ, Ziel-van der Made A, Dits NF, Boormans JL,
van der Kwast TH, van Dekken H, Bangma CH, et al: Truncated ETV1,
fused to novel tissue-specific genes, and full-length ETV1 in
prostate cancer. Cancer Res. 68:7541–7549. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clark JP and Cooper CS: ETS gene fusions
in prostate cancer. Nat Rev Urol. 6:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shin S, Kim TD, Jin F, van Deursen JM,
Dehm SM, Tindall DJ, Grande JP, Munz JM, Vasmatzis G and Janknecht
R: Induction of prostatic intraepithelial neoplasia and modulation
of androgen receptor by ETS variant 1/ETS-related protein 81.
Cancer Res. 69:8102–8110. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jane-Valbuena J, Widlund HR, Perner S,
Johnson LA, Dibner AC, Lin WM, Baker AC, Nazarian RM, Vijayendran
KG, Sellers WR, et al: An oncogenic role for ETV1 in melanoma.
Cancer Res. 70:2075–2084. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Goueli BS and Janknecht R: Upregulation of
the catalytic telomerase subunit by the transcription factor ER81
and oncogenic HER2/Neu, Ras, or Raf. Mol Cell Biol. 24:25–35. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hiyama E and Hiyama K: Telomerase as tumor
marker. Cancer Lett. 194:221–233. 2003. View Article : Google Scholar
|
|
36
|
Janknecht R: On the road to immortality:
hTERT upregulation in cancer cells. FEBS Lett. 564:9–13. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bosc DG and Janknecht R: Regulation of
HER2/Neu promoter activity by the ETS transcription factor, ER81. J
Cell Biochem. 86:174–183. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Signoretti S, Montironi R, Manola J,
Altimari A, Tam C, Bubley G, Balk S, Thomas G, Kaplan I, Hlatky L,
et al: Her-2-neu expression and progression toward androgen
independence in human prostate cancer. J Natl Cancer Inst.
92:1918–1925. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Osman I, Mikhail M, Shuch B, Clute M,
Cheli CD, Ghani F, Thiel RP and Taneja SS: Serum levels of shed
Her2/neu protein in men with prostate cancer correlate with disease
progression. J Urol. 174:2174–2177. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nishio Y, Yamada Y, Kokubo H, Nakamura K,
Aoki S, Taki T, Honda N, Nakagawa A, Saga S and Hara K: Prognostic
significance of immunohistochemical expression of the HER-2/neu
oncoprotein in bone metastatic prostate cancer. Urology.
68:110–115. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Dowdy SC, Mariani A and Janknecht R:
HER2/Neu- and TAK1-mediated up-regulation of the transforming
growth factor beta inhibitor Smad7 via the ETS protein ER81. J Biol
Chem. 278:44377–44384. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Massague J: TGFbeta in cancer. Cell.
134:215–230. 2008. View Article : Google Scholar
|
|
43
|
Yan X and Chen YG: Smad7: not only a
regulator, but also a cross-talk mediator of TGF-beta signalling.
Biochem J. 434:1–10. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
De Haro L and Janknecht R: Functional
analysis of the transcription factor ER71 and its activation of the
matrix metalloproteinase-1 promoter. Nucleic Acids Res.
30:2972–2979. 2002.PubMed/NCBI
|
|
45
|
De Haro L and Janknecht R: Cloning of the
murine ER71 gene (Etsrp71) and initial characterization of its
promoter. Genomics. 85:493–502. 2005.PubMed/NCBI
|
|
46
|
Goueli BS and Janknecht R: Regulation of
telomerase reverse transcriptase gene activity by upstream
stimulatory factor. Oncogene. 22:8042–8047. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shin S and Janknecht R: Concerted
activation of the Mdm2 promoter by p72 RNA helicase and the
coactivators p300 and P/CAF. J Cell Biochem. 101:1252–1265. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim TD, Oh S, Shin S and Janknecht R:
Regulation of tumor suppressor p53 and HCT116 cell physiology by
histone demethylase JMJD2D/KDM4D. PLoS One. 7:e346182012.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kim TD, Shin S and Janknecht R: Repression
of Smad3 activity by histone demethylase SMCX/JARID1C. Biochem
Biophys Res Commun. 366:563–567. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rossow KL and Janknecht R: Synergism
between p68 RNA helicase and the transcriptional coactivators CBP
and p300. Oncogene. 22:151–156. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Ben-Levy R, Paterson HF, Marshall CJ and
Yarden Y: A single autophosphorylation site confers oncogenicity to
the Neu/ErbB-2 receptor and enables coupling to the MAP kinase
pathway. EMBO J. 13:3302–3311. 1994.PubMed/NCBI
|
|
52
|
Knebel J, De Haro L and Janknecht R:
Repression of transcription by TSGA/Jmjd1a, a novel interaction
partner of the ETS protein ER71. J Cell Biochem. 99:319–329. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Mooney SM, Goel A, D’Assoro AB, Salisbury
JL and Janknecht R: Pleiotropic effects of p300-mediated
acetylation on p68 and p72 RNA helicase. J Biol Chem.
285:30443–30452. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kim J, Shin S, Subramaniam M, Bruinsma E,
Kim TD, Hawse JR, Spelsberg TC and Janknecht R: Histone demethylase
JARID1B/KDM5B is a corepressor of TIEG1/KLF10. Biochem Biophys Res
Commun. 401:412–416. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Shin S and Janknecht R: Activation of
androgen receptor by histone demethylases JMJD2A and JMJD2D.
Biochem Biophys Res Commun. 359:742–746. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mooney SM, Grande JP, Salisbury JL and
Janknecht R: Sumoylation of p68 and p72 RNA helicases affects
protein stability and transactivation potential. Biochemistry.
49:1–10. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Shin S and Janknecht R: Diversity within
the JMJD2 histone demethylase family. Biochem Biophys Res Commun.
353:973–977. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shin S, Bosc DG, Ingle JN, Spelsberg TC
and Janknecht R: Rcl is a novel ETV1/ER81 target gene upregulated
in breast tumors. J Cell Biochem. 105:866–874. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Shin S, Rossow KL, Grande JP and Janknecht
R: Involvement of RNA helicases p68 and p72 in colon cancer. Cancer
Res. 67:7572–7578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Oh S and Janknecht R: Histone demethylase
JMJD5 is essential for embryonic development. Biochem Biophys Res
Commun. 420:61–65. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim TD, Shin S, Berry WL, Oh S and
Janknecht R: The JMJD2A demethylase regulates apoptosis and
proliferation in colon cancer cells. J Cell Biochem. 113:1368–1376.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ozaki I, Mizuta T, Zhao G, Yotsumoto H,
Hara T, Kajihara S, Hisatomi A, Sakai T and Yamamoto K: Involvement
of the Ets-1 gene in overexpression of matrilysin in human
hepatocellular carcinoma. Cancer Res. 60:6519–6525. 2000.PubMed/NCBI
|
|
63
|
Crawford HC, Fingleton B, Gustavson MD,
Kurpios N, Wagenaar RA, Hassell JA and Matrisian LM: The PEA3
subfamily of Ets transcription factors synergizes with
beta-catenin-LEF-1 to activate matrilysin transcription in
intestinal tumors. Mol Cell Biol. 21:1370–1383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wei GH, Badis G, Berger MF, Kivioja T,
Palin K, Enge M, Bonke M, Jolma A, Varjosalo M, Gehrke AR, et al:
Genome-wide analysis of ETS-family DNA-binding in vitro and in
vivo. EMBO J. 29:2147–2160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Janknecht R, Monte D, Baert JL and de
Launoit Y: The ETS-related transcription factor ERM is a nuclear
target of signaling cascades involving MAPK and PKA. Oncogene.
13:1745–1754. 1996.PubMed/NCBI
|
|
66
|
Holbro T, Civenni G and Hynes NE: The ErbB
receptors and their role in cancer progression. Exp Cell Res.
284:99–110. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cai C, Hsieh CL, Omwancha J, Zheng Z, Chen
SY, Baert JL and Shemshedini L: ETV1 is a novel androgen
receptor-regulated gene that mediates prostate cancer cell
invasion. Mol Endocrinol. 21:1835–1846. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kumar-Sinha C, Tomlins SA and Chinnaiyan
AM: Recurrent gene fusions in prostate cancer. Nat Rev Cancer.
8:497–511. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yu YP, Landsittel D, Jing L, Nelson J, Ren
B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al: Gene
expression alterations in prostate cancer predicting tumor
aggression and preceding development of malignancy. J Clin Oncol.
22:2790–2799. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pajouh MS, Nagle RB, Breathnach R, Finch
JS, Brawer MK and Bowden GT: Expression of metalloproteinase genes
in human prostate cancer. J Cancer Res Clin Oncol. 117:144–150.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hashimoto K, Kihira Y, Matuo Y and Usui T:
Expression of matrix metalloproteinase-7 and tissue inhibitor of
metalloproteinase-1 in human prostate. J Urol. 160:1872–1876. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Ouyang XS, Wang X, Lee DT, Tsao SW and
Wong YC: Up-regulation of TRPM-2, MMP-7 and ID-1 during sex
hormone-induced prostate carcinogenesis in the Noble rat.
Carcinogenesis. 22:965–973. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Asamoto M, Hokaiwado N, Cho YM, Takahashi
S, Ikeda Y, Imaida K and Shirai T: Prostate carcinomas developing
in transgenic rats with SV40 T antigen expression under probasin
promoter control are strictly androgen dependent. Cancer Res.
61:4693–4700. 2001.
|
|
74
|
Szarvas T, Becker M, Vom Dorp F, Meschede
J, Scherag A, Bankfalvi A, Reis H, Schmid KW, Romics I, Rubben H
and Ergun S: Elevated serum matrix metalloproteinase 7 levels
predict poor prognosis after radical prostatectomy. Int J Cancer.
128:1486–1492. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wilson CL, Heppner KJ, Labosky PA, Hogan
BL and Matrisian LM: Intestinal tumorigenesis is suppressed in mice
lacking the metalloproteinase matrilysin. Proc Natl Acad Sci USA.
94:1402–1407. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Rudolph-Owen LA, Chan R, Muller WJ and
Matrisian LM: The matrix metalloproteinase matrilysin influences
early-stage mammary tumorigenesis. Cancer Res. 58:5500–5506.
1998.PubMed/NCBI
|
|
77
|
Crawford HC, Scoggins CR, Washington MK,
Matrisian LM and Leach SD: Matrix metalloproteinase-7 is expressed
by pancreatic cancer precursors and regulates acinar-to-ductal
metaplasia in exocrine pancreas. J Clin Invest. 109:1437–1444.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hollenhorst PC, Ferris MW, Hull MA, Chae
H, Kim S and Graves BJ: Oncogenic ETS proteins mimic activated
RAS/MAPK signaling in prostate cells. Genes Dev. 25:2147–2157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Powell WC, Knox JD, Navre M, Grogan TM,
Kittelson J, Nagle RB and Bowden GT: Expression of the
metalloproteinase matrilysin in DU-145 cells increases their
invasive potential in severe combined immunodeficient mice. Cancer
Res. 53:417–422. 1993.
|
|
80
|
Wroblewski LE, Noble PJ, Pagliocca A,
Pritchard DM, Hart CA, Campbell F, Dodson AR, Dockray GJ and Varro
A: Stimulation of MMP-7 (matrilysin) by Helicobacter pylori
in human gastric epithelial cells: role in epithelial cell
migration. J Cell Sci. 116:3017–3026. 2003.PubMed/NCBI
|
|
81
|
Remy L, Trespeuch C, Bachy S, Scoazec JY
and Rousselle P: Matrilysin 1 influences colon carcinoma cell
migration by cleavage of the laminin-5 beta3 chain. Cancer Res.
66:11228–11237. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Lee SK, Han YM, Yun J, Lee CW, Shin DS, Ha
YR, Kim J, Koh JS, Hong SH, Han DC and Kwon BM: Phosphatase of
regenerating liver-3 promotes migration and invasion by
upregulating matrix metalloproteinases-7 in human colorectal cancer
cells. Int J Cancer. 131:E190–E203. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ii M, Yamamoto H, Adachi Y, Maruyama Y and
Shinomura Y: Role of matrix metalloproteinase-7 (matrilysin) in
human cancer invasion, apoptosis, growth, and angiogenesis. Exp
Biol Med (Maywood). 231:20–27. 2006.PubMed/NCBI
|
|
84
|
Egeblad M and Werb Z: New functions for
the matrix metalloproteinases in cancer progression. Nat Rev
Cancer. 2:161–174. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
85
|
Deryugina EI and Quigley JP: Matrix
metalloproteinases and tumor metastasis. Cancer Metastasis Rev.
25:9–34. 2006. View Article : Google Scholar
|
|
86
|
Wang Y, Wang L, Chen Y, Li L, Yang X, Li
B, Song S, Yang L, Hao Y and Yang J: ER81 expression in breast
cancers and hyperplasia. Pathology Res Int. 2011:9805132011.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Chi P, Chen Y, Zhang L, Guo X, Wongvipat
J, Shamu T, Fletcher JA, Dewell S, Maki RG, Zheng D, et al: ETV1 is
a lineage survival factor that cooperates with KIT in
gastrointestinal stromal tumours. Nature. 467:849–853. 2010.
View Article : Google Scholar : PubMed/NCBI
|