Introduction
Despite recent advances in surgical treatment,
chemotherapy and radiotherapy, gastric cancer remains a major
global health burden. The most recent estimates found that gastric
cancer is the fourth most common cancer and the second most common
cause of cancer mortality worldwide (1,2). The
majority of gastric cancer mortalities are a result of
complications associated with recurrence and metastasis, even
following curative resection. However, the invasion and metastasis
of gastric cancer is a complex multistep process, in which an
invasive capacity is required for gastric cancer cells to undergo
metastasis. The aberrant expression of specific genes is known to
contribute to this transformation (3).
T-cell lymphoma invasion and metastasis inducing
factor 1 (Tiam-1), an important member of the Dbl oncogene family,
was identified in T-lymphoma cells using proviral tagging in
combination with in vitro selection for invasiveness
(4). The human Tiam-1 gene maps to
the syntenic region (q22) on human chromosome 21 (21q22) and
encodes a 170 kDa transmembrane glycoprotein with intrinsic Rho
GTPase activity. The gene contains a Dbl homologous (DH) domain
adjacent to a pleckstrin homologous domain, a typical structure of
guanine nucleotide exchange factors (GEFs) (5). As a specific GEF for Ras-related C3
botulinum toxin substrate 1 (Rac-1), a member of the Rho oncogene
family, Tiam-l catalyzes the transition of Rac-l from an inactive
GDP-bound state to an active GTP-bound state. In its active
GTP-bound state, Rac-1 activates downstream signaling pathways
associated with a number of important cellular events, including
cytoskeletal reorganization, cell adhesion and migration (6). In addition, Tiam-1 has been
demonstrated to increase the invasion of T-lymphoma cells,
stimulate the cellular migration of fibroblasts and promote the
motility of specific neurocytes. An increasing number of studies
are focusing on Tiam-1 regulatory mechanisms, as well as its role
in tumor progression and metastasis. Increased Tiam-1 expression
has been revealed to correlate with breast cancer grade in humans
and the metastatic potential of human breast cancer cell lines in
nude mice (7), moreover Tiam1 may
represent a marker of progression and metastasis of hepatocellular
carcinomas and colon tumors (8–10).
In our previous study, negative staining for Tiam-1 protein was
observed in paracarcinoma gastric mucosa, obtained from patients
with gastric cancer. By contrast, another study reported that
positive staining for Tiam-1 protein was detected in gastric cancer
tissues (11). In addition, as the
degree of histological differentiation of gastric cancer decreased,
the depth of invasion was found to increase, TNM stage was upgraded
and lymph node metastasis was noted. Staining corresponding to
Tiam-1 protein levels in gastric cancer tissues was observed to
increase, indicating that Tiam-1 may represent a candidate
biomarker of the invasive and metastatic state of gastric cancer
and for patient prognosis; however, the intrinsic biological
mechanism via which Tiam-1 exerts it effects remains unclear.
The aim of the present study was to further define
the mechanism of gastric cancer cell invasion induced by Tiam-1.
Firstly, two subclones with high (MH) or low (ML) invasive and
metastatic potentials were isolated from the MKN-45 human gastric
cancer cell line (M0) by in vitro adhesion selection, as
described previously (12). Tiam-1
expression levels and cellular morphology were compared between the
subclones. In addition, the correlation between Tiam-1 expression
and the in vitro invasive potential of the three cell lines
was analyzed. Next, the effects of Tiam-1 downregulation on changes
in cytoskeletal structure and the invasiveness of MH cells were
investigated by in vitro transfection of antisense
oligodeoxynucleotides (ASODNs). Results of the present study are
likely to provide an improved insight into the mechanism of gastric
cancer cell invasion and metastasis induced by Tiam-1 and promote
the development of novel treatment strategies in gastric
cancer.
Materials and methods
Cell culture and adhesion selection
The parental cell line MKN-45 (M0) is a poorly
differentiated gastric adenocarcinoma cell line (13) and was acquired from Shanghai
Institute of Digestive Disease (Shanghai Second Medical University
Renji Hospital, Shanghai, China). Frozen cell stocks were thawed
and grown in RPMI-1640 medium (Hyclone Laboratories Inc., Logan,
UT, USA) supplemented with 10% fetal calf serum (Gibco-BRL,
Carlsbad, CA, USA), L-glutamine, sodium bicarbonate, essential
amino acids, antibiotics (100 U/ml penicillin and 100 μg/ml
streptomycin) and sodium pyruvate. The medium was replaced with
fresh medium every 2–3 days to maintain optimal growth at 37ºC in a
humidified atmosphere containing 5% CO2. At ~80%
confluence, cells were passaged at a ratio of 1:3 and detached
using a 0.25% (w/v) trypsin-EDTA solution every 5–6 days.
M0 cells were selected for adhesion on mouse
laminin-1-coated substrates (Sigma-Aldrich, St Louis, MO, USA), as
described previously (12).
Briefly, culture dishes (diameter, 35 mm) were coated with 50 μg
laminin-1 in phosphate buffered saline (PBS) and air-dried
overnight at 4ºC. Subconfluent M0 cells were incubated in 0.25%
trypsin-EDTA solution, washed and resuspended in serum-free medium.
The cell suspension (2.5×105 cells/dish) was incubated
with laminin-1 coated substrate for 1 h and attached and unattached
cells were collected separately. The attached cells were cultured
to subconfluence on laminin-1 coated dishes, then selected again
for adhesion. Unaffected cells were prepared following the same
protocol. Serial adhesion selection was performed 20 times, then
attached (MH) or unattached (ML) cells were maintained under normal
culture conditions. Two passages after the last adhesion selection,
the selected cells (MH and ML) were used for further
experiments.
Antisense treatment of MH cell
subclones
The design of one 18-mer ASODN was based on the
nucleotide sequence of human Tiam-1, targeted to the DH domain,
while sense oligonucleotide (SODN) was used as a control. Specific
primers against ASODN and SODN were derived from previously
published sequences (14) and
determined using a BLAST search of a current EMBL
database/NCBI-Genebank database (http://blast.ncbi.nlm.nih.gov/). The sequences used
were as follows: SODN, 5′-*G*AT CTG CGA GCT
CCT GG*A*-3′; and ASODN,
5′-*T*CC AGG AGC TCG CAG
AT*C*-3′ (*represents
phosphorothioate linkages). All ODNs were synthesized using the
Applied Biosystems 391 DNA synthesizer (Perkin Elmer Applied
Biosystems, Inc., Foster City, CA, USA).
MH cells in log-phase growth (2.5×105
cells) were plated onto culture dishes (diameter, 35 mm) and
incubated under standard culture conditions for 24 h, then
transfected with an ASODN or SODN of Tiam-1 using DOTAP liposomal
transfection reagent (Roche GmbH, Mannheim, Germany). The
preparation of liposome-ODN was performing according to the
manufacturer’s instructions. Prior to treatment, ODNs and liposomes
(ratio w/v, 5/15 μl; 1:3) were dissolved separately in
HEPES-buffered saline solution (20 mm HEPES, 150 mm NaCl; pH 7.4)
and mixed at room temperature for 15 min. The optimal working
conditions for lipofectin-ODN were determined previously in
preliminary experiments (data not shown). Experimental cells were
treated with liposome-ASODN at a final concentration of 0.43 μM.
Control treatment groups were serum-free culture medium only,
liposome only (15 μl/dish) and liposome-SODN (0.43 μM). The culture
medium was removed and cells were exposed to lipofection solution
for 8 h. At the end of the incubation with ODN-liposome, the medium
was replaced with fresh serum-containing culture medium (3 ml/dish)
and placed into an incubator. Analysis of Tiam-1 mRNA expression in
MH cells was performed 48 h following transfection.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was extracted using TriPure Isolation
Reagent (Boehringer Mannheim, Mannheim, Germany) according to the
manufacturer’s instructions, and its content and purity were
evaluated by ultra-violet spectrophotography. cDNA was synthesized
in 20-μl reaction volumes containing 1 μg total RNA, 0.2 μg/μl
random hexamer primer, 4 μl 5X reaction buffer, 20 U/μl
ribonuclease inhibitor, 10 mmol/l deoxyribonucleotide triphosphate
(dNTPs) and 20 units Moloney murine leukemia virus reverse
transcriptase (G-Biosciences & Geno Technology Co., St. Louis,
USA). Procedures were performed according to the manufacturer’s
instructions of the First Strand cDNA synthesis kit (Sangon
Biological Engineering Co., Shanghai, China).
For the semi-quantitative assay, a standard
calibration curve was created to determine the optimum number of
cycles (15–35 cycles); the most appropriate number of cycles was 30
(data not shown). PCR was performed using the following materials:
2 μl cDNA, 5 μl 10X buffer, 4 μl MgCl2 (25 mmol/l), 8 μl
dNTPs (2.5 mmol/l), 0.5 μl of each forward and reverse
amplification primer (25 μmol/l) and 0.5 μl Ex Taq DNA
polymerase (2.5 units; Takara Biotechnology Co., Ltd., Dalian,
China) in a 50 μl reaction volume. Reactions were performed using
the PTC-100TM PCR system (MJ Research Inc., Watertown, MA, USA)
under the following conditions: 30 cycles of 94ºC (45 sec), 54ºC (1
min) and 72ºC (1.5 min). The following primer pairs (synthesized by
Sangon Biological Engineering Co.) were used to amplify fragments
of Tiam-1 (818 bp) and GADPH (310 bp) cDNAs: Tiam-1, forward 5′-TTC
TCA CCA GTC TGT TCA GC-3′ and reverse 5′-CCA GAC TTG GAA TCC TCA
GA-3′; and GADPH, forward 5′-AGG TCC ACC ACT GAC ACG TT-3′ and
reverse 5′-GCC TCA AGA TCA TCA GCA AT-3′.
PCR products (5 μl) amplified from the Tiam-1 or
GADPH genes were mixed and underwent 2% ethidium bromide agarose
gel electrophoresis with molecular weight standards. The band
intensities for Tiam-1 and GADPH were estimated using a Bio-Rad gel
documentation 2000 system (Bio-Rad, Hercules, CA, USA). The
intensity of Tiam-1 gene expression were reported as the ratio
(RV) of Tiam-1 to GADPH (as an internal control).
Specimens were analyzed three times and values were averaged.
Quantitative cell-ELISA
MH cells (1×105 cells) in log-phase
growth were plated onto 96-well plates, incubated under standard
culture conditions for 24 h and transfected as described. Control
groups were prepared following the same protocol. Quantitative
cellular-ELISA of the Tiam-1 protein in gastric cancer cells was
performed following transfection for 48 h, as described previously
(15).
In brief, culture medium was removed from the
96-well plates and the cells were washed twice with PBS, fixed with
4% paraformaldehyde (w/v in PBS) for 1 h and removed. Following
three PBS washes, cells were blocked with 10% (w/v) goat serum
albumin in PBS at 4ºC for 15 min and then incubated at 37ºC for 2 h
with a 1:100 working dilution of rabbit polyclonal antibody (50
μl/well) against human Tiam-l (Santa Cruz Biotechnology, Inc.,
Santa Cruz, CA, USA). Next, cells were washed 3 times with 0.05%
Tween-20/PBS and incubated at 37ºC for 1 h with a 1:5,000 (50
μl/well) working dilution of horseradish peroxidase-conjugated goat
anti-rabbit secondary antibody (Zhongshan Biotechnology Co., Ltd.,
Beijing, China). Following four washes with 0.05% Tween-20/PBS,
equal amounts of A and B fluid (50 μl each) containing
3,3′,5,5′-tetramethylbenzidine peroxidase chromogenic reagent (KPL,
Inc., Gaithersburg, MD, USA) were added to react in the dark for 10
min. The reaction was terminated with 50 μl/well HCl (1 mol/l)
under oscillation for 30 sec and the absorbance at 450 nm was
measured using an ELISA plate reader (Sunrise; Tecan Group, Ltd.,
Männedorf, Switzerland). Wells were washed with distilled water,
followed by the addition of 0.08% (w/v) crystal violet (100
μl/well) and incubation at room temperature for 20 min. The wells
were then washed again with distilled water and the reaction was
terminated using 33% (w/v) glacial acetic acid (100 μl/well) at
room temperature for 30 min and the absorbance was measured at 550
nm. The intensity of Tiam-1 protein was reported as the ratio
(RD) of D450 to D550. Specimens were analyzed three
times and the values were averaged. Negative controls consisted of
omission of the primary antibody.
Morphological and ultrastructural
observations
To examine morphological changes, cells were
directly cultured on slides and washed twice with PBS. Next, the
cells were stained with hematoxylin and eosin (H&E) and
observed under a light microscope, and representative images were
captured.
For scanning electron microscopy (SEM), cells were
grown on cover slips and fixed with 2.5% (w/v) glutaraldehyde at
4ºC overnight. Following this, cells were washed with PBS, treated
with 1% (w/v) osmium tetroxide (TAAB Laboratories Equipment Ltd.,
Berkshire, UK) at 4ºC for 1 h, washed three times with PBS and
dehydrated using a gradient ethano1. SEM specimens were dried using
a critical point dryer (Hcp-2; Hitachi Ltd., Tokyo, Japan) and the
sputter was coated with Pt + Cd using an ion sputtering device
(Oie-102; Hitachi Ltd.). Cell surface alteration was observed under
an SEM microscope (AMRay 1000B, Bedford, MA, USA).
Cytoskeletal staining
Cytoskeletal staining was performed as described
previously (16). Following two
washes with PBS, cells cultured on slides were permeabilized with
0.1% Triton-X-100 3 times for 8 min, washed in M-buffered solution
(50 mM imidazole, 50 mM potassium chloride, 0.5 mM magnesium
chloride, 2 mM EGTA, 0.1 mM EDTA, 1 mM mercaptoethanol, 4 M
glycerol and distilled water to 1,000 ml) 3 times for 4 min and
then fixed with 3% (w/v) glutaraldehyde at room temperature for 10
min. Next, cells were washed in M-buffered solution 3 times for 3
min and stained with coomasssie brilliant blue solution (0.2 g
R-250, 46.5 ml methanol, 7 ml glacial acetic acid and 46.5 ml
distilled water) at room temperature for 40 min, followed by PBS
washing, air-drying, dimethyl benzene clearing and neutral gum
sealing. Cells were then viewed under a light microscope (LH50A;
Olympus Optical Co., Tokyo, Japan).
In vitro invasion assay
In vitro cell invasion was analyzed using
24-transwell units (Millipore, Billerica, MA, USA), as described
previously (17).
Polyvinylpyrrolidone-free polycarbonate filters (diameter, 12 μm;
pore size, 8 μm; Millipore) were coated with 30 μg Matrigel/filter
(BD Biosciences, Franklin Lakes, NJ, USA). NIH-3T3 cell-conditioned
media was used as a chemoattractant and was obtained by culturing
cells for 24 h in serum-free RPMI-1640 medium. Cells in serum-free
RPMI-1640 medium (400 μl containing 1×105 cells) were
added to the upper compartment of the chamber, while 600 μl
conditioned medium was added to the lower compartment. Following 72
h incubation at 37ºC in humidified 95% air with 5% CO2,
basement membrane Matrigel and cells on the upper side of the
filter were removed by wiping with a cotton swab. Cells that had
migrated to the lower surface of the filters were fixed and stained
with H&E. The invasive potential of cells was determined by
measuring the number of cells that migrated to the lower side of
the filters through the basement membrane matrigel and pores. The
number of cells that penetrated the filter was counted in 10
microscopic fields of each filter. Assays were performed in
triplicate.
Statistical analysis
Quantitative data are presented as the mean ± SD.
All data were analyzed statistically by one-way ANOVA followed by a
least significant difference test, using the SPSS 10.0 software
program (SPSS Inc., Chicago, IL, USA). P<0.05 was considered to
indicate a statistically significant difference. Correlations were
calculated using the Spearman correlation coefficient.
Results
In vitro invasive potential of M0, ML and
MH cells
MH and ML cells, which exhibit different invasive
and metastatic potentials, were subcloned from the MKN-45 human
gastric cancer cell line (M0) by adhesion selection. A transwell
assay was performed to compare the invasive capacity of the three
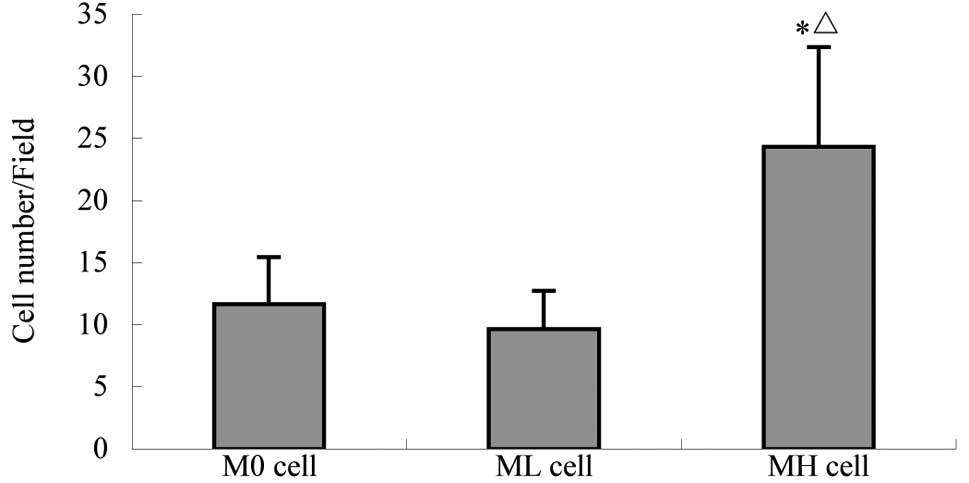
cell strains in vitro. As demonstrated in Table I and Fig. 1, MH cells exhibited a higher
invasive potential than M0 and ML cells in vitro. This is
consistent with results of a previous study on tumor-cell
transplantation and experimental metastasis in nude mice (12).
 | Table IIn vitro invasive potential of
gastric cancer cells. |
Table I
In vitro invasive potential of
gastric cancer cells.
| Variable | Cells (n)
| F-value |
|---|
| M0 | ML | MH |
|---|
| Cells/field | 11.67±3.79 | 9.67±3.06 | 24.33±8.02a,b | 6.470 |
Tiam-1 mRNA and protein expression levels
in M0, ML and MH cells
Since Tiam-l has been previously reported to be
associated with tumor invasion and metastasis, Tiam-l mRNA and
protein expression levels in M0, ML and MH cells were analyzed by
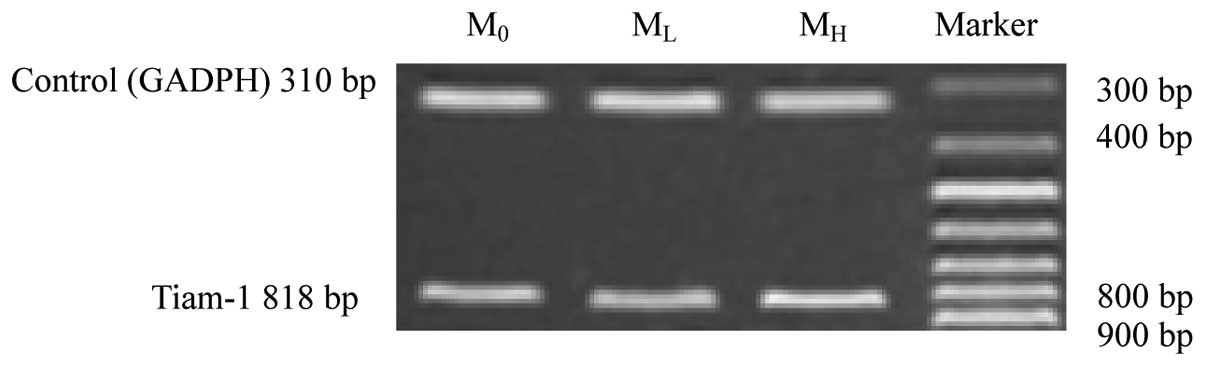
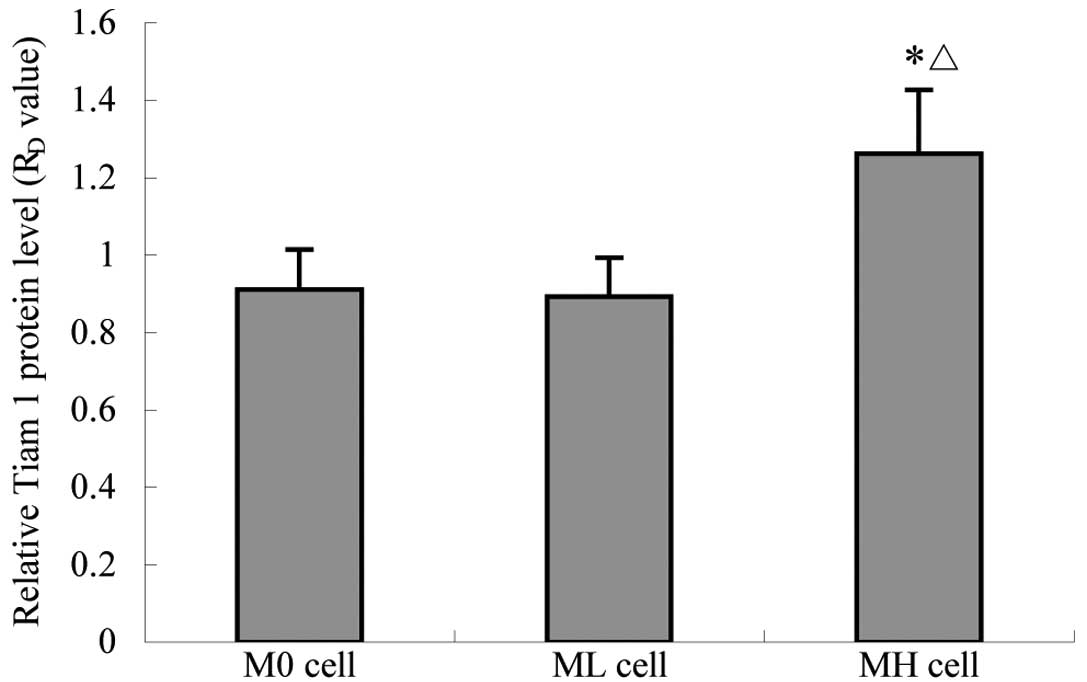
RT-PCR and quantitative cellular-ELISA. The expression of Tiam-1
mRNA and protein in the three strains was positive; however, levels
in MH cells were observed to be markedly higher than those of M0
and ML cells (Table II; Figs. 2–4)
 | Table IITiam-1 mRNA and protein expression
levels in gastric cancer cells. |
Table II
Tiam-1 mRNA and protein expression
levels in gastric cancer cells.
| Ratio | Cells | F-value |
|---|
|
|---|
| M0 | ML | MH |
|---|
| RV | 0.759±0.047 | 0.743±0.039 | 0.855±0.051a,b | 5.269 |
| RD | 0.911±0.104 | 0.892±0.101 | 1.262±0.165a,b | 13.429 |
These observations were analyzed further, revealing
a positive correlation between the expression levels of Tiam-l mRNA
or protein and the invasive capacity of the cells (Spearman
correlation coefficient, r=1.000; P<0.01).
Antisense treatment downregulates the
expression of Tiam-1 mRNA and protein in MH cells
To confirm the effect of Tiam-1 ASODN on MH cells,
RT-PCR and quantitative cell-ELISA were performed. The optimum
conditions for antisense treatment, determined in preliminary
experiments (data not shown), were 0.43 μM ODN, ratio (w/v) of
ODN/liposome=1:3, 4 h exposure and 48 h recovery. Following
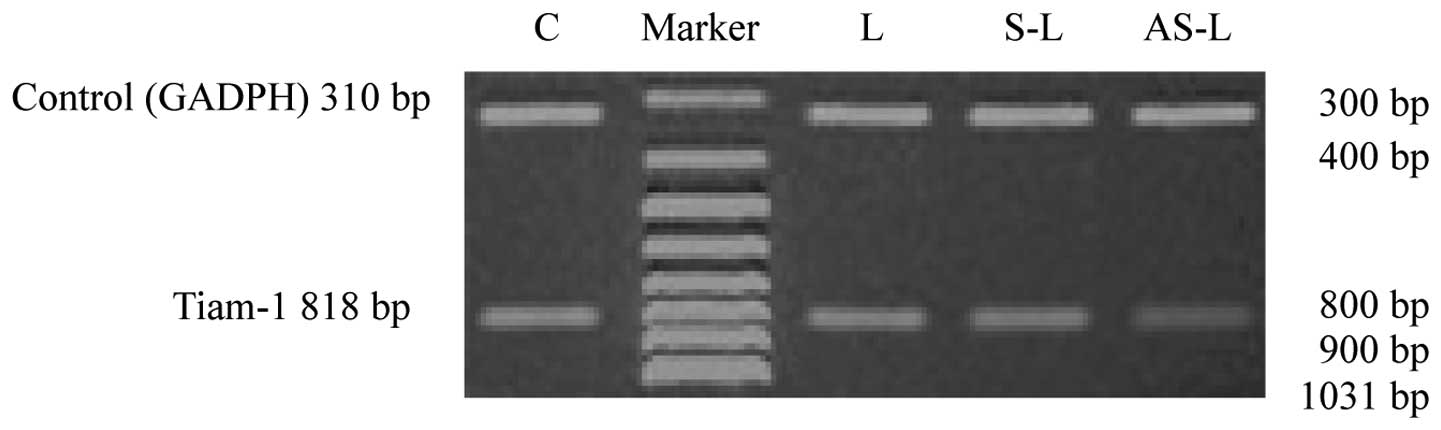
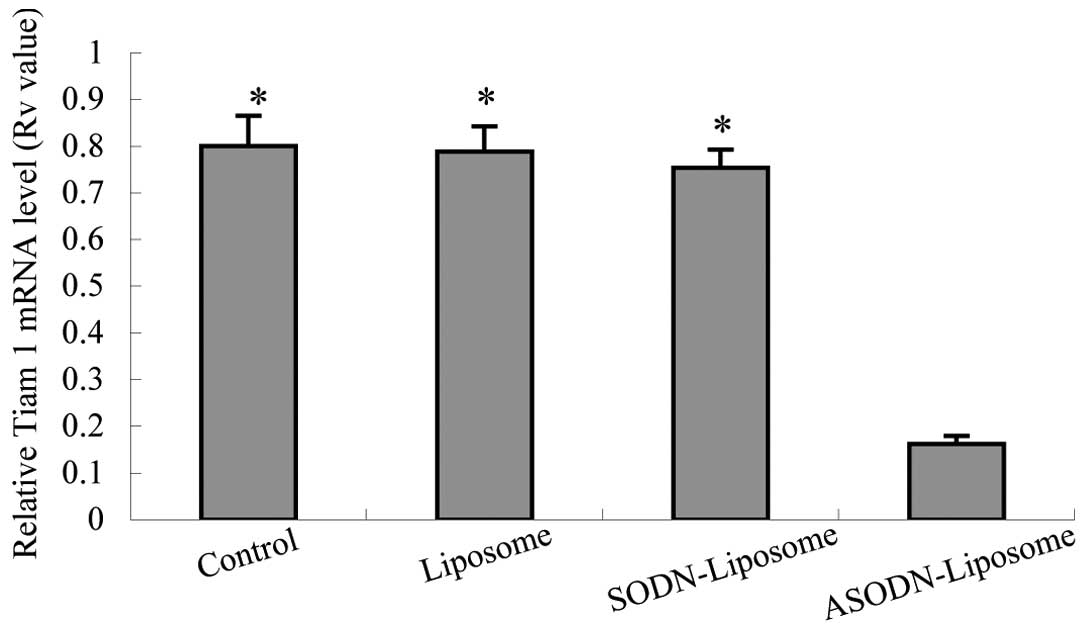
ASODN-liposome treatment, the expression of Tiam-1 in MH cells was
significantly inhibited compared with untreated (control),
liposome- or SODN-liposome-treated samples (Table III). RT-PCR revealed a faint
Tiam-1 band in MH cells following ASODN treatment. By contrast, the
band was significantly stronger in the other three groups. No
significant difference between the intensities of the stronger
bands was identified (Figs. 5 and
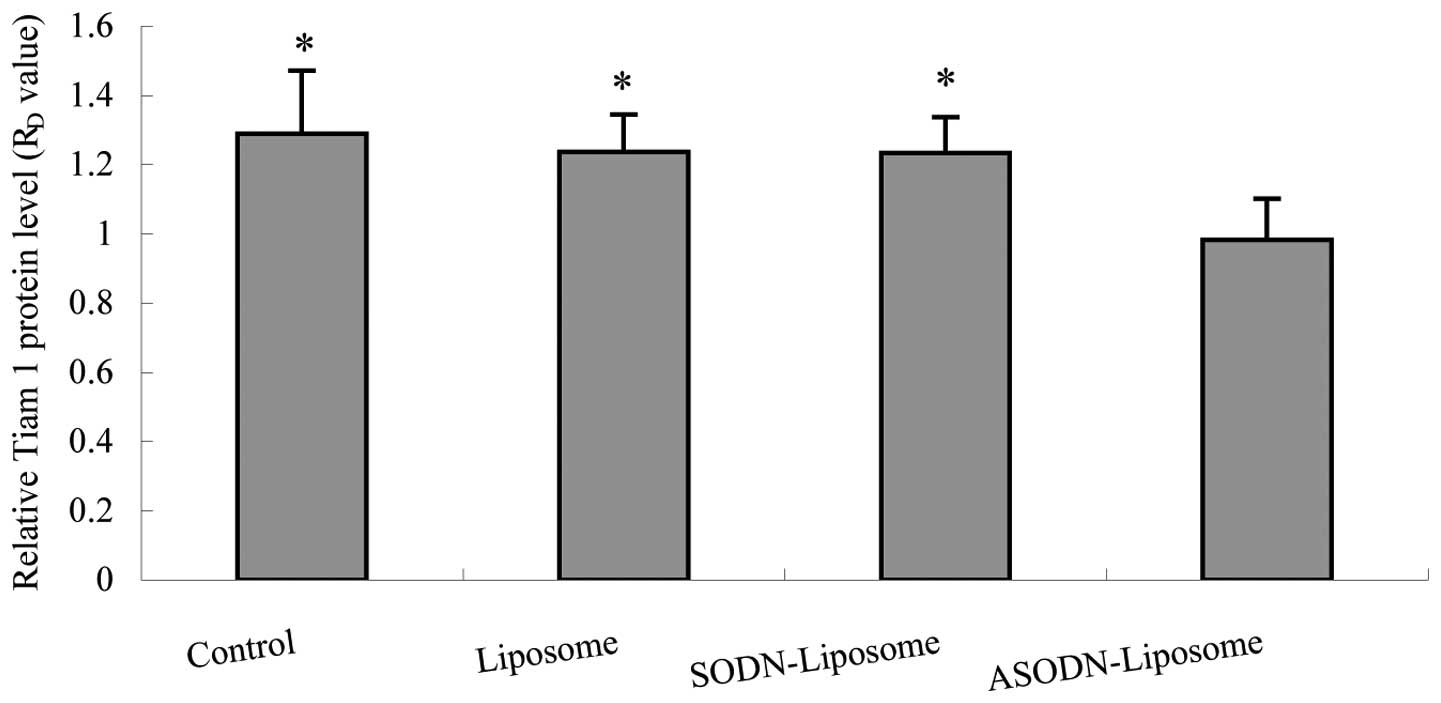
6). Quantitative cell-ELISA of MH
cells transfected with ASODN-liposome also revealed a significant
reduction in Tiam-1 protein levels compared with untreated,
liposome- or SODN-liposome treated groups (Fig. 7).
 | Table IIITiam-1 mRNA and protein expression
levels in gastric cancer cells. |
Table III
Tiam-1 mRNA and protein expression
levels in gastric cancer cells.
| Ratio | Control | Liposome | SODN-liposome | ASODN-liposome | F-value |
|---|
| RV | 0.801±0.065a | 0.789±0.054a | 0.754±0.039a | 0.162±0.018 | 130.160 |
| RD | 1.290±0.182a | 1.237±0.108a | 1.234±0.103a | 0.982±0.119 | 5.516 |
Alterations in cellular morphology and
ultrastructure following antisense treatment
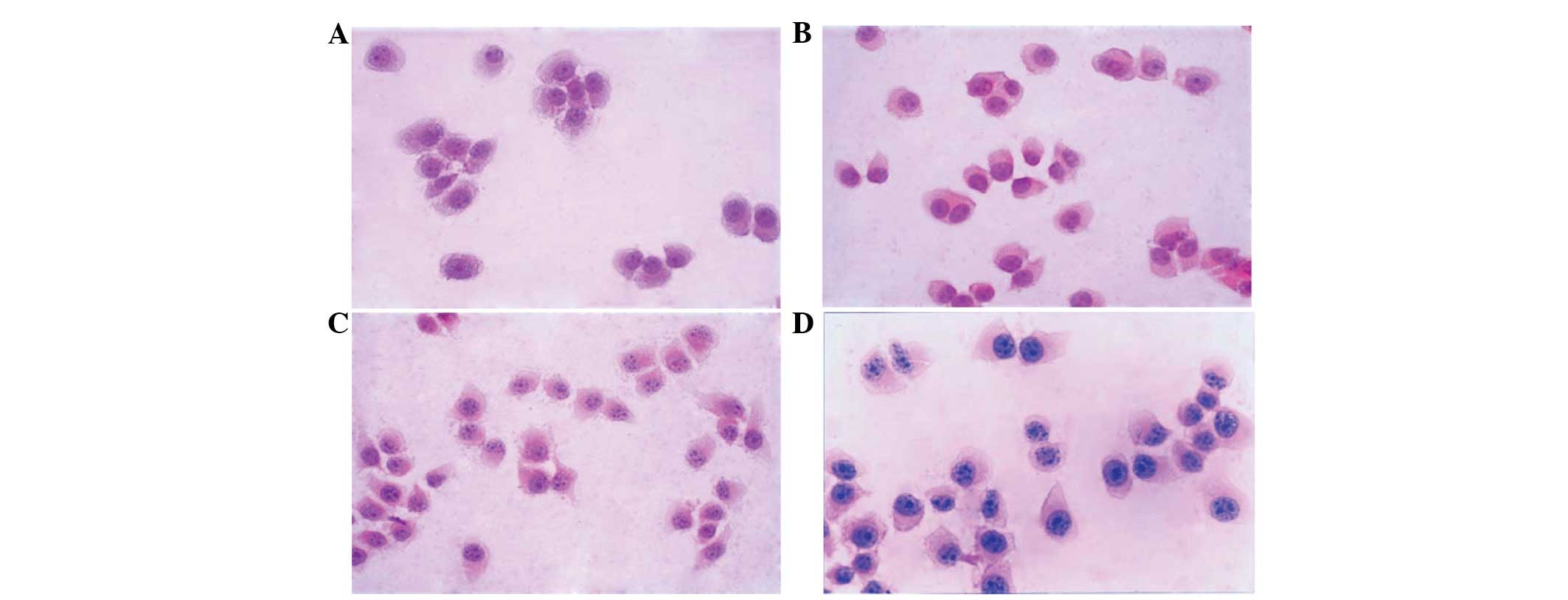
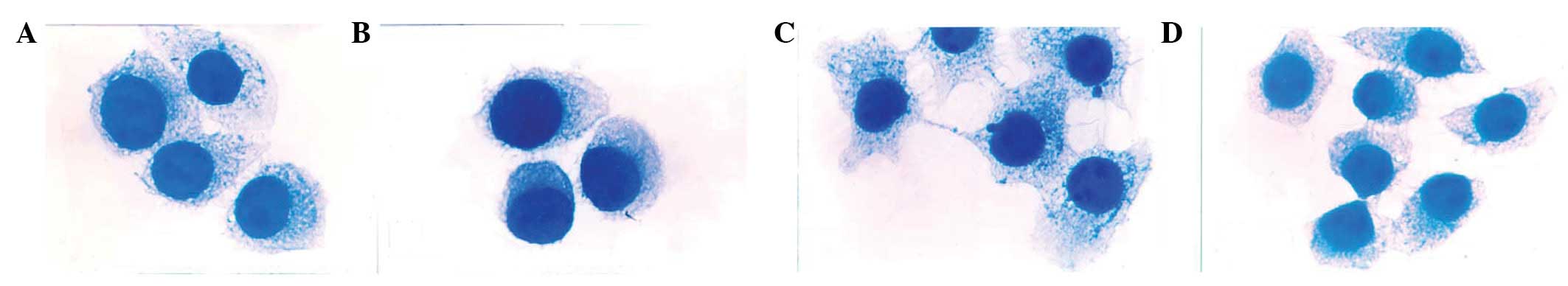
Following H&E staining, cells were observed
under a light microscope. M0 and ML cells were floating and
exhibited round epithelial-like morphologies with a large
nucleocytoplasmic ratio; no differences in morphology were found
between M0 and MH cells. However, MH cells were observed to be
multiform compared with the uniform appearance of M0 and ML cells.
A number of MH cells were observed to exhibit a polygonal
morphology with longer cytoplasmic processes (Fig. 8). Following Tiam-1 ASODN treatment,
the morphology of MH cells resembled that of M0 and ML cells.
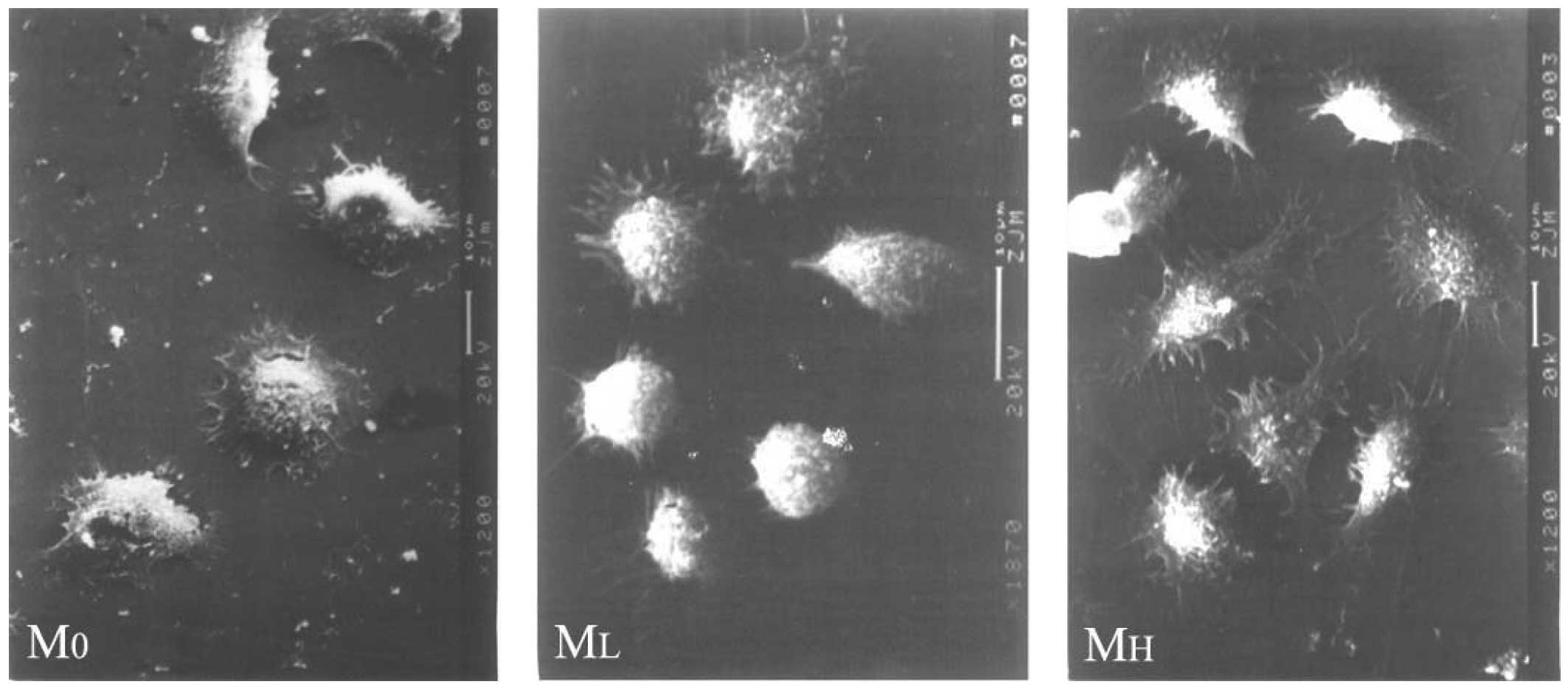
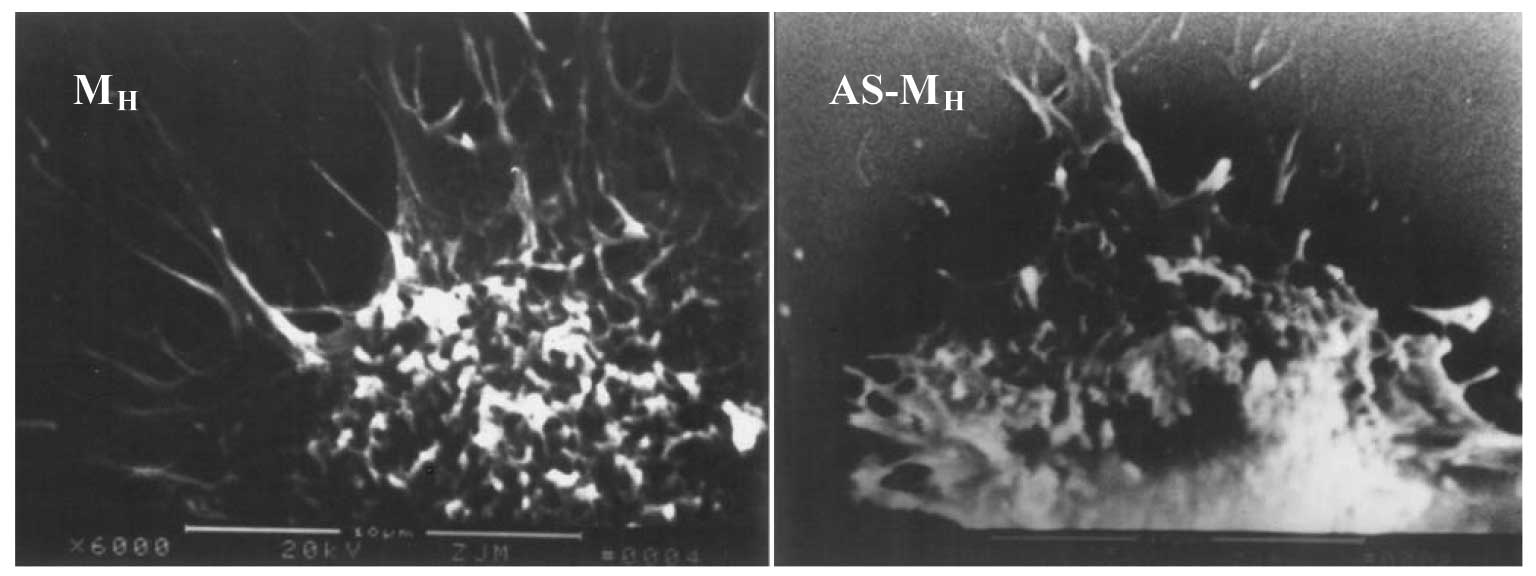
SEM revealed abundant microvilli, cytoplasmic
processes and large pseudopodia dotted on the surface of MH cells.
Certain processes extended a considerable distance, radiated in all
directions or formed bulges at their ends. By contrast, the
processes of M0 and ML cells were short and compact. No desmosomes,
tight conjunctions or other cell junction structures were observed
in the three cell types. Following antisense treatment, MH cells
exhibited a smooth surface with markedly reduced filopodia and
microspikes (Figs. 9 and 10).
Cytoskeletal structure and reorganization
following antisense treatment
Cells were stained with coomasssie brilliant blue
R-250 and viewed under an immersion objective. The cytoskeletal
structure of M0, ML and MH cells formed a complicated cytoskeletal
network in the cytoplasm and was markedly more concentrated in
perinuclear areas. Bundles of cytoskeletal filaments were stretched
and scattered intercellularly. Compared with M0 and ML cells,
cytoskeletal alignment in MH cells was disordered, accompanied with
more dotted actin bodies, cytoplasmic processes and long
filament-like structures. By contrast, no differences in
cytoskeletal alignment were observed between M0 and ML cells.
Following antisense treatment, cytoskeletal distribution in MH
cells changed from irregular to regular, with reduced long
filament-like structures on the cell surface and dotted actin
bodies in the cytoplasm, and resembled that of M0 and ML cells
(Fig. 11).
Tiam-1 ASODN treatment downregulates the
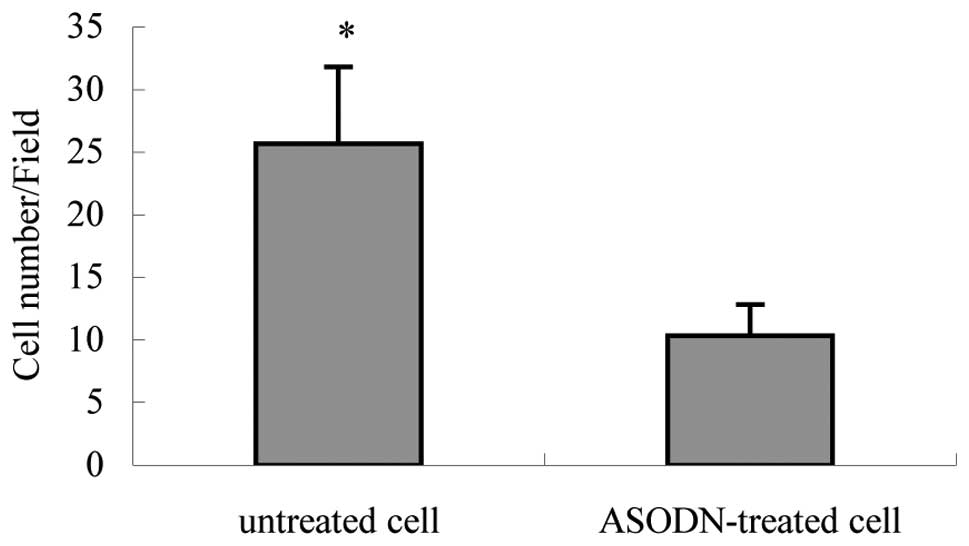
invasiveness of MH cells in vitro
An in vitro transwel1 assay revealed that the
number of antisense-treated MH cells penetrating the artificial
basement membrane was 10.33±2.52 cells/high power field, which was
significantly lower than that of untreated MH cells (25.67±6.11
cells/high power field; P<0.05; Table IV, Fig. 12).
 | Table IVEffect of antisense treatment on the
in vitro invasive potential of MH cells. |
Table IV
Effect of antisense treatment on the
in vitro invasive potential of MH cells.
| Variable | Cells (n)
| F-value |
|---|
| Untreated | ASODN-treated |
|---|
| Cells/field | 25.67±6.11 | 10.33±2.52a | 16.153 |
Discussion
Tumor cell invasion and metastasis depends on a
unique set of biological properties that enable malignant cells to
complete all steps of the metastatic cascade. A tumor mass commonly
consists of various cell subclones, a phenomenon known as tumor
heterogeneity (18). Since
subclones are from the same parental cell line, thus having the
same genetic background, the differences in their phenotypes are
likely to have a number of underlying molecular mechanisms
(19,20). In-depth study of the differences
between subclones may provide novel insight into the mechanisms of
tumor cell invasion and metastasis.
Members of the Rho GTPase family have emerged as key
players in the regulation of a diverse set of biological
activities, including actin organization, focal complex/adhesion
assembly, cell motility, cell polarity and gene transcription, the
importance of which is becoming evident in cancer progression,
particularly in the area of metastasis (21,22).
As an upstream regulator of Rho-GTPases, particularly Rac-1, the
potential role of Tiam-1 in tumor invasion and metastasis is
becoming increasingly apparent.
In the present study, two subclones with a high (MH)
or low (ML) invasive and metastatic potential were acquired from
the MKN-45 human gastric cancer cell line (M0) using in
vitro adhesion selection on laminin-1-coated dishes (12), and Tiam-1 expression levels were
compared. Results indicated that Tiam-1 mRNA and protein expression
levels in MH cells were markedly higher than that of M0 and ML
cells. In addition, a positive correlation between the expression
of Tiam-1 and the in vitro invasive potentials of M0, ML and
MH cells was observed. These results are consistent with previous
observations (11) and implicate a
role for Tiam-1 in the invasive process of human gastric
cancer.
Cytoskeletal components, including microtubules,
microfilaments and intermediate filaments, are responsible for the
maintenance of cell morphology, direction of cell locomotion, cell
spreading, cell migration and stability of cell-substrate contact
in normal cells (23). Alterations
in cytoskeletal structures, such as reduced microtubules,
disruption of stress fibers and redistribution of actin-filaments
in tumor cells, have been reported to be important determinants of
malignancy in tumor cells, enabling cells to transform and
transmigrate within tissues (24).
In the present study, analysis of the cellular morphology and
ultrastructure of gastric cancer cells revealed that M0 and ML
cells were floating and exhibited round epithelial-like structures.
By contrast, MH cells were multiform and a number of cells were
polygonal with long cytoplasmic processes. SEM also revealed
abundant microvilli, cytoplasmic processes and large microhills
located on the MH cell surface. Specific processes were observed to
extend considerable distances, radiate in all directions or form
bulges at their ends. By contrast, M0 and ML cells were observed to
exhibit short and compact processes. Following staining with
coomassie brilliant blue, the cytoskeletal structure of M0, ML and
MH cells was observed to form a complicated cytoskeletal network in
the cytoplasm. The network was particularly concentrated in
perinuclear areas and bundles of cytoskeletal filaments passed
through the cell wall to interact with cytoskeletal arrays of
adjacent cells. Compared with M0 and ML cells, cytoskeletal
alignment in MH cells was disordered, accompanied with increased
dotted actin bodies, cytoplasmic processes and long filament-like
structures, whereas no marked differences were observed between M0
and ML cells. The differences in cytoskeletal structure may be due
to cytodynamic activities associated with dissemination and
implantation, based on the invasive and metastatic potentials of
M0, ML and MH cells. As Tiam-1 is known to regulate the
reorganization of cytoskeletal structure, differences in the
morphological characteristics of gastric cancer cells with
different invasive and metastatic potentials may correlate with
Tiam-1 expression.
ASODNs bind and inactivate specific RNA sequences
and represent one of the best tools for studying gene function, the
regulation of gene expression and interactions between gene
products. Intracellular delivery of ASODNs results in the
downregulation of target gene expression by inhibiting
transcription or translation without affecting other cellular
functions. To validate the role of Tiam-1 in gastric cancer cell
invasion, a 18-mer ASODN was used to block Tiam-1 expression in MH
cells and the subsequent changes in cellular morphology and the
in vitro invasiveness of MH cells were analyzed. As
unmodified phosphodiester oligomers are susceptible to nucleases,
partial phosphorothioate oligomers were used, which are relatively
nuclease-resistant. Results demonstrated that the treatment of MH
cells with ASODNs (0.43 μM, 48 h) decreased Tiam-1 mRNA
transcription and protein expression by 70 [(0.801–0.162)/0.801]
and 65% [(1.290–0.982)/1.290] respectively, compared with untreated
controls. In addition, the in vitro invasive potential of
ASODN-treated MH cells was suppressed by 75% [(25.67–10.33)/25.67];
however, cells treated with SODN or lipofectin alone were not
affected. Morphological and ultrastructural observations also
revealed that ASODN-treated MH cells exhibited a smooth surface
with markedly reduced filopodia and microspikes, which resembled M0
and ML cells. Cytoskeletal distribution in ASODN-treated MH cells
was markedly altered from disordered to regular, with reduced long
filament-like structures, projections, pseudopodia at the cell
surface and reduced actin bodies in the cytoplasm. Our findings
showed that Tiam-1 induces gastric cancer cell invasion by
regulating cytoskeletal reorganization, which caused the
depolymerization of microtubules and microfilaments, leading to
disordered, actively motile properties, cell detachment and a
metastatic potential in gastric cancer cells. Previous observations
showed that Tiam-1 contributed to the cytoskeletal reorganization
required during cell migration and neurite extension by activation
of Rac 1 (25). Whether this is
the case in gastric cancer cell invasion it is not yet known.
In addition, abnormalities in the expression and
functional activity of cell adhesion molecules are implicated in
the invasive and metastatic progression of tumor cells.
Intercellular and cell-matrix adhesion molecules have been shown to
have roles in addition to acting as cementing substances, such as
regulating cell polarity, differentiation, invasion and migration
(26). A number of these cellular
events are mediated through direct associations with the
cytoskeletal network and interactions between the cytoskeletal
network. Therefore, the manipulation of molecules associated with
the cytoskeleton requires further investigation (27). The in vitro cell model of
laminin-1 adhesion selection used in the present study represents a
simple and time-effective method to screen a specific phenotypic
cell subpopulation and is likely to be useful for future analysis
of the cytoskeleton.
Laminin-1, a major basement membrane glycoprotein,
promotes the malignant phenotype and expression of specific laminin
receptors, and has been found to correlate with the malignant
characteristics of tumor cells (28). The adhesion of cells to laminin-1
induces collagenase IV production, which is required for tumor
cells to metastasize. Previous studies, as well as the present
study, have demonstrated that the adherent subclone (MH), isolated
from the MKN-45 gastric cancer cell line by in vitro
adhesion selection on laminin-1, is more invasive and metastatic
in vitro and in vivo compared with parental (M0) and
non-adherent subclone (ML) cells, indicating that the properties of
these cells are closely associated with adhesion to the basement
membrane and extracellular matrix (12). In addition, other studies have
demonstrated that a laminin-adhesive subclone of a human colon
cancer cell line revealed higher invasive and metastatic potentials
in vitro and in vivo than a laminin-non-adhesive cell
line. These observations indicate that the increased invasive and
metastatic potentials of the laminin adhesion-selected
subpopulation may be due to an alteration in the membrane
distribution and/or affinities of multiple laminin receptors,
including the 67 kDa laminin receptor and β1 integrin (29,30).
The cancer cell population, either as a solid tumor mass in
vivo or as a continuous cell line in vitro, is an
ever-changing entity due to their genetic instability and selective
environmental pressure (31).
Tiam-1 expression is associated with the invasive potential of
gastric cancer cells. Therefore, we hypothesized that Tiam-1
expression, the rearrangement of cytoskeletal structure and the
expression and/or redistribution of cell surface adhesion molecules
are linked. Additionally, the associated mechanisms of cellular
signal transduction remain unclear and require investigation.
Results of the present study demonstrated that
Tiam-1 induces gastric cancer cell invasion and may represent a
candidate target for biotherapy. However, additional studies are
required to determine the molecular and biological mechanisms
associated with these observations.
Acknowledgements
The authors thank Professor Zheng Jiang and his
personnel at the Molecular Biological Center of Xinan Hospital
(Chongqing, China) for their excellent technical assistance.
References
|
1
|
Mahar AL, McLeod RS, Kiss A, Paszat L and
Coburn NG: A systematic review of the effect of institution and
surgeon factors on surgical outcomes for gastric cancer. J Am Coll
Surg. 214:860–868. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee JH, Kim KM, Cheong JH and Noh SH:
Current management and future strategies of gastric cancer. Yonsei
Med J. 53:248–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lin LL, Huang HC and Juan HF: Discovery of
biomarkers for gastric cancer: a proteomics approach. J Proteomics.
75:3081–3097. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Habets GG, Scholtes EH, Zuydgeest D, van
der Kammen RA, Stam JC, Berns A and Collard JG: Identification of
an invasion-inducing gene, Tiam-1, that encodes a protein with
homology to GDP-GTP exchangers for Rho-like proteins. Cell.
77:537–549. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Habets GG, van der Kammen RA, Jenkins NA,
Gilbert DJ, Copeland NG, Hagemeijer A and Collard JG: The
invasion-inducing TIAM1 gene maps to human chromosome band 21q22
and mouse chromosome 16. Cytogenet Cell Genet. 70:48–51. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mertens AE, Roovers RC and Collard JG:
Regulation of Tiam1-Rac signalling. FEBS Lett. 546:11–16. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Minard ME, Kim LS, Price JE and Gallick
GE: The role of the guanine nucleotide exchange factor Tiam1 in
cellular migration, invasion, adhesion and tumor progression.
Breast Cancer Res Treat. 84:21–32. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen B, Ding Y, Liu F, et al: Tiam1,
overexpressed in most malignancies, is a novel tumor biomarker. Mol
Med Rep. 5:48–53. 2012.PubMed/NCBI
|
|
9
|
Ding Y, Chen B, Wang S, et al:
Overexpression of Tiam1 in hepatocellular carcinomas predicts poor
prognosis of HCC patients. Int J Cancer. 124:653–658. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Minard ME, Ellis LM and Gallick GE: Tiam1
regulates cell adhesion, migration and apoptosis in colon tumor
cells. Clin Exp Metastasis. 23:301–313. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu JM, Yu PW and Zhao YL: Relationship
between the expression of Tiam-1, Rac 1 and the pathobiological
behavior of gastric cancer. Chin J Gen Surg. 14:168–172. 2005.
|
|
12
|
Chen XR, Ren WP, Dong JF, Xiao SD and
Sloane BF: Screening of gastric cancer cell sublines by adhesion
method in vitro. Chin J Gastroenterol. 2:121–124. 2001.
|
|
13
|
Yokozaki H: Molecular characteristics of
eight gastric cancer cell lines established in Japan. Pathol Int.
50:767–777. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li ZZ, Zhang L, Mao HT, Wang Y, Li DH and
Gui SL: Effect of Tiam-1 antisense oligodeoxynucleotides (ASODNS)
on antimetastasis of tumor. Chin J Cancer Biother. 50:767–777.
2000.
|
|
15
|
Zhang XQ, Zhang R, Gu CH and Wang YM:
Expression level of ICAM-1 on cultured cells measured
quantitatively with cellular ELISA. Acta Acad Med Mil Tert.
23:117–118. 2001.
|
|
16
|
Zhang JH, Ding YQ and Yang XJ:
Introduction of a reforming staining method for cytoskeletal
proteins. J Diag Pathol. 3:235–236. 1996.
|
|
17
|
Albini A, Iwamoto Y, Kleinman HK, Martin
GR, Aaronson SA, Kozlowski JM and McEwan RN: A rapid in vitro assay
for quantitating the invasive potential of tumor cells. Cancer Res.
47:3239–3245. 1987.PubMed/NCBI
|
|
18
|
Lleonart ME, Martin-Duque P,
Sanchez-Prieto R, Moreno A and Ramon y Cajal S: Tumor
heterogeneity: morphological, molecular and clinical implications.
Histol Histopathol. 15:881–898. 2000.PubMed/NCBI
|
|
19
|
Marian AJ: Molecular genetic studies of
complex phenotypes. Transl Res. 159:64–79. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Prasun P, Pradhan M and Agarwal S: One
gene, many phenotypes. J Postgrad Med. 53:257–261. 2007. View Article : Google Scholar
|
|
21
|
Etienne MS and Hall A: Rho GTPases in cell
biology. Nature. 420:629–635. 2002. View Article : Google Scholar
|
|
22
|
Malliri A and Collard JG: Role of
Rho-family proteins in cell adhesion and cancer. Curr Opin Cell
Biol. 15:583–589. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hirohashi S and Kanai Y: Cell adhesion
system and human cancer morphogenesis. Cancer Sci. 94:575–581.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ben-Ze’ev A: The cytoskeleton in cancer
cells. Biochim Biophys Acta. 780:197–212. 1985.
|
|
25
|
Ehler E, van Leeuwen F, Collard JG and
Salinas PC: Expression of Tiam-1 in the developing brain suggests a
role for the Tiam-1-Rac signaling pathway in cell migration and
neurite outgrowth. Mol Cell Neurosci. 9:1–12. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Okegawa T, Pong RC, Li Y and Hsieh JT: The
role of cell adhesion molecule in cancer progression and its
application in cancer therapy. Acta Biochim Pol. 51:445–457.
2004.PubMed/NCBI
|
|
27
|
Marhaba R and Zoller M: CD44 in cancer
progression: adhesion, migration and growth regulation. J Mol
Histol. 35:211–231. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ekblom P, Lonai P and Talts JF: Expression
and biological role of laminin-1. Matrix Biol. 22:35–47. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim WH, Lee BL, Jun SH, Song SY and
Kleinman HK: Expression of 32/67-kDa laminin receptor in laminin
adhesion-selected human colon cancer cell lines. Br J Cancer.
77:15–20. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim WH, Jun SH, Kibbey MC, Thompson EW and
Kleinman HK: Expression of beta 1 integrin in
laminin-adhesion-selected human colon cancer cell lines of varying
tumorigenicity. Invasion Metastasis. 14:147–155. 1995.PubMed/NCBI
|
|
31
|
Brábek J, Mierke CT, Rösel D, Veselý P and
Fabry B: The role of the tissue microenvironment in the regulation
of cancer cell motility and invasion. Cell Commun Signal.
8:222010.PubMed/NCBI
|