Introduction
Glioblastoma multiforme (GBM) is the most aggressive
form of astrocytoma, with a low mean survival time after diagnosis
(1). Despite aggressive treatment
and recently developed clinical and targeted therapies, the overall
survival time for glioblastoma patients has not improved
significantly over the last twenty years, despite objective initial
responses (2). Thus, the
definition of novel biological characteristics is required for
informed diagnosis and treatment.
MicroRNAs (miRNAs) are small, non-coding single-
stranded RNAs ~19–25 nt long, which regulate genes at the
translational level by binding loosely to complimentary sequences
in the 3′-untranslated regions (UTRs) of target mRNAs, and are
involved in cell growth, differentiation, cytokine activities and
angiogenesis (3,4). Mounting evidence has demonstrated
that miRNAs are essential in regulating various pathways involved
in tumor pathogenesis, functioning as either oncogenes or tumor
suppressors (5–7).
Previous studies have shown that there is a
difference in the expression of miRNAs in glioblastoma tissues
compared to that in normal brain tissues, for example, miR-21 is
overexpressed in glioblastoma tissues, which inhibits cell
proliferation by the mitogen-activated protein kinase (MAPK) and
AKT pathways (5,6). miR-381 levels were increased in GBM.
By directly targeting leucine-rich repeat-containing protein 4
(LRRC4), miR-381 was regulated by LRRC4 via a feedback loop
involving the MAPK pathway (8).
The miR-17–92 cluster has been found to be upregulated in GBM and
directly targets the connective tissue growth factor (CTGF)
(9). miR-26a and miR-214 have been
shown to target PTEN and appeared to be upregulated in gliomas
(10–12). Conversely, levels of miR-7 were
found to be lower in GBM, the targets of which include the
epidermal growth factor receptor (EGFR), and the overexpression of
miR-7 reduces proliferation, survival and invasiveness in cultured
glioma cells. miR-124 and miR-137, which are both downregulated in
GBM, induce neuronal differentiation and inhibit glioma cell growth
in vitro.
Son of sevenless 1 (SOS1) is a dual guanine
nucleotide exchange factor (GEF) for Ras and Rac1 that converts
inactive Ras-GDP into active Ras-GTP in many EGF-stimulated cells
(13,14). SOS1 has two binding sites for Ras,
one of which is an allosteric site distal to the active site
(15). RTK activation results in
the translocation of SOS1, which mediates Ras activation. The
Ras-specific GEF activity of SOS1 is conferred by the Cdc25 domain
in the central region of the protein, which also contains a
Ras-binding region designated as the Ras exchanger motif (16). Ras is a critical signaling molecule
that is important in regulating cell growth (17). MAPK pathways are involved in a
variety of cellular functions including growth, proliferation,
differentiation, migration and apoptosis (18). The activation of ERK by growth
factors and mitogens leads to a series of phosphorylation reactions
involving Ras, Raf and ERK, and is particularly important in
understanding the pathogenesis of cancer. Active ERK signaling
results in the upregulation of transcriptional products, some of
which allow entry into the cell cycle, and some of which repress
the expression of genes that inhibited cell proliferation and the
cell cycle (19).
In this study we focused on miRNA-124, a
brain-enriched miRNA that has been broadly investigated in order to
understand physiological neural development (20,21).
We detected that miR-124 is significantly downregulated in glioma
cell lines, and that the overexpression of miR-124 induced cell
proliferation inhibition, which is associated with SOS1 signaling
in the MAPK pathway.
Materials and methods
Cell lines and culture
Human glioma cell lines (U87, U373, SW1088 and
SW1783) and HEK293T cells were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA). U87 and U373 cells
were cultured in minimal essential medium (MEM), and SW1088 and
SW1783 cells were cultured in Leibovitz’s L-15 medium (Invitrogen,
Carlsbad, CA, USA). All media were supplemented with 10% fetal
bovine serum (FBS) (Invitrogen), 100 U/ml of penicillin and 100
μg/ml of streptomycin (Gibco, Grand Island, NY, USA). Human
astrocytes (HA) and all growth media were obtained from ScienCell
Research Laboratories (Carlsbad, CA, USA). The cells were cultured
in a humidified 5% CO2 atmosphere.
Quantification of mRNA using real-time
qRT-PCR
Total RNA, including small RNA, was extracted from
cells using TRIzol (Invitrogen) according to the manufacturer’s
instructions. From each sample, 1 μg of RNA was reverse-transcribed
using the SuperScript™ III first-strand synthesis system and
oligo(dT) primers (Invitrogen) were synthesized according to the
manufacturer’s instructions. Real-time PCR (qRT-PCR) was performed
with the ABI 7500 (Applied Biosystems, Carlsbad, CA, USA). The
cycling parameters were 95°C for 10 min followed by 40 cycles of
95°C (15 sec) and 60°C (60 sec), followed by melting curve
analysis. The primers used were: SOS1, F:
5′-CAAGAACACCGTTAACACCTC-3′ and R: 5′-GGACAGGCACTTCATCAGTG-3′;
GAPDH, F: 5′-GGA GTCCACTGGCGTCTT-3′, and R: 5′-GAGTCCTTCCA
CGATACCAA-3′. For qRT-PCR of miR-124, 50 ng total RNA was
reverse-transcribed with a miRNA-specific stem-loop primer
(5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGAGGCATT-3′), and the
specific primers for U6 were sequenced as described previously
(5′-CGCTTCACGAATTTGCGTGTC-3′) (22). All reactions were performed in
triplicate with GAPDH as a reference (internal control) and the
median Ct (cycle threshold) value was used for analysis.
SOS1-3′UTR and miR-124 reporter
assays
There are three predicted target sites for miR-124
in the entire 3′UTR of SOS1 (www.targetscan.org). SOS1-3′UTR reporter assays were
performed in 293T cells. pCS2-Luc vector harboring SOS1-3′UTR
sequences with wild-type (WT) miR-124 binding sites or mutated
(MUT) miR-124 binding sites were generated by cloning the
subsequent 3′UTR of SOS1 into the EcoRI and XhoI
sites. The 293T cells were cultured in Dulbecco’s modified Eagle’s
medium (DMEM) supplemented with 10% FBS. Cells were transfected
with miR-124 or control mimics (50 ng), pMIR-REPORT vectors
containing WT or MUT miR-124 binding sites (100 ng) and pRL-SV40
(Promega, Madison, WI, USA) expressing Renilla luciferase (30 ng)
for normalization. Luciferase measurements were performed 48 h
post-transfection using the Dual-Luciferase Reporter Assay System
(Promega).
Vector constructs
The pri-miR-124 sequence was amplified and cloned
into the pcDN3.1 vector and then subcloned (BamHI +
EcoRI) into the pCDH-CMV-MCS-EF1-copGFP vector (SBI) to
generate pCDH-miR-124. A 2500 bp fragment of the 3′UTR of SOS1 was
obtained by PCR amplification of human genomic DNA subcloned into
pCS2-Luc vector using the primers: F: 5′-CGTGAATTCGCTGCAACATGGTGGG
AAC-3′ and R: 5′-TCACTCGAGGTGGGCTATGTAAGGCA TTTTTC-3′ (reverse).
The underlined sequences are the introduced EcoRI and
XhoI sites, respectively.
The mutated versions, mut-1, mut-2, mut-3 and mut,
were generated utilizing the SOS1-UTR plasmid as a template and
modifying the miR-124 seed binding site using the QuikChange II XL
site-directed mutagenesis kit. The mutagenic primers used were:
Mut1: 5′-GUUUAGUAAAUUCCA CCGGCCA-3′, Mut2: 5′-CAGUAGCUGCCAAAUGCCGGC
CU-3′ and Mut3: 5′-AAUAAUAAAGAAAAACCGGCCAC-3′. The underlined
sequences indicate the mutated bases. All constructs were sequenced
for verification.
Lentivirus production and
transduction
Virus particles were harvested 48–60 h after
pCDH-miR-124 transfection with the packaging plasmid pRSV/pREV,
pCMV/pVSVG and pMDLG/pRRE transfected into HEK293T cells using
Fugene® HD Transfection Reagent (Roche, Mannheim,
Germany). U87 and U373 cells were infected with recombinant
lentivirus-transducing units plus 8 μg/ml polybrene (Sigma, St
Louis, MO, USA).
Cell proliferation assay
Cell proliferation was measured using the Cell
Counting Kit-8 (CCK-8) assay kit (Dojindo Corp., Kunamoto, Japan).
Cells were seeded into a 96-well plate at a density of
3×103 cells in each well with 100 μl culture medium,
then 10 μl CCK-8 was added. The cells were subsequently incubated
for 1 h at 37°C and the absorbance was measured at 450 nm. Three
independent experiments were performed.
SDS-PAGE and western blotting
Cells were lysed in RIPA buffer, the lysate was
sonicated and centrifuged for 10 min at 13,000 × g to remove cell
debris. Protein concentrations were determined using a
bicinchoninic acid assay (BCA) (Thermo Scientific, Waltham, MA,
USA). Equal amounts of proteins (30–40 μg/lane) were separated
using SDS-PAGE and transferred to a nitrocellulose membrane
(Bio-Rad, Hercules, CA, USA). The membrane was probed with an
appropriate primary antibody and a secondary antibody conjugated to
horseradish peroxidase. The following antibodies were utilized:
GAPDH (1:1,000 dilution, Cell Signalling Technology Inc., Danvers,
MA, USA), SOS1 (1:1,000 dilution, Cell Signalling Technology Inc.),
Ras (1:1,000 dilution, Millipore, Billerica, MA, USA), p-Raf
(1:1,000 dilution, Epitomics, Burlingame, CA, USA), p-ERK (1:1,000
dilution, Epitomics) and ERK (1:5,000 dilution, Epitomics).
Proteins were visualized with enhanced chemiluminescence
(Millipore). Three independent experiments were performed and one
representative result is shown.
Statistical analysis
Data were presented as the means ± SE. Quantified
data represent an average of at least triplicate samples or as
otherwise indicated. Error bars represent SE. Statistically
significant differences were determined by the Student’s t-test.
P<0.05 was taken to indicate a statistically significant
difference.
Results
miR-124 is downregulated in GBM cell
lines and the stable overexpression of miR-124 inhibits cell
growth
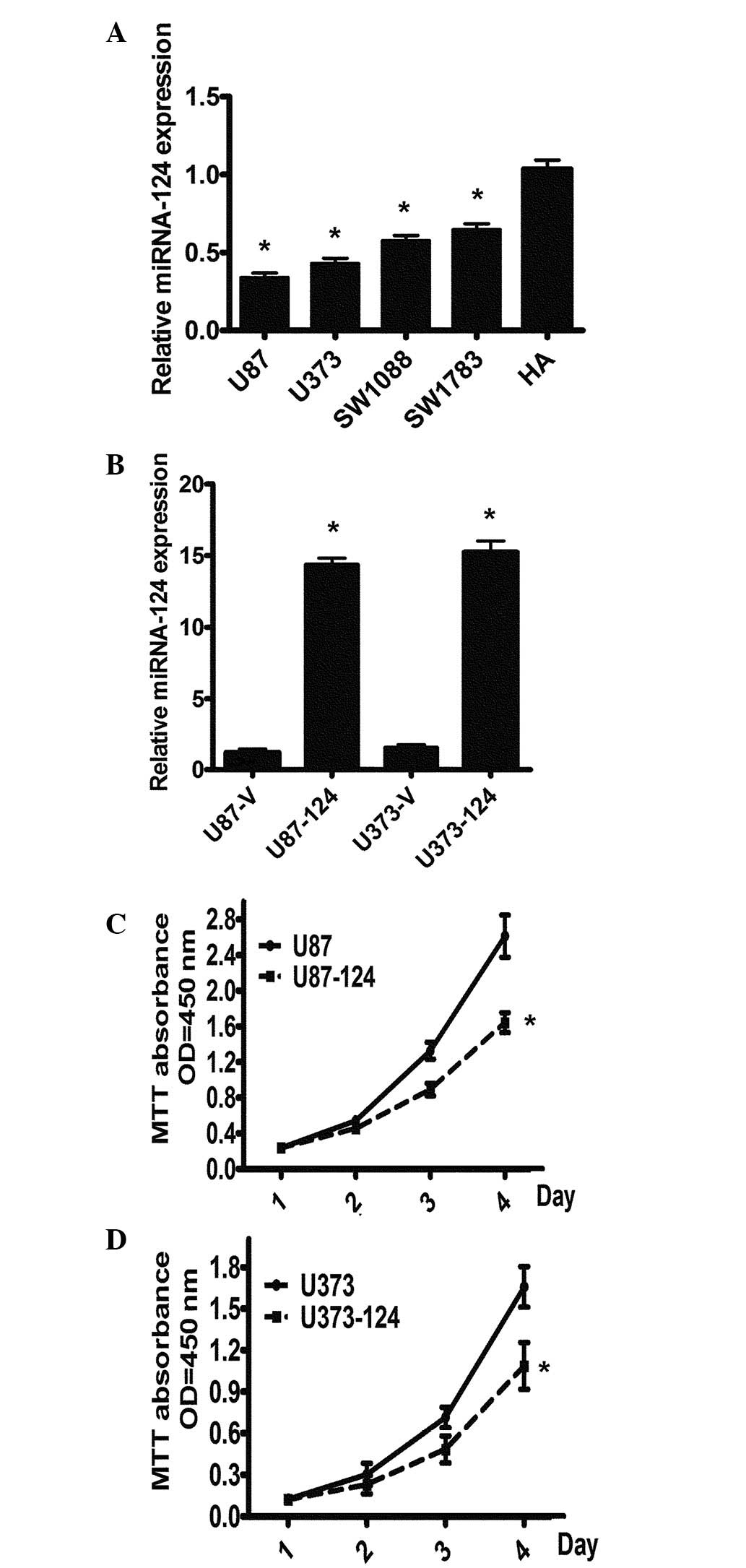
To explore the functional role of miR-124 in glioma
carcinogenesis, we first detected the expression levels of miR-124
using real-time PCR in five GBM cell lines. The results revealed
that all four GBM cell lines (U87, U373, SW1088 and SW1783) had
significantly lower levels of miR-124 expression than those of the
HA cell line (Fig. 1A). To further
explore the theory that miR-124 is important for cell
proliferation, we constructed a miR-124 overexpression model in U87
and U373 cells infected with miR-124 by the lentivirus pCDH-CMV
system, designated as U87-miR-124 or U373-miR-124, respectively,
and cells infected with an empty virus vector were used as a
control. The overexpression of miR-124 in U87-miR-124 and
U373-miR-124 cells was confirmed using qRT-PCR (Fig. 1B). The cell proliferation assays
revealed that the overexpression of miR-124 suppresses the
proliferation of GBM cells (Fig. 1C
and D). The data indicate that a decrease in miR-124 expression
exerts a growth-inhibiting function in human GBM.
miR-124 directly targets SOS1 in human
GBM cell lines and miR-124 negatively regulates endogenous SOS1
expression in GBM
It is generally accepted that miRNAs regulate
expression of their downstream gene targets in order to exert their
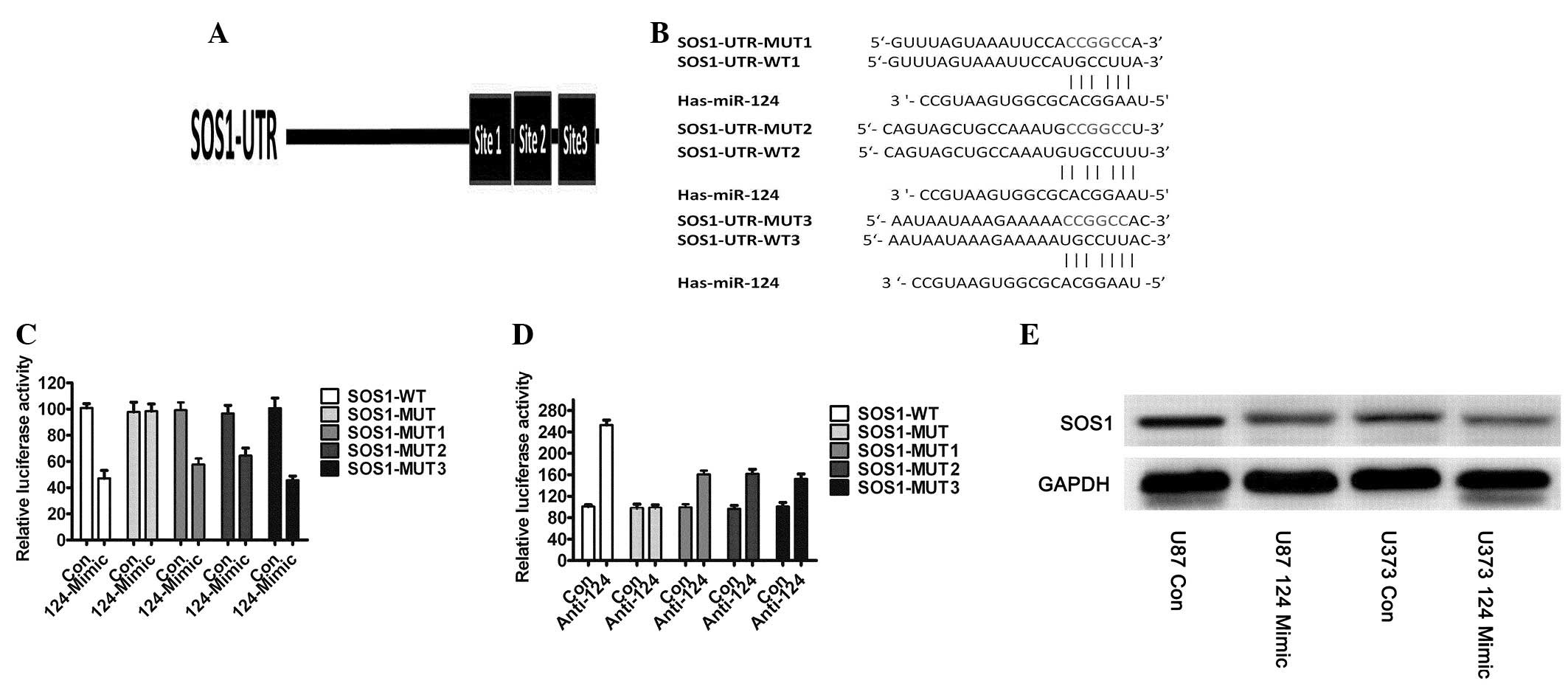
function. To clarify the molecular mechanisms by which miR-124
inhibits glioblastoma cell growth, we predicted its downstream
targets using the algorithms: TargetScan (23) and PicTar (24). Among the candidate target genes
commonly predicted by the algorithms was SOS1. To validate that
SOS1 is targeted by miR-124, we subcloned segments of the 3′UTRs of
SOS1 into a pCS2-Luc reporter vector. There are three binding sites
which miR-124 was predicted to target (Fig. 2A), and we constructed mutant
reporters, respectively (Fig. 2B).
We used 293T cells, which have very low or undetectable levels of
miR-124, for the reporter assays, and tested the effects of miRNA
mimics on the relative Firefly luciferase ratio. miR-124
overexpression reduced the expression of a luciferase reporter
containing wild-type SOS1 3′UTR, i.e., a mutant of all binding
sites did not affect luciferase activity, and every binding site
wound affected luciferase activity (Fig. 2C). We then co-transfected
anti-miR-124 and SOS1-3′UTR-WT or SOS1-3′UTR-MUT into
miR-124-overexpressing U87 (U87-miR-124) cells and observed that
the anti-miR-124 inhibitor rescued the luciferase activities of the
reporter containing the wild-type SOS1 3′UTR (Fig. 2D), but the mutant did not. We
detected the endogenous protein expression of SOS1 and discovered
that the protein expression of SOS1 was significantly decreased in
U87-miR-124 and U373-miR-124 cells which had overexpressed miR-124
(Fig. 2E). Taken together, these
results demonstrated that SOS1 was a direct target of miR-124
action in GBM.
miR-124 negatively regulates endogenous
SOS1 expression
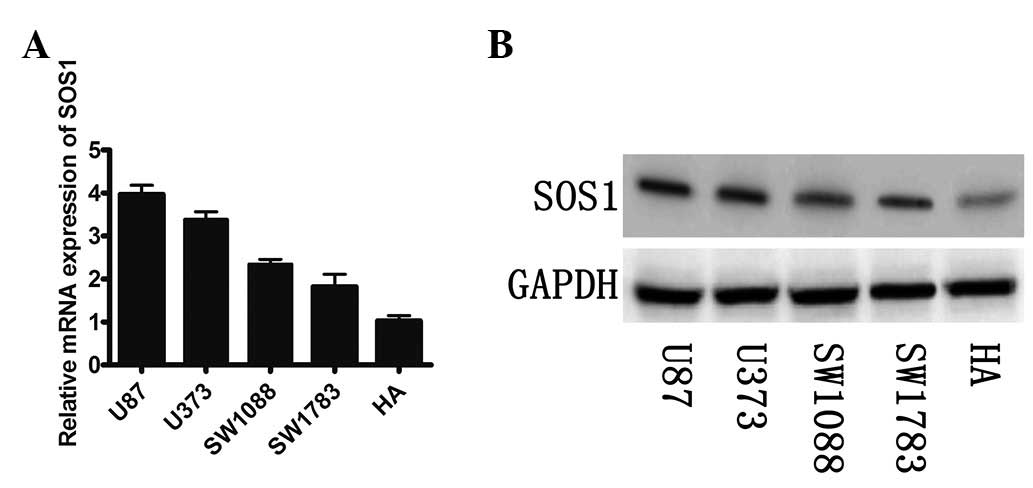
We examined the mRNA and protein expression of SOS1
in the GBM cell lines. A significant inverse correlation between
the mRNA and protein expression of SOS1 and levels of miR-124 was
observed (Fig. 3A and B). It was
demonstrated that low levels of miR-124 were more likely to be
observed in the GBM cell lines with a high expression of SOS1 mRNA
and protein.
miR-124 suppresses SOS1 and negatively
regulates the MAPK pathway in GBM cell lines
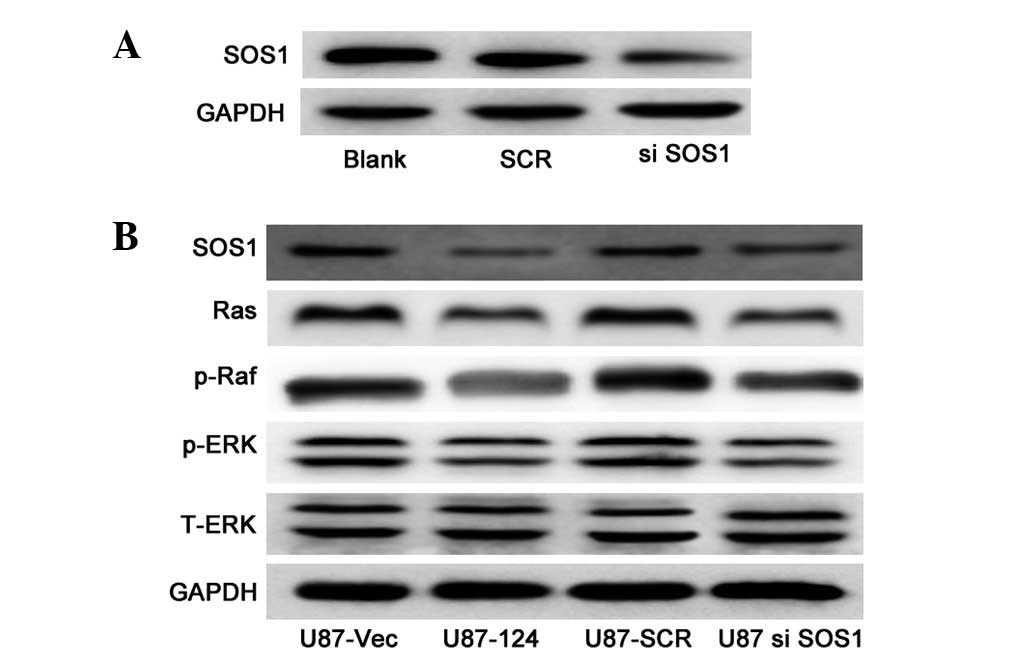
To investigate the molecular mechanism of
miR-124-mediated cell growth, we examined the expression of vital
components of the MAPK pathway, which are critical in the
regulation of cell growth in U87 cells with or without miR-124
overexpression. We performed loss-of-function experiments to
further verify that SOS1 targeting is involved in miR-124-mediated
growth inhibition in U87 cells. The SOS1 protein can be effectively
knocked down (Fig. 4A). Western
blot analysis revealed that the protein levels of Ras, p-Raf and
p-ERK were significantly decreased in the SOS1 knockdown cells
(Fig. 4B), compared to those in
the control cells. As expected, upregulation of miR-124 resulted in
downregulation of the protein levels of Ras, p-Raf and p-ERK
(Fig. 4B). The data indicate that
miR-124 is capable of suppressing the growth of U87 cells by
targeting SOS1 via the MAPK pathway.
Discussion
Over the last twenty years, miRNA has been proven to
play in the regulation of a wide variety of biological processes
and various studies have shown that miRNAs regulate gene expression
(25–28). Studies have has been undertaken in
order to understand miRNA expression, the impact it has on
malignant tumors and patient prognosis, as well as potential
strategies for individualized therapy. In this study, we selected
miR-124 for a detailed investigation into downregulation in
glioblastoma patient samples (29,30).
The detailed mechanism(s) surrounding the role of miR-124 in GBM
development requires further clarification.
We identified the differential expression of miR-124
in GBM cell lines and explored the molecular mechanism by which
miR-124 suppressed glioma cell growth. To identify genes that may
be regulated by miR-124, we used algorithms designed to search for
matching base pairs in miRNAs and mRNA targets. SOS1 was identified
as a direct and functional target of miR-124, a conclusion
supported by the following reasons: three complementary sequences
of miR-124 were identified in the 3′UTR of SOS1 mRNA; miR-124
overexpression suppresses SOS1 3′UTR luciferase report activity and
this effect was eliminated by mutation of the miR-124 seed binding
site; overexpression of miR-124 led to a significant reduction in
SOS1 at the protein level. SOS1 knockdown induced cell growth
inhibition similar to the phenotypes induced by miR-124
upregulation. These findings indicate that miR-124 inhibits glioma
cell growth by repressing SOS1 post-transcriptionally.
SOS1 is known to be overexpressed in various types
of cancer and plays an important role in signaling to the Ras/ERK
cascade (31–33). Grb2/Sos is central to signal
transduction following growth factor engagement of receptor
tyrosine kinases (RTKs). Upon further examination of the molecular
mechanisms of growth inhibition induced by miR-124, we observed the
expression of key components of the MAPK pathway. The results are
consistent with SOS1 downregulation by siRNA, indicating that the
protein levels of Ras, p-Raf and p-ERK are significantly suppressed
in cells with an overexpression of miR-124. The major implication
of these findings is that miR-124 is downregulated in GBM cells and
directly targets SOS1 to inhibit cell growth by the MAPK
pathway.
In conclusion, our study demonstrates that
downregulated miR-124 is responsible for the upregulation of SOS1
in GBM, and miR-124 has an important role in inhibiting cell growth
by regulating the SOS1/Raf/ERK signaling pathway. Our findings
bring new insights on the targeted delivery of miR-124 to GBM cells
as a potential therapeutic treatment for GBM.
References
|
1
|
Karsy M, Arslan E and Moy F: Current
progress on understanding microRNAs in glioblastoma multiforme.
Genes Cancer. 3:3–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Omuro AM, Faivre S and Raymond E: Lessons
learned in the development of targeted therapy for malignant
gliomas. Mol Cancer Ther. 6:1909–1919. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garofalo M and Croce CM: microRNAs: master
regulators as potential therapeutics in cancer. Annu Rev Pharmacol
Toxicol. 51:25–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kwak HJ, Kim YJ, Chun KR, et al:
Downregulation of Spry2 by miR-21 triggers malignancy in human
gliomas. Oncogene. 30:2433–2442. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou X, Ren Y, Moore L, et al:
Downregulation of miR-21 inhibits EGFR pathway and suppresses the
growth of human glioblastoma cells independent of PTEN status. Lab
Invest. 90:144–155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhu XC, Dong QZ, Zhang XF, et al:
microRNA-29a suppresses cell proliferation by targeting SPARC in
hepatocellular carcinoma. Int J Mol Med. 30:1321–1326.
2012.PubMed/NCBI
|
|
8
|
Tang H, Liu X, Wang Z, et al: Interaction
of hsa-miR-381 and glioma suppressor LRRC4 is involved in glioma
growth. Brain Res. 1390:21–32. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ernst A, Campos B, Meier J, et al:
De-repression of CTGF via the miR-17–92 cluster upon
differentiation of human glioblastoma spheroid cultures. Oncogene.
29:3411–3422. 2010.PubMed/NCBI
|
|
10
|
Huse JT, Brennan C, Hambardzumyan D, et
al: The PTEN-regulating microRNA miR-26a is amplified in high-grade
glioma and facilitates gliomagenesis in vivo. Genes Dev.
23:1327–1337. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang H, Kong W, He L, et al: MicroRNA
expression profiling in human ovarian cancer: miR-214 induces cell
survival and cisplatin resistance by targeting PTEN. Cancer Res.
68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kim H, Huang W, Jiang X, Pennicooke B,
Park PJ and Johnson MD: Integrative genome analysis reveals an
oncomir/oncogene cluster regulating glioblastoma survivorship. Proc
Natl Acad Sci USA. 107:2183–2188. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gureasko J, Galush WJ, Boykevisch S, et
al: Membrane-dependent signal integration by the Ras activator Son
of sevenless. Nat Struct Mol Biol. 15:452–461. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nimnual AS, Yatsula BA and Bar-Sagi D:
Coupling of Ras and Rac guanosine triphosphatases through the Ras
exchanger Sos. Science. 279:560–563. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Margarit SM, Sondermann H, Hall BE, et al:
Structural evidence for feedback activation by Ras. GTP of the
Ras-specific nucleotide exchange factor SOS. Cell. 112:685–695.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bar-Sagi D: The Sos (Son of sevenless)
protein. Trends Endocrinol Metab. 5:165–169. 1994. View Article : Google Scholar
|
|
17
|
Vetter IR and Wittinghofer A: The guanine
nucleotide-binding switch in three dimensions. Science.
294:1299–1304. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dhillon AS and Kolch W: Untying the
regulation of the Raf-1 kinase. Arch Biochem Biophys. 404:3–9.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamamoto T, Ebisuya M, Ashida F, Okamoto
K, Yonehara S and Nishida E: Continuous ERK activation
downregulates antiproliferative genes throughout G1 phase to allow
cell-cycle progression. Curr Biol. 16:1171–1182. 2006. View Article : Google Scholar
|
|
20
|
Cheng LC, Pastrana E, Tavazoie M and
Doetsch F: miR-124 regulates adult neurogenesis in the
subventricular zone stem cell niche. Nat Neurosci. 12:399–408.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yoo AS, Sun AX, Li L, et al:
MicroRNA-mediated conversion of human fibroblasts to neurons.
Nature. 476:228–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen C, Ridzon DA, Broomer AJ, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krek A, Grün D, Poy MN, et al:
Combinatorial microRNA target predictions. Nat Genet. 37:495–500.
2005. View
Article : Google Scholar
|
|
25
|
Cordes KR, Sheehy NT, White MP, et al:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009.PubMed/NCBI
|
|
26
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Makeyev EV, Zhang J, Carrasco MA and
Maniatis T: The MicroRNA miR-124 promotes neuronal differentiation
by triggering brain-specific alternative pre-mRNA splicing. Mol
Cell. 27:435–448. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
King IN, Qian L, Liang J, et al: A
genome-wide screen reveals a role for microRNA-1 in modulating
cardiac cell polarity. Dev Cell. 20:497–510. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Silber J, Lim DA, Petritsch C, et al:
miR-124 and miR-137 inhibit proliferation of glioblastoma
multiforme cells and induce differentiation of brain tumor stem
cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xia H, Cheung WK, Ng SS, et al: Loss of
brain-enriched miR-124 microRNA enhances stem-like traits and
invasiveness of glioma cells. J Biol Chem. 287:9962–9971. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Timofeeva OA, Zhang X, Ressom HW, et al:
Enhanced expression of SOS1 is detected in prostate cancer
epithelial cells from African-American men. Int J Oncol.
35:751–760. 2009.PubMed/NCBI
|
|
32
|
Daniels MA, Teixeiro E, Gill J, et al:
Thymic selection threshold defined by compartmentalization of
Ras/MAPK signalling. Nature. 444:724–729. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roose JP, Mollenauer M, Ho M, Kurosaki T
and Weiss A: Unusual interplay of two types of Ras activators,
RasGRP and SOS, establishes sensitive and robust Ras activation in
lymphocytes. Mol Cell Biol. 27:2732–2745. 2007. View Article : Google Scholar : PubMed/NCBI
|