Introduction
Translation initiation is one of the key regulatory
steps for protein synthesis in mammalian cells, and is performed by
eukaryotic translation initiation factors (eIFs). Eukaryotic
translation initiation factor 3 (eIF3) mediates the interaction
between the 43S pre-initiation complex and the eIF4F-bound mRNA
clients (1). eIF3 is comprised of
13 subunits (eIF3a-m; ~700–800 kDa total mass), and each of these
contributes to the function (2).
Similarly, each subunit of eIF3 may exert functions in biological
processes other than translation initiation. For example, the eIF3a
subunit is associated with cell cycle regulation, and the eIF3e
subunit may be involved in mitosis and segregation (2). The key position of eIF3 in the
mechanism of translation initiation and other functions of its
subunits contributes to the important roles of eIF3 and its
subunits in various aspects of cellular activities under
physiological and pathological conditions, including cancer
(2,3). The aberrant expression of certain
subunits of eIF3 in several types of cancer has been associated
with the phenotypes of cancer cells, including malignant growth,
proliferation, invasion, metastasis and clinical parameters,
including disease staging and prognosis (4–9).
eIF3g is a core eIF3 subunit, which is known to be
involved in the process of translation reinitiation (2). Previous studies have reported that
eIF3g is also involved in the cytoskeletal network and
caspase-mediated apoptosis (10,11).
Our previous study demonstrated that eIF3g was overexpressed in an
adriamycin-resistant human erythroleukemia cell line (12). In addition, a positive correlation
was observed between the over-expression of eIF3g and lymph node
metastasis in a pilot study performed to evaluate the clinical
significance of eIF3g over-expression in breast cancer (data not
shown). These results led to the hypotheses that the aberrant
expression of eIF3g may be involved in the development and/or the
progression of cancer by mediating aberrant apoptosis and multidrug
resistance. It is important to test these hypotheses and to examine
the possible mechanisms.
Our previous bioinformatics investigation to
identify functional domains of eIF3g other than those for
translation initiation revealed that eIF3g may have a nuclear
localization sequence. Therefore, the present study aimed to
examine the nuclear localization of eIF3g and to investigate the
possible functions of nuclear eIF3g, as well as its interacting
nuclear proteins, in order elucidate the mechanisms underlying the
effect of eIF3g on the development/progression of breast
cancer.
Materials and methods
Bioinformatics analysis
Several online bioinformatics tools were used to
predict the subcellular distribution of eIF3g, including WoLF PSORT
(http://wolfpsort.org), PROST II Prediction
(http://psort.hgc.jp/form2.html),
Subnuclear Compartments Prediction System (http://array.bioengr.uic.edu/subnuclear.htm) and
PredictProtein (https://www.predictprotein.org). The sequence-based
protein function was predicted using the online tool,
PredictProtein (https://www.predictprotein.org). Protein sequence data
were obtained from the National Center for Biotechnology
Information Protein database (http://www.ncbi.nlm.nih.gov/protein). Gene Ontology
terms are organized in two different ontolo-gies, Molecular
Function Ontology and Biological Process Ontology (http://geneontology.org/).
Overexpression of eIF3g in Bcap37 human
breast cancer cells
The human breast cancer cell line, Bcap37 (The Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences,
Shanghai, China), with doxycycline-inducible overexpression of
eIF3g (Bcap37/Tet-on-eIF3g) was used as previously described
(13). Briefly, the human breast
cancer Bcap37 cells were cultured in RPMI 1640 medium (Boster
Biological Technology) supplemented with 10% fetal bovine serum
(FBS; Gbico; Thermo Fisher Scientific, Inc., Waltham, MA, USA) or
10% Tet-approved FBS (Clontech Laboratories, Mountain View, CA,
USA) in a humidified 37°C incubator. The full length eIF3g cDNA in
the pLVX-Tight-Puro vector (Clontech Laboratories) was used to
produce the lentivirus, following packaging with a Lenti-X™ Tet-On
Avanced Inducible Expression System (Clontech Laboratories),
according to the manufacturer's protocol. HEK293T cells
(8×106 per dish; American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's
medium (DMEM) with high glucose (Boster Biological Technology),
supplemented with 10% FBS, and used for transfection. The
lentivirus-containing supernatant was collected. The Bcap37 cells
(6×105 per dish) were infected with the lentivirus in a
6-well plate. Geneticin (250 µg/ml; Amresco LLC, Solon, OH,
USA) and puromycin (0.25 µg/ml; Sigma-Aldrich, St. Louis,
MO, USA) were used to select and maintain stable transfectants.
Doxycycline (1 µg/ml; Sigma-Aldrich) was used to induce the
overexpression of eIF3g.
Rapid, efficient and practical method for
cell fractionation
Fractionation was performed to separate the
cytosolic fraction and nuclear fraction of the cells (14). Briefly, the trypsinized cells were
washed once for 1 min with ice-cold phosphate-buffered saline (PBS)
and lysed with 1 µl ice-cold 0.1% NP-40 in PBS. A small
volume (300 µl) of the lysate was saved as 'whole cell
lysate'. The remaining lysate was centrifuged at 15,000 × g (4°C)
for 5 min, and the supernatant was collected as the 'cytosolic
fraction'. The pelleted 'nuclear fraction' was further lysed in
radioimmunoprecipitation assay (RIPA) buffer [50 mM Tris-HCl (pH
7.4), 1% NP-40, 0.5% sodium deoxycholate, 150 mM NaCl and 0.1%
SDS], containing protease inhibitor cocktail tablets (Roche
Diagnostics, Basel, Switzerland). For crosslinking experiments, the
modified RIPA buffer (1X PBS, 1% NP-40, 0.5% sodium deoxycholate,
150 mM NaCl and 0.1% SDS) was used. The lysate was centrifuged at
15,000 × g (4°C) for 15 min and the supernatant was collected as
the 'nuclear fraction'.
Western blot analysis
The cells were lysed in RIPA buffer and the lysate
was centrifuged at 15,000 × g for 15 min at 4°C. The protein
supernatant was quantified using a DC Protein Assay kit (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The cell lysates and
protein samples, prepared in reducing sample buffer [50 mM Tris-HCl
(pH 6.8), 2% SDS, 0.01% Bromophenol Blue, 10% glycerol and 50 mM
DTT] were subjected to SDS-PAGE and transferred onto polyvinylidene
fluoride (0.2 µm PVDF) membranes. Following blocking with
10% nonfat milk for 1 h, the membranes were incubated with the
following antibodies: Polyclonal rabbit anti-human eIF3g (1:10,000;
Bethyl Laboratories, Inc., Montgomery, TX, USA; cat. no.
A301-757A), polyclonal rabbit anti-human hnRNP U (1:1,000; Abcam
Cambridge, UK; cat. no. ab20666), polyclonal goat anti-human
HSZFP36 (1:1,000; Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA; cat. no. sc-247189), polyclonal goat anti-human β-actin
(1:1,000; Santa Cruz Biotechnology, Inc.; cat. no. sc-1615),
polyclonal rabbit anti-human Histone H4 (cat. no. 07-108; 1:1,000;
EMD Millipore, Billerica, MA, USA) overnight at 4°C. This was
followed by incubation with secondary peroxidase-conjugated goat
anti-rabbit IgG (1:5,000; ZB-2301) and rabbit anti-goat IgG
(1:5,000; ZB-2306) (Zhongshan Golden Bridge Biotechnology, Co, Ltd,
Beijing, China) or horseradish peroxidase-conjugated polyclonal
rabbit anti-human GAPDH (cat. no. sc-25778; 1:1,000; Santa Cruz
Biotechnology, Inc.) for 2 h at room temperature. Enhanced
chemiluminescence substrates (EMD Millipore) were then applied and
signals were detected using a chemiluminescence imaging system
(ChemiDoc™ MP Imaging System; Bio-Rad Laboratories, Inc.).
Co-immunoprecipitation (co-IP)
The nuclear protein (200 µg) was incubated
with the primary antibodies mentioned above (5 µg
anti-eIF3g, 5 µg anti-hnRNPU, 5 µg anti-HSZFP36 or 5
µg anti-β-actin) and 50 µl protein A/G PLUS-Agarose
(Santa Cruz Biotechnology, Inc.) on a rotating device (Tube Tumbler
Rotating Mixer SBS550-2; Select BioProducts, Edison, NJ, USA) at
4°C overnight. Normal rabbit serum (2 µl) was used as a
control group. Following centrifugation at 5,000 × g for 10 min at
4°C, the co-immunoprecipitated proteins were washed with PBS four
times (5 min each time), treated with reducing sample buffer and
subjected to 10% SDS-PAGE. Following electrophoresis of the
proteins, the gel was stained using 50 ml Bio-safe Coomassie blue
stain (Bio-Rad Laboratories, Inc.) for 1 h at room temperature with
gentle agitation, followed by destaining in distilled water for
>2 h until the protein bands were clearly visible. The protein
bands were excised and placed into microtubes, and were sent for
mass spectrometry characterization of proteins by service provider
(Institute of Biomedical Sciences, Fudan University, Shanghai,
China).
In vitro crosslinking
The separated nuclear fraction was lysed with
modified RIPA buffer. DSS (Thermo Fisher Scientific, Inc.; spacer
arm, 11.4 Å) was added to the nuclear fraction to a final
concentration of 5 mM and incubated at room temperature for 30 min.
To quench the reaction, quenching buffer (1 M Tris-Cl; pH 7.5) was
added to a final concentration of 20–50 mM Tris (Bio-Rad
Laboratories, Inc.) at room temperature for 15 min. The mixtures
were then prepared in SDS-PAGE loading buffer for western blot
analysis.
Glutathione S-transferase (GST) pull-down
assay
The full length coding sequence of eIF3g was
inserted into the pGEX-5X-2 vector (Pharmacia Biotech, Tokyo,
Japan), in-frame with the GST open reading frame, which led to the
production of soluble GST-eIF3g fusion protein when transformed
into Escherichia coli DH5α (Takara Bio Inc., Otsu, Shiga,
Japan) and induced by 0.1 mM isopropyl-β-D-1 -thiogalactopyranoside
(Amresco LLC) for 4 h at 37°C. For the GST pull-down assays, the
GST or GST-eIF3g proteins expressed in E. coli were purified
using magnetic glutathione-beads (Promega Corp., Madison, WI, USA),
according to the manufacturer's protocol. Briefly, bacteria were
collected following culture by centrifugation at 5,000 × g for 15
min at 4°C, resuspension in 3 ml ice-cold PBS and lysis by
sonication on ice. The lysate was centrifuged at 5,000 × g, for 10
min at 4°C to remove the cell debris, and 100 µl magnetic
glutathione beads were added to the supernatant for binding of the
GST/GST-eIF3g protein. The beads were washed three times (5 min
each time) with PBS and then incubated with the nuclear lysates
from the Bcap37 cells with rotation for 30 min at room temperature.
The beads were then washed at least three times with PBS (5 min
each time); bound proteins were resolved by 8% SDS-PAGE and
detected by western blotting.
Immunofluorescent staining
The Bcap37/Tet-on-eIF3g and Bcap37 cells were seeded
separately into a 6-well plate with a glass-bottomed dish at a
density of 2×104 cells/dish and treated with doxycycline
for 3 days at 37°C. The cells were then washed with PBS three times
(10 min each time), fixed with 4% paraformaldehyde (Shanghai Ling
Feng Chemicals Co., Ltd., Shanghai, China) for 10 min, and
permeabilized with 0.1% Triton X-100 (Bio Basic Inc., Markham, ON,
Canada) for 10 min. Following washing with PBS three times, the
cells were blocked with 5% goat serum (Zhongshan Golden Bridge
Biotechnology Co., Ltd.) and incubated with anti-eIF3g (1:400),
anti-hnRNP U (1:200), anti-HSZFP36 (1:200) or anti-β-actin (1:200)
primary antibodies overnight at 4°C. The cells were then washed
with PBS three times (10 min each time) and further incubated with
following secondary antibodies: Fluorescein
isothiocyanate-conjugated donkey anti-rabbit IgG (1:500;
711-095-152) and/or tetramethylrhodamine-conjugated donkey
anti-goat IgG (1:500; 705-025-003) (Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA). The nuclei were stained
with 200 µg/ml DAPI (Sigma-Aldrich). The stained cells were
observed under a confocal microscope (LSM710; Carl Zeiss, Jena,
Germany).
Results
Nuclear localization of eIF3g
The present study hypothesized that eIF3g may link
other biological processes to the initiation of translation. To
confirm this, in silico analysis and subcellular
fractionation was performed to examine the functional domains and
subcellular distribution of eIF3g. As shown in Tables I and II, bioinformatics analyses using
state-of-art online tools predicted that eIF3g has a nuclear
distribution for potential functions.
 | Table IPredicted subcellular distribution of
eIF3g using online bioinformatics tools. |
Table I
Predicted subcellular distribution of
eIF3g using online bioinformatics tools.
| Bioinformatics
tool | Distribution |
|---|
| WoLF PSORT | Cytoplasm and
nucleus |
| PROST II
Prediction | Nucleus |
| Subnuclear
Compartments Prediction System | Nucleus |
| PredictProtein | Nucleus |
 | Table IIPredicted function of eukaryotic
translation initiation factor 3 subunit g in the nucleus. |
Table II
Predicted function of eukaryotic
translation initiation factor 3 subunit g in the nucleus.
| GO ID | Molecular
function/biological process | Reliability
(%) |
|---|
| GO:0003677 | DNA binding | 2 |
| GO:0003697 | Single-stranded DNA
binding | 2 |
| GO:0003690 | Double-stranded DNA
binding | 1 |
| GO:0006396 | RNA processing | 14 |
| GO:0051028 | mRNA transport | 9 |
| GO:0045449 | Regulation of
transcription | 8 |
| GO:0060211 | Regulation of
nuclear-transcribed mRNA poly (A) tail shortening | 8 |
| GO:0048255 | mRNA
stabilization | 8 |
| GO:0031047 | Gene silencing by
RNA | 8 |
| GO:0000184 | Nuclear-transcribed
mRNA catabolic process, nonsense-mediated decay | 8 |
| GO:0060212 | Negative regulation
of nuclear-transcribed mRNA poly (A) tail shortening | 8 |
| GO:0000398 | Nuclear mRNA
splicing, via spliceosome | 8 |
| GO:0006350 | Transcription | 8 |
To verify this prediction, the present study
examined the expression of eIF3g in Bcap37 breast cancer cells by
western blotting in a preliminary experiment, and the results
showed a weak, but distinct band of eIF3g in the nuclear protein
fraction (data not shown). The Bcap37/Tet-on-eIF3g cells with
inducible overexpression of eIF3g were then used in subsequent
investigations. The cells were harvested through subcellular
fractionation of the cytosolic and the nuclear proteins of breast
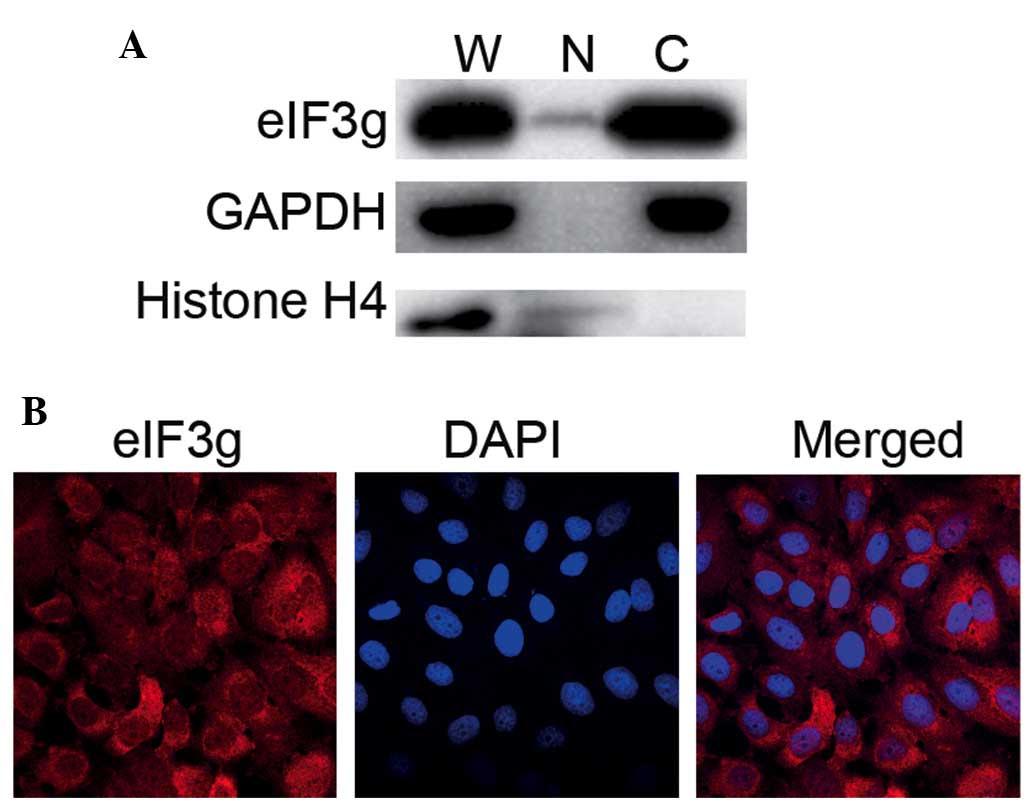
cancer cells expressing eIF3g (Bcap37/Tet-on-eIF3g). As shown in
Fig. 1A, the use of this method
successfully separated the cytosolic and the nuclear fractions,
confirmed by detection of nuclear protein, Histone H4, in the whole
cell lysate and nuclear fraction, but not in the cytoplasmic
fraction. By contrast, the cytoplasmic protein, GAPDH, was detected
in the whole cell lysate and cyto-solic fraction, but not in the
nuclear fraction. Although eIF3g was abundant in the cytosolic
fraction, it was also detected in the nuclear fraction.
Immunofluorescent staining confirmed the nuclear distribution of
eIF3g (Fig. 1B). This was a novel
finding, considering eIF3g was originally identified as a
cytoplasmic protein and as a component of the translation
initiation machinery (2).
Identification of nuclear proteins
potentially interacting with eIF3g
Functional nuclear proteins exert their functions
via binding directly to DNA or RNA to regulate gene expression or
RNA processing, or by forming a protein complex to cooperate with
other nuclear proteins (15). The
present study focused primarily on identifying the possible nuclear
proteins, which may interact with eIF3g. The nuclear fraction of
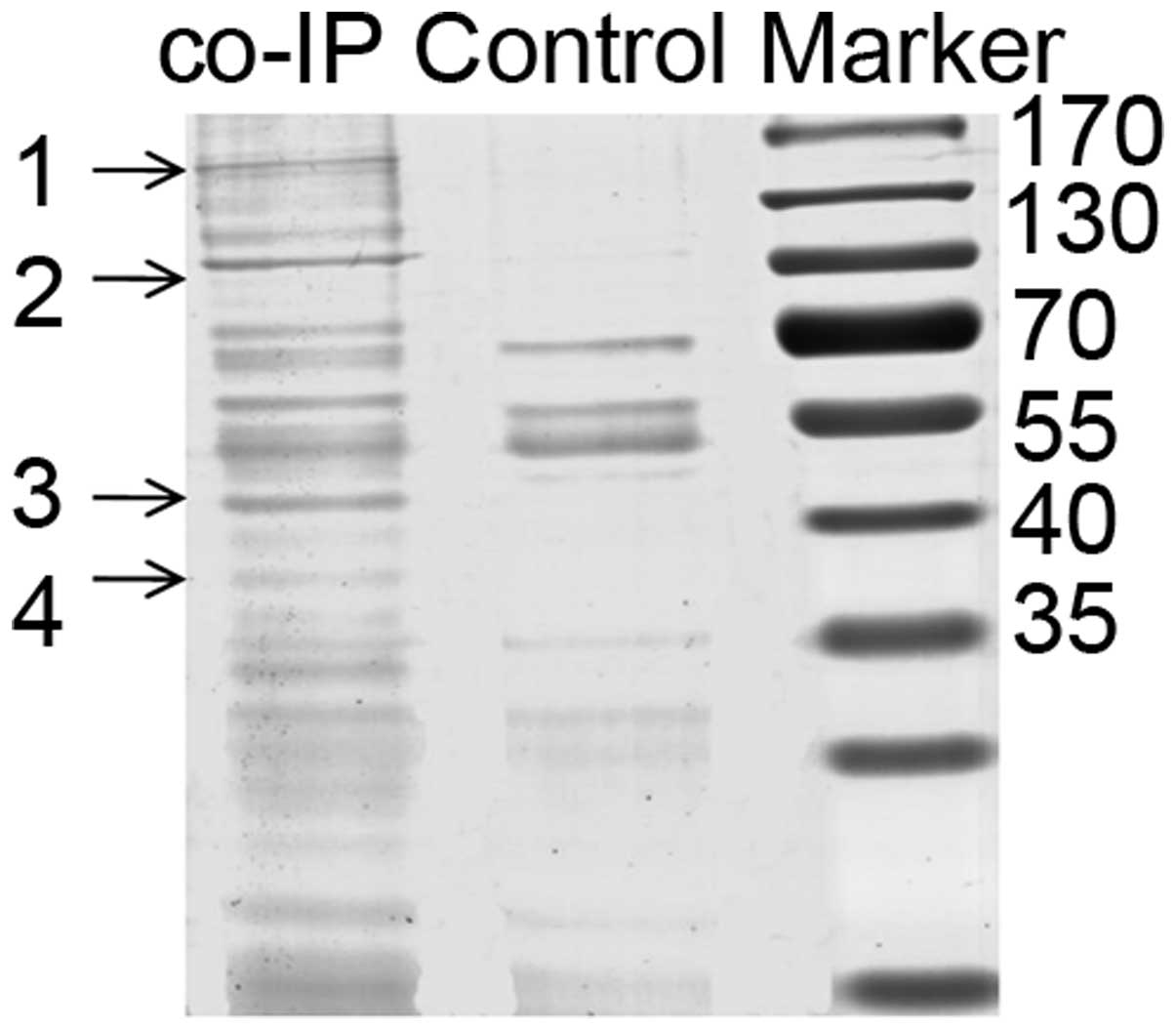
the eIF3g-expressing cells was isolated, and co-IP was formed with
anti-eIF3g antibody to pull down proteins that physically
interacted with eIF3g. The immunoprecipitated proteins were
separated by SDS-PAGE and visualized by Coomassie blue staining
(Fig. 2), and four bands were
selected for subsequent mass spectrometry for protein
characterization, excluding non-specific bands. As shown in
Table III, the proteins were
heterogeneous nuclear ribonucleoprotein U/scaffold attachment
factor A (hnRNP U/SAF-A), HSZFP36/zinc finger protein 823 (ZFP823),
β-actin and eIF3a, respectively.
 | Table IIIMass spectrometry data of the
proteins pulled down by eukaryotic translation initiation factor 3
subunit g. |
Table III
Mass spectrometry data of the
proteins pulled down by eukaryotic translation initiation factor 3
subunit g.
| Spot | Accession
number | Protein | Predicted mass
(Da) | Score | Function in
nucleus |
|---|
| 1 | gi|6685537 | Eukaryotic
translation initiation factor 3 subunit A (Homo
sapiens) | 166,468 | 547 | Not reported |
| 2 | gi|3202000 | Scaffold attachment
factor A (Homo sapiens) | 90,423 | 320 | DNA repair,
telomere length regulation, chromatin remodeling, transcription
regulation |
| 3 | gi|148887464 | Zinc finger protein
823 (Homo sapiens) | 70,225 | 53 | Transcriptional
regulation |
| 4 | gi|14250401 | β-actin (Homo
sapiens) | 40,978 | 193 | Chromatin
remodeling, transcription regulation nucleocytoplasmic
trafficking |
Confirmation of the interaction between
eIF3g and hnRNP U, HSZFP36 and β-actin
The present study confirmed the physical interaction
between eIF3g and its potential partners, hnRNP U, HSZFP36 and
β-actin, by performing co-IP and in vitro crosslinking in
combination with western blotting. As shown in Fig. 3A, eIF3g was co-immunoprecipitated
from the nuclear fractions using antibodies against hnRNP U,
HSZFP36 and β-actin, respectively. These proteins were also
co-immunoprecipitated using antibody against eIF3g (Fig. 3B–D).
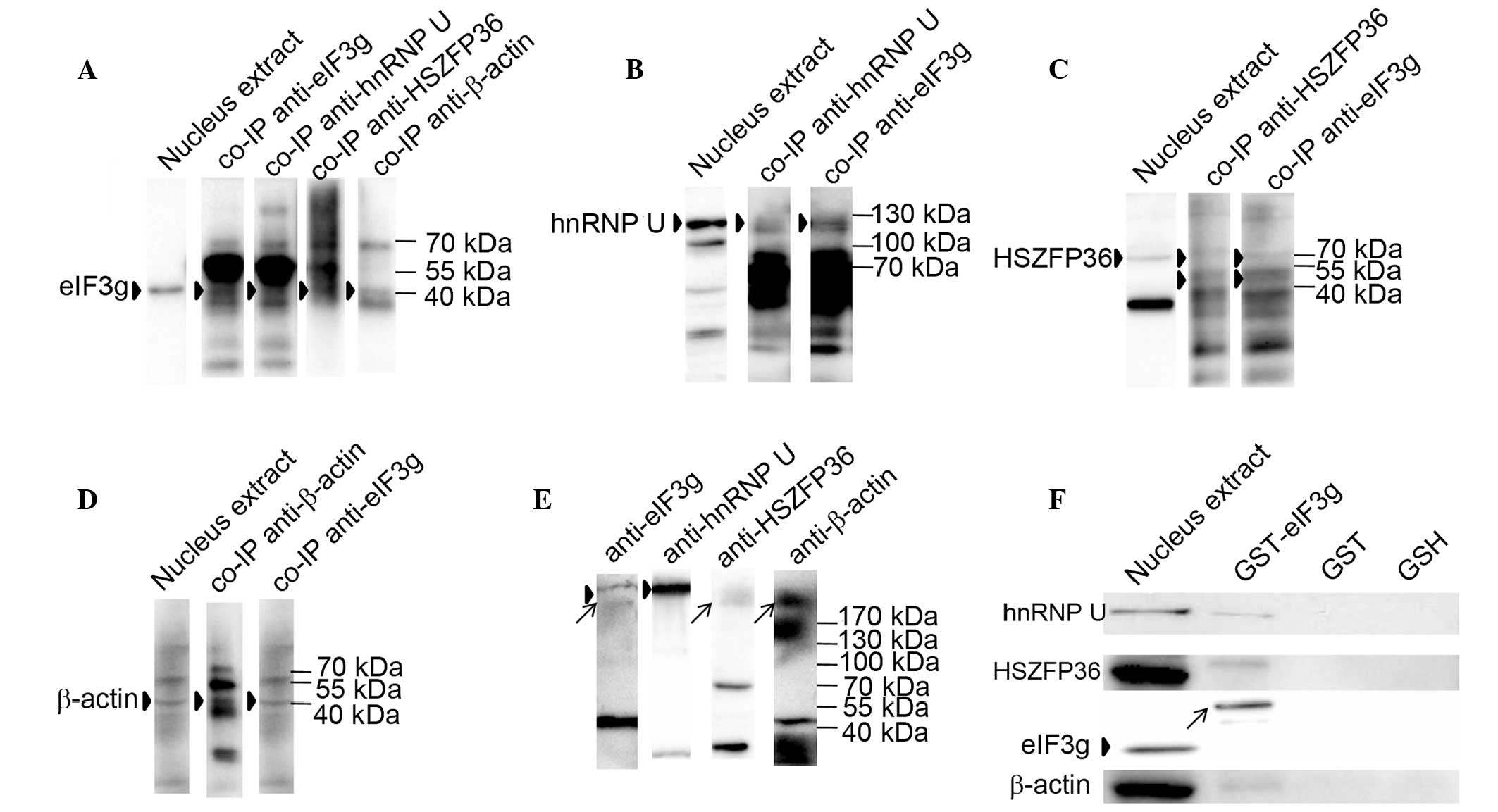 | Figure 3eIF3g interacts with hnRNP U, HSZFP36
and β-actin. (A) Nuclear extracts of Bcap37/Tet-on-eIF3g cells were
subjected to immunoprecipitation with anti-eIF3g (5 µg),
anti-hnRNP U (5 µg), anti-HSZFP36 (5 µg) and
anti-β-actin (5 µg) antibodies, respectively. Precipitated
proteins were resolved by 10% SDS-PAGE and analyzed by western
blotting with anti-eIF3g antibody. Arrowheads indicate eIF3g. (B)
Anti-eIF3g antibody-immunoprecipitated proteins were analyzed by
western blotting with anti-hnRNP U antibody. Arrowheads indicate
hnRNP U. (C) Anti-eIF3g (5 µg) and anti-hnRNP U (5
µg) antibody-immunoprecipitated proteins were analyzed by
western blotting with anti-HSZFP36 antibody. Arrowheads indicate
HSZFP36. (D) Anti-eIF3g (5 µg) and anti-β-actin (5
µg) antibody-immunoprecipitated proteins were analyzed by
western blotting with anti-β-actin antibody. Arrowheads indicate
β-actin. (E) Results of western blotting from the in vitro
crosslinking. The nuclear extract from Bcap37/Tet-on-eIF3g cells
was crosslinked by DSS, subject ed to 8% SDS-PAGE and
electrotransferred onto a polyvinylidene fluoride membrane,
followed by incubation with antibodies specific for eIF3g, hnRNP U,
HSZFP36 and β-actin. Arrowheads indicate larger complex bands;
arrows indicate smaller complex bands. (F) GST-pulldown results.
GST or GST-eIF3g proteins were incubated with nuclear lysates of
Bcap37 cells and glutathione beads; bound proteins were resolved by
10% SDS-PAGE and detected by western blotting. hnRNP U, HSZFP36 and
β-actin were pulled down by GST-eIF3g, with GST protein and GSH
beads alone used as negative controls. Arrowheads indicate eIF3g in
the Bcap37 cells; arrows indicate the GST-eIF3g protein band.
eIF3g, eukaryotic translation initiation factor 3 subunit g; hnRNP
U, heterogeneous nuclear ribonucleoprotein U; GST, glutathione
S-transferase; GSH, glutathione. |
The results of the western blotting for the
crosslinked proteins are shown in Fig.
3E, in which two specific bands were detected by the anti-eIF3g
antibody; these bands were >170 kDa in molecular weight. The
larger band was also detected by the anti-hnRNP U antibody, whereas
the smaller band was detected by the anti-HSZFP36 or anti-β-actin
antibodies. The results of the GST-pulldown assay showed that hnRNP
U, HSZFP36 and β-actin were pulled down by the GST-eIF3g proteins
(Fig. 3F). These results supported
the interaction between eIF3g and hnRNP U, HSZFP36 or β-actin, and
suggested that eIF3g, HSZFP36 and β-actin may form a protein
complex.
Co-localization of eIF3g and its
interacting proteins, HSZFP36 and β-actin, by immunofluorescent
staining
The subcellular distributions of eIF3g, hnRNP U,
HSZFP36 and β-actin were examined by immunofluorescent staining and
confocal microscopy. The cells incubated with anti-eIF3g antibody
showed strong fluorescent signals in the cytoplasm and weak
fluorescent signals in the nucleus. Similar patterns of
fluorescence were observed in the cells incubated with anti-HSZFP36
and anti-β-actin antibodies. In parallel, fluorescence was
predominantly observed in the nuclei of cells incubated with
anti-hnRNP U antibody (Fig. 4A).
The subcellular distributions of these four proteins were also
confirmed by western blotting with the fractionated cytosolic and
nuclear protein samples (Fig.
4B).
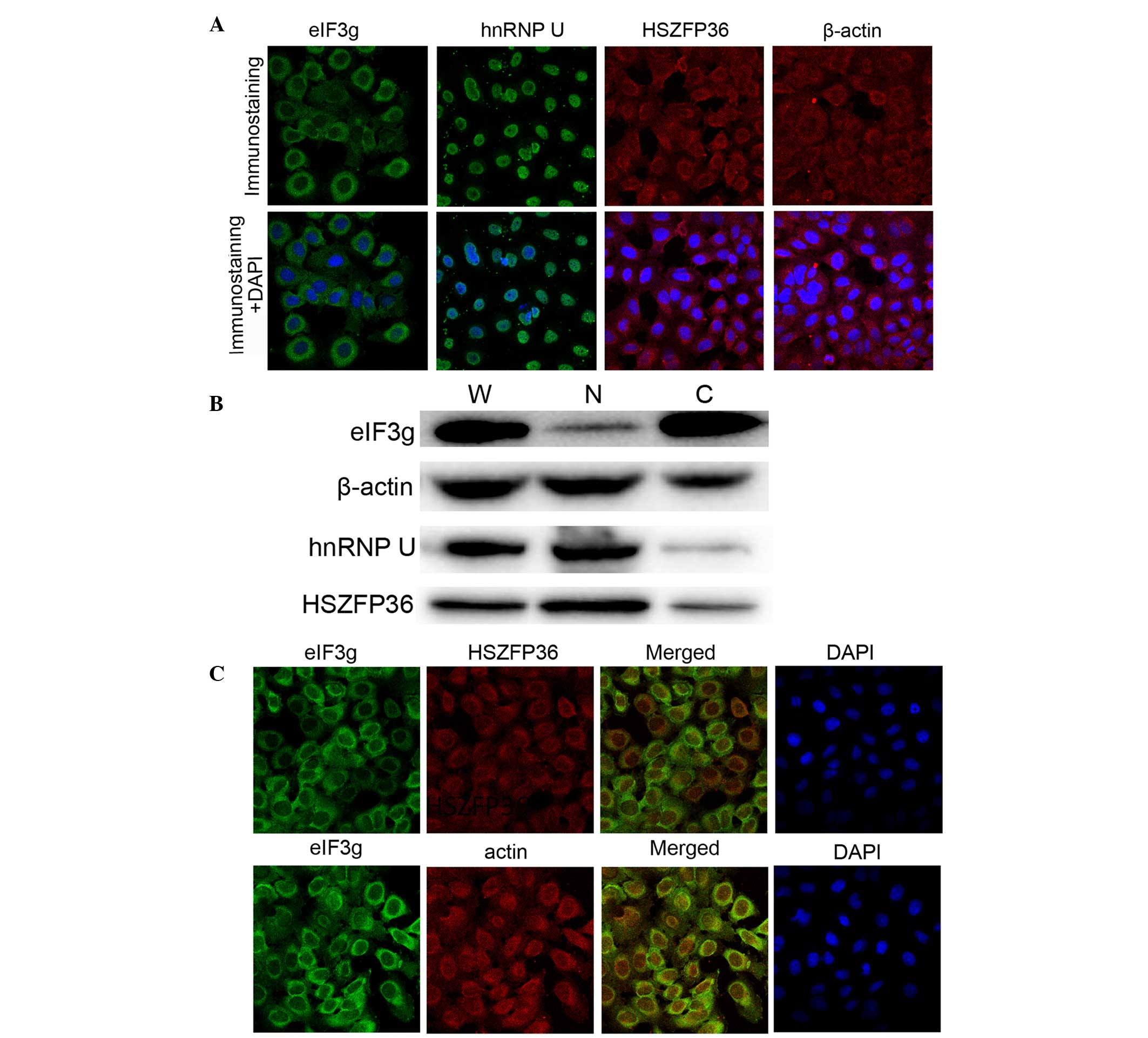 | Figure 4Co-localization of eIF3g, hnRNP U,
HSZFP36 and β-actin. (A) Bcap37/Tet-on-eIF3g cells were incubated
with anti-eIF3g, anti-hnRNP U, anti-HSZFP36 and anti-β-actin
antibodies, respectively (magnification, ×100). (B) Results of
western blot analysis. Extracts from Bcap37/Tet-on-eIF3g cells were
probed with anti-eIF3g, anti-hnRNP U, anti-HSZFP36 and anti-β-actin
antibodies, respectively. (C) Co-localization of eIF3g, HSZFP36 and
β-actin in the nuclei (magnification, ×100). eIF3g, eukaryotic
translation initiation factor 3 subunit g; hnRNP U, heterogeneous
nuclear ribonucleoprotein U; W, whole cell lysate; C, cytoplasmic
fraction; N, nuclear fraction. |
As shown in Fig.
4C, confocal microscopy revealed the co-localization of eIF3g
with HSZFP36 and β-actin in the nucleus, although the intensities
of the signals varied due to the different abundance of the
proteins in the nucleus.
Discussion
The results of the present study demonstrated for
the first time, to the best of our knowledge, the nuclear location
of eIF3g, and its interactions with nuclear and cytoskeleton
proteins. EIF3g is known to contain an RNA recognition motif (RRM)
in its C-terminal region (320 amino acids). This mediates the
interaction between the 43S pre-initiation complex and the
eIF4F-bound mRNA, leading to the formation of the translation
initiation factor complex (2). It
has been demonstrated that eIF3g is involved in the complex
formation by interacting with eIF3b, eIF3i, eIF4B and RNA. There is
also evidence showing that eIF3g (p44) interacts with eIF3a (p170)
and binds to rRNA via the RRM domain (16). In addition, eIF3g has been
suggested to be involved in the translational reinitiation process
of cauliflower mosaic virus (CaMV) polycistronic mRNA and the yeast
GCN4 mRNA (17,18). In CaMV-infected plant cells,
transactivator/viroplasmin (TAV) binds to the plant eIF3 via the
eIF3g subunit to form TAV-eIF3-40S and TAV-eIF3-80S complexes,
allowing the translation of polycistronic mRNAs by reinitiation
(17). The centre of eIF3g, a
fragment spanning amino acid residues 66–173, interacts with TAV or
eIF4B, thus eIF4B can preclude the formation of the TAV/eIF3
complex via competition with TAV for eIF3g binding (19,20).
In addition to its role in translation
initiation/reinitiation, eIF3g may be involved in the cytoskeletal
network and apoptosis. The cytoskeletal protein, erythroid protein
4.1 (4.1R) is reported to bind to eIF3g (p44) (21), and the evolutionary conserved
protein, pelota, is reported to interact with eIF3g in the
cytoskeleton (10). The
involvement of eIF3g (p42) in apoptosis was observed in a proteomic
study and confirmed using a knockdown assay, demonstrating that
RNAi-induced eIF3g suppression inhibits the apoptotic process
(22). The interaction between the
N-terminus of eIF3g and apoptosis-inducing factor, a
caspase-independent apoptotic factor, leads to the inhibition of
protein synthesis during apoptosis (23). Previously, eIF3g was found to be
cleaved by caspases during apoptosis, and the cleaved N-terminus
was translocated to the nucleus, showing a high level of DNase
activity (11).
In our preliminary study, it was found that the
increased expression of eIF3g in tumor cells correlates with lymph
node metastasis and drug resistance in patients with breast cancer,
and that the enforced expression of eIF3g in breast cancer cells
significantly enhances proliferation, migration and invasion in
vitro. In the present study, it was demonstrated that eIF3g can
translocate into the nuclei of breast cancer cells, and that hnRNP
U, HSZFP36 and β-actin can interact with eIF3g in the nucleus. As
nuclear factors may exert a marked effect on the modulation of gene
expression, which may lead to significant phenotypic changes
including cell growth, even in small quantities, trace quantities
of such nuclear eIF3g may be important to the behavior of the
cells. These data provided clues to further investigate the
molecular mechanisms underlying the tumor-promoting properties of
overexpressed eIF3g in cancer cells.
Studies on the nuclear matrix-associated hnRNP U
protein have shown that it is important in DNA repair, telomere
length regulation, chromatin remodeling and gene expression
regulation at the transcriptional and post-transcriptional levels
(24–26). hnRNP U is involved in the
transcriptional activation of certain genes when cooperating with
DNA topoisomerase II (27). It is
also a potent regulator of nuclear ribonucleoprotein particles in
diverse gene expression pathways (28), and enhances the expression of
specific genes by stabilizing mRNA (29). Notably, compared with normal
colonic epithelium, hnRNP U shows increased nuclear expression in
colorectal cancer tissue (30).
HSZFP36 is a member of the zinc finger protein family and is
located predominantly in the nucleus, acting as a transcriptional
regulator (31). Its roles in
physiological/pathological conditions has not been investigated,
however, studies on the functions of nuclear actin have shown that
it is implicated in diverse nuclear activities, including chromatin
remodeling, transcription regulation and nucleocytoplasmic
trafficking (32). In HeLa cells,
actin and hnRNP U cooperate with histone acetyltransferase PCAF in
promoting transcription by RNA polymerase II (33,34).
Whether the interactions between eIF3g and these proteins have
direct or indirect effects on the expression of genes responsible
for the development and/or progression of breast cancer remains to
be elucidated, however, nuclear eIF3g may act as an important
molecule in assisting or inhibiting the functions of these
interacting proteins. In the present study, further functional
analysis of the proteins using PredictProtein revealed the
possibility that eIF3g regulates gene expression at the
transcriptional and post-transcriptional levels (Table II). Further investigations are
required to identify the binding sites of eIF3g to hnRNP U, HSZFP36
and β-actin, to determine whether, and in what manner, eIF3g binds
to DNA and to elucidate how nuclear eIF3g promotes tumor
growth.
In conclusion, the data in the present study
revealed the novel finding that eIF3g interacted with hnRNP U,
HSZFP36 and β-actin in the nucleus. The present study identified
these proteins as eIF3g-interacting proteins, however, whether the
interaction between eIF3g and these proteins is direct or indirect
remains to be elucidated. A detailed analysis of the eIF3g
interaction with hnRNP U, HSZFP36 and β-actin may provide novel
insight into the functions of eIF3g. The present study provided a
basis for further investigation on the association between the
altered expression of eIF3g in breast cancer cells and the
malignant phenotypes of breast cancer cells, including invasion and
metastasis, and may also provide more potential novel markers
and/or therapeutic targets for breast cancer.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (grant no. 81172516) and the Science
and Technology Project (grant no. 2013B02) of Hangzhou Municipal
Health Bureau, China. The authors would like to thank Dr Hao Zhu of
the Department of Clinical Laboratory Sciences, University of
Kansas Medical Center (Kansas City, KS, USA) for his assistance,
comments and suggestions in manuscript preparation.
Abbreviations:
|
CaMV
|
cauliflower mosaic virus
|
|
co-IP
|
co-immunoprecipitation
|
|
eIF
|
eukaryotic translation initiation
factor
|
|
FBS
|
fetal bovine serum
|
|
GAPDH
|
glyceraldehyde-3-phosphate
dehydrogenase
|
|
hnRNP U/SAF-A
|
heterogeneous nuclear
ribonucleoprotein U/scaffold attachment factor A
|
|
PBS
|
phosphate buffer saline
|
|
PVDF
|
polyvinylidene fluoride
|
|
RRM
|
RNA recognition motif
|
|
TAV
|
transactivator/viroplasmin
|
|
ZFP823
|
zinc finger protein 823
|
|
4.1R
|
erythroid protein 4.1
|
References
|
1
|
Silvera D, Formenti SC and Schneider RJ:
Translational control in cancer. Nat Rev Cancer. 10:254–266. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dong ZZ and Zhang JT: Initiation factor
eIF3 and regulation of mRNA translation, cell growth, and cancer.
Crit Rev Oncol Hematol. 59:169–180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hershey JW: Regulation of protein
synthesis and the role of eIF3 in cancer. Braz J Med Biol Res.
43:920–930. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Buttitta F, Martella C, Barassi F,
Felicioni L, Salvatore S, Rosini S, D'Antuono T, Chella A, Mucilli
F, Sacco R, et al: Int6 expression can predict survival in
early-stage non-small cell lung cancer patients. Clin Cancer Res.
11:3198–3204. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cappuzzo F, Varella-Garcia M, Rossi E,
Gajapathy S, Valente M, Drabkin H and Gemmill R: MYC and EIF3H
coamplification significantly improve response and survival of
non-small cell lung cancer patients (NSCLC) treated with gefitinib.
J Thorac Oncol. 4:472–478. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Savinainen KJ, Helenius MA, Lehtonen HJ
and Visakorpi T: Overexpression of EIF3S3 promotes cancer cell
growth. Prostate. 66:1144–1150. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lei YX, Wei L, Wang M, Wu GR and Li M:
Malignant transformation and abnormal expression of eukaryotic
initiation factor in bronchial epithelial cells induced by cadmium
chloride. Biomed Environ Sci. 21:332–338. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang L, Pan X and Hershey JW: Individual
overexpression of five subunits of human translation initiation
factor eIF3 promotes malignant transformation of immortal
fibroblast cells. J Biol Chem. 282:5790–5800. 2007. View Article : Google Scholar
|
|
9
|
Umar A, Kang H, Timmermans AM, Look MP,
Meijer-van Gelder ME, den Bakker MA, Jaitly N, Martens JW, Luider
TM, Foekens JA and Pasa-Tolić L: Identification of a putative
protein profile associated with tamoxifen therapy resistance in
breast cancer. Mol Cell Proteomics. 8:1278–1294. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Burnicka-Turek O, Kata A, Buyandelger B,
Ebermann L, Kramann N, Burfeind P, Hoyer-Fender S, Engel W and
Adham IM: Pelota interacts with HAX1, EIF3G and SRPX and the
resulting protein complexes are associated with the actin
cytoskeleton. BMC cell Biol. 11:282010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim JT, Lee SJ, Kim BY, Lee CH, Yeom YI,
Choe YK, Yoon DY, Chae SK, Kim JW, Yang Y, et al: Caspase-mediated
cleavage and DNase activity of the translation initiation factor 3,
subunit G (eIF3g). FEBS Lett. 587:3668–3674. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu F, Wang Y, Zeng S, Fu X, Wang L and
Cao J: Involvement of annexin A1 in multidrug resistance of
K562/ADR cells identified by the proteomic study. Omics.
13:467–476. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li C, Chen L, Ye J, Zhang X and Cao J:
Establishment of breast cancer cell models with inducible
differential eIF3g expressions. Xi Bao Sheng Wu Xue Za Zhi.
34:1226–1231. 2012.In Chinese.
|
|
14
|
Suzuki K, Bose P, Leong-Quong RY, Fujita
DJ and Riabowol K: REAP: A two minute cell fractionation method.
BMC Res Notes. 3:2942010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cokol M, Nair R and Rost B: Finding
nuclear localization signals. EMBO Rep. 1:411–415. 2000. View Article : Google Scholar
|
|
16
|
Block KL, Vornlocher HP and Hershey JW:
Characterization of cDNAs encoding the p44 and p35 subunits of
human translation initiation factor eIF3. J Biol Chem.
273:31901–31908. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park HS, Himmelbach A, Browning KS, Hohn T
and Ryabova LA: A plant viral 'reinitiation' factor interacts with
the host translational machinery. Cell. 106:723–733. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cuchalova L, Kouba T, Herrmannová A, Dányi
I, Chiu WL and Valásek L: The RNA recognition motif of eukaryotic
translation initiation factor 3g (eIF3g) is required for resumption
of scanning of posttermination ribosomes for reinitiation on GCN4
and together with eIF3i stimulates linear scanning. Mol Cell Biol.
30:4671–4686. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park HS, Browning KS, Hohn T and Ryabova
LA: Eucaryotic initiation factor 4B controls eIF3-mediated
ribosomal entry of viral reinitiation factor. EMBO J. 23:1381–1391.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ryabova L, Park HS and Hohn T: Control of
translation reini-tiation on the cauliflower mosaic virus (CaMV)
polycistronic RNA. Biochem Soc Trans. 32:592–596. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hou CL, Tang Cj, Roffler SR and Tang TK:
Protein 4.1R binding to eIF3-p44 suggests an interaction between
the cytoskeletal network and the translation apparatus. Blood.
96:747–753. 2000.PubMed/NCBI
|
|
22
|
Machuy N, Thiede B, Rajalingam K, Dimmler
C, Thieck O, Meyer TF and Rudel T: A global approach combining
proteome analysis and phenotypic screening with RNA interference
yields novel apoptosis regulators. Mol Cell Proteomics. 4:44–55.
2005. View Article : Google Scholar
|
|
23
|
Kim JT, Kim KD, Song EY, Lee HG, Kim JW,
Kim JW, Chae SK, Kim E, Lee MS, Yang Y and Lim JS:
Apoptosis-inducing factor (AIF) inhibits protein synthesis by
interacting with the eukaryotic translation initiation factor 3
subunit p44 (eIF3g). FEBS Lett. 580:6375–6383. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carpenter B, MacKay C, Alnabulsi A, MacKay
M, Telfer C, Melvin WT and Murray GI: The roles of heterogeneous
nuclear ribonucleoproteins in tumour development and progression.
Biochim Biophys Acta. 1765:85–100. 2006.
|
|
25
|
Ford LP, Wright WE and Shay JW: A model
for heterogeneous nuclear ribonucleoproteins in telomere and
telomerase regulation. Oncogene. 21:580–583. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Göhring F and Fackelmayer FO: The
scaffold/matrix attachment region binding protein hnRNP-U (SAF-A)
is directly bound to chromosomal DNA in vivo: A chemical
cross-linking study. Biochemistry. 36:8276–8283. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kawano S, Miyaji M, Ichiyasu S, Tsutsui KM
and Tsutsui K: Regulation of DNA Topoisomerase IIbeta through
RNA-dependent association with heterogeneous nuclear
ribonucleoprotein U (hnRNP U). J Biol Chem. 285:26451–26460. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiao R, Tang P, Yang B, Huang J, Zhou Y,
Shao C, Li H, Sun H, Zhang Y and Fu XD: Nuclear matrix factor hnRNP
U/SAF-A exerts a global control of alternative splicing by
regulating U2 snRNP maturation. Mol Cell. 45:656–668. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yugami M, Kabe Y, Yamaguchi Y, Wada T and
Handa H: hnRNP-U enhances the expression of specific genes by
stabilizing mRNA. FEBS Lett. 581:1–7. 2007. View Article : Google Scholar
|
|
30
|
Hope NR and Murray GI: The expression
profile of RNA-binding proteins in primary and metastatic
colorectal cancer: Relationship of heterogeneous nuclear
ribonucleoproteins with prognosis. Hum Pathol. 42:393–402. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huebner K, Druck T, Croce CM and Thiesen
HJ: Twenty-seven nonoverlapping zinc finger cDNAs from human T
cells map to nine different chromosomes with apparent clustering.
Am J Hum Genet. 48:726–740. 1991.PubMed/NCBI
|
|
32
|
Bettinger BT, Gilbert DM and Amberg DC:
Actin up in the nucleus. Nat Rev Mol Cell Biol. 5:410–415. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kukalev A, Nord Y, Palmberg C, Bergman T
and Percipalle P: Actin and hnRNP U cooperate for productive
transcription by RNA polymerase II. Nat Struct Mol Biol.
12:238–244. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Obrdlik A, Kukalev A, Louvet E, Farrants
AK, Caputo L and Percipalle P: The histone acetyltransferase PCAF
associates with actin and hnRNP U for RNA polymerase II
transcription. Mol Cell Biol. 28:6342–6357. 2008. View Article : Google Scholar : PubMed/NCBI
|