Introduction
MicroRNAs (miRNAs) are a class of small non-coding
RNAs (length, ~22 nt) that regulate gene expression at the
post-transcriptional levels. miRNAs are involved in the regulation
of the majority of important biological events, including
differentiation, growth, proliferation, survival, signal
transduction and immune response (1–3).
However, the roles of miRNAs in regulating the activation of human
bone marrow-derived mesenchymal stromal cells (BM-MSCs) remain to
be elucidated.
The BM-MSCs are multipotent cells that differentiate
into osteoblasts, adipocytes, chondrocytes and other tissue cells
(4–7). It has been demonstrated that
toll-like receptors (TLRs) are expressed in MSCs to modulate their
proliferation, cytokine secretion, differentiation,
hematopoiesis-supporting functions and immunosuppressive capacity
(8–10). Notably, it is well established that
TLRs induce multiple miRNAs, which regulate TLR-signaling responses
at multiple levels. For example, TLR2 and TLR4 are repressed by
expression of miR-105, miR-146 and the let-7 miRNA family (11–14).
However, miR-155 and miR-146b directly target numerous TLR
signaling proteins (15,16). Regulatory molecules, TLR-induced
transcription factors and the inflammatory cytokines are also
regulated by miRNAs, including miR-155 (17).
It has previously been verified that pre-stimulation
with the TLR2 agonist, PAM3CSK4 (PM) or the TLR4
agonist, lipopolysaccharides (LPS) enables BM-MSCs to enhance
CD34+ cell proliferation and differentiation towards the
myeloid lineage (10). To further
elucidate the roles of miRNAs in mediating TLR-induced BM-MSC
activation, the present study aimed to determine the miRNA
expression profiles in unstimulated BM-MSCs and PM/LPS-stimulated
BM-MSCs, using high-throughput Illumine HiSeq 2000 technology.
Bioinformatic methods were also used to predict the potential
target genes of the abundant known miRNAs. Furthermore, data from
the present study indicated that miRNAs are involved in various
important functions in human BM-MSCs, including the TLR signaling
pathway.
Materials and methods
Isolation and culture of BM-MSCs
BM-MSCs were isolated from fresh bone marrow by
sterile puncture of a healthy donor. The present study was approved
by the Anhui Medical University Ethics Committee, and informed
written consent was obtained from the donor. The isolation,
expansion and identification of BM-MSCs were performed as described
previously (10). Briefly,
following density gradient centrifugation over Ficoll-Hypaque
(1.077 g/ml; Beijing Solarbio Science &Technology Co., Ltd.,
Beijing, China), BM-MSCs were cultured (1×106 cells/ml)
in Dulbecco's High Glucose Modified Eagles Medium (GE Healthcare
Life Sciences, Logan, UT, USA), supplemented with 10% fetal calf
serum (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and
containing 5% CO2 at 37°C. Non-adherent cells were
removed after 24 h and the culture medium was changed twice a week.
Adherent cells were cultured until 80–90% confluence was reached,
after which trypsin (0.25%; Gibco) digestion was conducted on
passage three cells. BM-MSCs were analyzed by flow cytometry
(FACSCalibur™; BD Biosciences, Franklin Lakes, NJ, USA) following
staining with monoclonal antibodies, as follows: Fluorescein
isothiocyanate (FITC)-conjugated anti-cluster of differentiation
(CD) 90 (1:10; cat. no. 555595); FITC-anti-CD14 (1:10; cat. no.
555397), phycoerythrin (PE)-anti-CD29 (cat. no. 555443),
cyanine5-PE-anti-CD34 (cat. no. 555823), PE-anti-CD166 (cat. no.
559263), FITC-anti-CD44 (1:10; cat. no. 555478), PE-anti-CD31
(1:10; cat. no. 560983), FITC-anti-CD45 (1:10; cat. no. 555482),
PE-anti-CD13 (1:10; cat. no. 560998) and FITC-anti-CD105 (1:10;
cat. no. 561443; all: BD Biosciences). BM-MSCs were confirmed by
negative staining of hematopoietic and endothelial lineage markers
(CD14, CD34, CD31 and CD45) and positive staining of CD90, CD105,
CD166, CD29, CD44 and CD13.
PM and LPS stimulation
LPS (100 ng/ml) and PM (100 ng/ml) were purchased
from R&D Systems, Inc. (Minneapolis, Minnesota, USA). The
BM-MSC feeder layers in 12-well plates were stimulated with
(experimental group) or without (control) TLR2 agonist, PM, or TLR4
agonist, LPS, for 24 h. The supernatants were removed and the
feeder layers were washed twice with fresh culture medium.
Total RNA extraction and small RNA
sequencing library preparation
Total RNA was isolated from confluent MSCs with
TRIzol (Promega Corporation, Madison, WI, USA). The purity and
concentration of total RNA samples were determined with NanoDrop
ND-1000 (Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
Total RNA of each sample was used to prepare the miRNA sequencing
library using the TruSeq Small RNA Library Prep kit (RS-200-0012;
Illumina, Inc., San Diego, CA, USA), according to the
manufacturer's protocol. This included the following steps: i)
3′-Adapter ligation with truncated T4 RNA ligase 2 (New England
BioLabs, Inc., Ipswich, MA, USA); ii) 5′-adapter ligation with T4
RNA ligase (Ambion; Thermo Fisher Scientific, Inc.); iii) cDNA
synthesis with reverse transcription primers (Bioligo Biological
Technology, Co., Ltd., Shanghai, China); iv) polymerase chain
reaction (PCR) amplification; and v) extraction and purification of
PCR amplified fragments [length,~138–158 bp; corresponding to small
RNAs (length,~15–35 nt)] from the agarose gel. Subsequently, the
completed libraries were quantified with an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Small RNA sequencing
The samples were diluted to a final concentration of
8 pM and cluster generation was performed on the Illumina cBot
using TruSeq Rapid SR cluster kit (Illumina, Inc.), following the
manufacturer's protocols. Sequencing was performed on an Illumina
HiSeq 2000 using TruSeq Rapid SBS kits (Illumina, Inc.), according
to the manufacturer's protocols. The DNA fragments in the libraries
were denatured with 0.1 M NaOH to generate single-stranded DNA
molecules, captured on flow cells (Illumina, Inc.), amplified in
situ and finally sequenced for 36 cycles on Illumina HiSeq
2000.
Analysis of sequencing data and length
distribution
Using high throughput sequencing with an Illumina
Hiseq 2000, total clean reads from the control and trial libraries
were obtained, and the length distribution of the clean reads was
summarized. Image analysis and base calling were performed using
Off-Line Basecaller software (v1.8.0; Illumina, San Diego, CA,
USA). Subsequently, the 3′-adapter sequence was trimmed from the
clean reads [that had passed the Solexa CHASTITY quality filter
(Illumina)] and the reads of length <15 nt were discarded.
Remaining reads (length, ≥15 nt) were aligned to the latest known
human reference miRNA precursor set [Sanger miRBase 19 (http://www.mirbase.org/)] using Novoalign software
(v2.07.11; http://www.novocraft.com/products/novoalign/). Reads
(<2 counts) were discarded when calculating the miRNA
expression. In order to characterize the isomiR variability, any
sequence that matched the miRNA precursors in the mature miRNA
region ±4 nt (with ≤1 mismatch) were accepted as mature miRNA
isomiRs, which were grouped according to the 5-prime (5p) or
3-prime (3p) arm of the precursor hairpin.
Prediction of novel miRNAs
miRDeep2 (http://www.mdc-berlin.de/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/miRDeep)
was used to predict novel miRNAs. For novel miRNA prediction, all
sequence data was pooled from the following 3′-adapter trimmed
files: LPS, PM and contrimmed_tags.fa, all adapter trimmed
sequences of length <17 bp and mismatch >1 were excluded from
the prediction pipeline. The higher the novel miRNA score of the
miRDeep2, the more reliable the novel miRNA was considered to
be.
Target gene prediction
Three online search algorithms, TargetScan version
6.2 (http://www.targetscan.org/vert_60/) and miRanda
(http://www.Microrna.org/microrna/home.do) and
MicroCosm Targets version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)
were used to predict the target genes of differentially expressed
(DE) miRNAs among the BM-MSCs activated with the TLR2 and TLR4
agonists, and BM-MSCs in the absence of agonists. The annotated
miRNA target genes that were selected from all the algorithms were
considered to be the target genes.
Gene ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway analysis of the predicted miRNA
target genes
In order to further realize the functions, the
predicted target genes were subjected to the analysis of GO project
(http://www.geneontology.org). Fisher's
exact test was used to find whether there was increased overlap
between the DE list and the GO annotation list than would be
expected by chance. The P-value denotes the significance of GO term
enrichment in the DE genes; the lower the P-value, the more
significant the GO term (P<0.05 is recommended). Furthermore,
pathway analysis was performed for these target genes. Pathway
analysis is a functional analysis that maps genes to the KEGG
pathways. The P-value (EASE-score, Fisher's method P-value or
hypergeometric P-value) indicates the significance of the pathway
correlated to the conditions. A lower P-value, indicates a more
significant pathway (P<0.05 is recommended).
Validation of miRNA expression by
quantitative PCR (qPCR)
A random selection of DE miRNAs between the
experimental and control groups from the sequencing data was
validated by qPCR. Corroboration of the six novel miRNAs was
performed according to the previously described conditions using
qPCR assays. Total RNA was isolated from each sample using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The miRNA quantification was performed by qPCR using Applied
Biosystems StepOne Real-Time PCR system (Thermo Fisher Scientific,
Inc.) and SYBR premix Ex Taq II kit (Takara Bio, Inc.) according to
the manufacturer's protocols. U6 served to normalize the levels of
all miRNA transcripts, the primer sequences were forward, 5′-GCT
TCG GCA GCA CAT ATA CTA AAAT-3′ and reverse, 5′-CGC TTC ACG AAT TTG
CGT GTCAT-3′ for U6. Relative expression of miRNA was evaluated by
the 2−ΔΔCq method (18). All miRNA samples and the internal
reference U6 were run in a qPCR reaction. Thermal cycling
conditions of the qPCR were 95°C for 10 min; 40 cycles of 95°C for
10 sec; and 40 cycles of 60°C for 60 sec.
Results
Overview of LPS and PM-induced miRNA
expression profiling
To obtain a comprehensive view of the miRNA profile
of BM-MSCs stimulated with LPS and PM, Illumine HiSeq 2000
technology was used to detect the global expression level annotated
in Sanger miRBase 19.0. A total of 67 miRNAs demonstrated different
expression patterns between the LPS and control groups (BM-MSCs
without stimulation), with 32 downregulated miRNAs (Table I)and 35 upregulated miRNAs
(Table II). In PM-treated cells,
the downregulation (Table III)
and upregulation (Table IV) were
observed for 51 and 46 miRNAs, respectively. To assess the
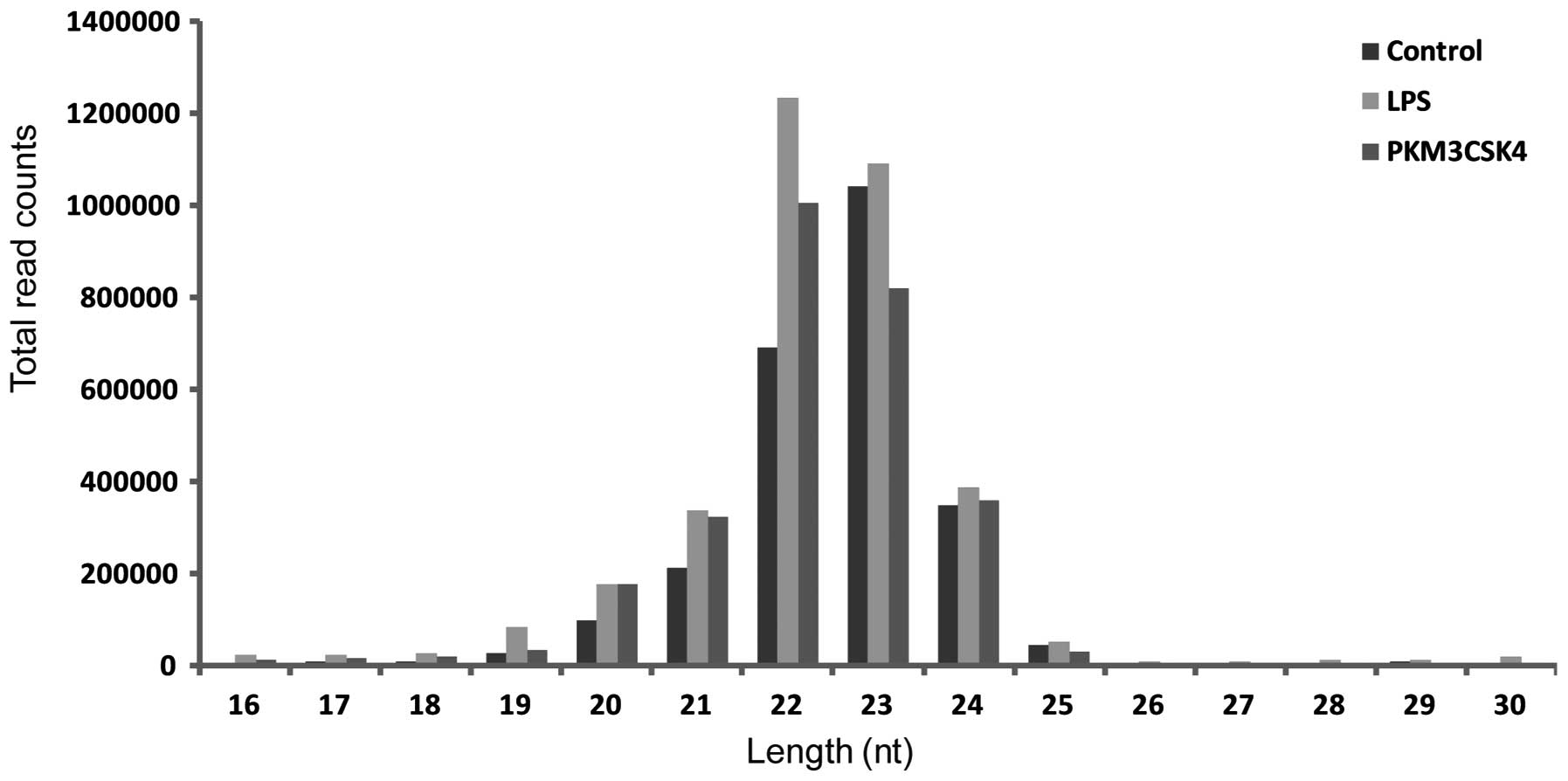
sequencing quality, the length distribution of the clean reads was
summarized. Few differences were observed in the length
distribution of the sequences from the control and trial libraries
(Fig. 1). The most abundant
sequence reads were 22, 23, 21 and 24 nt in length, which is
consistent with the typical small RNA distribution of mammals.
 | Table IDownregulated miRs in bone
marrow-derived mesenchymal stem cells stimulated with
lipopolysaccharide. |
Table I
Downregulated miRs in bone
marrow-derived mesenchymal stem cells stimulated with
lipopolysaccharide.
| miR name | Fold change |
|---|
| hsa-let-7a-3p | 0.333333333 |
| hsa-miR-1 | 0.157248157 |
|
hsa-miR-125a-5p | 0.403608737 |
| hsa-miR-132-5p | 0.161290323 |
| hsa-miR-137 | 0.253012048 |
| hsa-miR-154-3p | 0.153846154 |
| hsa-miR-15a-5p | 0.359223301 |
|
hsa-miR-181a-5p | 0.287637856 |
|
hsa-miR-181c-3p | 0.450000000 |
| hsa-miR-186-5p | 0.128352490 |
|
hsa-miR-199a-3p | 0.309404320 |
|
hsa-miR-199b-3p | 0.309404320 |
| hsa-miR-221-3p | 0.280632896 |
| hsa-miR-222-3p | 0.374696936 |
| hsa-miR-24-3p | 0.333686441 |
| hsa-miR-29a-3p | 0.187765957 |
| hsa-miR-30d-5p | 0.437901499 |
| hsa-miR-30e-5p | 0.300376648 |
|
hsa-miR-323a-3p | 0.182186235 |
|
hsa-miR-323b-3p | 0.094527363 |
| hsa-miR-335-5p | 0.413793103 |
| hsa-miR-337-3p | 0.482758621 |
| hsa-miR-361-5p | 0.454545455 |
| hsa-miR-369-3p | 0.404193782 |
|
hsa-miR-374a-5p | 0.271111111 |
| hsa-miR-380-3p | 0.400000000 |
| hsa-miR-411-3p | 0.083179298 |
| hsa-miR-425-5p | 0.016032811 |
|
hsa-miR-450a-5p | 0.183206107 |
| hsa-miR-708-5p | 0.258064516 |
| hsa-miR-941 | 0.131578947 |
| hsa-miR-99b-5p | 0.476327116 |
 | Table IIUpregulated miRs in bone
marrow-derived mesenchymal stem cells stimulated with
lipopolysaccharide. |
Table II
Upregulated miRs in bone
marrow-derived mesenchymal stem cells stimulated with
lipopolysaccharide.
| miR name | Fold change |
|---|
| hsa-let-7b-5p | 2.918778428 |
| hsa-miR-100-5p | 10.847709700 |
|
hsa-miR-106a-5p | 2.083333333 |
|
hsa-miR-106b-5p | 5.405405405 |
|
hsa-miR-125a-3p | 3.090909091 |
| hsa-miR-126-3p | 2.282608696 |
| hsa-miR-128 | 10.200000000 |
| hsa-miR-136-3p | 2.000000000 |
| hsa-miR-138-5p | 15.218750000 |
|
hsa-miR-146a-5p | 2.125000000 |
| hsa-miR-155-5p | 19.623376620 |
| hsa-miR-17-5p | 5.828767123 |
| hsa-miR-18a-5p | 4.888888889 |
|
hsa-miR-199a-5p | 32.703703700 |
| hsa-miR-20a-5p | 4.294478528 |
| hsa-miR-218-5p | 4.400000000 |
| hsa-miR-22-5p | 2.133333333 |
| hsa-miR-224-5p | 12.050000000 |
| hsa-miR-26a-5p | 8.077169132 |
| hsa-miR-26b-5p | 2.234290470 |
| hsa-miR-32-3p | 2.750000000 |
| hsa-miR-34a-5p | 2.252148997 |
|
hsa-miR-3591-5p | 3.214285714 |
| hsa-miR-361-3p | 4.136363636 |
| hsa-miR-369-5p | 2.222222222 |
| hsa-miR-424-3p | 9.666666667 |
| hsa-miR-425-3p | 3.461538462 |
| hsa-miR-455-3p | 6.421052632 |
| hsa-miR-486-5p | 3.418918919 |
| hsa-miR-497-5p | 2.230769231 |
|
hsa-miR-548d-5p | 2.800000000 |
| hsa-miR-574-5p | 7.185185185 |
| hsa-miR-7-5p | 4.600000000 |
| hsa-miR-941 | 2.250000000 |
| hsa-miR-99a-5p | 10.116279070 |
 | Table IIIDownregulated miRs in bone marrow
derived mesenchymal stem cells stimulated with
PAM3CSK4. |
Table III
Downregulated miRs in bone marrow
derived mesenchymal stem cells stimulated with
PAM3CSK4.
| miR name | Fold change |
|---|
| hsa-let-7a-3p | 0.339080460 |
| hsa-miR-1 | 0.090909091 |
| hsa-miR-101-3p | 0.413533835 |
|
hsa-miR-103a-3p | 0.169556840 |
|
hsa-miR-106b-3p | 0.383177570 |
| hsa-miR-107 | 0.200000000 |
| hsa-miR-10a-5p | 0.263522618 |
| hsa-miR-10a-3p | 0.406250000 |
| hsa-miR-10b-5p | 0.276625387 |
|
hsa-miR-125b-5p | 0.449112979 |
|
hsa-miR-125b-1-3p | 0.409638554 |
| hsa-miR-132-5p | 0.322580645 |
| hsa-miR-134 | 0.266187050 |
| hsa-miR-137 | 0.409638554 |
| hsa-miR-140-5p | 0.369565217 |
|
hsa-miR-146b-5p | 0.250539957 |
|
hsa-miR-148a-5p | 0.385714286 |
|
hsa-miR-148b-5p | 0.458333333 |
|
hsa-miR-151a-3p | 0.343228200 |
| hsa-miR-154-3p | 0.1111111110 |
|
hsa-miR-181c-3p | 0.316666667 |
| hsa-miR-186-5p | 0.223180077 |
| hsa-miR-190a | 0.284424379 |
| hsa-miR-191-5p | 0.262476895 |
|
hsa-miR-199a-3p | 0.255945887 |
|
hsa-miR-199b-3p | 0.255945887 |
| hsa-miR-21-3p | 0.239819005 |
| hsa-miR-210 | 0.234042553 |
| hsa-miR-221-3p | 0.215810131 |
| hsa-miR-222-3p | 0.383072515 |
| hsa-miR-27a-5p | 0.405405405 |
| hsa-miR-27b-3p | 0.278292181 |
| hsa-miR-29a-3p | 0.392021277 |
|
hsa-miR-30c-2-3p | 0.431818182 |
| hsa-miR-30d-5p | 0.423982869 |
| hsa-miR-30e-5p | 0.284369115 |
|
hsa-miR-323a-3p | 0.287449393 |
|
hsa-miR-323b-3p | 0.074626866 |
| hsa-miR-335-3p | 0.438016529 |
| hsa-miR-34c-5p | 0.138211382 |
| hsa-miR-369-3p | 0.149674620 |
|
hsa-miR-374a-5p | 0.182222222 |
| hsa-miR-379-5p | 0.346481876 |
| hsa-miR-411-3p | 0.118299445 |
| hsa-miR-425-5p | 0.026099925 |
|
hsa-miR-450a-5p | 0.114503817 |
| hsa-miR-493-5p | 0.460829493 |
| hsa-miR-671-5p | 0.326530612 |
| hsa-miR-708-5p | 0.370967742 |
| hsa-miR-92a-3p | 0.430016863 |
| hsa-miR-99b-5p | 0.199426112 |
 | Table IVUpregulated miRs in bone
marrow-derived mesenchymal stem cells stimulated with
PAM3CSK4. |
Table IV
Upregulated miRs in bone
marrow-derived mesenchymal stem cells stimulated with
PAM3CSK4.
| miR name | Fold change |
|---|
| hsa-let-7a-5p | 4.209545028 |
| hsa-let-7b-5p | 6.022742040 |
| hsa-let-7c | 3.173728814 |
| hsa-let-7d-5p | 11.85106383 |
| hsa-let-7e-5p | 2.807106599 |
| hsa-let-7f-5p | 2.787247317 |
| hsa-let-7g-5p | 2.138705416 |
| hsa-miR-100-5p | 9.835217133 |
|
hsa-miR-106b-5p | 4.783783784 |
| hsa-miR-127-3p | 2.286982249 |
|
hsa-miR-1307-5p | 2.181818182 |
|
hsa-miR-130a-3p | 2.319148936 |
| hsa-miR-136-3p | 2.102564103 |
|
hsa-miR-146a-5p | 22.854166670 |
| hsa-miR-155-5p | 5.000000000 |
| hsa-miR-15b-5p | 3.800000000 |
| hsa-miR-17-3p | 2.000000000 |
| hsa-miR-185-5p | 2.1111111110 |
| hsa-miR-18a-5p | 2.222222222 |
|
hsa-miR-193b-3p | 4.820512821 |
| hsa-miR-195-5p | 16.080000000 |
|
hsa-miR-196a-5p | 2.655948553 |
| hsa-miR-214-3p | 84.462585030 |
| hsa-miR-23a-3p | 3.709433962 |
| hsa-miR-23b-3p | 5.725806452 |
| hsa-miR-23c | 6.500000000 |
| hsa-miR-24-3p | 29.494703390 |
| hsa-miR-27b-5p | 5.000000000 |
| hsa-miR-28-5p | 2.307692308 |
| hsa-miR-320a | 2.166666667 |
| hsa-miR-335-5p | 2.379310345 |
|
hsa-miR-374b-3p | 4.285714286 |
|
hsa-miR-376c-3p | 2.636363636 |
| hsa-miR-377-3p | 2.133333333 |
| hsa-miR-421 | 3.884615385 |
| hsa-miR-423-5p | 2.112903226 |
| hsa-miR-424-5p | 2.350000000 |
| hsa-miR-431-5p | 3.600000000 |
| hsa-miR-4510 | 6.818181818 |
| hsa-miR-484 | 2.535714286 |
| hsa-miR-487b | 3.000000000 |
| hsa-miR-493-3p | 2.100000000 |
| hsa-miR-598 | 2.300000000 |
| hsa-miR-92b-3p | 3.000000000 |
| hsa-miR-941 | 5.400000000 |
| hsa-miR-99a-5p | 2.523255814 |
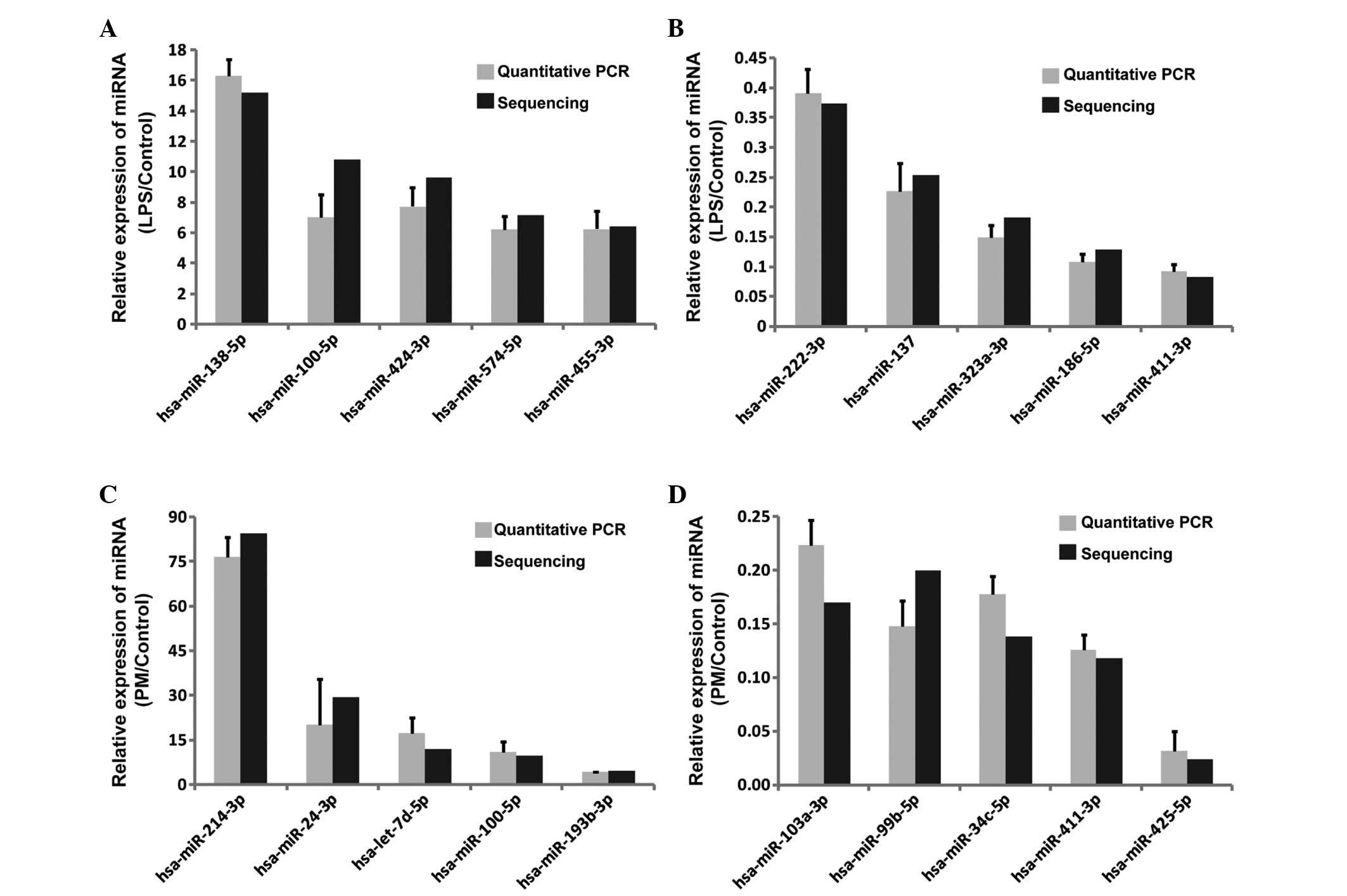
qPCR verification of known miRNAs
For identification of the authenticity of the miRNAs
detected by Illumine HiSeq 2000 technology, 40 known miRNAs from
Tables I–IV were randomly selected for further
validation with additional samples by qPCR. As presented in
Fig. 2, the qPCR data were highly
consistent with Illumine HiSeq 2000 technology, confirming the
reliability of the HiSeq 2000 sequencing data.
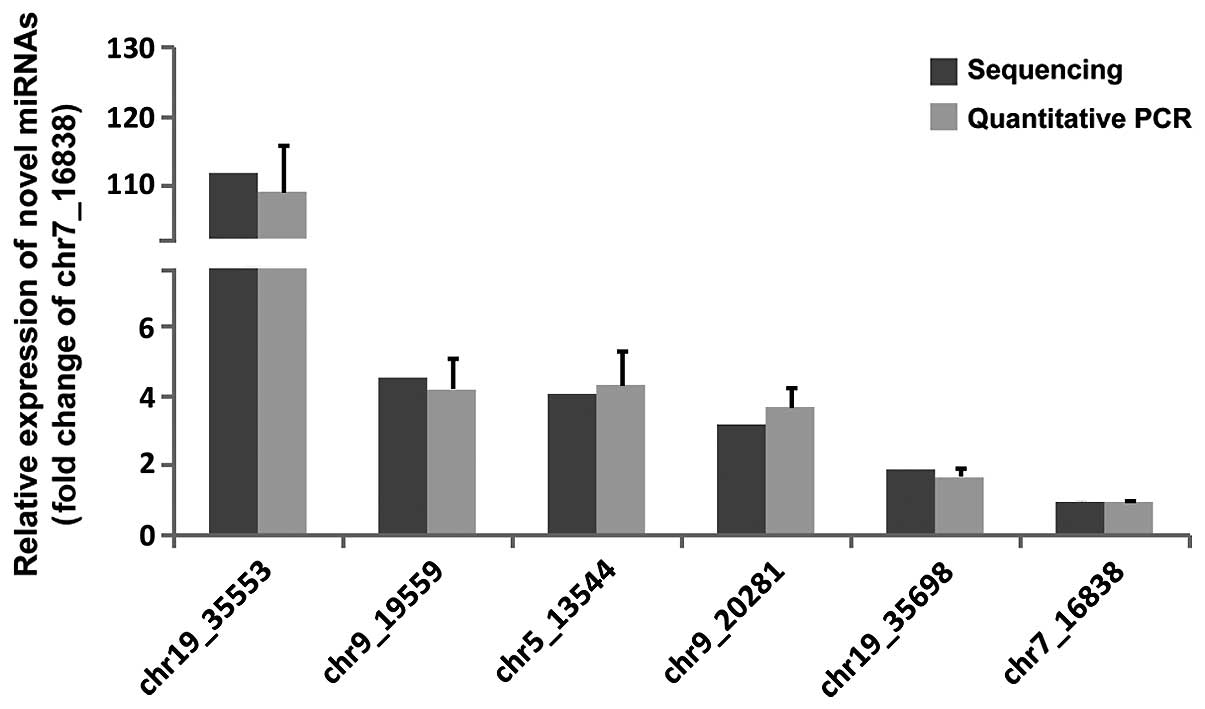
Novel miRNA identification
To identify the novel, unannotated miRNAs in human
BM-MSCs, the unlabeled reads were analyzed by miRDeep2. The present
study focused on six potential novel miRNAs with the most frequent
appearance among all the samples and the highest miRDeep2 score.
The expression levels of these six miRNAs were further validated by
qPCR. Compared with Illumine HiSeq 2000 sequencing, qPCR indicated
the same trend of expression levels for the six novel miRNA
candidates (Fig. 3).
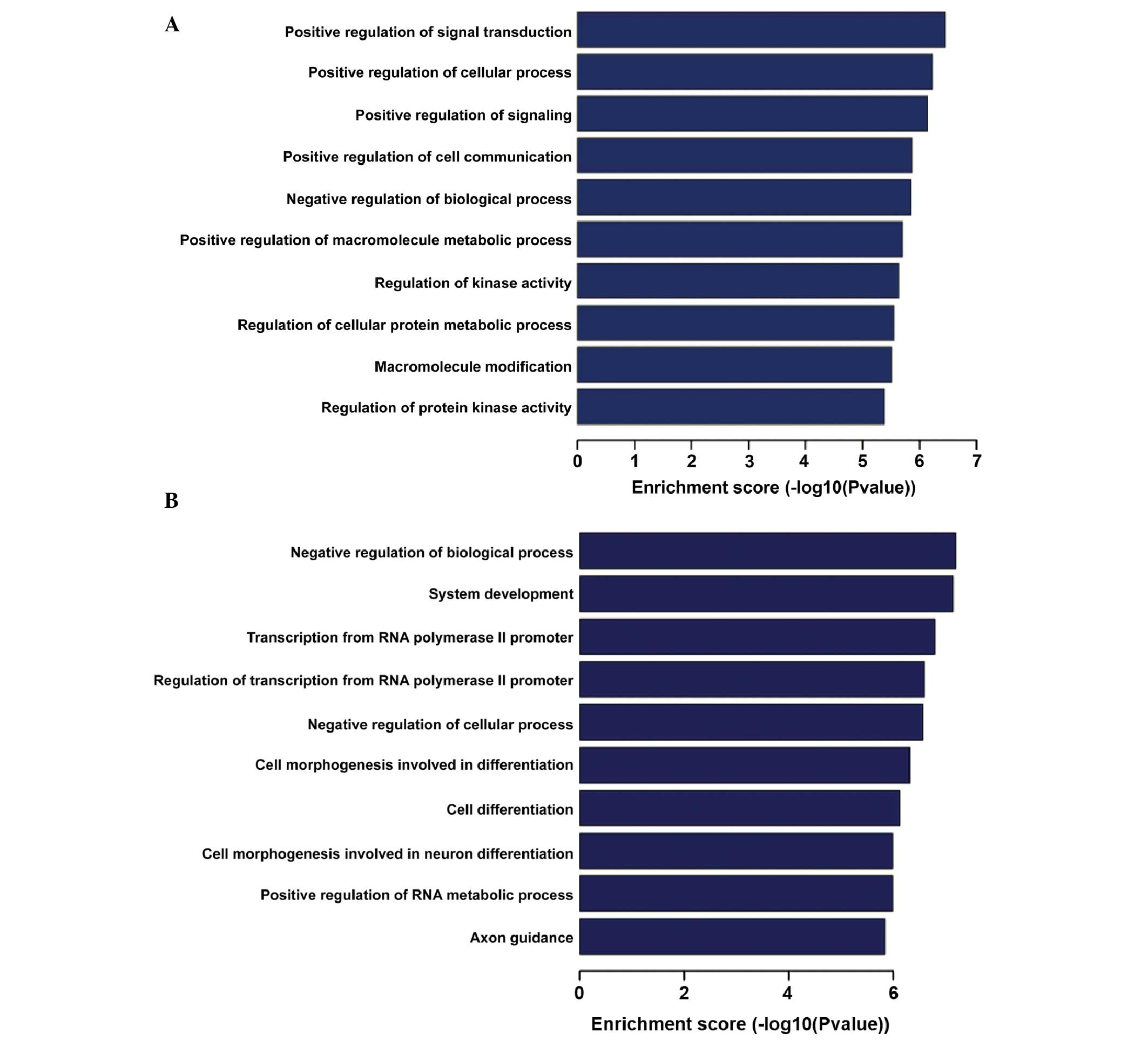
Predicted target genes of LPS- and
PM-responsive miRNAs
To characterize the DE miRNAs between LPS, PM and
the controls, the target genes of the DE miRNAs were predicted
using three different miRNA target prediction algorithms
(TargetScan, MicroCosm and miRanda). Subsequently, the target genes
that were identified in all three databases underwent GO analysis.
In the LPS group, the targets genes of downregulated miRNAs were
significantly enriched in transcripts from the RNA polymerase II
promoter and included regulation of macromolecule metabolic
processes, gene expression and nucleobase-containing compound
metabolic processes; the most enriched GO terms of targets genes of
upregulated miRNAs included anatomical structure morphogenesis,
regulation of cellular processes, biological processes and cell
development. In the PM group, significantly enriched GO terms of
targets genes of down-regulated miRNAs included regulation of
biological process, system development, transcription from RNA
polymerase II promoter and cell differentiation; the most enriched
GO terms of targets genes of downregulated miRNAs included
regulation of signal transduction, cellular process, cell
communication and biological process (Fig. 4). KEGG pathway analysis indicated
that the predicted target genes were significantly enriched in a
wide range of pathways. LPS-regulated miRNAs predominantly target
mitogen activated protein kinases (MAPKs), estrogen, T cell
receptors, phosphatidylinositol-4,5-bisphosphate 3-kinase
(PI3)-Akt, the mammalian target of rapamycin signaling pathway and
cancer-associated signaling pathways, while PM-regulated miRNAs
target the P13-Akt, ras-related protein 1, neurotrophin, MAPK and
Ras signaling pathways (Fig.
5).
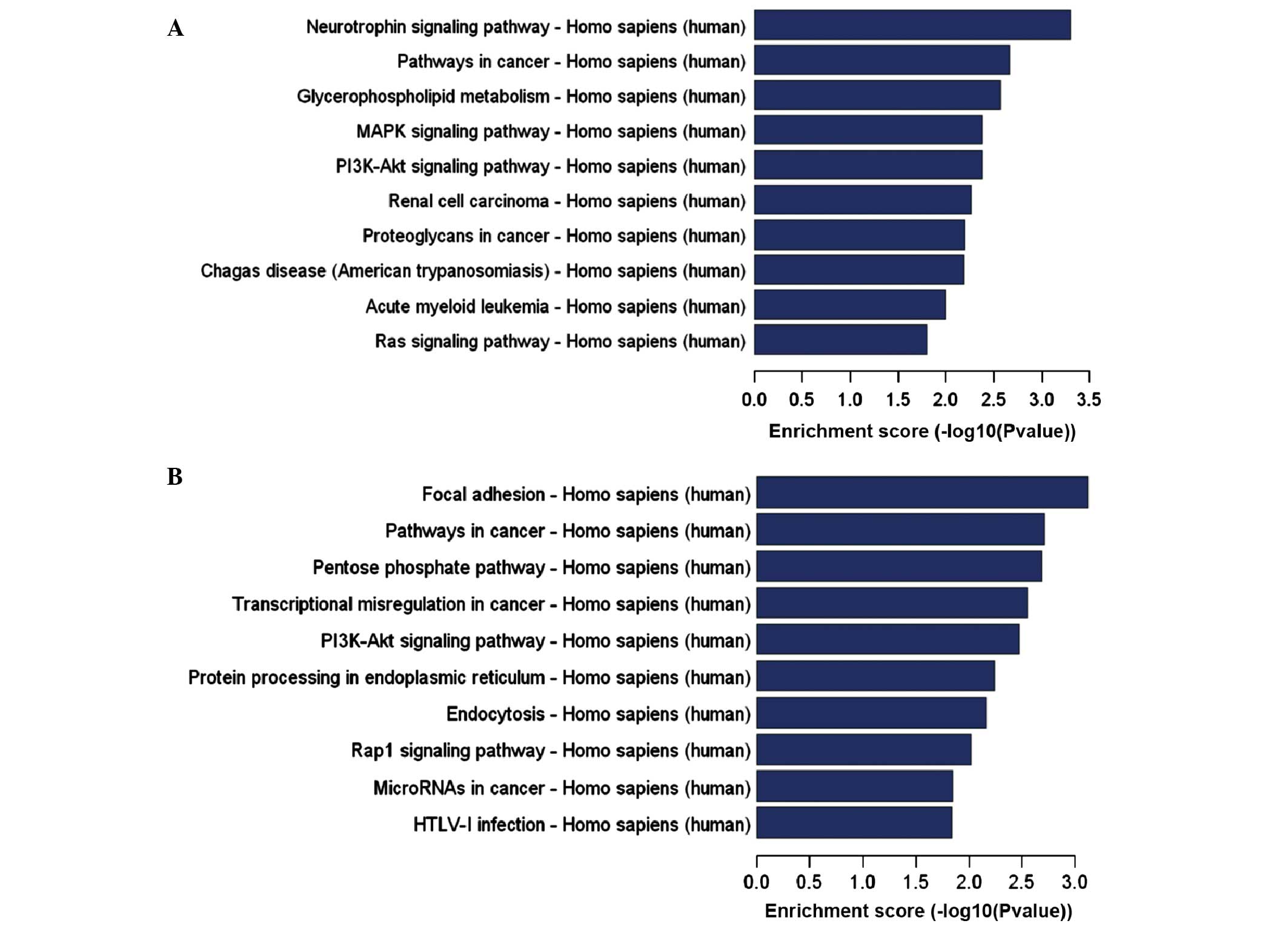 | Figure 5KEGG pathways of target genes
demonstrating the greatest enrichment for differentially expressed
miRNAs. The KEGG pathways enriched for target genes regulated by
(A) increased and (B) decreased expression of miRNAs in the
PAM3CSK4 group. KEGG, Kyoto Encyclopedia of Genes and
Genomes; miRNA, microRNA; PI3K,
phosphatidylinositol-4,5-bisphosphate 3-kinase; MAPK,
mitogen-activated protein kinase; mTOR, mammalian target of
rapamycin; Rap-1, ras-related protein 1; HTLV, human T-lymphotropic
virus; FoxO, forkhead box O. KEGG pathways of target genes
demonstrating the greatest enrichment for differentially expressed
miRNAs. The KEGG pathways enriched for target genes regulated by
(C) increased and (D) decreased expression of miRNAs in the
lipopolysaccharide group. KEGG, Kyoto Encyclopedia of Genes and
Genomes; miRNA, microRNA; PI3K,
phosphatidylinositol-4,5-bisphosphate 3-kinase; MAPK,
mitogen-activated protein kinase; mTOR, mammalian target of
rapamycin; Rap-1, ras-related protein 1; HTLV, human T-lymphotropic
virus; FoxO, forkhead box O. |
Discussion
A number of studies have reported the expression of
TLRs upon MSC and TLR activation in MSCs regulates their functions
in immunomodulation, migration, repair and regeneration of damaged
tissues/organs (19–24). In addition, a previous study
reported that TLR2 agonists (PM) and/or TLR4 agonists (LPS) were
able to activate MSCs, thus modulating their
hematopoiesis-supporting role in vitro (10). The present study hypothesized that
the miRNAs induced by TLR activation may control MSC processes,
including immunomodulation, migration, and repair and regeneration
of damaged tissues/organs. The present study analyzed the
differential miRNA expression profiles in the MSCs derived from
human fresh bone marrow and stimulated with LPS or PM. By
high-throughput sequencing, compared with the expression levels in
unstimulated MSCs, 67 miRNAs were identified with 35 miRNAs
upregulated and 32 downregulated following LPS stimulation; 97
miRNAs with 46 miRNAs upregulated and 51 downregulated following PM
stimulation. Data from the present study demonstrated that the
responses to LPS or PM prompted quick and specific in vitro
alterations to the miRNA expression profile of BM-MSCs.
miR-425, which has been demonstrated to be DE and
the most markedly downregulated, was observed in the LPS and
PM-stimulated BM-MSCs. It was reported that upregulated expression
of miR-425 was associated with a variety of types of cancer,
including renal and gastric cancers (25,26).
A recent study demonstrated that nuclear factor-κB
(NF-κB)-dependent miR-425 upregulation promotes gastric cancer cell
growth by targeting phosphatase and tensin homolog in response to
IL-1β treatment (25,26). Furthermore, this indicates that the
action of miR-425 may mediate cell proliferation. However, to the
best of our knowledge, there are no reports investigating the
effects of miR-425 in BM-MSCs. Compared with previous studies, the
marked difference in expression level of miR-425 indicates that
miRNAs may respond differently in LPS or PM-stimulated BM-MSCs and
exert different effects in the different groups.
In accordance with the miRNA results demonstrating
upregulation of miR-199a in human BM-MSCs during osteoblastic
differentiation and chondrogenesis (27), the most abundantly expressed miRNA
in LPS-stimulated BM-MSCs was miR-199a, which was also upregulated
in LPS-stimulated leukocytes derived from cord blood (CB) (28). Laine et al (27) demonstrated that cells transfected
with pre-miR-199a reduced the proliferation of human MSCs and
miR-199a restricts chondrogenic differentiation by suppressing the
expression of SRY-box 9, which is a positive regulator of
chondrogenesis (29). In addition,
miR-199a has been identified as a negative regulator of
chondrogenesis (30). Wu et
al (31) inferred that the
overexpression of miR-199a-5p inhibits the proliferation of keloid
fibroblasts. However, Shi et al (32) demonstrated that overexpression of
miR-199a-5p promoted the proliferation of porcine preadipocytes and
suppressed adipogenic differentiation. These findings indicate that
increased expression of miR-199a in LPS-stimulated BM-MSCs may be a
key regulator for MSC differentiation and proliferation. As miR-199
was observed to exert opposing effects on proliferation in
different cells, this is likely explained by miRNA-mRNA targeting,
and the different interactions among tissue and cell types and
physiological/pathological conditions (33). Further studies are required to
investigate the exact underlying mechanism of miR-199a action in
BM-MSCs.
The miRNA that was most abundantly expressed in
human PM-stimulated BM-MSCs was miR-214. Previous studies reported
that miR-214 may be important in promoting myoblast proliferation
and differentiation (34);
overexpression of miR-214 was connected to gastric cancer tissue,
and deletion of miR-214 inhibited the proliferation, migration and
invasion of gastric cancer cells (35). The results are consistent with
recent findings by Zhang et al (36), that miR-214 is overexpressed in
nasopharyngeal carcinoma tissues and cell lines, and that knockdown
of miR-214 suppressed cell proliferation and induced apoptosis.
However, Derfoul et al (37), demonstrated that overexpression of
miR-214 inhibited the proliferation and invasion of breast cancer
cells. Thus, the abnormal expression of miR-214 may be closely
associated with certain types of cancer by affecting cell growth,
differentiation and migration.
However, whether the TLR signaling pathway is
regulated by the above-mentioned miRNAs, which are expressed most
differentially, and whether their target genes are involved in TLR
signal transduction, requires further investigation.
To investigate the function of the LPS- or
PM-induced DE miRNAs further, the target genes were predicted, and
GO and KEGG pathway analyses were applied to these predicted target
genes. The GO term annotation suggests that the presumed target
genes of these DE miRNAs in BM-MSCs are involved in a broad range
of physiological processes in response to stimuli. Consistent with
the GO assay, among those pathways, the MAPK and PI3-Akt signaling
pathway are notable, as they have been reported to be important in
promoting cell proliferation, growth and differentiation (38).
Notably, in the present study, a number of the
verified TLR signaling pathway-associated miRNAs were markedly DE
between the control and trial groups. For example, in the TLR4
agonist group, the associated markedly DE miRNAs were miR-146a,
miR-155, miR-132, miR-29, miR-199a-5p, let-7b, miR-24, miR-221,
miR-181, miR-106a/b, miR-20a, miR-26a and miR-34a (14,15,39-48).
In the TLR2 agonist group, the associated markedly DE miRNAs were
miR-146a/b, miR-155, miR-132, miR-29, miR-199a-3p, let-7b, miR-24,
miR-221, miR-181, miR-106b, miR-92a, miR-210, miR-27b, miR-21,
miR-125b, miR-101 and miR-148a/b (12,15,16,38-45,49-54).
These miRNAs in BM-MSCs may suggest a potentially important role in
TLR signaling regulation. Furthermore, miR-146b was observed to be
upregulated in CB leukocytes following LPS stimulation and may
control LPS-stimulated neonatal early phases of inflammation via
negative feedback loops (28). Ma
et al (55) reported that
miR-301a was upregulated in umbilical cord-MSCs stimulated with
LPS, while miR-301a was not regulated by LPS and PM in the dataset
produced in the present study, this notable difference is likely
due to the different experimental methods and sources of
material.
TLR activation triggers myeloid differentiation
primary response gene 88-dependent and -independent downstream
signaling cascades, leading to the activation of a number of
transcription factors and genes (56–61).
These downstream signaling molecules predominantly include
transforming growth factor-β-activated kinase, IL-1R-associated
kinases, MAPKs, NF-κB and tumor necrosis factor-receptor-associated
factor 6 (59). It has been
demonstrated that following stimulation with ligands specific to
TLRs, the NF-κB, MAPK, and PI3K signaling pathways were activated
with a subsequent induction of multiple genes and cytokines
(62). Furthermore, combined with
KEGG pathways analysis, the results of the present study indicate a
critical role for miRNAs, via the TLR signaling pathway, in the
growth, differentiation, and migration of LPS and PM-induced
BM-MSCs.
During comparison of these two sets of experiments,
a number of the TLR signaling pathway-associated miRNAs (miR-146a,
miR-155, let-7b, miR-106b, miR-132, miR-29, miR-221 and miR-181)
were found to be the same, and the up- or downregulation of
specific miRNAs was also consistent, suggesting that LPS- or
PM-induced miRNAs may have common targets and exert similar effects
in BM-MSCs. Future studies are required to validate these
predictions.
Six novel miRNAs that demonstrated marked changes in
response to TLR activation were observed in the present study,
which were not similar to any known miRNAs. Future investigations
are required to predict and functionally validate these DE miRNA
targets.
In conclusion, the results of the current study
identify the global expression change of miRNAs in BM-MSCs
stimulated with LPS and PM, providing a framework for further
analysis of miRNAs and their role in mediating TLR signals to
regulate the functions of BM-MSCs.
Acknowledgments
The authors would like to thank Dr Jun Liu for
reviewing the manuscript. The present study was supported by the
National Natural Science Foundation (grant no. 81270573) and the
Natural Science Foundation of Anhui Higher Education Institutions
(grant no. KJ2012Z188).
Abbreviations:
|
BM-MSCs
|
bone marrow-derived mesenchymal
stromal cells
|
|
GO
|
gene ontology
|
|
LPS
|
lipopolysaccharides
|
|
MAPKs
|
mitogen-activated protein kinases
|
|
miRNAs
|
microRNAs
|
|
NF-κB
|
nuclear factor-κB
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
TLRs
|
toll-like receptors
|
References
|
1
|
Yates LA, Norbury CJ and Gilbert RJ: The
long and short of microRNA. Cell. 153:516–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pittenger MF, Mackay AM, Beck SC, Jaiswal
RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S and
Marshak DR: Multilineage potential of adult human mesenchymal stem
cells. Science. 284:143–147. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang Y, Jahagirdar BN, Reinhardt RL,
Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund
T, Blackstad M, et al: Pluripotency of mesenchymal stem cells
derived from adult marrow. Nature. 418:41–49. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Charbord P: Bone marrow mesenchymal stem
cells: Historical overview and concepts. Hum Gene Ther.
21:1045–1056. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oreffo RO, Cooper C, Mason C and Clements
M: Mesenchymal stem cells: Lineage, plasticity and skeletal
therapeutic potential. Stem Cell Rev. 1:169–178. 2005. View Article : Google Scholar
|
|
8
|
Hwa Cho H, Bae YC and Jung JS: Role of
toll-like receptors on human adipose-derived stromal cells. Stem
Cells. 24:2744–2752. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van den Berk LC, Jansen BJ,
Siebers-Vermeulen KG, Netea MG, Latuhihin T, Bergevoet S, Raymakers
RA, Kögler G, Figdor CC, Adema GJ and Torensma R: Toll-like
receptor triggering in cord blood mesenchymal stem cells. J Cell
Mol Med. 13:3415–3426. 2009. View Article : Google Scholar
|
|
10
|
Wang X, Cheng Q, Li L, Wang J, Xia L, Xu X
and Sun Z: Toll-like receptors 2 and 4 mediate the capacity of
mesenchymal stromal cells to support the proliferation and
differentiation of CD34+cells. Exp Cell Re. 318:196–206. 2012.
View Article : Google Scholar
|
|
11
|
Benakanakere MR, Li Q, Eskan MA, Singh AV,
Zhao J, Galicia JC, Stathopoulou P, Knudsen TB and Kinane DF:
Modulation of TLR2 protein expression by miR-105 in human oral
keratinocytes. J Biol Chem. 284:23107–23115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Quinn EM, Wang JH, O'Callaghan G and
Redmond HP: MicroRNA-146a is upregulated by and negatively
regulates TLR2 signaling. PLoS One. 8:e622322013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
O'Hara SP, Splinter PL, Gajdos GB,
Trussoni CE, Fernandez-Zapico ME, Chen XM and LaRusso NF: NFkappaB
p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated
transcriptional repression of microRNA let-7i following microbial
infection. J Biol Chem. 285:216–225. 2010. View Article : Google Scholar :
|
|
14
|
Yang K, He YS, Wang XQ, Lu L, Chen QJ, Liu
J, Sun Z and Shen WF: MiR-146a inhibits oxidized low-density
lipoprotein-induced lipid accumulation and inflammatory response
via targeting toll-like receptor 4. FEBS Lett. 585:854–860. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tili E, Michaille JJ, Cimino A, Costinean
S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA and
Croce CM: Modulation of miR-155 and miR-125b levels following
lipopolysaccharide/TNF-alpha stimulation and their possible roles
in regulating the response to endotoxin shock. J Immunol.
179:5082–5089. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Curtale G, Mirolo M, Renzi TA, Rossato M,
Bazzoni F and Locati M: Negative regulation of Toll-like receptor 4
signaling by IL-10-dependent microRNA-146b. Proc Natl Acad Sci USA.
110:11499–11504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ceppi M, Pereira PM, Dunand-Sauthier I,
Barras E, Reith W, Santos MA and Pierre P: MicroRNA-155 modulates
the interleukin-1 signaling pathway in activated human
monocyte-derived dendritic cells. Proc Natl Acad Sci USA.
106:2735–2740. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Hwa Cho H, Bae YC and Jung JS: Role of
toll-like receptors on human adipose-derived cells. Stem Cells.
24:2744–2752. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pevsner-Fischer M, Morad V, Cohen-Sfady M,
Rousso-Noori L, Zanin-Zhorov A, Cohen S, Cohen IR and Zipori D:
Toll-like receptors and their ligands control mesenchymal stem cell
functions. Blood. 109:1422–1432. 2007. View Article : Google Scholar
|
|
21
|
Liotta F, Angeli R, Cosmi L, Filì L,
Manuelli C, Frosali F, Mazzinghi B, Maggi L, Pasini A, Lisi V, et
al: Toll-like receptors 3 and 4 are expressed by human bone
marrow-derived mesenchymal stem cells and can inhibit their T cell
modulatory activity by 634 impairing Notch signaling. Stem Cells.
26:279–289. 2008. View Article : Google Scholar
|
|
22
|
Opitz CA, Litzenburger UM, Lutz C, Lanz
TV, Tritschler I, Köppel A, Tolosa E, Hoberg M, Anderl J, Aicher
WK, et al: Toll-like receptor engagement enhances the
immunosuppressive properties of human bone marrow-derived
mesenchymal stem cells by inducing indoleamine-2,3-dioxygenase-1
via interferon-beta and protein kinase R. Stem Cells. 27:909–919.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tomchuck SL, Zwezdaryk KJ, Coffelt SB,
Waterman RS, Danka ES and Scandurro AB: Toll-like receptors on
human mesenchymal stem cells drive their migration and
immunomodulating responses. Stem Cells. 26:99–107. 2008. View Article : Google Scholar
|
|
24
|
Romieu-Mourez R, François M, Boivin MN,
Bouchentouf M, Spaner DE and Galipeau J: Cytokine modulation of TLR
expression and activation in mesenchymal stromal cells leads to a
proinflammatory phenotype. J Immunol. 182:7963–7973. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
White NM, Bao TT, Grigull J, Youssef YM,
Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R, et
al: miRNA profiling for clear cell renal cell carcinoma: Biomarker
discovery and identification of potential controls and consequences
of miRNA dysregulation. J Urol. 186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma J, Liu J, Wang Z, Gu X, Fan Y, Zhang W,
Xu L, Zhang J and Cai D: NF-kappaB-dependent MicroRNA-425
upregulation promotes gastric cancer cell growth by targeting PTEN
upon IL-1β induction. Mol Cancer. 13:402014. View Article : Google Scholar
|
|
27
|
Laine SK, Alm JJ, Virtanen SP, Aro HT and
Laitala-Leinonen TK: MicroRNAs miR-96, miR-124 and miR-199a
regulate gene expression in human bone marrow-derived mesenchymal
stem cells. J Cell Biochem. 113:2687–2695. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen J, Liu Z and Yang Y: In vitro
screening of LPS-induced miRNAs in leukocytes derived from cord
blood and their possible roles in regulating TLR signals. Pediatr
Res. 75:595–602. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Akiyama H: Control of chondrogenesis by
the transcription factor Sox9. Mod Rheumatol. 18:213–219. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin EA, Kong L, Bai XH, Luan Y and Liu CJ:
miR-199a, a bone morphogenic protein 2-responsive MicroRNA,
regulates chondrogenesis via direct targeting to Smad1. J Biol
Chem. 284:11326–11335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu ZY, Lu L, Liang J, Guo XR, Zhang PH and
Luo SJ: Keloid microRNA expression analysis and the influence of
miR-199a-5p on the proliferation of keloid fibroblasts. Genet Mol
Res. 13:2727–2738. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shi XE, Li YF, Jia L, Ji HL, Song ZY,
Cheng J, Wu GF, Song CC, Zhang QL, Zhu JY and Yang GS:
MicroRNA-199a-5p affects porcine preadipocyte proliferation and
differentiation. Int J Mol Sci. 15:8526–8538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Feng Y, Cao JH, Li XY and Zhao SH:
Inhibition of miR-214 expression represses proliferation and
differentiation of C2C12 myoblasts. Cell Biochem Funct. 29:378–383.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer Cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang ZC, Li YY, Wang HY, Fu S, Wang XP,
Zeng MS, Zeng YX and Shao JY: Knockdown of miR-214 promotes
apoptosis and inhibits cell proliferation in nasopharyngeal
carcinoma. PLoS One. 9:e861492014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Derfoul A, Juan AH, Difilippantonio MJ,
Palanisamy N, Ried T and Sartorelli V: Decreased microRNA-214
levels in breast cancer cells coincides with increased cell
proliferation, invasion and accumulation of the Polycomb Ezh2
methyltransferase. Carcinogenesis. 32:1607–1614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Brzezianska E and Pastuszak-Lewandoska D:
A minireview: The role of MAPK/ERK and PI3K/Akt pathways in thyroid
follicular cell-derived neoplasm. Front Biosci (Landmark Ed).
16:422–439. 2011. View
Article : Google Scholar
|
|
39
|
Nahid MA, Yao B, Dominguez-Gutierrez PR,
Kesavalu L, Satoh M and Chan EK: Regulation of TLR2-mediated
tolerance and cross-tolerance through IRAK4 modulation by miR-132
and miR-212. J Immunol. 190:1250–1263. 2013. View Article : Google Scholar :
|
|
40
|
Ahmed F, Shiraishi T, Vessella RL and
Kulkarni P: Tumor necrosis factor receptor associated factor-4: An
adapter protein overexpressed in metastatic prostate cancer is
regulated by microRNA-29a. Oncol Rep. 30:2963–2968. 2013.PubMed/NCBI
|
|
41
|
Chen R, Alvero AB, Silasi DA, Kelly MG,
Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T and Mor G:
Regulation of IKKB by miR-199a affects NF-kappB activity in ovarian
cancer cells. Oncogene. 27:4712–4723. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhou R, O'Hara SP and Chen XM: MicroRNA
regulation of innate immune responsesin epithelial cells. Cell Mol
Immunol. 8:371–379. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang Z, Chen X, Yu B and Chen D: Cloning
and functional characterization of rat stimulator of interferon
genes (STING) regulated by miR-24. Dev Comp Immunol. 37:414–420.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lai NS, Yu HC, Chen HC, Yu CL, Huang HB
and Lu MC: Aberrant expression of microRNAs in T cells from
patients with ankylosing spondylitis contributes to the
immunopathogenesis. Clin Exp Immunol. 173:47–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhao J, Gong AY, Zhou R, Liu J, Eischeid
AN and Chen XM: Downregulation of PCAF by miR-181a/b provides
feedback regulation to TNF-α-induced transcription of
proinflammatory genes in liver epithelial cells. J Immunol.
188:1266–1274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sharma A, Kumar M, Aich J, Hariharan M,
Brahmachari SK, Agrawal A and Ghosh B: Posttranscriptional
regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl
Acad Sci USA. 106:5761–5766. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang M, Liu Q, Mi S, Liang X, Zhang Z, Su
X, Liu J, Chen Y, Wang M, Zhang Y, et al: Both miR-17-5p and
miR-20a alleviate suppressive potential of myeloid-derived
suppressor cells by modulating STAT3 expression. J Immunol.
186:4716–4724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Witwer KW, Sisk JM, Gama L and Clements
JE: MicroRNA regulation of IFN-beta protein expression: Rapid and
sensitive modulation of the innate immune response. J Immunol.
184:2369–2376. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lai L, Song Y, Liu Y, Chen Q, Han Q, Chen
W, Pan T, Zhang Y, Cao X and Wang Q: MicroRNA-92a negatively
regulates Toll-like receptor (TLR)-triggered inflammatory response
in macrophages by targeting MKK4 kinase. J Biol Chem.
288:7956–7967. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Qi J, Qiao Y, Wang P, Li S, Zhao W and Gao
C: MicroRNA-210 negatively regulates LPS-induced production of
proinflammatory cytokines by targeting NF-κB1 in murine
macrophages. FEBS Lett. 586:1201–1207. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jennewein C, von Knethen A, Schmid T and
Brüne B: MicroRNA-27b contributes to lipopolysaccharide-mediated
peroxisome proliferator-activated receptor gamma (PPARgamma) mRNA
destabilization. J Biol Chem. 285:11846–11853. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lu TX, Munitz A and Rothenberg ME:
MicroRNA-21 is up-regulated in allergic airway inflammation and
regulates IL-12p35 expression. J Immunol. 182:4994–5002. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhu QY, Liu Q, Chen JX, Lan K and Ge BX:
MicroRNA-101 targets MAPK phosphatase-1 to regulate the activation
of MAPKs in macrophages. J Immunol. 185:7435–7442. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liu X, Zhan Z, Xu L, Ma F, Li D, Guo Z, Li
N and Cao X: MicroRNA-148/152 impair innate response and antigen
presentation of TLR-triggered dendritic cells by targeting CaMKIIα.
J Immunol. 185:7244–7251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ma F, Chen D, Chi Y, Chen F, Li X and Han
Z: The expression and role of miR-301a in human umbilical
cord-derived mesenchymal stromal cells. Cytotherapy. 15:1511–1516.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Akira S, Uematsu S and Takeuchi O:
Pathogen recognition and innate immunity. Cell. 124:783–801. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ozinsky A, Underhill DM, Fontenot JD,
Hajjar AM, Smith KD, Wilson CB, Schroeder L and Aderem A: The
repertoire for pattern recognition of pathogens by the innate
immune system is defined by cooperation between toll-like
receptors. Proc Natl Acad Sci USA. 97:13766–13771. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Pasare C and Medzhitov R: Toll-like
receptors: Linking innate and adaptive immunity. Adv Exp Med Biol.
560:11–18. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hoebe K, Jiang Z, Georgel P, Tabeta K,
Janssen E, Du X and Beutler B: TLR signaling pathways:
Opportunities for activation and blockade in pursuit of therapy.
Curr Pharm Des. 12:4123–4134. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kawai T and Akira S: TLR signaling. Semin
Immunol. 19:24–32. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
O'Neill LA and Bowie AG: The family of
five: TIR-domain-containing adaptors in Toll-like receptor
signalling. Nat Rev Immunol. 7:353–364. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
DelaRosa O and Lombardo E: Modulation of
adult mesenchymal stem cells activity by toll-like receptors:
Implications on therapeutic potential. Mediators Inflamm.
2010:8656012010. View Article : Google Scholar : PubMed/NCBI
|