Introduction
Dengue virus (DENV), a plus-strand RNA virus with an
enveloped icosahedral nucleocapsid, can be transmitted through
mosquito vectors, and is the causative agent of dengue fever,
dengue hemorrhagic fever and dengue shock syndrome (1). Dengue hemorrhagic fever and dengue
shock syndrome are potentially life threatening, and the risk of
developing these diseases is correlated with infection by one of
the four DENV serotypes (DENV1-4) and the carrying of antibodies to
another DENV serotype from a previous infection (2). Currently, no specific treatment or
vaccine for DENV is available (3).
There has been a significant increase in the number
of reports of DENV-associated infections and DENV-associated
mortality (1). Notably, over the
last 50 years, the incidence of DENV-associated infections has
increased by 30-fold, and the World Health Organization estimate
that there are currently 50,000,000 cases per annum worldwide
(3). Infections due to DENV are
now of serious concern worldwide, particularly in subtropical
areas, including Southeast Asia (4). DENV was first isolated by Kimura and
Hotta from blood samples obtained during the 1943 dengue epidemic,
which was predominantly confined to the Japanese port cities of
Nagasaki, Kobe and Osaka (5). On
the 28th August 2014, the Ministry of Health, Labour and Welfare in
Japan reported the country's first domestically acquired case of
dengue fever for almost 70 years (6). Therefore, the implementation of
procedures to monitor for any further potential outbreak of dengue
fever in the country are required.
Consequently, the development of effective methods
for the surveillance of DENV are urgently required. One approach to
achieve sensitive detection of DENV is to establish a method to
concentrate the viral particles. Several approaches to concentrate
virus have been suggested, including ultracentrifugation and
polyethylene glycol (PEG)-mediated precipitation. Although these
methods are applicable to a number of viruses, they have
significant practical limitations. Specifically,
ultracentrifugation is time-consuming and can increase the
false-positive rate when combined with polymerase chain reaction
(PCR) analysis (7). Although
PEG-mediated precipitation is simple and easy to perform, PEG
interferes with the subsequent PCR procedure (8,9). An
alternative to these conventional methods is the use of magnetic
beads coated with molecules, which efficiently bind to the virus,
which allows the capture and concentration of the viral particles
by applying a magnetic field. A potential approach to the capture
of a target virus is to use magnetic beads coated with an antibody
specific to the particular virus of interest.
Magnetic nanoparticles (MNPs), including iron,
nickel or cobalt, have been widely investigated for biomedical and
environmental applications due to their high specific surface area
and the ease of magnetic collection of target materials adsorbed by
the MNPs (10,11). However, a significant problem with
these beads is their inherent chemical instability, which can limit
their application in the field of biological and environmental
science (12). To overcome this
limitation, the MNPs are typically encapsulated with a protective
shell of graphite, silica or polymer (12).
Graphite-encapsulated MNPs (GrMNPs) are usually
hydrophobic, which is a limitation for several biomedical
applications. However, appropriate surface modification can improve
the properties of the GrMNPs, allowing them to efficiently
recognize and bind to molecular targets, including antibodies,
antigens and receptors (13).
Amino group functionalization, which is a desirable functionality
for graphite, improves the reactivity and hydrophilic nature of the
GrMNPs (12). A promising method
for the amino functionalization of GrMNPs is to use inductively
coupled radiofrequency (RF) plasma, which is environmentally
friendly and requires a short duration for the reaction to reach
completion. Using this approach, it is possible to introduce amino
groups effectively (14).
Furthermore, the degree of surface derivatization with amino groups
of GrMNPs can be optimized by altering the plasma discharge
conditions (15). With these
amino-modified GrMNPs, the efficient surface immobilization of
sugar chains has been demonstrated, including dextran, as well as
antibodies against several pathogens, including anti-influenza
virus and anti-Salmonella antibodies (12,16,17).
On the basis of this background, the present study
was performed to expand on previous results examining the influenza
virus to investigate DENV via the immobilization of anti-DENV
antibody onto the functionalized surface of GrMNPs. The modified
GrMNPs were then assessed for their ability to capture DENVs, and
the concentrated virus was then detected in combination with a
PCR-based amplification procedure.
Materials and methods
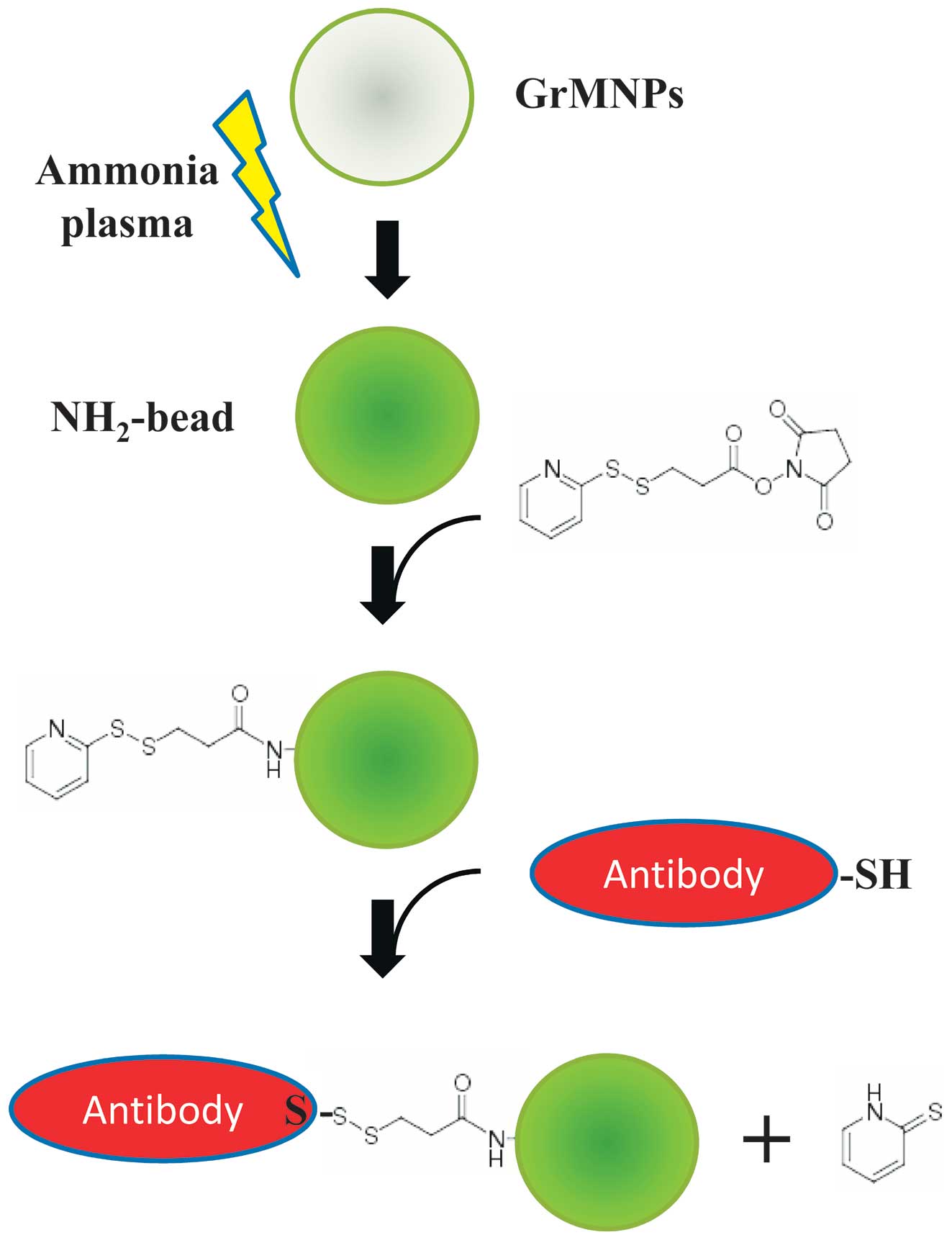
Plasma-functionalized GrMNPs and
production of antibody-integrated magnetic beads
The graphite-encapsulated iron compound
nanoparticles were prepared using an arc discharge method by
applying a 150–200 A direct current at ~20 V between an anode and
cathode, as described previously (15). A graphite electrode, molded using
graphibond-551R with Fe2O3 powder, was used
as the anode. On the opposite side, a graphite rod (50 mm•Ø10 mm;
99.9%) was used as the cathode. The resulting graphite-encapsulated
iron compound nanoparticles were then exposed to plasma, which was
produced using an RF power supply (18,19)
in an atmosphere containing ammonia at 13.56 MHz and 80 W via a
matching network (18,19). Initial pretreatment was performed
for 10 min using Ar plasma, followed by 2 min of ammonia plasma
post-treatment for amino group introduction. During the
experiments, the gas pressure was maintained at 50 Pa. The amino
groups on the surface of the magnetic beads were then further
labelled with 0.3 µM of the coupling agent,
N-succinimidyl 3-(2-pyridyldithio) propionate (SPDP; Dojindo
Laboratories, Kumamoto, Japan) at pH 7–8. A human mono-clonal
antibody (clone no. D23-1G7C2) recognizing the first domain II
fusion region of the DENV envelope glycoprotein (E) (20) was reduced using dithiothreitol
(DTT), resulting in breakage of the S-S bonds and generation of S-H
groups. The D23-1G7C2 antibody was produced from hybridomas using
peripheral blood mononuclear cells from patients in the acute phase
of dengue fever 5 days following the onset of illness, and exhibits
neutralizing activity against DENV1-4 (20). The S-H groups on the antibody were
then reacted with the SPDP-NH2-magnetic beads, resulting
in covalent crosslinking of the antibody onto the surface of the
beads. The resulting magnetic beads were termed antibody-integrated
magnetic beads (Fig. 1).
Cell culture and virus
A C6/36 cell culture (American Type Culture
Collection, Manassas, VA, USA), derived from Aedes
albopictus, was maintained in Leibovitz L15 medium (Thermo
Fisher Scientific, Inc., Waltham, MA, USA) containing 0.3% tryptose
phosphate broth (TPB) and 10% fetal calf serum (FCS; Wako Pure
Chemical Industries, Ltd., Osaka, Japan). The laboratory DENV
strains (21), DENV1 (Mochizuki
strain), DENV2 (16681 strain), DENV3 (80-2 strain) and DENV4 (H241
strain), were used to infect the C6/36 cell cultures. The C6/36
cells were cultured to ~80% confluence, then infected with the
DENVs at a multiplicity of infection of 0.1 in Leibovitz L15 medium
containing 0.3% TPB and 2% FCS, and were incubated for 3 days at
28°C. The medium was then collected and used for viral capture
experiments.
DENV capture
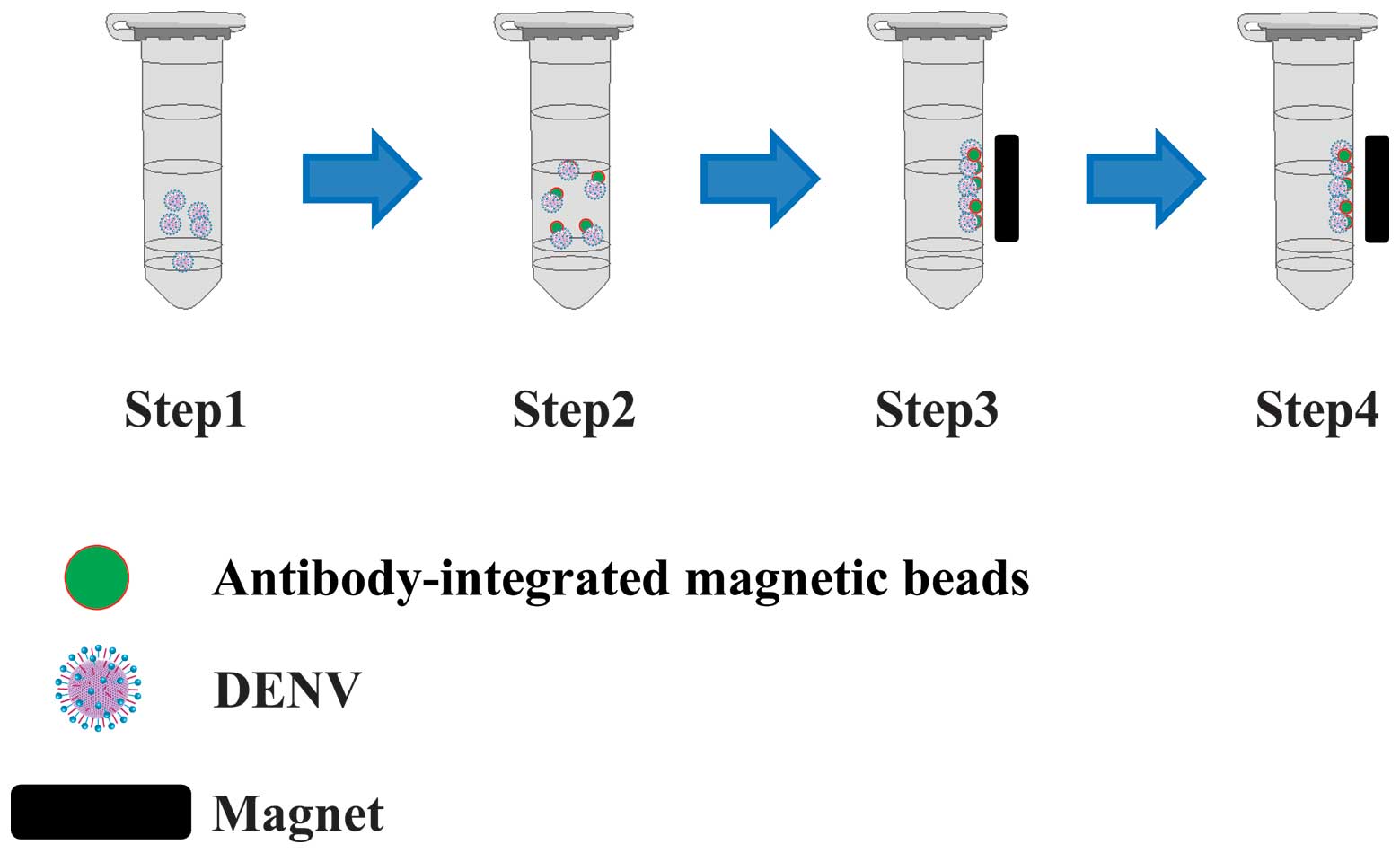
The capture of DENV1-4 was performed as follows.
Briefly, 10 µl of the magnetic beads were washed twice with
phosphate-buffered saline (PBS). A 10 µl sample of medium
from uninfected (Mock) or DENV-infected cell cultures were added to
the washed beads with 1 ml PBS, and the tube was incubated for 15
min at room temperature. The tubes containing the mixtures were
then set in a magnetic field for 5 min using an Adem-Mag SV
magnetic device (Ademtech, Pessac, France). Following magnetic
separation, the supernatant was removed, and the beads were washed
three times with PBS and resuspended in 10 µl PBS. This
procedure produced two fractions: Bead fraction (BD) and
supernatant fraction (SP). A 10 µl sample of each fraction
was used for reverse transcription (RT)-PCR analysis.
RT-PCR analysis
The DENV genomic RNA from each fraction was
extracted using a QIAamp Viral RNA Mini kit (Qiagen, Hilden,
Germany), according to the manufacturer's protocol. The RT
reactions were performed using a PrimeScriptII First UStrand cDNA
synthesis kit (Takara Bio, Inc., Otsu, Japan) with random primers.
Following incubation at 65°C for 5 min, the viral RNA was reverse
transcribed at 42°C for 60 min. The resultant cDNA (2 µl)
was amplified by PCR, in a reaction mixture (20 µl)
containing primers (0.2 µl each, 100 pmol/µl),
MgCl2 (1.6 µl, 25 mM), dNTP mixture (1.6
µl, 2.5 mM each), Ex Taq (0.1 µl, 5 U/µl;
Takara Bio, Inc.) and 10X Ex Taq buffer (2 µl) under
conditions of 35 cycles at 94°C for 30 sec, 55°C for 30 sec and
72°C for 1 min. The primers used were as follows: D1, forward
5′-tcaatatgctgaaacgcgggagaaaccg-3′ and D2, reverse
5′-ttgcaccaacagtcaatgtcttcaggttc-3′ (22). The amplified DNA fragments were
purified and analyzed by DNA sequencing on an ABI PRISM3100 genetic
analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.) to
verify the identity of the amplified product. Then, the product
sequences were compared with the sequences in the Genbank database
(ncbi.nlm.nih.gov/genbank/).
Amino group detection using
fluorescamine
Fluorescamine reacts with amino groups and forms
blue-green fluorescent derivatives. In the present study,
fluorescence intensity increased in a dose-dependent manner with
bovine serum albumin (BSA) concentrations following incubation with
fluorescamine (Wako Pure Chemical Industries, Ltd.) at an
excitation of 400 nm and emission of 490 nm. The amino groups on
the GrMNPs treated with ammonia plasma were measured using a
fluorometric method with fluorescamine, in accordance with a
previous study with modification (23). Briefly, 5 µl of the
plasma-treated nanoparticles (2 mg/ml) were solubilized with 100
µl 50 mM borate buffer, following which 4 µl 0.075%
fluorescamine in acetonitrile was added to each sample. The
fluorescence, at an emission of 490 nm and excitation of 400 nm,
was measured using a SpectraMax M2e HK fluorometer (Molecular
Devices, Sunnyvale, CA, USA).
Amino group detection using
2,4,6-trinitrobenzenesulfonic acid (TNBS)
TNBS reacts with amino groups and shows an increase
in absorbance at ~350 nm (24).
When 10 mg/ml BSA was reacted with TNBS (Wako Pure Chemical
Industries, Ltd.), the maximal peak of the spectra was 345 nm. In
addition, a dose-dependent increase of the corresponding peak in
BSA was observed following the TNBS reaction at 345 nm. The amino
groups on the GrMNPs treated with ammonia plasma were measured
using a colorimetric method using TNBS in accordance with a
previous study with modification (24). Briefly, 20 µl of the
plasma-treated GrMNPs (2 mg/ml) in 50 mM borate buffer was
incubated for 16 h at 4°C following the addition of 0.5% TNBS.
Subsequently, 250 µl of 5% sodium dodecyl sulfate and 2 N
HCl were added, and the absorbance at 345 nm was measured using a
spectrophotometer (UVmini-1240; Shimadzu Corporation, Kyoto,
Japan).
RT-PCR for DENV
Viral genomic RNA was extracted from the samples
prior to and following magnetic separation using a QIAamp Viral RNA
mini kit (Qiagen). The RNA was reverse transcribed using a
PrimeScriptII first strand cDNA synthesis kit (Takara Bio, Inc.)
using the following temperature regime: 65°C for 5 min, 4°C for 5
min and 42°C for 60 min. The resulting cDNAs were subjected to PCR
using SYBR Premix Ex TaqII (Tli RNase H Plus; Takara Bio, Inc.).
The primers for DENV were as follows: forward
5′-ct(a,t)tcaatatgctgaaacgcg-3′ and reverse
5′-tctatcca(g,a)aat(t,c)cctgctgtt-3′ (22). The temperature cycling conditions
used for PCR were as follows: 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec and 60°C for 1 min.
Statistical analysis
Statistical analyses were performed using Prism 4
software (GraphPad Software, Inc., San Diego, CA, USA). The TNBS
and fluorescamine data were subjected to a non-repeated measures
analysis of variance followed by the Bonferroni correction test.
The PCR data were subjected to Student's t-test (unpaired).
P<0.05 is considered to indicate a statistically significantly
difference.
Results
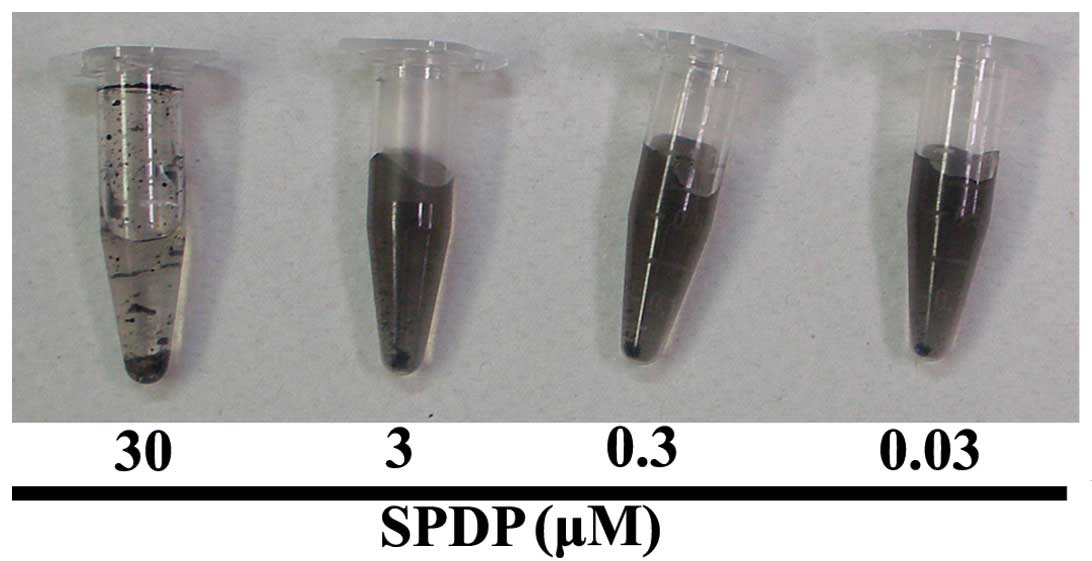
In order to analyze the hydrophilicity of the
ammonia plasma-treated GrMNPs, the resultant NH2-beads
were incubated with various concentrations of SPDP (Fig. 2). The results showed that the beads
aggregated following incubation with 30 µM SPDP, whereas the
beads incubated with 0.03, 0.3 and 3 µM SPDP remained in
suspension. These observations suggested that the ammonia
plasma-treated GrMNPs possessed hydrophilic properties, possibly
due to the introduction of amino groups to the bead surface.
To further confirm the presence of amino groups on
the surface of the beads, the ammonia plasma-treated GrMNPs were
incubated with TNBS or fluorescamine, which react with an amine
functionality (Tables I and
II). BSA, which has a surface
exposed amino groups, was also reacted with TNBS or fluorescamine.
As expected, an increase in the absorbance of TNBS at 345 nm and in
the fluorescent intensity of fluorescamine (excitation 400 nm;
emission 490 nm) was observed following incubation of BSA with the
respective reagents. Subsequently, the ammonia plasma-treated
GrMNPS were exposed to either TNBS or fluorescamine. The ammonia
plasma-treated GrMNPs exhibited increased absorbance at 345 nm and
increased fluorescent intensity (excitation 400 nm; emission 490
nm) following incubation with TNBS and fluorescamine, respectively.
Following incubation with TNBS, the samples were centrifuged for 5
min at 20,000 × g and room temperature, and the resultant
supernatant also showed increased absorbance at 345 nm, indicating
that the increased absorbance was not due to turbidity. Taken
together, these results suggested that the ammonia plasma-treated
GrMNPs possessed surface-exposed amino groups.
 | Table IConfirmation of amino groups on the
surface of ammonia plasma-treated GrMNPs by reaction with TNBS. |
Table I
Confirmation of amino groups on the
surface of ammonia plasma-treated GrMNPs by reaction with TNBS.
| Treatment | Absorbance at 345
nm |
|---|
| Buffer + TNBS | 0.2973±0.002 |
|
Ammonia-plasma-treated GrMNPs + TNBS |
0.8153±0.004a |
| Supernatant of
ammonia-plasma-treated GrMNPs + TNBS (post-centrifugation) |
0.6833±0.003a |
| BSA (0 mg/ml) | 0.295 |
| BSA (0.1
mg/ml) | 0.327 |
| BSA (1 mg/ml) | 0.499 |
| BSA (10 mg/ml) | 1.745 |
 | Table IIConfirmation of amino groups on the
surface of ammonia plasma-treated GrMNPs by reaction with
fluorescamine. |
Table II
Confirmation of amino groups on the
surface of ammonia plasma-treated GrMNPs by reaction with
fluorescamine.
| Treatment | Fluorescence
(excitation 400 nm; emission 490 nm) |
|---|
|
Ammonia-plasma-treated GrMNPs | 112.49±7.71a |
| Antibody-integrated
magnetic beads | 101.00±7.29a |
| BSA (0 mg/ml) | 84.23±2.55 |
| BSA (0.1
mg/ml) |
178.28±15.88a |
| BSA (1 mg/ml) | 862.58±5.70a |
Subsequently, the present study examined the
functionality of the resultant antibody-integrated magnetic beads
by incubating them with a suspension of DENV prior to separation of
the beads by applying a magnetic field to confirm capture of the
viral particles (Fig. 3).
Specifically, media from C6/36 mosquito cells infected with DENV1-4
were diluted with PBS and mixed with the magnetic beads. The
mixture was then magnetically separated into the BD and supernatant
SP fractions (Fig. 4). Medium from
mock-infected cells was used to prepare the control fractions.
Finally, the fractions were analyzed using RT-PCR to determine the
extent of DENV capture by the beads.
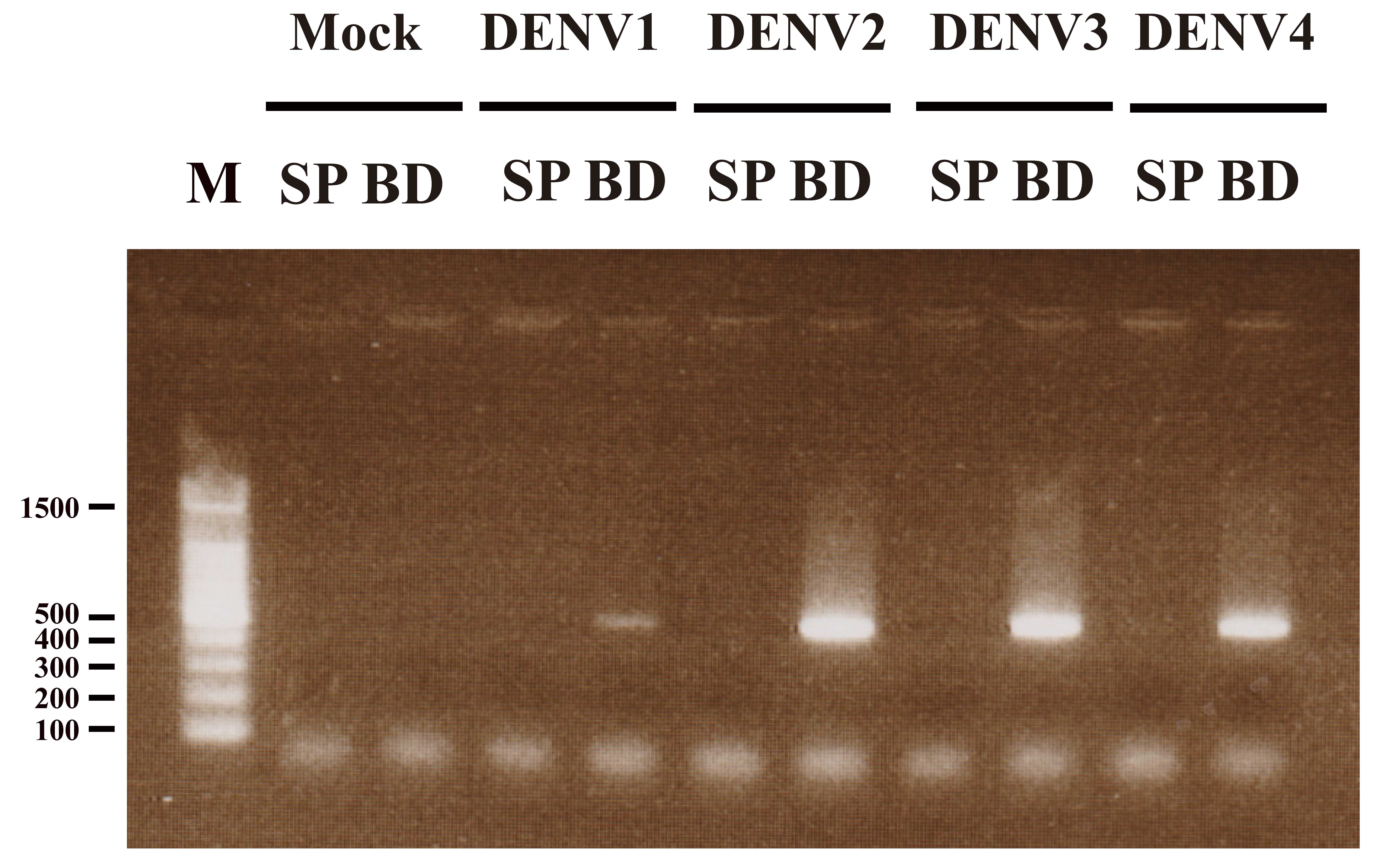
Using primers specific for a 511 bp section of DENV
RNA, RT-PCR was used to amplify a product of the expected size in
the BD fraction, but not in the SP fraction in DENV1-4 (Fig. 4). Two independent DNA sequence
analyses were performed to confirm the identity of the 511 bp band.
In each case, within the sequenced region, the following results
were obtained using the Genbank database: DENV1 (Mochizuki strain)
exhibited 98 and 99% sequence identity to Genbank accession no.
AB074760; DENV2 (16681 strain) exhibited 98 and 99% sequence
identity to Genbank accession no. U87411; DENV3 (80-2 strain)
exhibited 95 and 95% sequence identity to Genbank accession no.
AF317645; and DENV4 (H241 strain) exhibited 99 and 99% sequence
identity to Genbank accession no. AY947539.
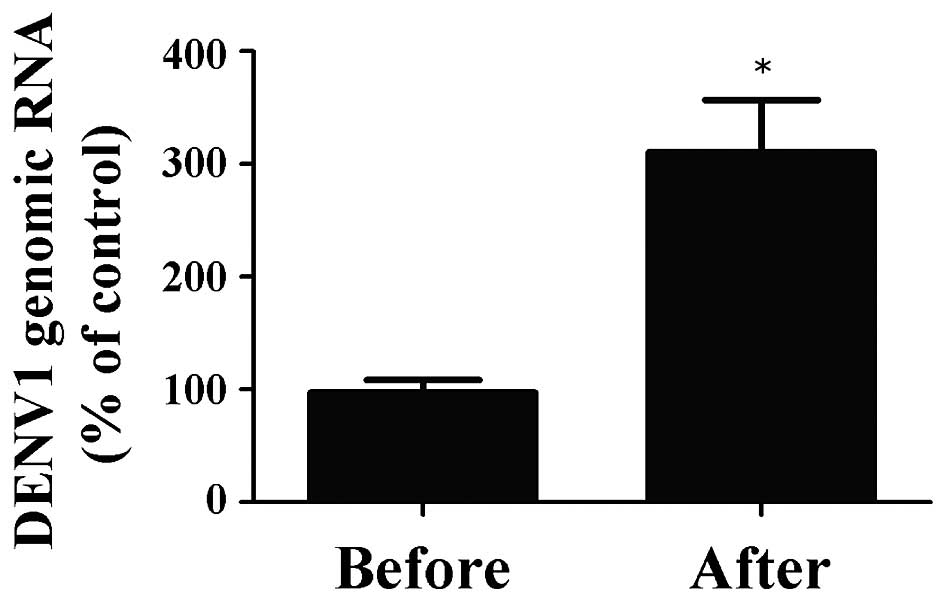
Subsequently, viral RNA was extracted, transcribed
and subjected to RT-PCR using specific forward and reverse primers
for DENV in order to investigate the quantity of DENV genomic RNA
prior to and following magnetic separation (Fig. 5). The percentage of genomic RNA in
DENV1 recovered following magnetic separation by the
antibody-integrated magnetic beads was 310.7±46.0%, compared with
the control. Of note, this sample was the same as the BD fraction.
By contrast, the percentage in the sample prior to incubation with
the beads was 97.3±11.2%, compared with the control. These findings
suggested that the antibody-integrated magnetic beads concentrated
the DENV1 by 3.2-fold.
The above results confirmed that the BD fraction
contained the corresponding DENV genomic RNAs, confirming that the
beads had successfully captured and concentrated the DENV
particles.
Discussion
Our previous studies showed that influenza virus, an
enveloped RNA virus, and Salmonella enterica, a pathogenic
bacterium associated with food poisoning can be efficiently
captured using amino modified GrMNPs, generated by an ammonia gas
plasma mediated strategy, coated with the corresponding antibody
(16,17). Therefore, the generic strategy of
using ammonia gas plasma to introduce amino groups into GrMNP, and
then covering the beads with antibodies directed against a specific
bacteria, virus or other pathogen, facilitates their efficient
capture from liquid samples. The targeted pathogen is then
pre-concentrated using immunomagnetic separation, enabling their
identification using a suitable detection procedure, for example
PCR. This method offers potential as an effective monitoring tool
for emerging viruses and other pathogens by facilitating their
rapid and sensitive detection. Therefore, the method described in
the present study contributed to controlling current and future
global infectious threats in the areas of food, medical and
environmental science.
The efficiency of introducing amino groups onto the
bead surface using the ammonia gas plasma technique may be
increased by carefully adjusting the treatment conditions. Although
the mechanism underlying the attachment of amino groups via plasma
treatment remains to be fully elucidated, its clarification may
further enhance the efficient introduction of these functional
units. For example, beads with a higher number of amino groups are
likely to adsorb a higher number of antibody molecules, resulting
in the enhanced capture and concentration of viruses. In addition,
the replacement of iron, which was used in the present study, with
alternative metals possessing stronger magnetic properties for the
fabrication of the beads may enhance their overall capture
efficiency. Furthermore, a combination of various antibodies and
beads may enable the multiplex detection of pathogens.
Following the initial emergence of DENV, its spread
is enhanced in a time-dependent manner. Although vaccination
strategies for DENV have not been established, it is likely that
infectious viruses are indispensable for future vaccine production.
Therefore, the viral isolation step is essential for vaccine
production. Early and efficient vaccine production is particularly
important during outbreaks of DENV-associated infections. Although
the early detection and isolation of DENV from mosquitoes is
crucial for preventing the potential spread of disease, progress
towards the development of methods for DENV detection and isolation
has been limited. Ultracentrifugation and PEG precipitation are
conventionally used to concentrate viruses (8,9).
However, these methods partially inactivate the viruses during the
concentration procedure and are unsuitable for the routine
monitoring of samples (8,9). The possibility of using magnetic
beads coated with bioadhesive molecules to concentrate viruses has
been suggested previously (25-42).
An example includes anionic magnetic beads coated with an anionic
polymer, poly (methyl vinyl ether-maleic anhydrate), termed poly
(MVE-MA), which can be used to concentrate DENV from infected
mosquito cells derived from patients with dengue fever (37). Notably, the magnetic capture of
other viruses using poly (MVE-MA) has been reported previously,
including human immunodeficiency virus (35), borna disease virus (38), respiratory syncytial virus
(39), influenza virus (40,41)
and adenovirus (42). The most
important aspects of this method are its simplicity and rapidity
(<30 min). Furthermore, the applicability of antibody-integrated
magnetic beads to the broad serotypes of DENV1-4 is another
promising feature of this approach. Therefore, the method for DENV
capture from solution, in combination with sensitive detection
methods, may contribute to preventing the spread of different
subtypes of DENV. In addition, the efficient capture of infectious
DENV may assist in the development of vaccines against dengue
fever. The magnetic bead-concentration method may facilitate the
isolation and sensitive detection of DENVs, and may contribute to
efficient surveillance and future vaccine production. The general
utility of the method described in the present study may be further
enhanced if it is found to be applicable to other emerging viruses
and bacterial pathogens.
In conclusion, the present study demonstrated that
antibody-integrated magnetic beads were useful for the capture of
DENVs. The capture of DENV1-4 using the antibody-integrated
magnetic beads was confirmed by the results of the RT-PCR analysis,
showing that the BD fraction contained DENV genomic RNA. Therefore,
this method may be used in combination with conventional PCR for
the detection of DENV, and may increase the sensitivity of viral
detection for the diagnosis of DENV.
Acknowledgments
This study was supported by a grant for the
Promotion of Basic Research Activities for Innovative Biosciences
from the Bio-oriented Technology Research Advancement Institution
(BRAIN), the Science and Technology Research Promotion Program for
Agriculture, Forestry, Fisheries and Food Industry, the Amano
Institute of Technology, and grants for Scientific Research on
Innovative Areas (grant nos. 21110010, 22110514 and 24110717) and
for Scientific Research (grant no. 25246029 and 16K04997) from the
Japan Society for the Promotion of Science.
References
|
1
|
World Health Organization: http://www.who.int/media-centre/factsheets/fs117/en/
Accessed July 8, 2015.
|
|
2
|
Morens DM: Antibody-dependent enhancement
of infection and the pathogenesis of viral disease. Clin Infect
Dis. 19:500–512. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
World Health Organization: Dengue
Hemorrhagic Fever: Diagnosis, Treatment and Control. 2nd edition.
World Health Organization; Geneva: 1997
|
|
4
|
Sakudo A, Onodera T, Shintani H and Ikuta
K: Dengue virus presence and surveillance in Okinawa (Review). Exp
Ther Med. 3:15–17. 2012.PubMed/NCBI
|
|
5
|
Hotta S: Experimental studies on dengue.
I. Isolation, identification and modification of the virus. J
Infect Dis. 90:1–9. 1952. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ministry of Health Labour and Welfare:
http://www.mhlw.go.jp/bunya/kenkou/kekkaku-kansenshou19/dengue_fever.html
Accessed July 8, 2015.
|
|
7
|
Roth WK, Weber M and Seifried E:
Feasibility and efficacy of routine PCR screening of blood
donations for hepatitis C virus, hepatitis B virus, and HIV-1 in a
blood-bank setting. Lancet. 353:359–363. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hamelin C and Lussier G: Concentration of
human cytomegalovirus from large volumes of tissue culture fluids.
J Gen Virol. 42:193–197. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Novotný J, Svobodová J, Ransnäs LA and
Kubistová K: A method for the preparation of purified antigens of
coxsackievirus B3 from a large volume of cell culture supernatant.
Acta Virol. 36:483–487. 1992.PubMed/NCBI
|
|
10
|
Safarikova M and Safarik I: The
application of magnetic techniques in biosciences. Magn Elect Sep.
10:223–252. 2001. View Article : Google Scholar
|
|
11
|
Pankhurst QA, Connolly J, Jones SK and
Dobson J: Applications of magnetic nanoparticles in biomedicine. J
Phys D Appl Phys. 36:R167–R181. 2003. View Article : Google Scholar
|
|
12
|
Saraswati TE, Ogino A and Nagatsu M:
Plasma-activated immobilization of biomolecules onto
graphite-encapsulated magnetic nanoparticles. Carbon. 50:1253–1261.
2012. View Article : Google Scholar
|
|
13
|
Poplawska M, Bystrzejewski M, Grudziński
IP, Cywińska MA, Ostapko J and Cieszanowski A: Immobilization of
gamma globulins and polyclonal antibodies of class IgG onto
carbon-encapsulated iron nanoparticles functionalized with various
surface linkers. Carbon. 74:180–194. 2014. View Article : Google Scholar
|
|
14
|
Saraswati TE, Matsuda T, Ogino A and
Nagatsu M: Surface modification of graphite encapsulated iron
nanoparticles by plasma processing. Diam Relat Mater. 20:359–363.
2011. View Article : Google Scholar
|
|
15
|
Saraswati TE, Tsumura S and Nagatsu M:
High-efficiency plasma surface modification of
graphite-encapsulated magnetic nanoparticles using a pulsed
particle explosion technique. Jpn J Appl Phys. 53:0102052014.
View Article : Google Scholar
|
|
16
|
Sakudo A, Chou H and Nagatsu M:
Antibody-integrated and functionalized graphite-encapsulated
magnetic beads, produced using ammonia gas plasma technology, for
capturing Salmonella. Bioorg Med Chem Lett. 25:1012–1016. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sakudo A, Chou H, Ikuta K and Nagatsu M:
Integration of antibody by surface functionalization of
graphite-encapsulated magnetic beads using ammonia gas plasma
technology for capturing influenza A virus. Bioorg Med Chem Lett.
25:1876–1879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagatsu M, Yoshida T, Mesko M, et al:
Narrow multi-walled carbon nanotubes produced by chemical vapor
deposition using graphene layer encapsulated catalytic metal
particles. Carbon. 44:3336–3341. 2006. View Article : Google Scholar
|
|
19
|
Saito Y, Yoshikawa T, Okuda M, et al: Iron
particles nesting in carbon cages grown by arc discharge. Chem Phys
Lett. 212:379–383. 1993. View Article : Google Scholar
|
|
20
|
Setthapramote C, Sasaki T, Puiprom O,
Limkittikul K, Pita ksajja kul P, Pipattanaboon C, Sasayama M,
Leuangwutiwong P, Phumratanaprapin W, Chamnachanan S, et al: Human
monoclonal antibodies to neutralize all dengue virus serotypes
using lymphocytes from patients at acute phase of the secondary
infection. Biochem Biophys Res Commun. 423:867–872. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Masrinoul P, Diata MO, Pambudi S,
Limkittikul K, Ikuta K and Kurosu T: Highly conserved region
141–168 of the NS1 protein is a new common epitope region of dengue
virus. Jpn J Infect Dis. 64:109–115. 2011.
|
|
22
|
Paudel D, Jarman R, Limkittikul K,
Klungthong C, Chamnanchanunt S, Nisalak A, Gibbons R and
Chokejindachai W: Comparison of real-time SYBR green dengue assay
with real-time taqman RT-PCR dengue assay and the conventional
nested PCR for diagnosis of primary and secondary dengue infection.
N Am J Med Sci. 3:478–485. 2011. View Article : Google Scholar
|
|
23
|
Chen X and Wang J: A sequential injection
fluorometric procedure for rapid determination of total protein in
human serum. Talanta. 69:681–685. 2006. View Article : Google Scholar
|
|
24
|
Plapp BV, Moore S and Stein WH: Activity
of bovine pancreatic deoxyribonuclease A with modified amino
groups. J Biol Chem. 246:939–945. 1971.PubMed/NCBI
|
|
25
|
Safaríková M and Safarík I: Immunomagnetic
separation of Escherichia coli O26, O111 and O157 from vegetables.
Lett Appl Microbiol. 33:36–39. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Safarík I, Safaríková M and Forsythe SJ:
The application of magnetic separations in applied microbiology. J
Appl Bacteriol. 78:575–585. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kobayashi S, Natori K, Takeda N and Sakae
K: Immunomagnetic capture rt-PCR for detection of norovirus from
foods implicated in a foodborne outbreak. Microbiol Immunol.
48:201–204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Clavet CR, Margolin AB and Regan PM:
Herpes simplex virus type-2 specific glycoprotein G-2
immunomagnetically captured from HEp-2 infected tissue culture
extracts. J Virol Methods. 119:121–128. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jothikumar N, Cliver DO and Mariam TW:
Immunomagnetic capture PCR for rapid concentration and detection of
hepatitis A virus from environmental samples. Appl Environ
Microbiol. 64:504–508. 1998.PubMed/NCBI
|
|
30
|
Satoh K, Iwata A, Murata M, Hikata M,
Hayakawa T and Yamaguchi T: Virus concentration using
polyethyleneimine-conjugated magnetic beads for improving the
sensitivity of nucleic acid amplification tests. J Virol Methods.
114:11–19. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Uchida E, Sato K, Iwata A, Ishii-Watabe A,
Mizuguchi H, Hikata M, Murata M, Yamaguchi T and Hayakawa T: An
improved method for detection of replication-competent retrovirus
in retrovirus vector products. Biologicals. 32:139–146. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Uchida E, Kogi M, Oshizawa T, Furuta B,
Satoh K, Iwata A, Murata M, Hikata M and Yamaguchi T: Optimization
of the virus concentration method using
polyethyleneimine-conjugated magnetic beads and its application to
the detection of human hepatitis A, B and C viruses. J Virol
Methods. 143:95–103. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iwata A, Satoh K, Murata M, Hikata M,
Hayakawa T and Yamaguchi T: Virus concentration using sulfonated
magnetic beads to improve sensitivity in nucleic acid amplification
tests. Biol Pharm Bull. 26:1065–1069. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hatano B, Kojima A, Sata T and Katano H:
Virus detection using Viro-Adembeads, a rapid capture system for
viruses and plaque assay in intentionally virus-contaminated
beverages. Jpn J Infect Dis. 63:52–54. 2010.PubMed/NCBI
|
|
35
|
Sakudo A and Ikuta K: A technique for
capturing broad subtypes and circulating recombinant forms of HIV-1
based on anionic polymer-coated magnetic beads. Int J Mol Med.
30:437–442. 2012.PubMed/NCBI
|
|
36
|
Sakudo A and Onodera T: Virus capture
using anionic polymer-coated magnetic beads. Int J Mol Med. 30:3–7.
2012.PubMed/NCBI
|
|
37
|
Sakudo A, Masrinoul P, Tanaka Y and Ikuta
K: Capture of dengue virus type 3 using anionic polymer-coated
magnetic beads. Int J Mol Med. 28:625–628. 2011.PubMed/NCBI
|
|
38
|
Sakudo A, Tanaka Y and Ikuta K: Capture of
infectious borna disease virus using anionic polymer-coated
magnetic beads. Neurosci Lett. 494:237–239. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sakudo A, Baba K, Tsukamoto M and Ikuta K:
Use of anionic polymer, poly(methyl vinyl ether-maleic
anhydride)-coated beads for capture of respiratory syncytial virus.
Bioorg Med Chem Lett. 19:4488–4491. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sakudo A, Baba K, Tsukamoto M, Sugimoto A,
Okada T, Kobayashi T, Kawashita N, Takagi T and Ikuta K: Anionic
polymer, poly(methyl vinyl ether-maleic anhydride)-coated
beads-based capture of human influenza A and B virus. Bioorg Med
Chem. 17:752–757. 2009. View Article : Google Scholar
|
|
41
|
Sakudo A and Ikuta K: Efficient capture of
infectious H5 avian influenza virus utilizing magnetic beads coated
with anionic polymer. Biochem Biophys Res Commun. 377:85–88. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sakudo A, Baba K and Ikuta K: Capturing
and concentrating adenovirus using magnetic anionic nanobeads. Int
J Nanomedicine. 11:1847–1857. 2016. View Article : Google Scholar : PubMed/NCBI
|