Introduction
The development of the mammalian kidney occurs via a
series of precisely controlled processes. Derived from a region of
the mesoderm, the kidney is gradually formed from the metanephric
mesenchymal (MM) cells and the ureteric bud (UB). Furthermore,
under the reciprocal induction of complex molecular regulatory
processes, the kidney with the entirety of its functions comes into
being (1–3). In the early development of the
kidney, Wilms' tumor suppressor 1 (WT1), a zinc-finger
transcription factor, is expressed in the MM cells, cap mesenchyme
(CM), and further during the course of nephron development. WT1
exerts an important role in maintaining the self-renewal of MM
cells, and the well-balanced development of the kidney (4–7).
Loss of WT1 results in embryonic lethality and hypoplasia of the
gonads, together with bilateral renal agenesis, which is
characterized by an incomplete formation of the UB and concurrent
apoptosis of the MM cells (7,8).
Therefore, these findings suggested that WT1 is intrinsically
required for the functioning of the MM cells, and is indispensable
for the completion of the kidney developmental program (9), and consequently, further research on
WT1 regulation is urgently required.
In addition to the regulation of coding genes, there
also exists non-coding RNA regulation. MicroRNAs (miRNAs) are a
class of endogenous non-coding RNAs that regulate gene expression
either through translational repression or via mRNA degradation by
binding to the 3′-untranslated region (3′-UTR) of the targeted
mRNAs (10,11). MiRNAs exert important roles in
development, proliferation, apoptosis and other biological
processes (12). Based on previous
studies, miRNA-dependent gene regulation serves an important role
in kidney development and disease (13). For example, miR-181 was
demonstrated to downregulate Six homeobox 2 (Six2) protein and
inhibit the proliferation of MM cells (14,15);
miR-135 mediated kidney podocyte injury through Wnt/β-catenin
signaling (16); miR-17 promoted
cell proliferation through polycystin-2 (PKD2) (17); and depletion of miR-150 suppressed
acute kidney injury induced by myocardial infarction (18). Of the miRNAs, miR-743a has rarely
been reported, and therefore its function has yet to be fully
elucidated. The present study therefore aimed to explore the
biological function of miR-743a in kidney development.
In the current study, miR-743a has been demonstrated
to directly target the 3′-UTR of WT1, and to downregulate the
expression of WT1 at the mRNA and the protein level. Furthermore,
miR-743a is revealed to inhibit the proliferation of MM cells.
These results have disclosed the functional effects of miR-743a in
kidney development, and opened up novel means for investigating
potential cures for kidney diseases in which WT1 exerts a
participatory role.
Materials and methods
Plasmid construction
The plasmids, pcDNA3.1-luciferase-WT1-3′-UTR-WT,
pcDNA3.1-luciferase-WT1-3′-UTR-MUT, pdsAAV-CB-EGFP-miR-743a and
pRL-SV40, were used in dual-luciferase assays, as described below.
The 3′-UTR of WT1, together with miR-743a, was cloned from C57BL/6
mouse genomic DNA using polymerase chain reaction (PCR). C57BL/6
mouse tail was digested with proteinase K at 55°C with vibration
overnight, and centrifuged at 13,800 × g for 5 min at room
temperature and then the supernatant was collected to obtain
genomic DNA. The mouse was obtained from the animal centre of
Chongqing Medical University (Chongqing, China), and the use of the
mouse was approved by the Animal Care and Use Committee of
Chongqing Medical University. The vector, pcDNA3.1(+)-WT1-CDS, was
obtained by PCR from the complementary DNA (cDNA) of mK3 cells (an
immortalized cell line of undifferentiated MM cells), and was
recombined at the restriction sites of BamHI and
EcoRI to construct pcDNA3.1 (+)-WT1-CDS. Details of the
primers used in the present study are provided in Table I.
 | Table I.Sequences of the primers used in
qPCR. |
Table I.
Sequences of the primers used in
qPCR.
| Gene | Primer name | Sense (5′→3′) | Antisense
(5′→3′) |
|---|
| mus.WT1 |
pcDNA3.1-luciferase-3′-UTR |
TACCGAGCTCGGATCCGCCATCACAACATGCATCAG |
GATATCTGCAGAATTCATCCACACAGTGATGCAGCT |
| mus.miR743a |
pdsAAV-CB-EGFP-miR-743a |
TTTCAGGTCCCGGATCATAGCCTACTGTAGGAATG |
TTGCACCACCACCGGAGGTAACAGATTAGGACTGG |
| mus.WT1 |
pcDNA3.1(+)-WT1-CDS |
ACCGAGCTCGGATCCGGTAAGGAGTTCAAGGCAGC |
CCCTCTAGACTCGAGATGCTGGACTGTCTCCGTGT |
| mus.WT1 |
pcDNA3.1-luciferase-3′-UTR-mutation |
TTGTTCTGATTTATTTTTTAGTTGTAATTAGGTACATCC |
AATAAATCAGAACAATTAAAAAAAGATCTCTGCTCTTAAAACAT |
| mus.WT1 | RT-qPCR |
CAGGATGTTCCCCAATGC |
GGTCCTCGTGTTTGAAGGAA |
| mus.miR743a | RT-qPCR |
TGCGAAAGACACCAAGCTG |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCTACTCA |
| mus.18s | RT-qPCR |
GTAACCCGTTGAACCCCATT |
CCATCCAATCGGTAGTAGCG |
| mus.U6 | RT-qPCR |
CTCGCTTCGGCAGCACA |
AACGCTTCACGAATTTGCGT |
Cell culture
293T cells, as well as mK3 cells, obtained from the
American Type Culture Collection (Manassas, VA, USA) were cultured
in Dulbecco's modified Eagle's medium [DMEM (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA)], with 10% FBS (Gemini 900–108)
and penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C with 5% CO2.
Transfection
The plasmids, pcDNA3.1-luciferase-WT1-3′-UTR-WT,
pcDNA3.1-luciferase-WT1-3′-UTR-MUT, pdsAAV-CB-EGFP-miR-743a and
pRL-SV40 were transfected in 293T cells via the calcium phosphate
transfection method. When 293T cells reached 50% confluence, 2.5 µl
2.5 M CaCl2 was mixed with 500 ng
pcDNA3.1-luciferase-WT1-3′-UTR-WT or 500 ng
pcDNA3.1-luciferase-WT1-3′-UTR-MUT and 500 ng pdsAAV-CB-EGFP or 500
ng pdsAAV-CB-EGFP-miR-743a together with 10 ng pRL-SV40, then
isovolumetric 2X Hank's balanced salt solution (16.3 g NaCl, 0.74 g
KCl, 0.214 g Na2HPO4, 2.4 g glucose, 10 g
HEPES, adjusted to pH 7.05) was added to the mixture above nad
inubated for 2 min at room temperature. The mixture was
subsequently added to the cells in 24-well plates in serum-free
medium. The plasmids, pcDNA3.1 (+)-WT1-CDS and miR-743a mimic or
control mimic (50 nM) (Guangzhou RiboBio Co., Ltd., Guangzhou,
China) were transfected into mK3 cells using
Lipofectamine®-2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Following transfection, the medium was replaced
with fresh complete medium after 6 h and after 48 h, luciferase
activity was measured using the Dual-Luciferase Reporter assay
system (Promega Corporation, Madison, WI, USA).
Dual-luciferase assays
Luciferase assays were performed following the
method described in a previous study (14). The plasmids,
pdsAAV-CB-EGFP-miR-743a and pRL-SV40, were co-transfected with
pcDNA3.1-luciferase or pcDNA3.1-luciferase-WT1-3′-UTR or
pcDNA3.1-luciferase-WT1-3′-UTR-mutant using the calcium phosphate
method. After 48 h, luciferase activity was measured using the
Dual-Luciferase® Reporter Assay system, following the
manufacturer's protocol.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total mK3 RNA was collected with TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The cDNA was
reverse-transcribed using the RevertAid First-Strand cDNA Synthesis
kit (cat. no. K1622; Thermo Fisher Scientific, Inc.), and RT-qPCR
experiments were performed using the SYBR Premix Ex Taq™ II kit
(Takara Biotechnology Co., Ltd., Dalian, China) and cycled at 95°C
for 10 min followed by 39 cycles of 95°C for 15 sec and 60°C for 1
min. The expression level of WT1 mRNA was normalized against the
internal control (18S), whereas the expression of miR-743a was
normalized against U6. The primers are provided in Table I. The expression levels were
normalized using the ΔΔCq method (19).
Western blot analysis
MiR-743a mimic (50 nM) or control mimic (50 nM) was
transfected into mK3 cells, and, after 48 h, the cells were rinsed
with 1X phosphate-buffered saline three times and subsequently
lysed with radioimmunoprecipitation assay (RIPA) lysis buffer
(Beyotime Institute of Biotechnology, Beijing, China). The cells
were incubated on ice for 15 min and then ruptured by
ultrasonication. The lysates were centrifuged at 2–8°C, 13,800 ×
g for 20 min, and subsequently boiled with 5X SDS loading
buffer at 95°C for 10 min. The proteins were separated using 12%
SDS-PAGE. The proteins were transferred to polyvinylidene fluoride
membrane and then blocked with 5% non-fat powdered milk in
TBS-Tween (1.21 g Tris, 5.84 g NaCl, adjusted to pH 7.4, in 1 L
ddH2O, with 0.1% Tween-20) at room temperature for 2 h.
The membranes were incubated with rabbit anti-WT1 (cat. no. sc-192;
1:1,000 dilution, Santa Cruz Biotechnology, Inc., Santa Cruz, CA,
USA) and mouse anti-β-actin (cat. no. HC201-02; 1:2,000 dilution,
Beijing Transgen Biotech Co., Ltd., Beijing, China) at 4°C
overnight then washed with TBS-Tween three times for 10 min. The
blots were incubated with peroxidase-conjugated Affinipure goat
anti-rabbit immunoglobulin G [(IgG), H+L, cat. no. 10285-1-AP;
1:5,000 dilution, ProteinTech Group, Inc., Chicago, IL, USA) for 2
h at room temperature. Images were detected using the ChemiDoc™
XRS+ system of Bio-Rad (Bio-Rad Laboratories, Inc., Hercules, CA,
USA).
Cell proliferation assay
At 36 h after transfection, the extent of
proliferation of the mK3 cells was determined on a glass slide
using a Cell-Light EdU Apollo 567 in vitro kit (Guangzhou
RiboBio Co., Ltd., Guangzhou, China), according to the
manufacturer's protocol. The visual fields were observed under a
fluorescence inverted microscope (Nikon, Japan). The ratio of red
cells (EdU-staining-positive cells) to the blue cells (total cells
labeled by DAPI) was identified as the proliferation rate.
Bioinformatic analysis
The evolutionary conservation of WT1 3′UTR was
obtained from The University of California Santa Cruz Genome
Browser. The potential miRNAs that target WT1 were predicted by the
cited prediction programs using miRWalk (www.microwalk.org), microRNA.org
and TargetScan (/www.targetscan.org).
Statistical analysis
All the experiments were performed in triplicate,
and the results and analyses are presented as the means ± standard
deviation and as a paired t-test. GraphPad Prism5 software
(GraphPad, San Diego, CA, USA) was used to analyze the data and to
construct the bar charts. P<0.05 was considered to indicate a
statistically significant difference.
Results
Bioinformatics analysis of WT1
3′-UTR
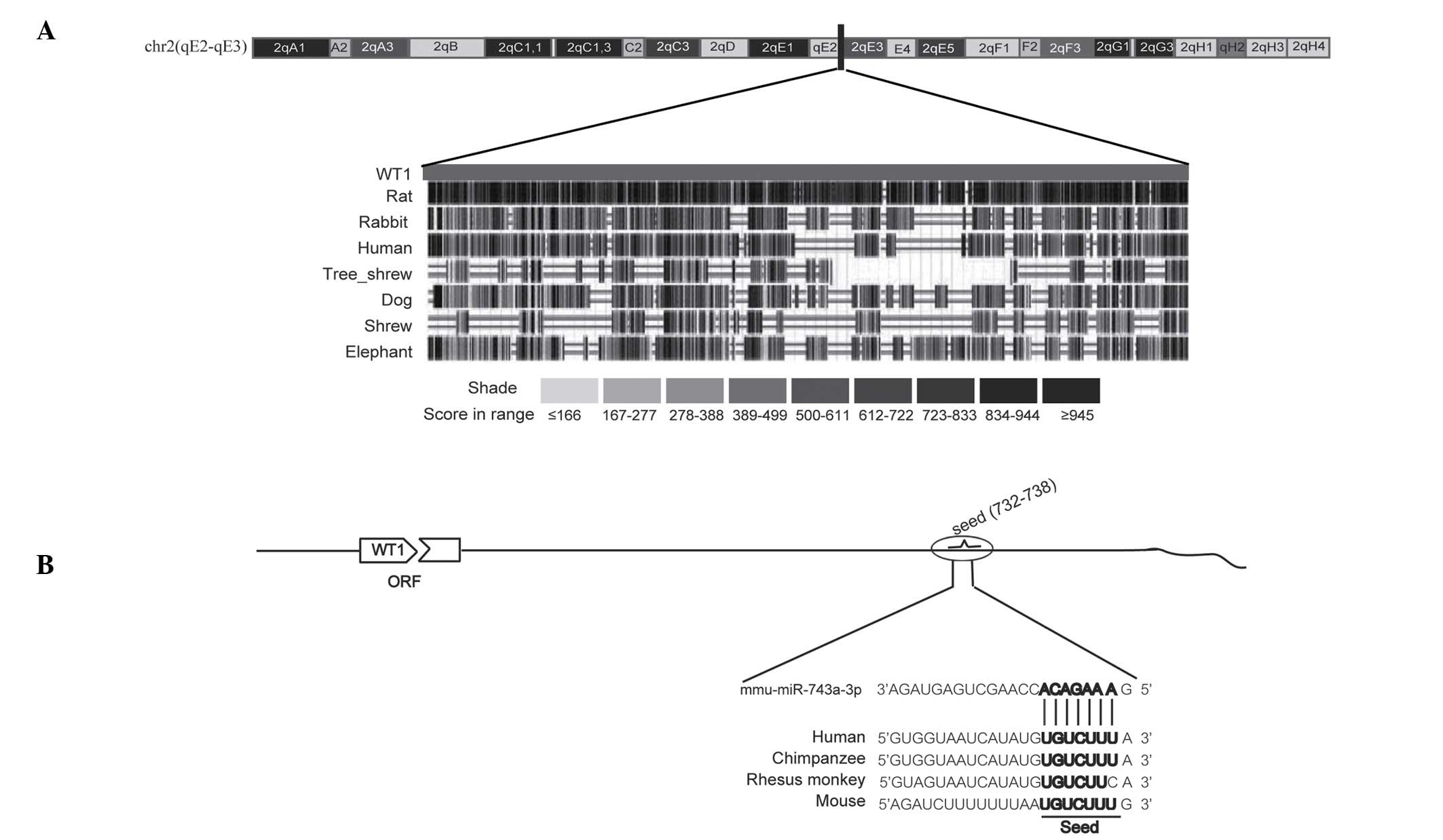
To analyze the conservation of the 3′-UTR of WT1, a
BLAST search was performed on the sequences across a variety of
species through the online bioinformatics site [The University of
California Santa Cruz Genome Browser]. As shown in Fig. 1A, the 3′-UTR of WT1 is reasonably
well conserved across different species, including rat, human,
rabbit and dog. Subsequently, the potential miRNAs were screened
using three online bioinformatics tools (miRWalk, microRNA.org and TargetScan). The preliminary results
revealed that miR-743a is likely to bind the 3′-UTR of WT1, and the
binding sites are also conserved to a certain extent (Fig. 1B). To the best of our knowledge, up
to this point, the role of miR-743a in, and its possible
association with, kidney development has rarely been reported.
These data suggested that the 3′-UTR of WT1 is conserved among
vertebrates to a certain extent, and that miR-743a is potentially
able to target it.
miR-743a directly targets the 3′-UTR
of WT1
To investigate whether miR-743a is able to directly
target the 3′-UTR of WT1, the plasmids,
pcDNA3.1-luciferase-WT1-3′-UTR and pdsAAV-CB-EGFP-miR-743a, were
constructed (Fig. 2A). The
plasmids, pcDNA3.1-luciferase-WT1-3′-UTR and
pdsAAV-CB-EGFP-miR-743a or pdsAAV-CB-EGFP were co-transfected into
293T cells. At the same time, the plasmid pRL-SV40 was included as
an internal reference. Compared with the control transfected with
pdsAAV-CB-EGFP, overexpression of miR-743a led to a downregulation
in relative luciferase activity (Fig.
2B). When the binding sites of miR-743a on the 3′-UTR of WT1
were scrambled (Fig. 2A), no
significant difference in the activity of luciferase was identified
following the transfection with miR-743a (Fig. 2B). These data indicated that
miR-743a is able to directly target the 3′-UTR of WT1, leading to a
decrease in the reporter gene activity.
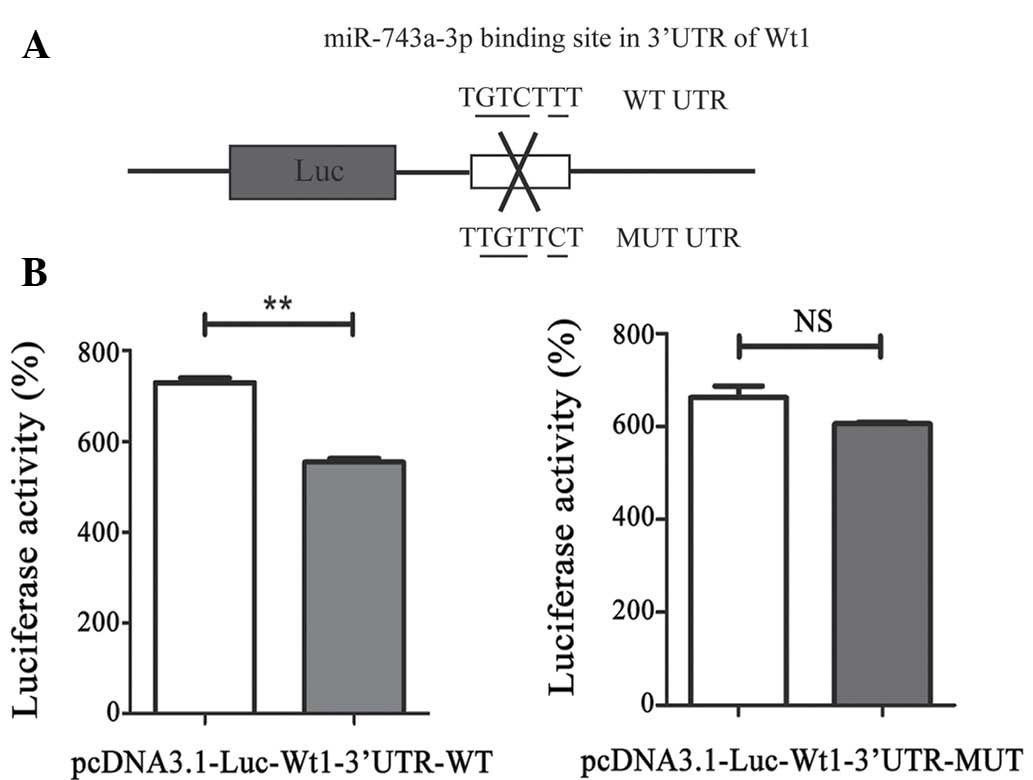 | Figure 2.miR-743a directly targets the 3′-UTR
of WT1. (A) The reporter vector contains the 3′-UTR of WT1. Two
fragments of WT1 3′-UTR, including wild-type as well as the mutant
type, are formed. (B) Transfection of the plasmids,
pcDNA3.1-luciferase-3′-UTR and pdsAAV-CB-EGFP-miR-743a, together
with pRL-SV40 reporter, into 293T cells is shown (the control
group: pcDNA3.1-luciferase-3′-UTR, pdsAAV-CB-EGFP and pRL-SV40).
After 48 h, the cells were lysed and the fluorescence was detected
in order to assess the activity, normalized against Renilla
activity. P<0.01 compared with the control. 3′-UTR,
3′-untranslated region; WT1, Wilms' tumor suppressor 1; Luc,
luciferase; WT, wild type; NS, not significant. |
miR-743a suppresses the expression of
WT1
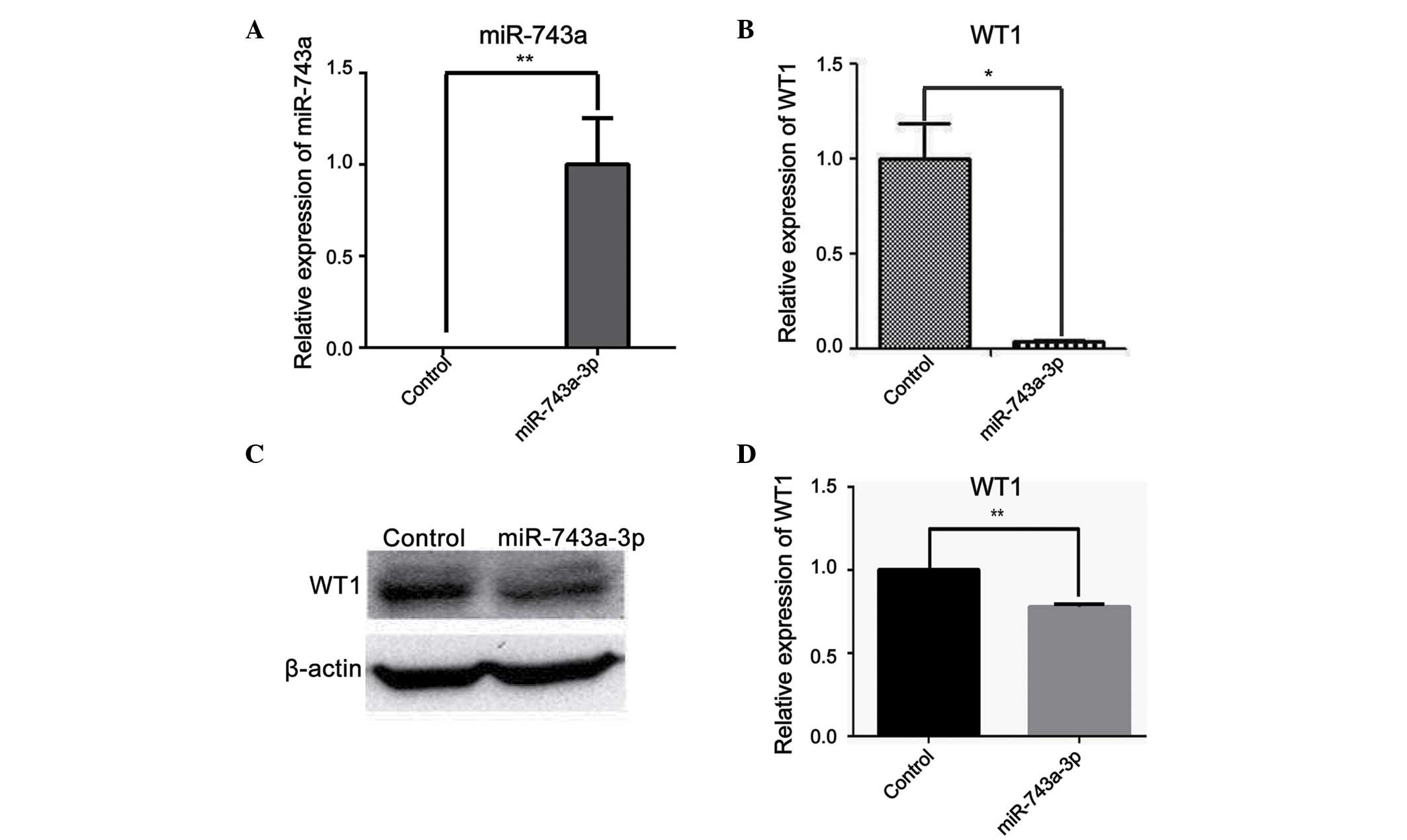
To further explore the effects of miR-743a on WT1 in
mK3 cells (an immortalized cell line of undifferentiated MM cells),
control mimic or miR-743a mimic (50 nM) was transfected into mK3
cells. After 48 h, the cells were collected in order to determine
the mRNA and protein expression levels of WT1. The cells
transfected with miR-743a mimic revealed a marked increase in
miR-743a expression at the mRNA level (Fig. 3A). However, the expression of WT1
was clearly decreased when transfected with miR-743a compared with
the control group, as demonstrated using RT-qPCR (Fig. 3B). Furthermore, the protein level
of WT1 also revealed a marked decrease, as revealed by western blot
analysis (Fig. 3C and D). Taken
together, these data suggested that miR-743a is able to
downregulate the expression of WT1 at the mRNA and the protein
level.
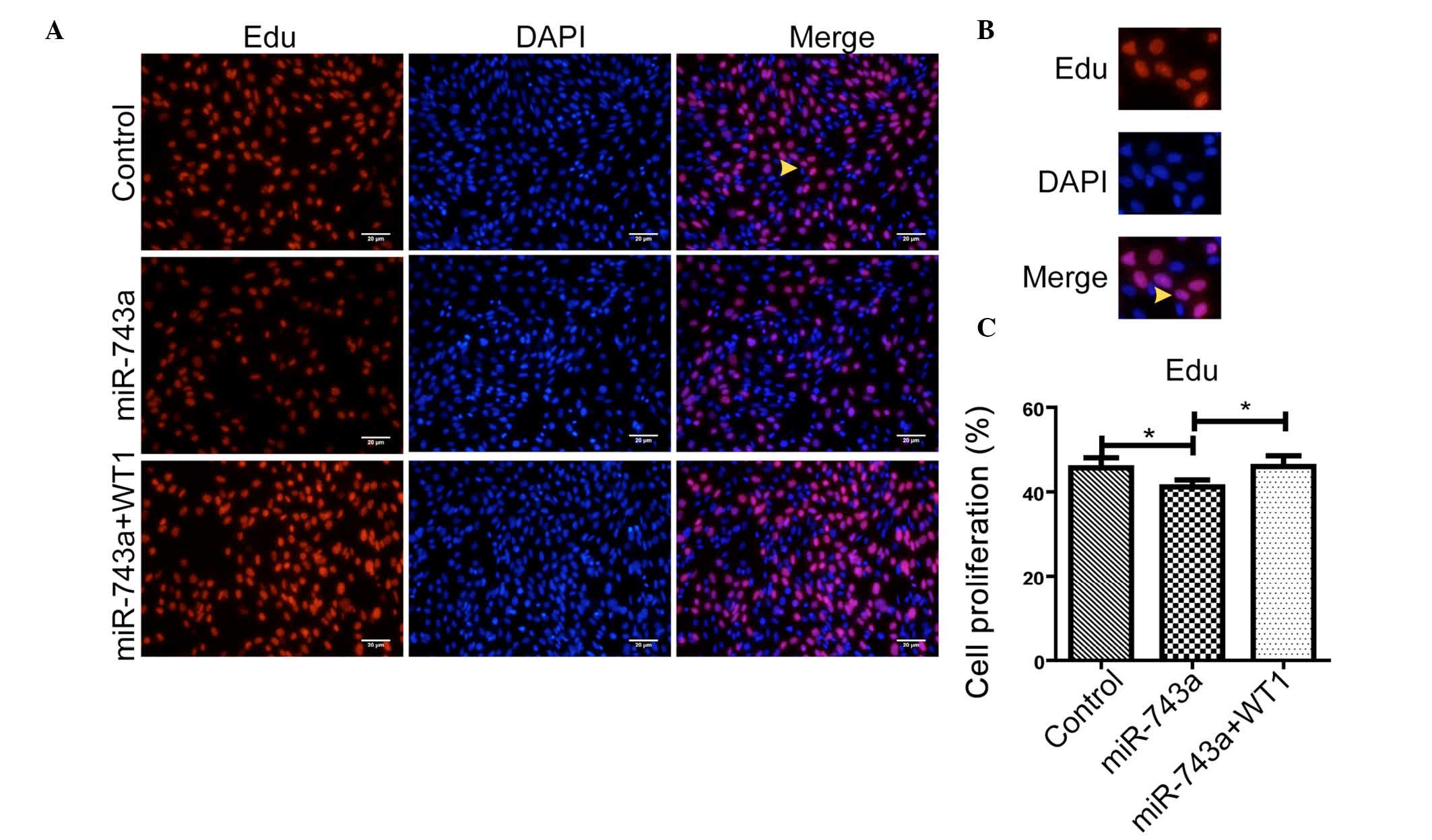
Proliferation of MM cells may be
attenuated by miR-743a, and rescued in part by WT1
To verify whether miR-743a is able to affect the
proliferation of mK3 cells via WT1, miR-743a mimics or negative
control mimics (50 nM) were transfected into mK3 cells. After 36 h,
the proliferation was measured using an EdU kit. As shown in
Fig. 4A-C, the proliferation rate
was markedly decreased in cells transfected with miR-743a together
with pCNDA3.1(+). Furthermore, as shown in Fig. 4A-C, the proliferation rate was
rescued when the cells overexpressing miR-743a were transfected
with WT1. These data suggested that miR-743a is able to inhibit the
proliferation of MM cells, and that this may be partially rescued
by WT1.
Discussion
Three important findings are associated with the
present study: i) miR-743a is able to directly target WT1; ii)
miR-743a is able to decrease the expression of WT1 at the mRNA and
the protein level; and iii) miR-743a inhibits the proliferation of
MM cells via WT1. To the best of our knowledge, these mechanistic
details have been established for the first time in the search to
identify the important functions of miR-743a in kidney
development.
WT1 was originally described in Wilm's tumor, a
condition that is characterized by undifferentiated blastema,
stromal cells, and even structures such as epithelium in developing
kidneys (20). In addition to
Wilm's tumors, mutation of WT1 can result in other severe kidney
diseases. WT1 fulfills important roles in maintaining the
proliferation of MM cells and in the balance of kidney development
(21). In addition, WT1 controls a
series of progenitor-specific genes in the kidney, including Sall1,
Pax2, Wnt8b, and so forth (4,21). A
proportion of reported kidney failures are caused by WT1 mutations,
which consequently creates public health problems worldwide
(22). Abnormal WT1 protein
results in the dysfunction of either renal progenitors or
podocytes, which ultimately lead to renal agenesis (22). Therefore, an investigation of the
upstream regulation of WT1 becomes a priority.
Other than code gene regulation in kidney
development, regulation of non-coding RNA is also functionally
important in the kidney. To the best of our knowledge, there is a
scarcity of reports on miR-743a, and particularly regarding its
function in the kidney In the present study, it has been
demonstrated that miR-743a may directly target the 3′-UTR of WT1.
For the first time, the important function of miR-743a has been
identified in the kidney, and the foundations have been laid for
further study of the role of miR-743a in kidney development and
disease.
In conclusion, in the current study it has been
identified that miR-743a targets WT1 in kidney MM cells.
Furthermore, the mechanism underpinning miR-743a inhibition of the
proliferation of MM cells through WT1 has been partly elucidated.
Future studies will focus on the possible role of miR-743a in
kidney diseases, with a view to elucidating new leads in curing
kidney diseases caused by WT1.
Acknowledgements
We would like to thank all the members of the
Division of Molecular Nephrology and the Creative Training Center
for Undergraduates, The M.O.E. Key Laboratory of Laboratory Medical
Diagnostics, The College of Laboratory Medicine, Chongqing Medical
University, for their assistance. This work was funded by the
National Basic Research Program of China (grant no. 2011CB944002 to
Professor Qin Zhou) and The National Natural Science Foundation of
China (grant nos. 31271563 and 81572076 to Professor Qin Zhou).
References
|
1
|
Dressler GR: The cellular basis of kidney
development. Annu Rev Cell Dev Biol. 22:509–529. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Little MH and McMahon AP: Mammalian kidney
development: Principles, progress, and projections. Cold Spring
Harb Perspect Biol. 4(pii): a0083002012.PubMed/NCBI
|
|
3
|
Lindström NO, Lawrence ML, Burn SF,
Johansson JA, Bakker ER, Ridgway RA, Chang CH, Karolak MJ, Oxburgh
L, Headon DJ, et al: Integrated β-catenin, BMP, PTEN, and Notch
signalling patterns the nephron. Elife. 3:e040002015.PubMed/NCBI
|
|
4
|
Motamedi FJ, Badro DA, Clarkson M, Lecca
MR, Bradford ST, Buske FA, Saar K, Hübner N, Brändli AW and Schedl
A: WT1 controls antagonistic FGF and BMP-pSMAD pathways in early
renal progenitors. Nat Commun. 5:44442014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Armstrong JF, Pritchard-Jones K, Bickmore
WA, Hastie ND and Bard JB: The expression of the Wilms' tumour
gene, WT1, in the developing mammalian embryo. Mech Dev. 40:85–97.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pritchard-Jones K, Fleming S, Davidson D,
Bickmore W, Porteous D, Gosden C, Bard J, Buckler A, Pelletier J,
Housman D, et al: The candidate Wilms' tumour gene is involved in
genitourinary development. Nature. 346:194–197. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Berry RL, Ozdemir DD, Aronow B, Lindström
NO, Dudnakova T, Thornburn A, Perry P, Baldock R, Armit C, Joshi A,
et al: Deducing the stage of origin of Wilms' tumours from a
developmental series of Wt1-mutant mice. Dis Model Mech. 8:903–917.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kirschner KM, Braun JF, Jacobi CL,
Rudigier LJ, Persson AB and Scholz H: Amine oxidase
copper-containing 1 (AOC1) is a downstream target gene of the Wilms
tumor protein, WT1, during kidney development. J Biol Chem.
289:24452–24462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Donovan MJ, Natoli TA, Sainio K, Amstutz
A, Jaenisch R, Sariola H and Kreidberg JA: Initial differentiation
of the metanephric mesenchyme is independent of WT1 and the
ureteric bud. Dev Genet. 24:252–262. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cannell IG, Kong YW and Bushell M: How do
microRNAs regulate gene expression? Biochem Soc Trans.
36:1224–1231. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Z: miRNA in the regulation of ion
channel/transporter expression. Compr Physiol. 3:599–653.
2013.PubMed/NCBI
|
|
12
|
Alvarez-Garcia I and Miska EA: MicroRNA
functions in animal development and human disease. Development.
132:4653–4662. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ho J and Kreidberg JA: The long and short
of microRNAs in the kidney. J Am Soc Nephrol. 23:400–404. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lyu Z, Mao Z, Wang H, Fang Y, Chen T, Wan
Q, Wang M, Wang N, Xiao J, Wei H, et al: MiR-181b targets Six2 and
inhibits the proliferation of metanephric mesenchymal cells in
vitro. Biochem Biophys Res Commun. 440:495–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lv X, Mao Z, Lyu Z, Zhang P, Zhan A, Wang
J, Yang H, Li M, Wang H, Wan Q, et al: miR181c promotes apoptosis
and suppresses proliferation of metanephric mesenchyme cells by
targeting Six2 in vitro. Cell Biochem Funct. 32:571–579. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang X, Wang X, Nie F, Liu T, Yu X, Wang
H, Li Q, Peng R, Mao Z, Zhou Q and Li G: miR-135 family members
mediate podocyte injury through the activation of Wnt/β-catenin
signaling. Int J Mol Med. 36:669–677. 2015.PubMed/NCBI
|
|
17
|
Sun H, Li QW, Lv XY, Ai JZ, Yang QT, Duan
JJ, Bian GH, Xiao Y, Wang YD, Zhang Z, et al: MicroRNA-17
post-transcriptionally regulates polycystic kidney disease-2 gene
and promotes cell proliferation. Mol Biol Rep. 37:2951–2958. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ranganathan P, Jayakumar C, Tang Y, Park
KM, Teoh JP, Su H, Li J, Kim IM and Ramesh G: MicroRNA-150 deletion
in mice protects kidney from myocardial infarction-induced acute
kidney injury. Am J Physiol Renal Physiol. 309:F551–F558. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hastie ND: The genetics of Wilms' tumor -
a case of disrupted development. Annu Rev Genet. 28:523–558. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong L, Pietsch S and Englert C: Towards
an understanding of kidney diseases associated with WT1 mutations.
Kidney Int. 88:684–690. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Eckardt KU, Coresh J, Devuyst O, Johnson
RJ, Köttgen A, Levey AS and Levin A: Evolving importance of kidney
disease: From subspecialty to global health burden. Lancet.
382:158–169. 2013. View Article : Google Scholar : PubMed/NCBI
|