Introduction
Abdominal aortic aneurysm (AAA) is a common disease
that is correlated with dilatation of the aorta to ≥5 mm in
diameter, and occurs below the renal arteries (1). Vascular degradation is closely
associated with advanced age, being female, smoking and a family
history of the condition (2). AAA
is responsible for ~15,000 mortalities in the USA per annum,
despite the recent reduction in prevalence and mortality due to
decreased smoking rates, increased early detection and improved
surgical procedures (3,4). The incidence of AAA is 6-fold higher
in males >60 years of age and females >65 years of age. AAA
develops due to extensive vascular inflammation combined with
maladaptive remodeling of the aortic wall (5,6).
Although various clinical and laboratory studies have provided
insights into the pathogenesis of AAA, the underlying mechanisms
remain to be elucidated (7).
The pathogenesis of AAA is very complex; however,
increasing evidence suggests that vascular smooth muscle cells
(VSMC) are associated with the development of AAA. VSMCs provide a
source of elastin, which serves an important role in maintaining
the elasticity of the aortic wall (8,9).
However, during the development of AAA, proteolytic processes
induce the degradation of the media layer, which leads to a
reduction of elastin content and expansion of the aortic wall. The
deficiency in elastin content subsequently leads to a compensatory
increase in collagen synthesis, which contributes to remodeling of
the AAA wall (10). This decrease
in elastin has been attributed to the induction of senescence and
apoptosis of VSMCs (11). AAA is
characterized by degradation of the extracellular matrix, a potent
inflammatory response, and increased oxidative stress in the
abdominal aortic wall (12,13).
In addition, infiltration by inflammatory cells may accelerate
apoptosis of VSMCs. Inhibition of apoptosis and SMC regeneration
are considered to be desirable endpoints for the treatment of AAA.
Through the identification of a large number of miRNA sequences
with differences in expression levels between AAA samples and those
derived from abdominal aortic tissues from age and sex-matched
controls, a number of previous reports have indicated that
microRNAs (miRNAs) may be involved in AAA (14–17).
miRNAs are a class of small non-coding RNAs that
bind preferentially to the 3′-untranslated region (3′-UTR) of
target mRNA sequences and transport them to the RNA-induced
silencing complex, which results in the downregulation of gene
expression by mRNA degradation and inhibition of translation
(18). This process contributes to
the regulation of crucial biological activities, including cell
proliferation, apoptosis and differentiation (19,20).
Despite the identification of <1,000 human miRNAs (21), it is now evident that >30% of
the genome is regulated by miRNAs (22). Previous studies have confirmed that
miRNAs serve crucial roles in tumorigenesis, as well as the
invasion and apoptosis of cancer cells in various human
malignancies (23,24). In addition, miRNAs have been
demonstrated to target tumor suppressor genes inhibiting smooth
muscle cell proliferation and migration, thus influencing
angiogenesis (25,26).
Although a considerable number of studies
investigating miRNAs have been conducted, a limited number of miRNA
expression profiling studies involving VSMCs from patients with AAA
have been performed to date. Therefore, the present study
investigated the expression patterns of miRNAs in aortic SMCs from
patients with AAA, in order to identify potential miRNAs that are
associated with the disease. A microarray-based genome-wide
screening study was first performed, followed by analysis of the
expression of individual miRNAs by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Bioinformatic analyses were conducted to predict gene targets of
the identified miRNAs and determine their putative roles in
AAA.
Materials and methods
Human aortic SMC samples
Aortic SMC specimens were obtained from patients
undergoing AAA repair operations (n=60) at the Department of
Cardiology, The Fourth Affiliated Hospital of Harbin Medical
University (Harbin, China) from January 2013 to December 2014.
Aortic wall tissues and matched non-aneurysmal aortic samples were
snap-frozen in liquid nitrogen and stored at −80°C until RNA was
extracted. Written informed consent was obtained from all study
percipients. The present study received ethical approval from the
Independent Ethics Committee of Shanghai First People's Hospital
(Shanghai, China), and all experiments were conducted in accordance
with the Declaration of Helsinki.
Cell culture and RNA isolation
SMCs were propagated in SmGM-2 Smooth Muscle Growth
Medium-2 (Lonza Group, Basel, Switzerland) containing 10% fetal
bovine serum (FBS) according to the manufacturer's instructions.
Cells were maintained in a humidified atmosphere at 37°C and 5%
CO2. After 48 h, cells were harvested and total RNA
isolated using the mirVana™ miRNA Isolation kit (Ambion; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's instructions.
Preparation of miRNA and total RNA and
RT-qPCR analysis
The quality of RNA and miRNA in samples was assessed
using a NanoDrop spectrophotometer (NanoDrop; Thermo Fisher
Scientific, Pittsburgh, PA, USA), and verified using the Agilent
2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA;
all sample RNA integrity values were >9). For quantification of
gene transcription, 10 µl of cDNA containing 1010 copies
of RNA was generated using the Moloney murine leukemia virus
reverse transcriptase (primers were sense: 5′-NNG/T; anti-sense:
5′-A/CNN), and then amplified on the ABI PRISM 7900HT Sequence
Detection System (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using TaqMan primers (TM001973; forward ACC CTG GTC TGC ACT
CTA TCT and reverse TGC CCT CTG TAT GGG AAA CC; Applied Biosystems;
Thermo Fisher Scientific, Inc.). The expression of miRNA sequences
was quantified using TaqMan. miRNA Reverse Transcription kits
(SM-10153) and TaqMan miRNA assays (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Thermal cycling parameters were 94°C for
30 sec, 57°C for 30 sec and 72°C for 30 sec. For miRNAs
quantification, 10 fmol was diluted to 0.01 amol and spiked into a
constant amount of carrier RNA, and the Cq values were
measured.
miRNA array hybridization
RNA was purified and pooled from samples of
treatments using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. This
method yielded an average of 30 µg total RNA from 106
cells. Hybridized microarray was performed by the Agilent Human
miRNA 8×15k Array 2.0 (cat. no. G4470B; Agilent Technologies, Inc.)
according to manufacturer's protocol, and the quantity of RNA which
was used array hybridization was 5 µg. The microarray contained 830
miRNA probes from the Sanger database (version 10.1, http://www.sanger.ac.uk/). Arrays were scanned using
the Agilent Microarray Scanner System and Feature Extractor
Software (version 9.5.1; Agilent Technologies, Inc.).
miRNA array analysis
Quality control (QC) reports were generated using
the Agilent Feature Extraction software, and the array data were
analyzed using GeneSpring GX software (version 10.0.2; Agilent
Technologies, Inc.). Arrays were examined by QC metrics and
principal components analysis, as well as hierarchical
agglomerative clustering. Two chips clustered separately and were
therefore excluded. The remaining arrays were subject to one-way
analysis of variance (threshold, P<0.05), with a
Benjamini-Hochberg post hoc correction for multiple testing.
Significant miRNAs were required to exhibit a >2-fold increase
or decrease in expression compared with the control at the 72
h-time point. Pairwise analysis was performed between the control
and the 72 h-time point using statistical analysis of microarrays
with false detection ratio of <1. TargetScan (version 5.1,
http://www.targetscan.org/worm_12/docs/help.html.)
and PicTar version 4.0.24 (http://www.pictar.org/.) software programs were used
to identify putative miRNA gene targets (27–29).
Validation of miRNA array results
In order to validate the expression levels of miRNA
sequences identified by microarray analysis, further RT-qPCR was
performed. Briefly, samples were incubated for 60 min at 42°C,
followed by heat inactivation of the reverse transcriptase for 5
min at 95°C prior to storage at 4°C. To convert mRNA to cDNA, total
RNA was reverse transcribed using the High-Capacity cDNA reverse
transcription kit (cat. no. 4368813; Applied Biosystems; Thermo
Fisher Scientific, Inc.). Reaction mixtures were incubated at 25°C
for 5 min, followed by 37°C for 120 min then 85°C for 5 min, prior
to storage at −20°C. Standard thermocycling conditions were used
and reactions were performed in triplicate with the corresponding
positive and negative controls. Primer sequences for Let-7c were
GCT GAC TGA AGA TAT GAT AAGG and ATG ACA CAT TAC CTT CTT GC; for
miR-449a CAC GAG AAT CGG CAG TGAC and AAC ACT GTC TAC TCC GAA AGA
CTCC; for miR-144-3p GCT GGG ATA TCA TCA TAT ACTG and CGG ACT AGT
ACA TCA TCT ATA CTG; for miR-192-5p CTG ACC TAT GAA TTG ACA GCC and
TGA CCT ATG GAA TTG GCAG; for miR-504 TCT ACA CAG GTT GGG ATC GG
and CGG GAC AAG TGC AAT ACC ATA; for miR-542-3p GAG CTA GCA CTT CCC
GAG C and TAG CTG TCT GCC CCT TGT CT, and the primer for endogenous
control was TTG CGG GTC TAA TCA CCG ATT and TCG GTG ATT AGA CCC GCA
ATT. The thermal cycling parameters were as follows: 95°C for 5
min, 45 cycles of 10 sec at 95°C, 20 sec at 60°C and 15 sec at
72°C; followed by a melting curve of 10 sec at 95°C, 60 sec at 60°C
and continued melting. A SYBR-Green Master mix (Thermo Fisher
Scientific, Inc.) was used. Target gene expression was quantified
using the 2−ΔΔCq method (30). All RT-qPCR analyses for the
quantification of miRNA expression levels were completed within 24
h of the reverse transcription step.
miRNA transfection
The DNA fragments encoding miRNA-504 or control
miRNA sequences and the miRNA-504 inhibitor or control small
interfering-RNA sequences (NC) were purchased from GeneCopoeia,
Inc. (Rockville, MD, USA), and were inserted into the pMSCV vector
(Clontech Laboratories Inc., Mountain View, CA, USA). Transfection
was performed with Lipofectamine 2000 transfection reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to
manufacturer protocols (PanVera; Thermo Fisher Scientific, Inc.).
The number of cells transfected was 1.5×104
cm−2, and the quantity of plasmid DNA was 1.5 mg in a
volume of 10 µl. Aortic SMCs were cultured in medium containing 10%
FBS for 24 h prior to transfection with miRNA sequences. The viral
vectors were added to the medium for transfection. At 48 h
following transfection, cells were cultured in medium containing 1
µg/ml puromycin to select for positive transfection clones. In
order to maintain stable expression of the inserted sequence, the
positive cells were cultured in medium containing 0.5 µg/ml
puromycin. The primer sequence for miRNA-504 was
5′-GGTGAAGGTCGGTGTGAACG-3′ (forward), and
5′-TGGAGGCCATGTAGGCCATG-3′ (reverse).
Luciferase reporter assay
In order to determine the effect of miRNA-504 on the
3′-UTR of the p53 mRNA sequence, the Dual-Luciferase Reporter 1000
assay system (Promega Corporation, Madison, WI, USA) was utilized.
Briefly, 1,000 to 15,000 cells were cultured in 24-well cell
culture plate until the cells reached 70% confluence. Cells were
co-transfected using a co-transfected reagent from Qiagen Sciences,
Inc. (Gaithersburg, MD, USA) with hsa-miRNA-504 or control miRNA
sequences and the 3′-UTR of p53 (mutant of p53 3′-UTR was
5′-ACUUGUUUUAUAGCAUCUACU-3′). The quantities of hsa-miR-504 and
control miRNA were calculated using 2−∆∆Cq method
(30). At 48 h following
transfection, cells were harvested and firefly and Renilla
luciferase activities were assayed. Lipofectamine 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) and a Dual-Luciferase reporter
assay system (Promega Corporation) were used for sequential
measurement of firefly and Renilla luciferase activities.
Quantification of luciferase activities and calculation of relative
ratios were carried out using a luminometer (TD-20/20; Turner
Designs, Sunnyvale, CA, USA); at least three independent
transfections were performed in triplicate. The results were
normalized to Renilla luciferase activity.
Western blot analysis
After 24 h, whole aortic cell lysates were extracted
using radioimmunoprecipitation assay lysis buffer (150 mM Tris-HCl,
50 mM NaCl, 1% Nonidet P-40 and 0.1% Tween-20) and the protein
concentration was determined using the Bradford assay (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Protein extracts (30 µg)
were resolved by SDS-PAGE (6 or 7.5% w/v) and transferred
electrophoretically to polyvinylidene difluoride membranes.
Following blocking of membranes with dried non-fat milk (5% w/v; 1
h at room temperature), membranes were probed with the following
primary antibodies overnight at 4°C: Anti-proliferating cell
nuclear antigen (anti-PCNA; cat. no. sc-25280; 1:200, Santa Cruz
Biotechnology, Inc., Dallas, TX, USA), anti-replication factor C
subunit 4 (RFC4) (cat. no. sc-28301; 1:1,000, Santa Cruz
Biotechnology, Inc.), anti-caspase-3 (cat. no. sc-7272; 1:500;
Santa Cruz Biotechnology, Inc.), anti-caspase-9 (cat. no. sc-81589;
1:500; Santa Cruz Biotechnology, Inc.), anti-B-cell lymphoma-2
(Bcl-2) (cat. no. sc-23960; 1:1,000; Santa Cruz Biotechnology,
Inc.), the anti-p53 rabbit monoclonal antibody (53 kD cat. no.
8712; 1:1,000; Cell Signaling Technology, Inc., Danvers, MA, USA),
the anti-p21 rabbit monoclonal antibody (21 kD; cat. no. 2947;
1:1,000; Cell Signaling Technology, Inc.) and anti-bcl-2-like
protein 4 (Bax; cat. no. 2772; 1:1,000; Cell Signaling Technology,
Inc). Membranes were subsequently incubated with the
peroxidase-conjugated goat anti-rabbit IgG secondary antibody (cat.
no. A0208; Beyotime Institute of Biotechnology, Shanghai, China) at
1:2,000 for 1 h at room temperature. Immunoreactive bands were
normalized to β-actin and visualized using an ECL kit (Amersham
Pharmacia Biotech Inc., Piscataway, NJ USA) then quantified using
ImageQuant software version 5.2 (GE Healthcare Life Sciences,
Chalfont, UK).
Determination of cell
proliferation
Treated and untreated aortic cells were seeded at
3.7×104 cells/cm3 in 96-well plates and
incubated for 24, 48, 72, 96 and 120 h. A CCK-8 assay (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) was performed by
adding 10% CCK-8 solution (v/v) to each well and incubating cells
for 1 h at 37°C. The optical density values for each well were
measured at a wavelength of 450 nm using a microplate reader
(Bio-Rad Laboratories, Inc.). All experiments were performed in
triplicate and repeated at least three times.
Cell apoptosis assay
Apoptosis was analyzed using the Annexin
V-fluorescein isothiocyanate (FITC; 5 µl)/propidium iodide (PI; 5
µl; both from Invitrogen; Thermo Fisher Scientific, Inc.) staining
method. The reagents were mixed with 500 µl buffer and flow
cytometry analysis was preformed according to manufacturer's
instructions. Cells (2–4×105 cells/well) were cultured
on 60-mm diameter dishes and infected with the aforementioned viral
vectors. At 48 h following transfection, cells were harvested by
trypsinization, washed with ice-cold phosphate-buffered saline
(PBS), and fixed with ice-cold 70% ethanol for >2 h at −20°C.
The fixed cells were then washed with PBS and incubated with
ribonuclease A (20 µg/ml at final concentration; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) and PI (0.05 mg/ml; Sigma-Aldrich;
Merck KGaA) at room temperature for 30 min. Cells were then labeled
with Annexin V-FITC (final concentration 0.25 µg/ml) and PI (final
concentration 12.5 µg/ml) in the dark for 15 min at room
temperature and analyzed by flow cytometry (BD Biosciences, San
Jose, CA, USA). At least 2×104 cells were analyzed for
each sample and the distribution of cells was analyzed using
CellQuest™ software (version 3.3; BD Biosciences). The experiments
were performed in triplicate.
Statistical analysis
Data are expressed as the mean ± standard error of
the mean from at least three repeated experiments. Differences
between groups were analyzed using one-way analysis of variance
followed by Tukey's test. All statistical analyses were conducted
using SPPS software (version 20.0; IBM SPSS, Armonk, NY, USA).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miRNA expression profiling of clinical
samples
In order to identify miRNA sequences associated with
AAA, the expression levels of 84 miRNAs that have been previously
implicated in angiocardiopathy and cancer in AAA were investigated.
Following analysis of the miRNAs that demonstrated differential
expression patterns in AAA samples compared with control samples, a
number of miRNAs that were reproducibly up- or downregulated in the
aortic cells were identified. The criteria for differentially
expressed miRNAs was based on the following two conditions: miRNA
sequences should demonstrate a >2-fold alteration in gene
expression when compared with the controls, and exclusion of miRNAs
with low expression levels (quantification cycle, >30), as
quantification by RT-qPCR analysis may produce unreliable results.
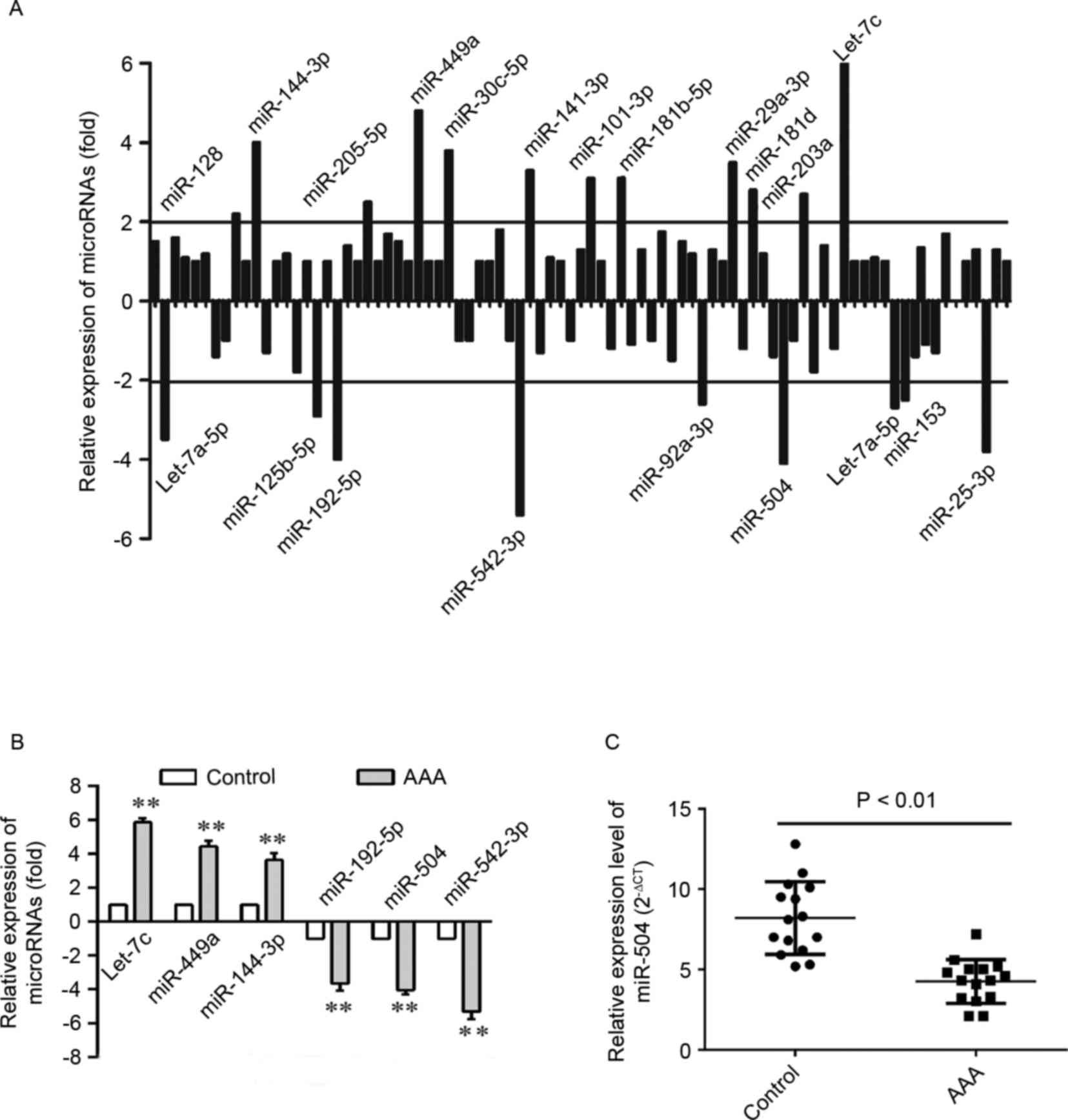
Accordingly, 21 differentially expressed miRNAs were identified in
aortic cells from patients with AAA (Fig. 1A). A total of 12 miRNAs were
upregulated, while 9 were downregulated in aortic cells. Among
these, the expression of 6 miRNAs, which were significantly altered
when compared with the remaining miRNAs, was verified by RT-qPCR
analysis (Fig. 1B). As shown in
Fig. 1C, the expression level of
miRNA-504 was significantly higher in the AAA aortic SMCs when
compared with matched normal controls (P<0.01). Therefore, the
results suggest that miRNA-504 expression was significantly
increased in AAA. In addition, previous studies have reported that
miRNA-504 exhibits anti-apoptotic functions, and AAA is closely
associated with apoptosis of smooth muscle cells (31,32).
The result indicated that miRNA-504 may be closely related with AAA
and therefore the functional role of miRNA-504 was investigated
further in the present study.
Effect of miRNA-504 on the
proliferation of aortic SMCs
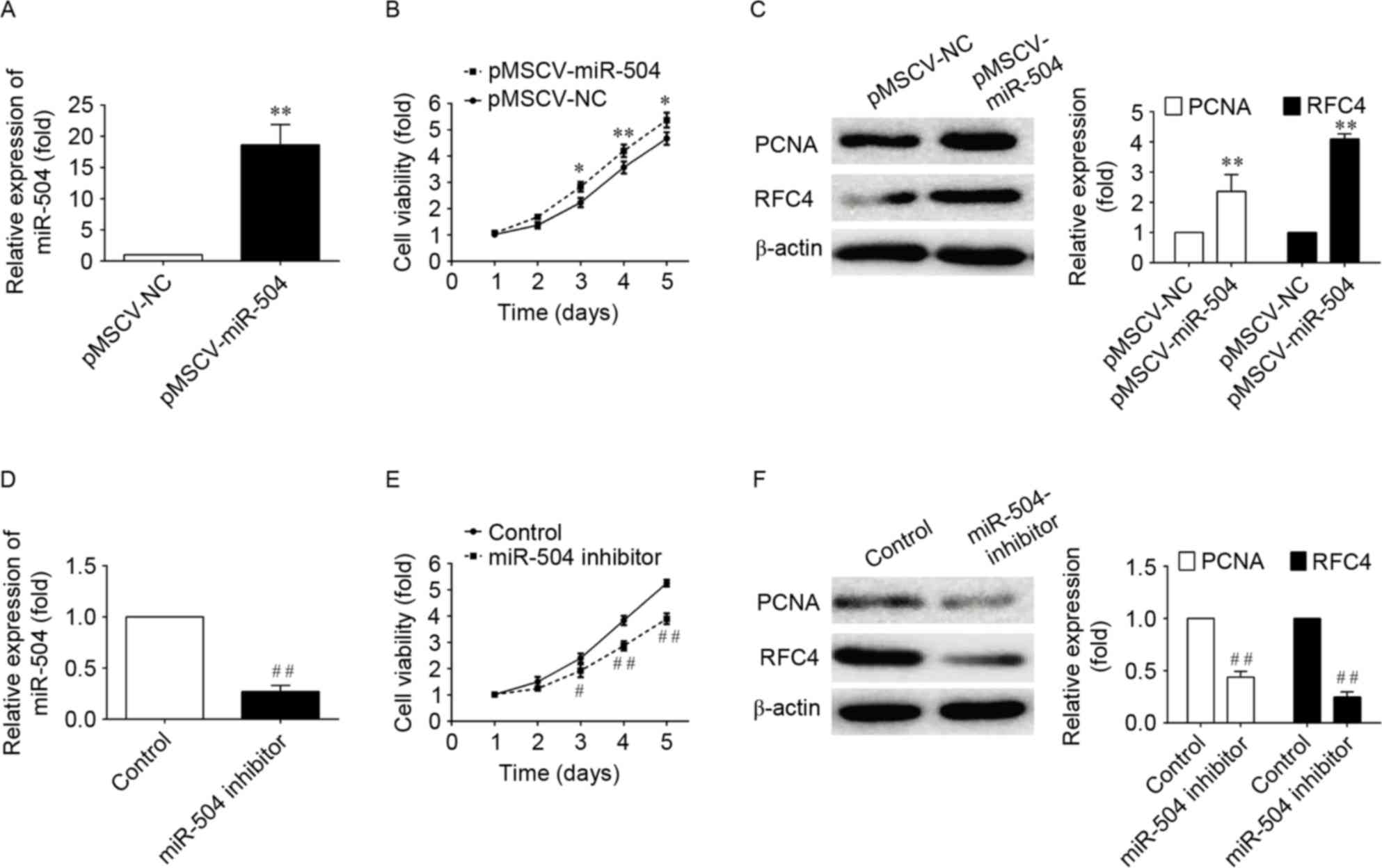
To examine the effect of miRNA-504 on the
proliferation of SMCs, the proliferation of aortic cells infected
with pMSCV vectors containing non-targeting control, miRNA-504 or
miRNA-504 inhibitor sequences was examined using the CCK-8 assay.
In the pMSCV-miRNA-504-transfected aortic cells, cell growth was
significantly increased within 5 days when compared with negative
controls (P<0.05; Fig. 2). By
contrast, transfection of cells with the miRNA-504 inhibitor,
significantly inhibited cell proliferation when compared with the
non-targeting controls (P<0.05; Fig. 2). These results indicate that
miRNA-504 may promote the proliferation of SMC cells.
In order to investigate the signaling pathways
associated with the miRNA-504-mediated increase in SMC
proliferation further, the expression levels of two genes involved
in DNA replication, PCNA and RFC4 (33,34),
which serve as checkpoints of DNA replication, were examined. The
western blotting results demonstrated that the PCMA and RFC4
expression was significantly increased in
pMSCF-miRNA-504-transfected cells, and significantly decreased
following transfection with the miRNA-504 inhibitor when compared
with controls (Fig. 2). These
results suggest that miRNA-504 may promote the proliferation of
SMCs by regulating the expression of DNA replication-associated
genes, PCMA and RFC4, which are involved in mediating the growth of
cells.
Inhibition of miRNA-504 induces
apoptosis of SMCs
Apoptosis is a form of cell death characterized by
cell shrinkage, chromatin margination, membrane blebbing and
nuclear condensation. A number of studies have demonstrated that
numerous miRNA sequences regulate apoptosis (35–38).
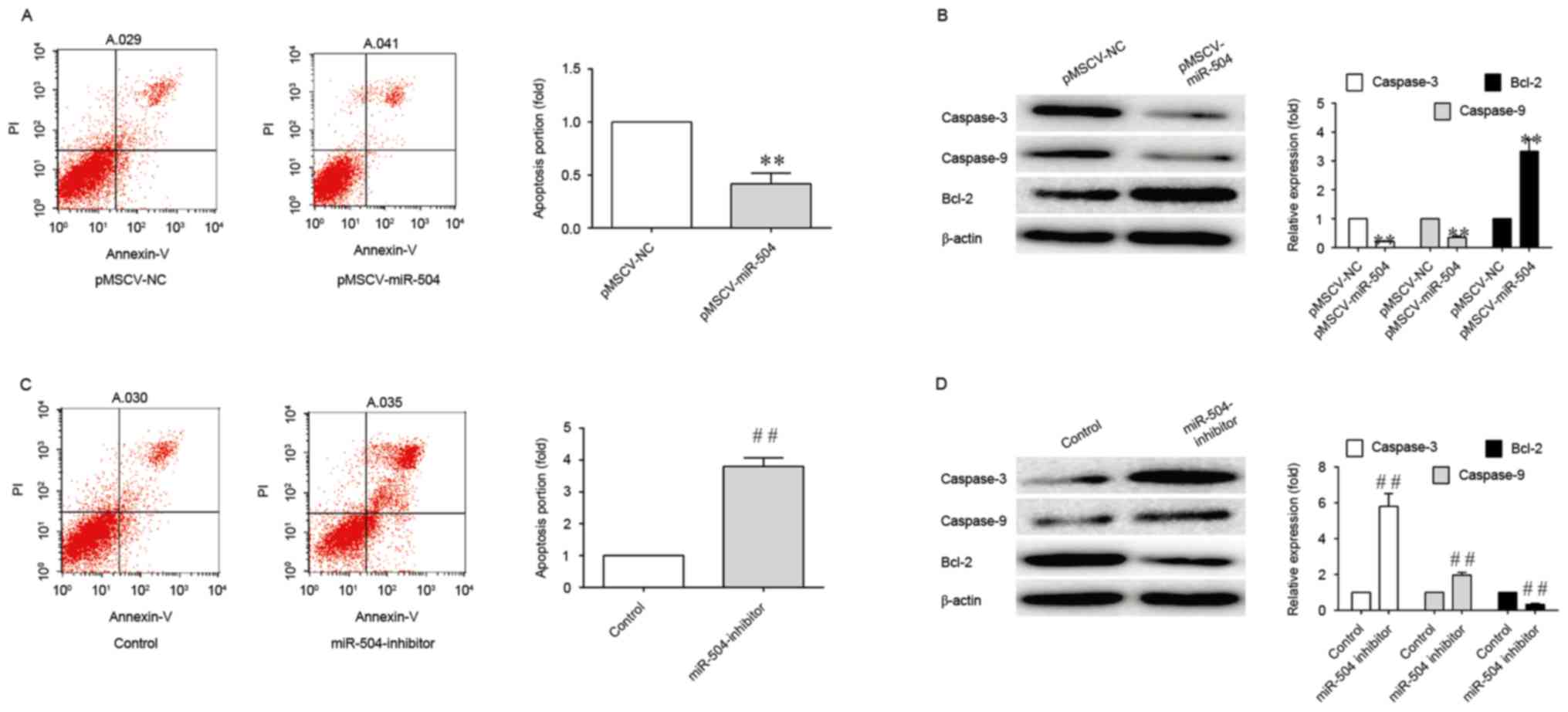
Therefore, in order to determine whether miRNA-504 may be involved
in the apoptosis of SMCs, the number of apoptotic cells in
pMSCV-miRNA-504 and miRNA-504 inhibitor-transfected SMCs was
ascertained using an Annexin V-FITC plus PI staining assay. As
shown in Fig. 3, miRNA-504
significantly inhibited the apoptosis of SMCs, as demonstrated by
the 4-fold reduction in apoptosis levels when compared with the
controls. By contrast, transfection with the miRNA-504 inhibitor
was associated with a 4-fold increase in the degree of apoptosis
when compared with controls (Fig.
2). The results indicated that miRNA-504 may regulate the
apoptosis of SMCs.
In order to investigate the association between
miRNA-504 and the expression of caspase-3, caspase-9 and Bcl-2, the
expression levels of these proteins were determined in SMCs
transfected with pMSCV-miRNA-504 and miRNA-504 inhibitors. As shown
in Fig. 3,
pMSCV-miRNA-504-transfected SMCs exhibited a significant reduction
in caspase-3 and caspase-9 protein expression levels, and a
significant increase in Bcl-2 expression, when compared with the
negative controls. This may have been responsible for the observed
increase in apoptosis levels. A significant increase in caspase-3
expression levels was observed in SMCs transfected with the
miRNA-504 inhibitor when compared with the controls (Fig. 3). By contrast, the expression
levels of the anti-apoptotic gene, Bcl-2 (39), were significantly decreased
following transfection with the miRNA-504 inhibitor (Fig. 3). These results indicate that
miRNA-504 may serve an anti-apoptotic role in SMCs.
miRNA-504 regulates SMC apoptosis via
an p53-dependent mechanism
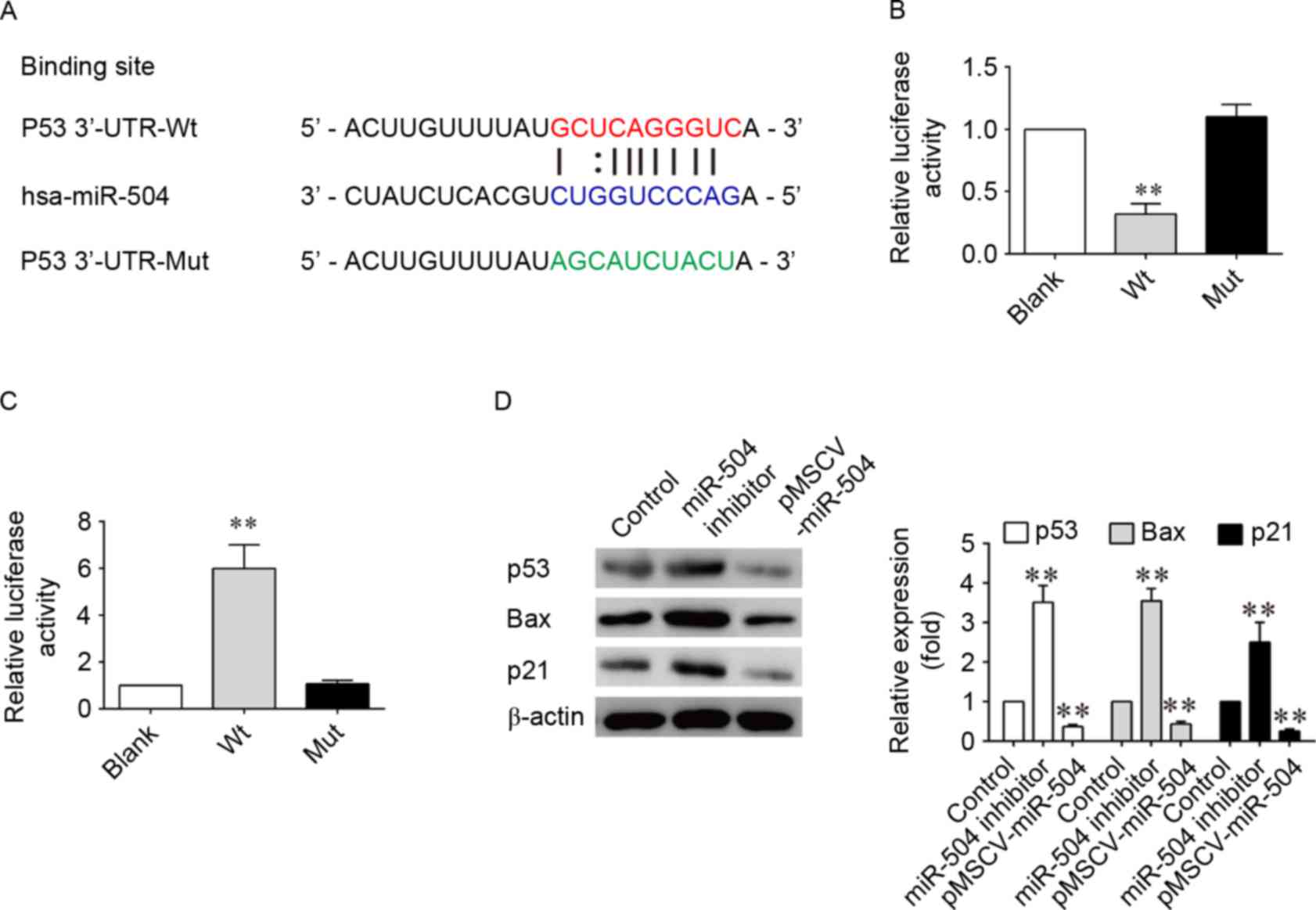
In order to determine the molecular mechanism
underlying the effects of miRNA-504 on SMCs, TargetScan software,
which is a bioinformatics tool for miRNA target screening, was
employed to search for putative target genes of miRNA-504. p53 was
identified as a potential target gene of miRNA-504. In addition, a
previous study has reported that overexpression of miRNA-504
inhibits the transcriptional activity of p53 and decreases
p53-induced apoptosis and cell-cycle arrest (40). Based on these previous
observations, a miRNA-504-based luciferase reporter assay in SMCs
was therefore performed in the present study. Comparison of the
miRNA-504 and p53 3′-UTR sequences indicated that p53 may be a
direct target of miRNA-504 (Fig.
4A). As shown in Fig. 4B and
C, the luciferase reporter activity of the wild-type 3′-UTR of
p53 was significantly decreased when compared with the luciferase
reporter activity of the mutant 3′-UTR sequence following
transfection of cells with hsa-miRNA-504 (P<0.001; Fig. 4B). By contrast luciferase reporter
expression of wild-type p53 3′-UTR sequences was significantly
increased in SMCs following inhibition of miRNA-504. Western
blotting analysis confirmed the decreased and increased expression
of p53, as well as its downstream factors, p21 and the
pro-apoptotic Bax gene, following transfection of SMCs with the
pMSCV-miRNA-504 and miRNA-504 inhibitors, respectively (Fig. 4D). Inhibition of miRNA-504 led to a
3-fold increase in the expression levels of p53, p21 and Bax when
compared with the controls (P<0.05; Fig. 4D). These results suggest that
miRNA-504 may regulate SMC growth and apoptosis by directly
targeting the 3′-UTR of p53, thereby altering its expression and
that of downstream genes involved in cell proliferation and
apoptosis.
Discussion
In normal aortic SMC cells, miRNA-504 is
upregulated. In the present study, however, results indicated that
miRNA-504 was downregulated in AAA samples of patient, and this
suggested that miRNA-504 may be correlated with heritable
cardiovascular risk. miRNA-504 may achieve this by regulating the
proliferation of SMCs; a critical determinant of vessel function.
The ability of SMCs to modulate their phenotype from a quiescent,
contractile state to a proliferative, synthetic profile has been
implicated in a number of conditions, including atherosclerosis,
post-angioplasty restenosis and aortic aneurysm formation (41–45).
In view of this important feature of SMCs that is associated with
AAA, an in-depth study was performed to investigate the mechanisms
underlying the effects of miRNA-504 on SMCs. According to previous
studies demonstrating that phenotypic alterations in SMCs are
regulated by miRNAs (46–48), miRNA microarray analysis of human
SMCs was performed to identify the miRNA sequences involved in AAA.
By employing an agnostic whole-genome approach, 12 upregulated and
9 downregulated miRNAs were identified to be significantly
differentially expressed in AAA samples. Among these miRNAs, 6
miRNAs exhibited 4- and 6-fold differences in expression in AAA
samples when compared with controls. Verification of the expression
of these miRNAs using RT-qPCR indicated that miRNA-504 was closely
associated with AAA. Subsequent functional investigation of
miRNA-504 demonstrated that this miRNA sequence promoted the
proliferation of SMCs.
Over the last several years, a number of studies
have provided extensive evidence that miRNAs serve important roles
in controlling numerous fundamental cellular processes, including
aneurysm formation (49). A number
of studies have detected frequent alterations in the expression of
miRNAs in a variety of human tumors, suggesting that miRNAs may
serve a role as a novel class of tumor promoters or suppressors
(50,51). Consistent with these observations,
the results of the present study demonstrated that miRNA-504 is an
essential miRNA that regulates the proliferation of SMCs in AAA.
The function of miRNA-504 was examined by transfecting
pMSCV-miRNA-504 into SMCs, which led to an increase in SMC
proliferation, as well as an increase in the expression levels of
the mRNA/DNA replication genes, PCNA and RFC4. Subsequent analysis
revealed that miRNA-504 may have promoted the proliferation of SMCs
by reducing the expression of caspase 3 and caspase 9. When
activated, these enzymes serve a key role in mediating apoptotic
signaling events (52). In
addition, miRNA-504 augmented the expression levels of the
anti-apoptotic gene, Bcl-2.
A number of different physiological death signals,
as well as pathological cellular insults, activate the genetically
programmed pathway of apoptosis (53). Apoptosis manifests as two major
execution programs downstream of the death signal, one of which is
the caspase and organelle dysfunction-induced pathway. As part of
this pathway, the mitochondrial dysfunction-induced pathway is the
best characterized (54). During
mitochondrial dysfunction, Bcl-2 resides upstream of irreversible
cellular damage and serves a pivotal role in determining cell fate
following induction of the mitochondrial dysfunction pathway. The
results of the present study suggest that miRNA-504 inhibited the
pathway of apoptosis induction, and enhanced the proliferation of
SMCs.
It is well known that the basic function of miRNAs
is to regulate target genes by facilitating the direct cleavage of
target mRNA sequences and/or by suppressing protein synthesis,
which depends on the degree of complementarity of miRNAs with the
3′-UTR of target gene mRNA (55).
In present study, p53 was selected as the target gene for miRNA-504
according to TargetScan software. Luciferase reporter assays
confirmed that miRNA-504 targets the 3′-UTR of p53, and that p53
expression was significantly downregulated by miRNA-504
overexpression. A number of previous studies have revealed that the
function of p53 as an anti-oncogene is short-lived, as it is
maintained at low levels by rapid degradation through the mouse
double minute 2 homolog, which is a p53-specific E3 ubiquitin
ligase (56,57). Activated p53 induces the
transcription of target genes, such as p21, to initiate various
cellular responses, including cell cycle arrest, apoptosis or
senescence. The results of the current study indicated that
overexpression of miRNA-504 inhibited the transcriptional activity
of p53 and potentially decreased p53-induced apoptosis.
In conclusion, the present study investigated the
function of miRNA-504 in SMCs from patients with AAA, and
demonstrated that miRNA-504 increased the growth of aortic SMCs and
inhibited apoptosis. Therefore, miRNA-504 may function as an
anti-neoplastic factor in AAA. Investigating the role of miRNA-504
in SMCs from patients with AAA further, may provide an improved
understanding of the molecular mechanisms underlying AAA
development. These results would provide important information and
a wider perspective on AAA intervention, prevention and treatment
strategies.
Acknowledgements
The current study was financially supported by the
Natural Science Foundation of Heilongjiang Province (grant no.
H201387) and Heilongjiang Postdoctoral Fund (LBH-Z16159).
References
|
1
|
Sakalihasan N, Limet R and Defawe OD:
Abdominal aortic aneurysm. Lancet. 365:1577–1589. 2005. View Article : Google Scholar
|
|
2
|
Boddy AM, Lenk GM, Lillvis JH, Nischan J,
Kyo Y and Kuivaniemi H: Basic research studies to understand
aneurysm disease. Drug News Perspect. 21:142–148. 2008.PubMed/NCBI
|
|
3
|
Curci JA and Thompson RW: Adaptive
cellular immunity in aortic aneurysms: Cause, consequence, or
context? J Clin Invest. 114:168–171. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Svensjo S, Björck M and Wanhainen A:
Update on screening for abdominal aortic aneurysm: A topical
review. Eur J Vasc Endovasc Surg. 48:659–667. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kuivaniemi H and Elmore JR: Opportunities
in abdominal aortic aneurysm research: Epidemiology, genetics, and
pathophysiology. Ann Vasc Surg. 26:862–870. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shimizu K, Mitchell RN and Libby P:
Inflammation and cellular immune responses in abdominal aortic
aneurysms. Arterioscler Thromb Vasc Biol. 26:987–994. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Daugherty A and Cassis LA: Mouse models of
abdominal aortic aneurysms. Arterioscler Thromb Vasc Biol.
24:429–434. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rowe VL, Stevens SL, Reddick TT, Freeman
MB, Donnell R, Carroll RC and Goldman MH: Vascular smooth muscle
cell apoptosis in aneurysmal, occlusive, and normal human aortas. J
Vasc Surg. 31:567–576. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Allaire E, Muscatelli-Groux B, Mandet C,
Guinault AM, Bruneval P, Desgranges P, Clowes A, Méllière D and
Becquemin JP: Paracrine effect of vascular smooth muscle cells in
the prevention of aortic aneurysm formation. J Vasc Surg.
36:1018–1026. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choke E, Cockerill G, Wilson WR, Sayed S,
Dawson J, Loftus I and Thompson MM: A review of biological factors
implicated in abdominal aortic aneurysm rupture. Eur J Vasc
Endovasc Surg. 30:227–244. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rotmans JI, Velema E, Verhagen HJ,
Blankensteijn JD, de Kleijn DP, Stroes ES and Pasterkamp G: Matrix
metalloproteinase inhibition reduces intimal hyperplasia in a
porcine arteriovenous-graft model. J Vasc Surg. 39:432–439. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuivaniemi H, Platsoucas CD and Tilson MD
III: Aortic aneurysms: An immune disease with a strong genetic
component. Circulation. 117:242–252. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Abdul-Hussien H, Hanemaaijer R, Kleemann
R, Verhaaren BF, van Bockel JH and Lindeman JH: The pathophysiology
of abdominal aortic aneurysm growth: Corresponding and discordant
inflammatory and proteolytic processes in abdominal aortic and
popliteal artery aneurysms. J Vasc Surg. 51:1479–1487. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lenk GM, Tromp G, Weinsheimer S, Gatalica
Z, Berguer R and Kuivaniemi H: Whole genome expression profiling
reveals a significant role for immune function in human abdominal
aortic aneurysms. BMC Genomics. 8:2372007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pahl MC, Derr K, Gäbel G, Hinterseher I,
Elmore JR, Schworer CM, Peeler TC, Franklin DP, Gray JL, Carey DJ,
et al: MicroRNA expression signature in human abdominal aortic
aneurysms. BMC Med Genomics. 5:252012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stather P, Sylvius N, Sidloff D, Dattani
N, Verissimo A, Wild JB, Butt HZ, Choke E, Sayers RD and Bown MJ:
Identification of microRNAs associated with abdominal aortic
aneurysms and peripheral arterial disease. Br J Surg. 102:755–766.
2015. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maegdefessel L, Spin JM, Raaz U, Eken SM,
Toh R, Azuma J, Adam M, Nakagami F, Heymann HM, Chernogubova E, et
al: miR-24 limits aortic vascular inflammation and murine abdominal
aneurysm development. Nature Commun. 5:52142014. View Article : Google Scholar
|
|
18
|
Angaji SA, Hedayati SS, Poor RH, Madani S,
Poor SS and Panahi S: Application of RNA interference in treating
human diseases. J Genet. 89:527–537. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kong W, Zhao JJ, He L and Cheng JQ:
Strategies for profiling microRNA expression. J Cell Physiol.
218:22–25. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gartel AL and Kandel ES: miRNAs: Little
known mediators of oncogenesis. Semin Cancer Biol. 18:103–110.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee KH, Chen YL, Yeh SD, Hsiao M, Lin JT,
Goan YG and Lu PJ: MicroRNA-330 acts as tumor suppressor and
induces apoptosis of prostate cancer cells through E2F1-mediated
suppression of Akt phosphorylation. Oncogene. 28:3360–3370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:2015. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Su Z, Yang Z, Xu Y, Chen Y and Yu Q:
MicroRNAs in apoptosis, autophagy and necroptosis. Oncotarget.
6:8474–8490. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Q, Morgan S, Ren J and Liu B:
Abstract 295: Macrophages induce apoptosis of arterial smooth
muscle cells via a monocyte chemoattractant protein-1 (MCP-1)
mediated cytotoxicity. Arteriosc Thromb Vasc Biol. 33:A2952013.
|
|
33
|
Dietrich DR: Toxicological and
pathological applications of proliferating cell nuclear antigen
(PCNA), a novel endogenous marker for cell proliferation. Crit Rev
Toxicol. 23:77–109. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Reynolds N, Fantes PA and MacNeill SA: A
key role for replication factor C in DNA replication checkpoint
function in fission yeast. Nucleic Acids Res. 27:462–469. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Maalouf SW, Liu WS and Pate JL: MicroRNA
in ovarian function. Cell Tissue Res. 363:7–18. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Singh DK, Bose S and Kumar S: Role of
microRNA in regulating cell signaling pathways, cell cycle, and
apoptosis in non-small cell lung cancer. Curr Mol Med. Apr
29–2016.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sherrard R, Luehr S, Holzkamp H, McJunkin
K, Memar N and Conradt B: miRNAs cooperate in apoptosis regulation
during C. elegans development. Genes Dev. 31:209–222. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Liu W, Wang H, Xu B, Li H and Cao
X: miRNA-126b regulated apoptosis of the human tongue carcinoma
cell line Tca8113-P60 via P38 signaling pathway. Int J Clin Exp
Med. 10:2654–2659. 2017.
|
|
39
|
Gross A, McDonnell JM and Korsmeyer SJ:
BCL-2 family members and the mitochondria in apoptosis. Genes Dev.
13:1899–1911. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Soutto M, Chen Z, Saleh MA, Katsha A, Zhu
S, Zaika A, Belkhiri A and El-Rifai W: TFF1 activates p53 through
down-regulation of miR-504 in gastric cancer. Oncotarget.
5:5663–5673. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Thompson RW, Liao S and Curci JA: Vascular
smooth muscle cell apoptosis in abdominal aortic aneurysms. Coron
Artery Dis. 8:623–631. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Inoue T and Node K: Molecular basis of
restenosis and novel issues of drug-eluting stents. Circ J.
73:615–621. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Doran AC, Meller N and McNamara CA: Role
of smooth muscle cells in the initiation and early progression of
atherosclerosis. Arterioscler Thromb Vasc Biol. 28:812–819. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cai X: Regulation of smooth muscle cells
in development and vascular disease: Current therapeutic
strategies. Expert Rev Cardiovasc Ther. 4:789–800. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hoshina K, Koyama H, Miyata T, Shigematsu
H, Takato T, Dalman RL and Nagawa H: Aortic wall cell proliferation
via basic fibroblast growth factor gene transfer limits progression
of experimental abdominal aortic aneurysm. J Vasc Surg. 40:512–518.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Scirocco A, Matarrese P, Carabotti M,
Ascione B, Malorni W and Severi C: Cellular and molecular
mechanisms of phenotypic switch in gastrointestinal smooth muscle.
J Cell Physiol. 231:295–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Charolidi N and Carroll VA: Hypoxia and
pulmonary hypertensionHypoxia and Human Diseases. InTech; 2017,
View Article : Google Scholar
|
|
48
|
Pei H, Tian C, Sun X, Qian X, Liu P, Liu W
and Chang Q: Overexpression of microRNA-145 promotes ascending
aortic aneurysm media remodeling through TGF-β1. Eur J Vasc
Endovasc Surg. 49:52–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kin K, Miyagawa S, Fukushima S, Shirakawa
Y, Torikai K, Shimamura K, Daimon T, Kawahara Y, Kuratani T and
Sawa Y: Tissue-and plasma-specific microRNA signatures for
atherosclerotic abdominal aortic aneurysm. J Am Heart Assoc.
1:e0007452012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Cohn DE, Fabbri M, Valeri N, Alder H,
Ivanov I, Liu CG, Croce CM and Resnick KE: Comprehensive miRNA
profiling of surgically staged endometrial cancer. Am J Obstet
Gynecol. 202:656.e1–e8. 2010. View Article : Google Scholar
|
|
51
|
Kwak PB, Iwasaki S and Tomari Y: The
microRNA pathway and cancer. Cancer Sci. 101:2309–2315. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Stadelmann C and Lassmann H: Detection of
apoptosis in tissue sections. Cell Tissue Res. 301:19–31. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Vaux DL and Korsmeyer SJ: Cell death in
development. Cell. 96:245–254. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Green DR and Reed JC: Mitochondria and
apoptosis. Science. 281:1309–1312. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chekulaeva M and Filipowicz W: Mechanisms
of miRNA-mediated post-transcriptional regulation in animal cells.
Curr Opin Cell Biol. 21:452–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Harris SL and Levine AJ: The p53 pathway:
Positive and negative feedback loops. Oncogene. 24:2899–2908. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Itahana K, Mao H, Jin A, Itahana Y, Clegg
HV, Lindström MS, Bhat KP, Godfrey VL, Evan GI and Zhang Y:
Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase
activity in the mouse reveals mechanistic insights into p53
regulation. Cancer Cell. 12:355–366. 2007. View Article : Google Scholar : PubMed/NCBI
|