Introduction
Blackcurrants (Ribes nigrum L.,
Grossulariaceae) contain a number of biochemical constituents,
including polyphenols, polyunsaturated fatty acids, organic acids
and phenolic acids (1–4). Blackcurrant flavonoids are a group of
polyphenolic compounds that include anthocyanins and flavonols.
Anthocyanins are natural plant pigments that are widely distributed
among flowers, fruits and vegetables. Blackcurrants are reported to
contain four anthocyanins: Delphinidin-3-glucoside;
delphinidin-3-rutinoside; cyanidin-3-glucoside; and
cyanidin-3-rutinoside. Delphinidin-3-rutinoside and
cyanidin-3-rutinoside are specific blackcurrant anthocyanins
(2). These blackcurrant
constituents have been associated with certain health benefits,
including improved blood flow (5),
decreased inflammatory marker levels (6) and protection against DNA damage
induced by hydrogen peroxide (7).
Previous studies have demonstrated that blackcurrant
phytochemicals exhibit anti-breast cancer effects. For example,
blackcurrant extract (BCE) or juice has been demonstrated to
inhibit breast cancer cell proliferation (8–10).
However, the effects of BCEs on normal human breast epithelial
cells have not been reported.
The objective of the present study was to
investigate whether an anthocyanin-rich BCE confers novel health
benefits to healthy breast epithelial cells. MCF10A healthy human
breast epithelial cells were exposed to BCE, and microarray and
Ingenuity® Pathway Analysis was performed to assess
alterations in gene expression. The regulation of the cell cycle
and the impact on cell viability was evaluated using
fluorescence-activated cell sorting. DNA damage and cell death was
examined by detecting apoptotic and necrotic cells using a comet
assay. mRNA expression of the DNA stability regulator
lysine-specific demethylase 5B (KDM5B) (11,12)
was investigated using the reverse transcription-quantitative
polymerase chain reaction (RT-qPCR).
Materials and methods
Materials and cell culture
The BCE powder, CaNZac-35, was purchased from Koyo
Mercantile Co., Ltd. (Tokyo, Japan). The BCR powder contained high
concentrations of polyphenols (37.6 g polyphenol/100 g BCE) and
anthocyanins (38.0 g anthocyacin/100 g BCE) (13). MCF10A breast cancer cells were
obtained from the American Type Culture Collection (Manassas, VA,
USA). Cells were maintained in a Mammary Epithelial Cell Growth
Medium kit (PromoCell GmbH, Heidelberg, Germany). The culture
experiments were conducted at 37°C in a humidified 5%
CO2 incubator.
Cell viability assay
MCF10A cells were seeded in 96-well plates at 5,000
cells/well and cultured overnight. Cells were then incubated with
0, 10, 25, 50, 100, 200 and 400 µg/ml BCE for 24 h, and observed
under a fluorescence microscope at ×200 magnification (FSX100;
Olympus Corporation, Tokyo, Japan). Cell viability was determined
using the PrestoBlue cell viability reagent (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA), according to the
manufacturer's protocol. PrestoBlue reagent was added to the cells
and cells were incubated for 6 h at 37°C. Absorbance was measured
on a Benchmark microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) at a wavelength of 570 nm. The absorbance was
assumed to be directly proportional to the number of cells. BCE
concentration-cell growth (dose effect) curves were drawn for each
treatment and approximate half-maximal response concentration
(EC50) values were determined using Excel software
(version 2013; Microsoft Corporation, Redmond, WA, USA).
Microarray gene expression
profiling
MCF10A cells were seeded in 21-cm2
culture dishes in mammary epithelial cell growth medium and grown
under the conditions described above until confluent, at which
point the medium was replaced with fresh medium with or without BCE
(50 µg/m). After 24 h incubation, the cells were washed twice with
PBS and total RNA was extracted using the RNeasy mini kit (Qiagen,
Inc., Valencia, CA, USA). Total RNA (100 ng) was used to produce
cyanine 3-CTP-labeled cRNA using a low input quick amp labeling kit
(Agilent Technologies, Inc., Santa Clara, CA, USA). Labeled and
fragmented cRNA was hybridized to a SurePrint G3 Human Gene
Expression microarray (8×60K; version 2; Agilent Technologies,
Inc.). Labeling, hybridization, image scanning and data analysis
was performed at Bio Matrix Research, Inc. (Chiba, Japan). The
MCF10A microarray dataset is available at www.ncbi.nlm.nih.gov/geo under accession code
GSE58304.
Genes with ≥2-fold upregulation following exposure
of MCF10A cells to 50 µg/ml BCE were analyzed using
Ingenuity® Pathway Analysis software (version 18030641;
Qiagen, Inc.).
Cell cycle analysis
Cell cycle analysis was performed as previously
described (13,14). MCF10A cells were seeded in
21-cm2 culture dishes at 5×105 cells/well and
cultured overnight. The medium was then replaced with fresh medium
with or without BCE (0, 50 or 100 µg/ml). The cells were cultured
for 24 h prior to DNA staining with propidium iodide at room
temperature for 2 h. Cell cycle analysis was performed by
fluorescence-activated cell sorting, which was conducted using a
Cell Cycle Phase Determination kit (Cayman Chemical Company, Ann
Arbor, MI, USA) and a Cytomics FC500 flow cytometer (Beckman
Coulter, Inc., Brea, CA, USA). Data were analyzed using CXP
analysis software (version 2.0; Beckman Coulter, Inc.).
Alkali comet assay
A Trevigen Comet Assay™ kit (Trevigen, Gaithersburg,
MD, USA) was used to assess DNA strand breaks. MCF10A cells were
treated with BCE or H2O2 as a positive
control for 24 h and resuspended in ice-cold PBS at a density of
1×105 cells/ml. A 50-µl aliquot of cell suspension
(containing 1×105 cells/ml) was mixed with 500 µl 1%
low-melting agarose maintained at 37°C. A total 50 µl mixture was
immediately obtained and evenly spread onto the comet slides. To
accelerate gelling of the agarose disc, the slides were incubated
at 4°C in the dark for 10 min. The slides were subsequently
transferred to a prechilled lysis solution and incubated for 60 min
at 4°C. The slides were then transferred to an alkali unwinding
solution [200 mM NaOH, 1 mM EDTA (pH>13.0)] and incubated at
room temperature for 20 min in the dark to perform denaturation.
Subsequently, the slides were transferred to a prechilled alkaline
electrophoresis solution and electrophoresed at 21 V for 30 min
followed by washing with distilled water. The slides were then
immersed in ice-cold 70% ethanol at room temperature for 5 min and
air-dried. The slides were incubated with 100 µl SYBR Green I dye
[Thermo Fisher Scientific, Inc.; in 10 mM Tris-HCl (pH 7.5), 1 mM
EDTA buffer] for 5 min at 4°C to stain the DNA and were immediately
analyzed using an FSX100 fluorescence microscope. The images were
captured at ×100 magnification. Quantitative analysis of tail
length was performed using the Comet Assay IV image analysis system
(Perceptive Instruments Ltd., Bury St Edmunds, UK).
Cell death detection
MCF10A cells were seeded in 9-cm2 culture
dishes and cultured overnight. The medium was then replaced with
fresh medium with or without BCE (0, 50 or 100 µg/ml). Apoptosis
and necrosis was detected using a Cell Meter™ Apoptotic and
Necrotic Detection kit (AAT Bioquest, Inc., Sunnyvale, CA, USA).
The cells were observed with an FSX100 fluorescence microscope. The
images were captured at ×42 magnification. Quantitative analysis of
apoptotic and necrotic cells was performed using FSX-BSW software
(version 03.01; Olympus Corporation).
RT-qPCR
MCF10A cells were seeded in 9-cm2 culture
dishes and cultured as described above until confluent. The used
medium was then replaced with fresh medium with or without BCE (0,
25 or 50 µg/ml). The cells were incubated for 24 h and washed twice
with PBS. Total RNA was extracted using the RNeasy mini kit
(Qiagen, Inc.). cDNA was reverse-transcribed from total RNA (0.5
µg) using the Omniscript RT kit (Qiagen, Inc.). A MiniOpticon
Real-Time PCR system (Bio-Rad Laboratories, Hercules, CA, USA) and
GoTaq Green Master Mix (Promega Corporation, Madison, WI, USA) were
used for the quantification of specific mRNA. PCRs were denatured
at 92°C for 2 min, and amplified at 92°C for 30 sec, 58°C for 30
sec and 72°C for 30 sec, for 40 cycles. Transcript levels were
normalized to that of GAPDH cDNA. The primers used were as follows
(5′-3′): KDM5B forward, ATT GCC TCA AAG GAA TTT GGC AGT G and
reverse, CAT CAC TGG CAT GTT GTT CAA ATT C; GAPDH forward, AGA AGG
CTG GGG CTC ATT TG and reverse, AGG GGC CAT CCA CAG TCT TC. PCR
specificity was confirmed by melting curve analysis. Relative gene
expression was calculated using the 2−ΔΔCq method
(15).
Statistical analysis
Results are presented as the mean ± standard error
of the mean of at least three independent experiments. Statistical
significance was determined using one-way analysis of variance
followed by Tukey's test. SPSS statistical software (version 16.0;
SPSS, Inc., Chicago, IL, USA) was used for statistical analysis.
P<0.05 was considered to indicate a statistically significant
difference.
Results
BCE decreases MCF10A cell
viability
To investigate BCE toxicity to MCF10A cells,
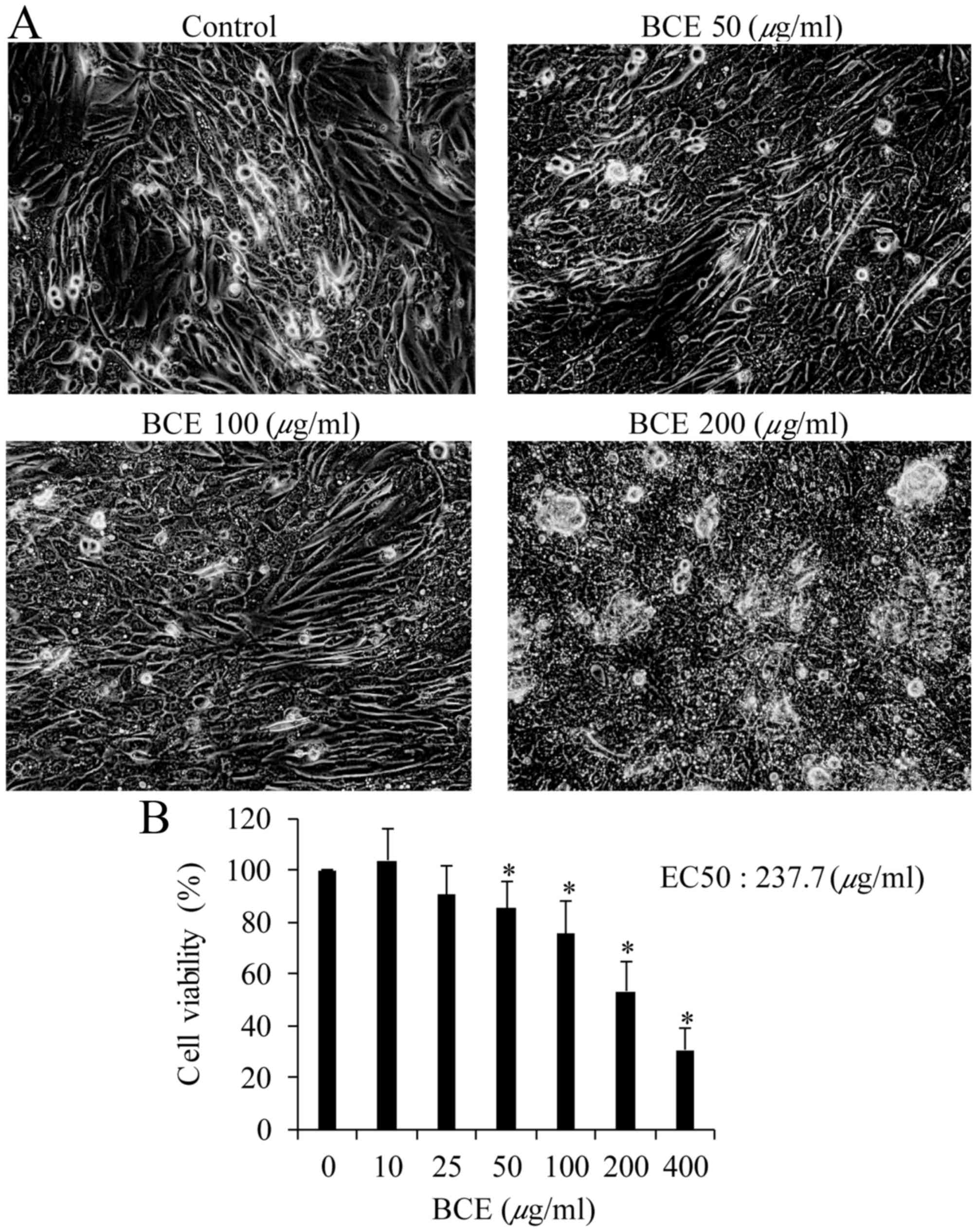
viability was determined using the PrestoBlue assay. Morphological
alterations were observed in cells exposed to 200 µg/ml BCE but not
in cells exposed to the lower concentrations of 50 and 100 µg/ml
(Fig. 1A). The percentages of
cells that remained viable following exposure to 50, 100, 200 and
400 µg/ml BCE were 85, 75, 53 and 31%, respectively (Fig. 1B). There was no significant
alteration in the viability of cells exposed to 10 or 25 µg/ml
BCE.
Ingenuity Pathway Analysis
To investigate the impact of BCE on MCF10A cells,
gene expression in the cells prior to and following BCE treatment
was compared using microarrays. Ingenuity Pathway Analysis was used
to investigate the functional associations between sets of genes
with modified expression levels. Given that 50 µg/ml BCE did not
affect MCF10A cell morphology and the EC50 value of BCE
was 237.7 µg/ml (Fig. 1B), a
concentration of 50 µg/ml was selected for microarray analysis. A
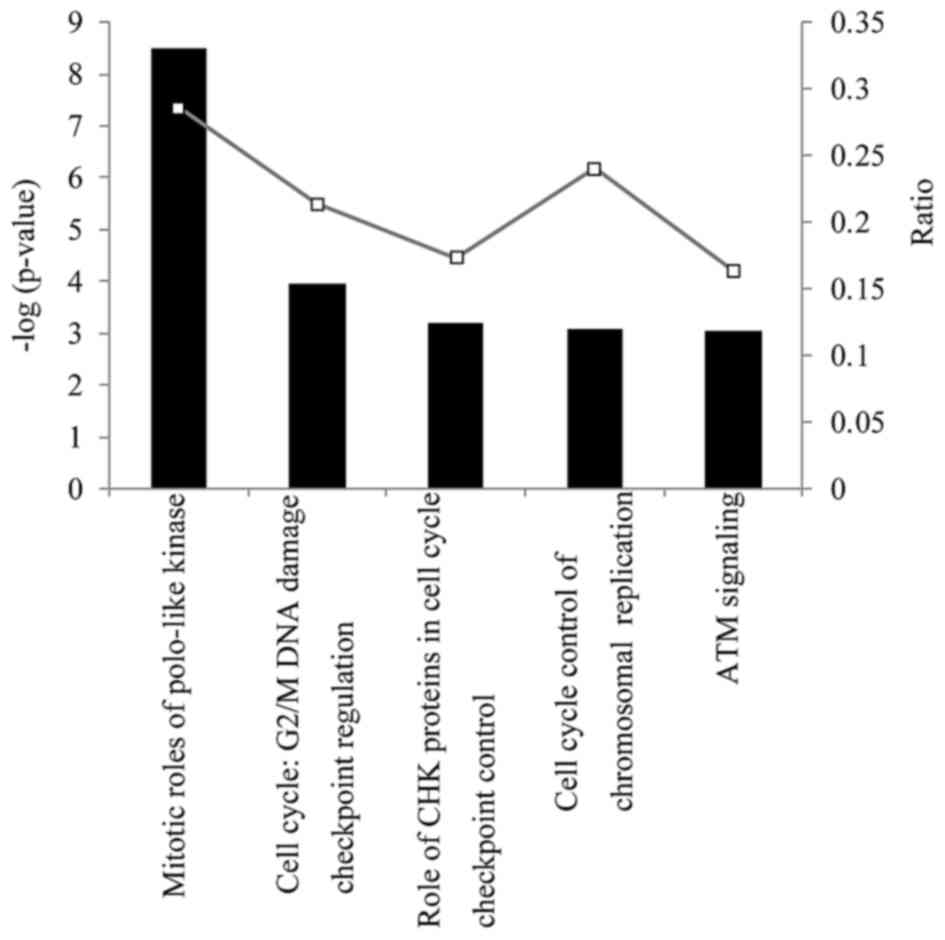
total of 147 significant canonical pathways were identified
(P<0.05), the majority of which were associated with cell cycle
signaling functions. The primary canonical pathways were ‘mitotic
roles of polo-like kinase’, ‘G2/M DNA damage checkpoint
regulation’, ‘the role of checkpoint kinase proteins in cell
cycle’, ‘cell cycle control of chromosomal replication checkpoint
control’ and ‘ataxia telangiectasia mutated signaling’ (Fig. 2). Detailed gene expression
alterations in each significantly affected canonical pathway are
presented in Table I. Mitotic
roles of polo-like kinase-related genes were affected the most.
Certain genes were common genes altered between mitotic roles of
polo-like kinase and other canonical pathways (G2/M DNA damage
checkpoint regulation, role of checkpoint kinase proteins in cell
cycle or ataxia telangiectasia mutated signaling) (Table I). These results indicate that BCE
is associated with significantly downregulated expression levels of
‘mitotic roles of Polo-like kinase’ and ‘cell cycle control of
chromosomal replication checkpoint control’ genes.
 | Table I.Details of altered gene expression in
canonical pathways. |
Table I.
Details of altered gene expression in
canonical pathways.
| Canonical
pathway | Gene symbol | Gene name | Fold-change |
|---|
| Mitotic roles of
polo-like kinase | AURKA | Aurora kinase
A | 0.46 |
| related genes | CCNB1 | Cyclin B1 | 0.40 |
|
| CCNB2 | Cyclin B2 | 0.37 |
|
| CDC20 | Cell division cycle
20 | 0.37 |
|
| CDC25A | Cell division cycle
25A | 0.44 |
|
| CDC25C | Cell division cycle
25C | 0.44 |
|
| CDK1 | Cyclin-dependent
kinase 1 | 0.43 |
|
| CHEK2 | Checkpoint kinase
2 | 0.48 |
|
| ESPL1 | Extra spindle pole
bodies homolog 1 | 0.45 |
|
| FBXO5 | F-box protein
5 | 0.43 |
|
| KIF11 | Kinesin family
member 11 | 0.48 |
|
| KIF23 | Kinesin family
member 23 | 0.41 |
|
| PKMYT1 | Protein kinase,
membrane associated tyrosine/threonine 1 | 0.42 |
|
| PLK1 | Polo-like kinase
1 | 0.42 |
|
| PLK4 | Polo-like kinase
4 | 0.41 |
|
| PRC1 | Protein regulator
of cytokinesis 1 | 0.39 |
| Cell cycle: G2/M
DNA damage | CCNB1 | Cyclin B1 | 0.40 |
| checkpoint
regulation | CCNB2 | Cyclin B2 | 0.37 |
|
| CDC25C | Cell division cycle
25C | 0.44 |
|
| CDK1 | Cyclin-dependent
kinase 1 | 0.43 |
|
| CHEK2 | Checkpoint kinase
2 | 0.48 |
|
| CKS2 | CDC28 protein
kinase regulatory subunit 2 | 0.49 |
|
| PKMYT1 | Protein kinase,
membrane associated 1 tyrosine/threonine | 0.42 |
|
| PLK1 | Polo-like kinase
1 | 0.42 |
|
| TOP2A | Topoisomerase (DNA)
IIα | 0.46 |
| Role of CHK
proteins in cell cycle | CDC25A | Cell division cycle
25A | 0.44 |
| checkpoint
control | CDC25C | Cell division cycle
25C | 0.44 |
|
| CDK1 | Cyclin-dependent
kinase 1 | 0.43 |
|
| CHEK2 | Checkpoint kinase
2 | 0.48 |
|
| CLSPN | Claspin | 0.50 |
|
| PLK1 | Polo-like kinase
1 | 0.42 |
|
| RFC3 | Replication factor
C subunit 3 | 0.37 |
|
| SLC19A1 | Solute carrier
family 19 (folate transporter), member 1 | 0.48 |
| Cell cycle control
of chromosomal | CDC45 | Cell division cycle
45 | 0.38 |
| replication | CDT1 | Chromatin licensing
and DNA replication factor 1 | 0.43 |
|
| CHEK2 | Checkpoint kinase
2 | 0.48 |
|
| MCM7 | Minichromosome
maintenance complex component 7 | 0.48 |
|
| ORC1 | Origin recognition
complex subunit 1 | 0.31 |
|
| ORC6 | Origin recognition
complex subunit 6 | 0.37 |
| Ataxia
telangiectasia mutated | BLM | Bloom syndrome,
recQ helicase-like | 0.44 |
| signaling | CCNB1 | Cyclin B1 | 0.40 |
|
| CCNB2 | Cyclin B2 | 0.37 |
|
| CDC25A | Cell division cycle
25A | 0.44 |
|
| CDC25C | Cell division cycle
25C | 0.44 |
|
| CDK1 | Cyclin-dependent
kinase 1 | 0.43 |
|
| CHEK2 | Checkpoint kinase
2 | 0.48 |
|
| RAD51 | RAD51
recombinase | 0.50 |
|
| TP73 | Tumor protein
p73 | 15.20 |
Cell cycle analysis
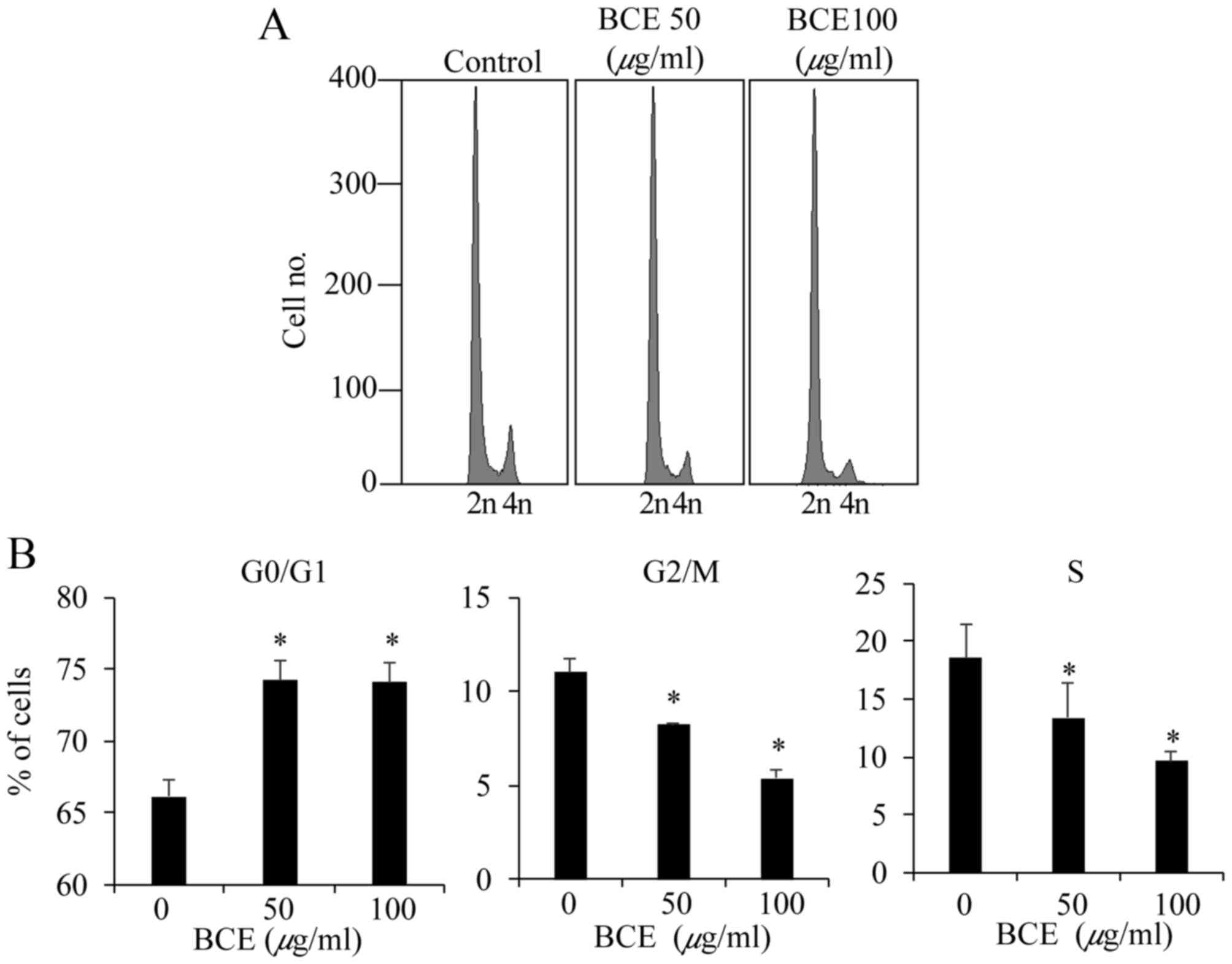
MCF10A cells were treated with BCE (0, 50 and 100
µg/ml) for 24 h prior to cell cycle analysis. Treatment with BCE
significantly increased the proportion of G0/G1 phase cells, and
decreased the proportions of G2/M phase and S phase cells (Fig. 3). These results indicate that BCE
inhibits cell cycle progression by inducing G0/G1 arrest in MCF10A
cells.
BCE-induced DNA damage
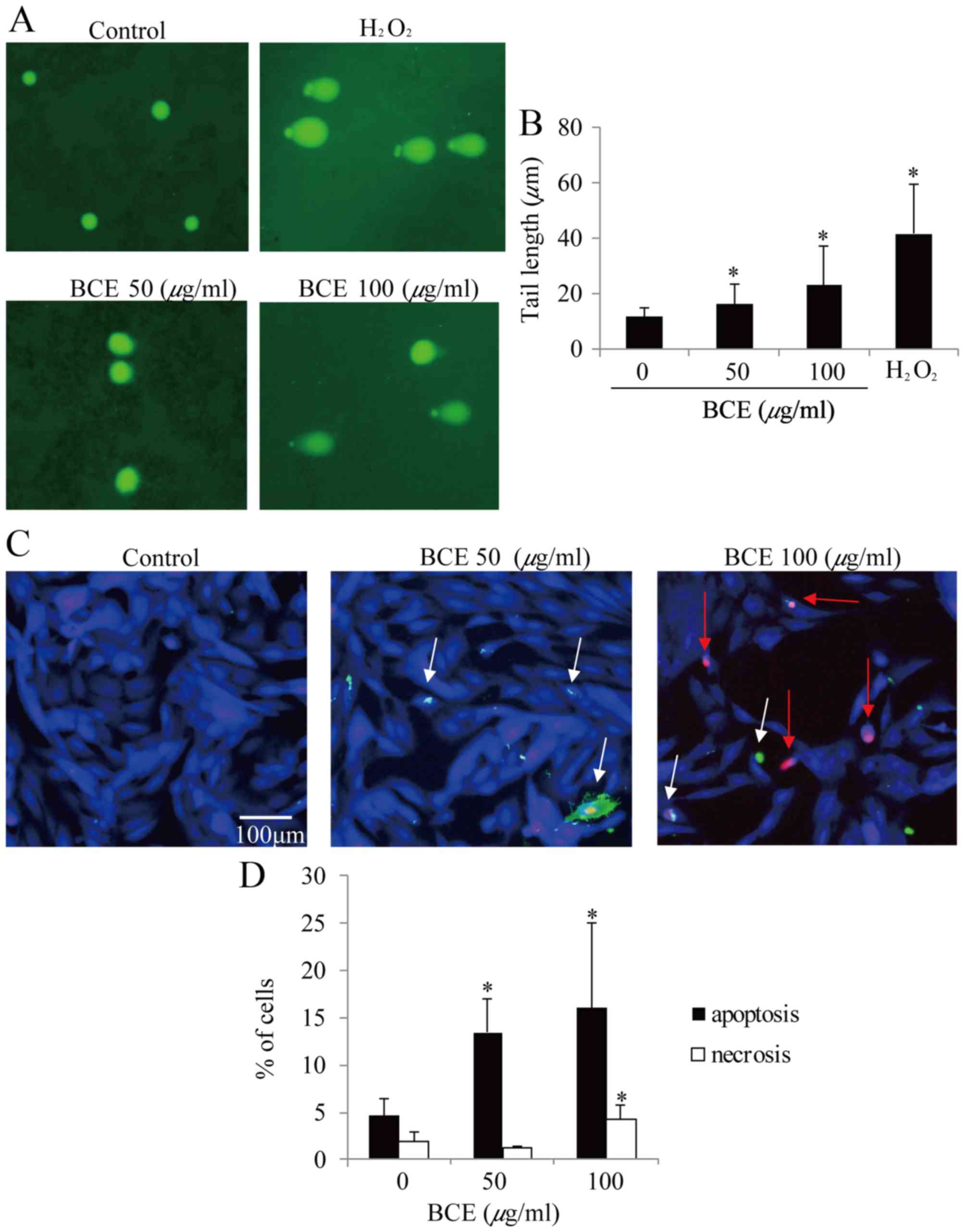
As DNA damage is an early event of cell cycle
arrest, it was further examined whether BCE induces DNA damage in
MCF10A cells. As presented in Fig. 4A
and B, 50 and 100 µg/ml BCE induced DNA damage in a
dose-dependent manner as measured by alkaline comet assay. The
comet tail lengths of 0, 50, 100 µg/ml BCE- and
H2O2-treated MCF10A cells were 11.4, 15.9,
22.9, and 41.6 µm, respectively. These results suggest that BCE
caused significant DNA damage.
Induction of apoptosis and
necrosis
As BCE decreased cell viability and induced G0/G1
phase arrest and DNA damage in MCF10A, it was hypothesized that BCE
may contribute to cell death. To examine this possibility, the
number of apoptotic and necrotic MCF10A cells were counted
following treatment with BCE. The cultures treated with 0, 50 and
100 µg/ml BCE contained 4.6, 13.4 and 16.0% apoptotic cells,
respectively (Fig. 4C and D). As
compared with the untreated cells (1.9%), the percentage of
necrotic cells increased in the 100 µg/ml BCE-treated culture
(4.3%) but not in the 50 µg/ml BCE-treated culture (Fig. 4C and D). These results indicate
that BCE acts as an apoptotic inducer in MCF10A cells.
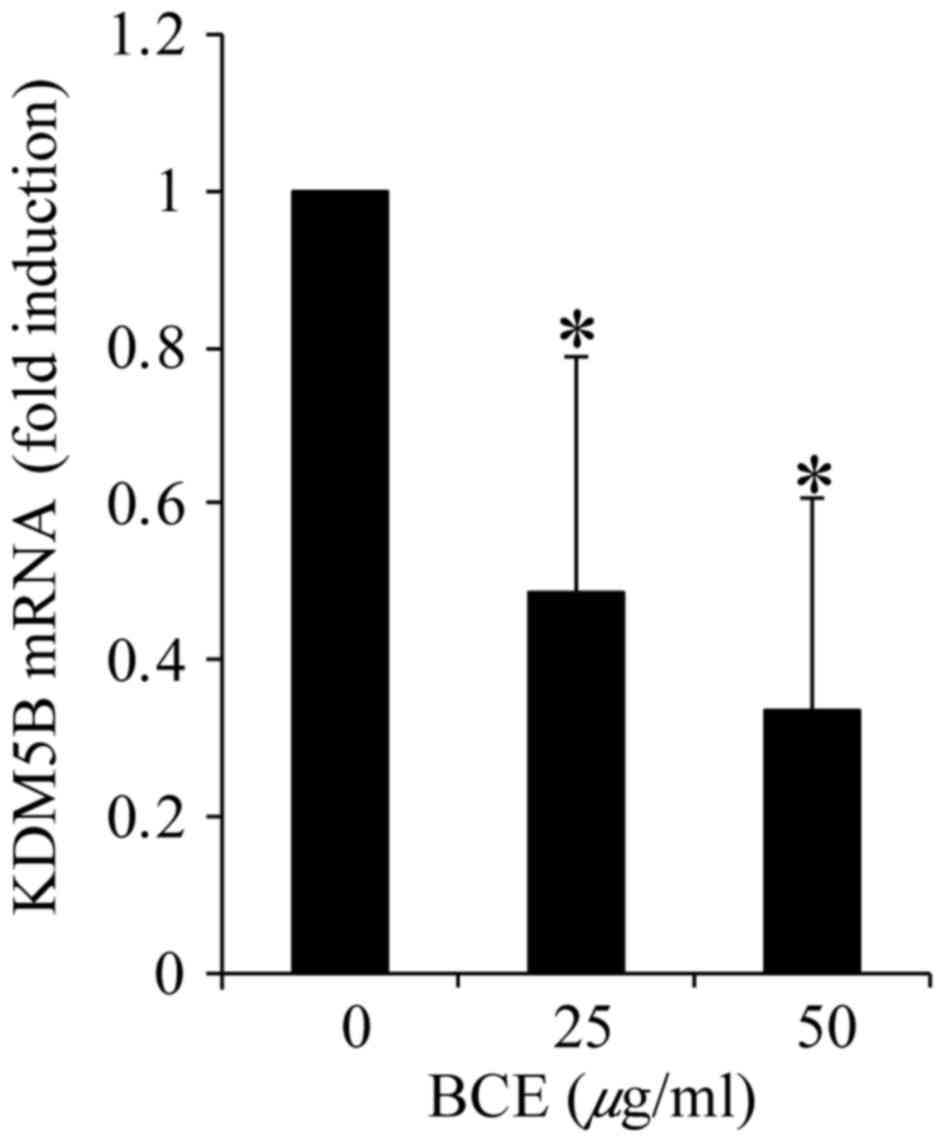
BCE decreases KDM5B expression
As KDM5B is known to be a DNA stability regulator
with high expression in breast cancer cells, its gene expression
was investigated by RT-qPCR. Treatment with BCE decreased KDM5B
expression in a dose-dependent manner (Fig. 5). These result indicate that BCE
induces KDM5B downregulation.
Discussion
In addition to the known health benefits of
blackcurrants, the present study investigated a novel function of
anthocyanin-rich BCE in the MCF10A healthy mammary epithelial cell
line. The present study demonstrated that exposure of MCF10A cells
to BCE reduces the expression of genes involved in cell signaling
pathways, including the mitotic roles of polo-like kinase signaling
and associated genes, and induces G0/G1 arrest and cell death.
Although canonical pathway analysis demonstrated that BCE reduced
certain signaling pathways, certain genes were common among
pathways. The important dysregulated signaling pathways appeared to
be the ‘mitotic roles of polo-like kinase’ and ‘cell cycle control
of chromosomal replication checkpoint control’ pathways.
Fluorescence-activated cell sorting analysis revealed that cell
cycle arrest reduced M and S phase cells, and the comet assay
revealed DNA damage following exposure to 50 or 100 µg/ml BCE.
Previous studies have demonstrated that polyphenols induce DNA
damage, and G2/M or G0/G1 cell cycle arrest (16–19).
BCE has been demonstrated to exert an
anti-proliferative effect on HT29 colon cancer cells, possibly
through the suppression of the cyclin-dependent kinase inhibitor 1
(p21WAF1) signaling pathway (20) and a potent cytotoxic effect against
hepatocellular carcinoma (21).
However, in the present study, microarray and Ingenuity Pathway
Analysis did not indicate suppression of the p21WAF1
signaling pathway (data not shown).
Microarray and Ingenuity Pathway Analysis revealed a
reduction in polo-like kinase signaling and associated genes. It is
known that almost every step of cell division, from entry into
mitosis to cytokinesis, is regulated by protein phosphorylation by
certain kinases, including aurora kinase A (AURKA), AURKB, the
cyclin-dependent kinase/cyclin B complex and polo-kinase 1 (PLK1)
(22,23). As the AURKs are frequently
overexpressed in cancer cells, it is hypothesized that AURKA may
cause the progression of gene instability observed in cancer.
Furthermore, AURKA and PLK1 cross-talk during mitotic entry and
spindle assembly (24). Therefore,
inhibitors of AURK activity may have the potential to be used as
novel anticancer agents. As BCE suppressed AURKA and PLK1
expression, the present study may lead to the development of a
treatment for the prevention of breast cancer.
Phytoestrogens are chemical compounds synthesized by
plants and are obtained naturally in a wide range of foods, and can
have estrogenic effects (25–28).
The authors previously reported that low-density blackcurrant
anthocyanins and BCE exhibit phytoestrogenic activity mediated via
estrogen receptor α signaling (13). As certain phytoestrogens were
demonstrated to inhibit breast cell proliferation and induce
apoptosis (29,30), it was hypothesized that BCE may be
effective for the prevention of the breast cancer. Anthocyanins or
polyphenols cause DNA damage and are known to cause apoptosis, by
activation of the ataxia telangiectasia-mutated signaling gene and
induction of p53 and p21 (31–33).
However, in the present study, ataxia telangiectasia-mutated
signaling was not activated and expression of the key regulator of
DNA stability, KDM5B, was decreased. These results are different
from previous reports.
KDM5B with methylation H3K4 is required for mammary
gland development and serves an important role in the proliferative
capacity of breast cancer cells (34,35).
The interaction of cyclin B1 and cyclin-dependent kinase 1 is
essential for control of the cell cycle at the G2/M phase.
Conversely, KDM5B has been reported to possibly be involved in the
regulation of the G2/M checkpoint and late M phase of the cell
cycle (36–39). Furthermore, interference of KDM5B
was demonstrated to increase the accumulation of G1 phase MCF7
breast cancer cells (40). As
treatment with BCE reduces KDM5B expression, and induces DNA
damage, G0/G1 cell cycle arrest and apoptosis in MCF10A cells, BCE
may prove to be useful as a breast cancer preventative food.
In conclusion, the present study demonstrated that
anthocyanin-rich BCE induces G0/G1 cell cycle arrest and apoptosis
in the normal breast epithelial cell line MCF10A. BCE dysregulated
polo-like kinase signaling and reduced breast cancer cell
proliferation-associated genes, including AURKA and KDM5B. These
results suggest that blackcurrant anthocyanins may be useful as a
component of breast cancer prevention.
Acknowledgements
The authors of the present study would like to thank
Ms. Yukimi Kato (Hirosaki University, Hirosaki, Japan) and Ms.
Chiaki Uehara (Hirosaki University) for their support, and Editage
(http://www.editage.jp) for their English language
editing. The present study was supported in part by a Hirosaki
University Institutional Research Grant for Young Scientists and a
Grant-in-Aid from the Japan Cassis Association.
Glossary
Abbreviations
Abbreviations:
|
AURKA
|
aurora kinase A
|
|
BCE
|
blackcurrant extract
|
|
KDM5B
|
lysine-specific demethylase 5B
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Delazar A, Khodaie L, Afshar J, Nahar L
and Sarker SD: Isolation and free-radical-scavenging properties of
cyanidin 3-O-glycosides from the fruits of Ribes biebersteinii.
Berl. Acta Pharm. 60:1–11. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gopalan A, Reuben SC, Ahmed S, Darvesh AS,
Hohmann J and Bishayee A: The health benefits of blackcurrants.
Food Funct. 3:795–809. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Määttä KR, Kamal-Eldin A and Törrönen AR:
High-performance liquid chromatography (HPLC) analysis of phenolic
compounds in berries with diode array and electrospray ionization
mass spectrometric (MS) detection: Ribes species. J Agric Food
Chem. 51:6736–6744. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nielsen IL, Haren GR, Magnussen EL,
Dragsted LO and Rasmussen SE: Quantification of anthocyanins in
commercial black currant juices by simple high-performance liquid
chromatography. Investigation of their pH stability and
antioxidative potency. J Agric Food Chem. 51:5861–5866. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matsumoto H, Takenami E, Iwasaki-Kurashige
K, Osada T, Katsumura T and Hamaoka T: Effects of blackcurrant
anthocyanin intake on peripheral muscle circulation during typing
work in humans. Eur J Appl Physiol. 94:36–45. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu Y, Xia M, Yang Y, Liu F, Li Z, Hao Y,
Mi M, Jin T and Ling W: Purified anthocyanin supplementation
improves endothelial function via NO-cGMP activation in
hypercholesterolemic individuals. Clin Chem. 57:1524–1533. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamamoto A, Nakashima K, Kawamorita S,
Sugiyama A, Miura M, Kamitai Y and Kato Y: Protective effects of
raw and cooked blackcurrant extract on DNA damage induced by
hydrogen peroxide in human lymphoblastoid cells. Pharm Biol.
52:782–788. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aiyer HS, Warri AM, Woode DR,
Hilakivi-Clarke L and Clarke R: Influence of berry polyphenols on
receptor signaling and cell-death pathways: Implications for breast
cancer prevention. J Agric Food Chem. 60:5693–5708. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Boivin D, Blanchette M, Barrette S,
Moghrabi A and Beliveau R: Inhibition of cancer cell proliferation
and suppression of TNF-induced activation of NFkappaB by edible
berry juice. Anticancer Res. 27:937–948. 2007.PubMed/NCBI
|
|
10
|
Olsson ME, Gustavsson KE, Andersson S,
Nilsson A and Duan RD: Inhibition of cancer cell proliferation in
vitro by fruit and berry extracts and correlations with antioxidant
levels. J Agric Food Chem. 52:7264–7271. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Catchpole S, Spencer-Dene B, Hall D,
Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG,
Burchell JM and Taylor-Papadimitriou J: PLU-1/JARID1B/KDM5B is
required for embryonic survival and contributes to cell
proliferation in the mammary gland and in ER+ breast cancer cells.
Int J Oncol. 38:1267–1277. 2011.PubMed/NCBI
|
|
12
|
Li X, Liu L, Yang S, Song N, Zhou X, Gao
J, Yu N, Shan L, Wang Q, Liang J, et al: Histone demethylase KDM5B
is a key regulator of genome stability. Proc Natl Acad Sci USA.
111:7096–7101. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nanashima N, Horie K, Tomisawa T, Chiba M,
Nakano M, Fujita T, Maeda H, Kitajima M, Takamagi S, Uchiyama D, et
al: Phytoestrogenic activity of blackcurrant (Ribes nigrum)
anthocyanins is mediated through estrogen receptor alpha. Mol Nutr
Food Res. 59:2419–2431. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nanashima N, Yamada T, Shimizu T and
Tsuchida S: Deletion of phospholipase A2 group IVc induces
apoptosis in rat mammary tumour cells by the nuclear
factor-κB/lipocalin 2 pathway. Biochem J. 469:315–324. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin CJ, Chang YA, Lin YL, Liu SH, Chang CK
and Chen RM: Preclinical effects of honokiol on treating
glioblastoma multiforme via G1 phase arrest and cell apoptosis.
Phytomedicine. 23:517–527. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu JJ, Cai YJ and Ding J: Curcumin induces
DNA damage and caffeine-insensitive cell cycle arrest in colorectal
carcinoma HCT116 cells. Mol Cell Biochem. 354:247–252. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ouyang G, Yao L, Ruan K, Song G, Mao Y and
Bao S: Genistein induces G2/M cell cycle arrest and apoptosis of
human ovarian cancer cells via activation of DNA damage checkpoint
pathways. Cell Biol Int. 33:1237–1244. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Z, Wang CZ, Du GJ, Qi LW, Calway T,
He TC, Du W and Yuan CS: Genistein induces G2/M cell cycle arrest
and apoptosis via ATM/p53-dependent pathway in human colon cancer
cells. Int J Oncol. 43:289–296. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu QK, Koponen JM, Mykkanen HM and
Törrönen AR: Berry phenolic extracts modulate the expression of p21
(WAF1) and Bax but not Bcl-2 in HT-29 colon cancer cells. J Agric
Food Chem. 55:1156–1163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bishayee A, Haznagy-Radnai E, Mbimba T,
Sipos P, Morazzoni P, Darvesh AS, Bhatia D and Hohmann J:
Anthocyanin-rich black currant extract suppresses the growth of
human hepatocellular carcinoma cells. Nat Prod Commun. 5:1613–1618.
2010.PubMed/NCBI
|
|
22
|
Strebhardt K: Multifaceted polo-like
kinases: Drug targets and antitargets for cancer therapy. Nat Rev
Drug Discov. 9:643–660. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ng WT Weng, Shin JS, Roberts TL, Wang B
and Lee CS: Molecular interactions of polo-like kinase 1 in human
cancers. J Clin Pathol. 69:557–562. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Asteriti IA, De Mattia F and Guarguaglini
G: Cross-talk between AURKA and Plk1 in mitotic entry and spindle
assembly. Front Oncol. 5:2832015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Guo D, Wang J, Wang X, Luo H, Zhang H, Cao
D, Chen L and Huang N: Double directional adjusting estrogenic
effect of naringin from Rhizoma drynariae (Gusuibu). J
Ethnopharmacol. 138:451–457. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee YM, Kim JB, Bae JH, Lee JS, Kim PS,
Jang HH and Kim HR: Estrogen-like activity of aqueous extract from
Agrimonia pilosa Ledeb. in MCF-7 cells. BMC Complement Altern Med.
12:2602012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Limer JL and Speirs V: Phyto-oestrogens
and breast cancer chemoprevention. Breast Cancer Res. 6:119–127.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mahmoud AM, Yang W and Bosland MC: Soy
isoflavones and prostate cancer: A review of molecular mechanisms.
J Steroid Biochem Mol Biol. 140:116–132. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li T, Zhu J, Guo L, Shi X, Liu Y and Yang
X: Differential effects of polyphenols-enriched extracts from
hawthorn fruit peels and fleshes on cell cycle and apoptosis in
human MCF-7 breast carcinoma cells. Food Chem. 141:1008–1018. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rice S and Whitehead SA: Phytoestrogens
oestrogen synthesis and breast cancer. J Steroid Biochem Mol Biol.
108:186–195. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Seo HS, Ju JH, Jang K and Shin I:
Induction of apoptotic cell death by phytoestrogens by
up-regulating the levels of phospho-p53 and p21 in normal and
malignant estrogen receptor α-negative breast cells. Nutr Res.
31:139–146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Prasad R and Katiyar SK: Polyphenols from
green tea inhibit the growth of melanoma cells through inhibition
of class I histone deacetylases and induction of DNA damage. Genes
Cancer. 6:49–61. 2015.PubMed/NCBI
|
|
33
|
Demoulin B, Hermant M, Castrogiovanni C,
Staudt C and Dumont P: Resveratrol induces DNA damage in colon
cancer cells by poisoning topoisomerase II and activates the ATM
kinase to trigger p53-dependent apoptosis. Toxicol In Vitro.
29:1156–1165. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamane K, Tateishi K, Klose RJ, Fang J,
Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P
and Zhang Y: PLU-1 is an H3K4 demethylase involved in
transcriptional repression and breast cancer cell proliferation.
Mol Cell. 25:801–812. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zou MR, Cao J, Liu Z, Huh SJ, Polyak K and
Yan Q: Histone demethylase jumonji AT-rich interactive domain 1B
(JARID1B) controls mammary gland development by regulating key
developmental and lineage specification genes. J Biol Chem.
289:17620–17633. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Barrett KL, Demiranda D and Katula KS:
Cyclin b1 promoter activity and functional cdk1 complex formation
in G1 phase of human breast cancer cells. Cell Biol Int. 26:19–28.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chang DC, Xu N and Luo KQ: Degradation of
cyclin B is required for the onset of anaphase in Mammalian cells.
J Biol Chem. 278:37865–37873. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ducommun B, Brambilla P, Félix MA, Franza
BR Jr, Karsenti E and Draetta G: cdc2 phosphorylation is required
for its interaction with cyclin. EMBO J. 10:3311–3319.
1991.PubMed/NCBI
|
|
39
|
Scibetta AG, Santangelo S, Coleman J, Hall
D, Chaplin T, Copier J, Catchpole S, Burchell J and
Taylor-Papadimitriou J: Functional analysis of the transcription
repressor PLU-1/JARID1B. Mol Cell Biol. 27:7220–7235. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Mitra D, Das PM, Huynh FC and Jones FE:
Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle
progression through epigenetic repression of microRNA let-7e. J
Biol Chem. 286:40531–40535. 2011. View Article : Google Scholar : PubMed/NCBI
|