Introduction
Progression has previously been made regarding the
diagnosis and treatment of various cancers associated with
mortality in women, however, breast cancer is still considered to
be of primary concern worldwide (1). In order to diagnose and treat breast
cancer during an earlier progressive stage, the underlying
molecular mechanisms of its development and progression need to be
elucidated (2).
It has been reported that microRNAs (miRNAs), small
RNAs composed of 18–25 nucleotides, are incapable of coding
protein, and dysregulated miRNA expression is present in multiple
cancers, including breast cancer (3). miRNA is important in cancer
progression via an influence on diverse cellular activities,
including cell growth (4),
apoptosis (5), metastasis
(6), invasion (7) and the cell cycle (8). MiR-455-5p as an oncogene or tumor
suppressor has been verified to be associated with several cancers,
including hepatocellular adenoma (9), head and neck squamous cell carcinoma
(10) and oral squamous cell
carcinoma (11). It has previously
been demonstrated that in hepatocellular adenoma tissues and cell
lines, miR-455-5p expression levels are deregulated (9). In oral cancer, transforming growth
factor (TGF)-β-mediated highly expressed miR-455-5p promotes cancer
progression via decreasing the expression of ubiquitin conjugating
enzyme E2 B (UBE2B) (11).
Currently, there are not many studies that have been conducted
regarding the expression and role of miR-455-5p. The present study
demonstrated that miR-455-5p was upregulated in breast cancer.
Further statistical analysis revealed that miR-455-5p was notably
associated with breast cancer patient prognosis. Cytological
experimental results demonstrated that miR-455-5p exerted a
positive influence on breast cancer cell migratory and invasive
abilities.
Previous studies have reported that miRNAs directly
interact with their targets to regulate tumor progression (12). In gastric cancer, miR-455-5p acts
as a tumor suppressor via targeting and downregulating RAB18,
member RAS oncogene family RAB18 (13). The present study demonstrated that
miR-455-5p targeted and negatively regulated programmed cell death
(PDCD)4, which has been identified as a tumor suppressor in breast
cancer, and inhibits breast cancer cell migration, invasion and
growth. Overall, the results of the present study suggest that
miR-455-5p acts as a biomarker that may be used in the prediction
of breast cancer patient prognosis, in addition to aiding in the
development of novel molecular targets and therapeutic strategies
to treat the disease.
Patients and methods
Patients and tissue samples
A total of 70 paired breast cancer tissue samples
and adjacent normal tissues were collected from patients (age
range, 24–76 years) following confirmation of written informed
consent. All the patients underwent surgical resections at the
Affiliated Tumor Hospital of Xinjiang Medical University (Urumqi,
China), during the period of January 2010-March 2016. The present
study gained the approval of the Ethics Committee of the Affiliated
Tumor Hospital of Xinjiang Medical University. Prior to mastectomy,
the 70 patients had not received radiotherapy or chemotherapy. The
extracted specimens were verified as breast cancer tissue with
pathological diagnosis according to the International Union against
Cancer. All fresh samples were immediately placed in a liquid
nitrogen container and then stored at −80°C for further
analysis.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Invitrogen TRIzol® reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) was used to isolate total RNA
samples from tissues or cells. In order to obtain cDNA, the
All-in-One miRNA qRT-PCR kit (GeneCopoeia, Inc., Rockville, MD,
USA) was used for conduction of reverse transcription and the PCR
reaction, according to the manufacturer's protocol. For reverse
transcription, the reaction was incubated at 42°C for 15 min;
heated to 95°C for 5 min and finally incubated at 5°C for 5 min.
Stem-loop primers used for reverse transcription of miR-455-5p and
U6 were 5′-GTCGTATCGAGTGGAGCGTCGAGCTATACGCACTCGATACGACACAAA-3′ and
5′-GTCCTATCCAGTGCAGGGTCCGAGGTGCACTGGATACGACAAAATATGGAAC-3′,
respectively. For RT-qPCR, the following thermocycling conditions
were used: Initial denaturation at 95°C for 5 min; followed by 40
cycles of denaturation at 95°C for 15 sec and annealing/elongation
at 60°C for 30 sec. The PCR primers for U6 and miR-455-5p were
obtained from GeneCopoeia, Inc. The PCR primer sequences were as
follows: U6 forward, 5′-CGCTTCGGCAGCACATATACTA-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCA-3′; miR-455-5p forward,
5′-CGAGCTTCCTTCTGCAGGT-3′ and reverse, 5′-CACCACTGCCATCCCACA-3′.
The 2−ΔΔCq method was used to quantify the level of
miRNA (14).
Cell culture and transfection
The breast cancer cell lines, including MDA-MB-453,
MCF-7, SK-BR-3, MDA-MB-231 and the normal breast cell line MCF-10A
were purchased from American Type Culture collection (Manassas, VA,
USA), then cultured with Dulbecco's modified Eagle medium (Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (GE Healthcare Life Sciences, Logan, UT,
USA). All of the cells were maintained in an incubator at 37°C, in
an environment containing 5% CO2. The miR-455-5p
inhibitors (5′-CGAUGUAGUCCAAAGGCACAUA-3′) and negative control (NC)
(5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from Shanghai
GenePharma Co., Ltd., (Shanghai, China). Cells underwent the
transfection procedure when the cell confluence reached 50–70%,
using Lipofectamine® 2000 (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The final
concentrations of the mimics and its corresponding NC were adjusted
to 50 nM (6-well plate), while inhibitors and its NC were modulated
to 200 nM (6-well plate). Total RNA and protein was extracted 24 h
following transfection, and the cells were harvested 48 h following
transfection for the subsequent experiments.
Wound-healing assay
The 6-well plates were used to culture cells
(5×105) and scratch wounds were created using a 200 µl
pipette tip. Following a culture period of 0 and 48 h, cells were
washed three times using sterile PBS to remove cellular debris,
then were imaged under an inverted microscope.
Transwell assay
The cell migratory and invasive abilities were
detected using non-Matrigel-coated/Matrigel-coated chambers,
respectively (BD Biosciences, Franklin Lakes, NJ, CA, USA).
Dulbecco's modified Eagle's medium supplemented with 10% FBS was
added into the lower chambers. MDA-MB-231 and MCF-7 cells
(1×105 cells/well), which had been transfected with
miR-455-5p inhibitors or NC, were plated into the upper chambers.
Following incubation for 24 or 48 h, the cells that remained in the
upper chambers were removed by PBS and cotton swabs. Following
this, 4% paraformaldehyde was used to fix cells at room temperature
for 30 min, which had passed through the filters, and cells were
stained with 0.1% crystal violet at room temperature for 5 min.
Cells were observed and imaged using a fluorescence microscope
(Olympus Corporation, Tokyo, Japan; magnification, ×100) and 5
separate fields were used for every filter to obtain an
average.
Western blotting
Total protein was extracted from breast cancer cells
using a radioimmunoprecipitation assay lysis buffer (Beyotime
Institute of Biotechnology, Haimen, China), according to the
manufacturer's protocol. The protein concentration was determined
using the bicinchonininc protein assay kit (Thermo Fisher
Scientific, Inc.). Following separation by 10% SDS-PAGE (25
µg/lane), the proteins were transferred to a polyvinlyidene
membrane. Then, the membrane was blocked using 5% non-fat milk at
room temperature for 2 h and incubated with PDCD4 (Cell Signaling
Technology, Inc., Danvers, MA, USA) or β-actin (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) primary antibodies, overnight at
4°C. The primary antibodies included anti-PDCD4 (1:1,000; cat. no.
ab51495; Abcam, Cambridge, MA, USA) and anti-β-actin (1:1,000; cat.
no. ab8226; Abcam). Following incubation with their respective
secondary antibodies at room temperature for 1 h the proteins were
visualized using an enhanced chemiluminescence chromogenic
substrate [Multi Sciences (Lianke) Biotech Co., Ltd., Hangzhou,
China]. The secondary antibodies included goat anti-mouse IgG2b
(1:8,000; cat. no. sc-2062; Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) and goat anti-rabbit IgG2b (1:8,000; cat. no. sc-2004;
Santa Cruz Biotechnology, Inc.). The immune complexes were
quantified using Image J (version 1.46; National Institutes of
Health, Bethesda, MA, USA).
Bioinformatics analysis
Bioinformatics tools TargetScan (www.targetscan.org), miRNA.org
(www.microrna.org) and miRbase (www.mirdb.org) were used to predict the targets of
miR-455-5p. Then the data from the three databases were combined to
obtain the most popular candidates in the three databases using
Microsoft Excel (version 2017; Microsoft Corporation, Redmond,
Washington) and SPSS software (version 22.0; IBM SPSS, Armonk, NY,
USA).
Statistical analysis
Data was analyzed using the Kaplan-Meier method,
unpaired Student's t-test, χ2 test, analysis of variance
followed by the least significant difference test and univariate
and multivariate Cox regression analysis. SPSS software, version
22.0 (IBM SPSS, Armonk, NY, USA) was used to analyze data. All the
presented data were expressed as the mean ± standard deviation.
Images were created using GraphPad Prism software (version 6;
GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of miR-455-5p in breast
cancer tissues increases
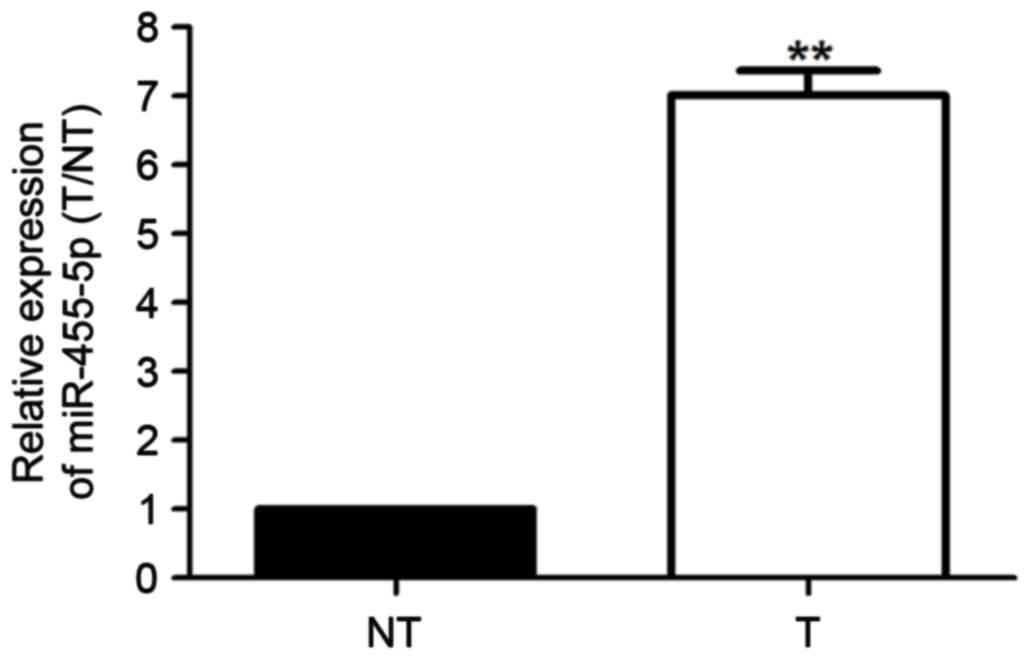
The present study explored miR-455-5p expression
levels in 70 paired breast cancer and adjacent non-cancerous tissue
samples. Results from RT-qPCR revealed that compared with control
group, miR-455-5p expression levels in breast cancer tissues were
notably increased (Fig. 1;
P<0.01). Therefore, miR-455-5p may be considered a valuable
oncogene in the progression of breast cancer.
Clinical significance of miR-455-5p
expression in breast cancer
Based on miR-455-5p average expression level (7.02;
range, 6.87–7.11), the 70 breast cancer patients were categorized
into two groups, low miR-455-5p and high miR-455-5p groups. Then,
the correlations between clinical characteristics and miR-455-5p
expression were assessed. The analysis revealed that miR-455-5p
expression was notably correlated with TNM stage, lymph node
metastasis and tumor differentiation (P<0.05; Table I). Additionally, univariate Cox
regression analysis (Table II)
and multivariate Cox regression analysis (Table III) were conducted for the
identification of prognostic factors for overall survival (OS) and
disease-free survival (DFS) time in patients with breast cancer.
Results revealed that TNM stage, lymph node metastasis, tumor
differentiation and miR-255-5p were prognostic factors of patients
with breast cancer. Therefore, it was hypothesized that miR-455-5p
may be associated with the survival rate of breast cancer and may
be used to predict the prognosis of breast cancer patients.
 | Table I.Association between miR-455-5p
expression level and clinical characteristics. |
Table I.
Association between miR-455-5p
expression level and clinical characteristics.
|
|
| miR455-5p
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
characteristics | Cases (n=70) | High (n=36) | Low (n=34) | P-value |
|---|
| Age, years |
|
|
|
|
|
<45 | 29 | 13 | 16 | 0.467 |
| ≥45 | 41 | 23 | 18 |
|
| Tumor size, cm |
|
|
|
|
|
<2 | 23 | 13 | 10 | 0.616 |
| ≥2 | 47 | 23 | 24 |
|
| Tumor location |
|
|
|
|
| Left | 34 | 16 | 18 | 0.633 |
|
Right | 36 | 20 | 16 |
|
| Differentiation |
|
|
|
|
|
Moderate/High | 26 | 7 | 19 | 0.003b |
| Low | 44 | 29 | 15 |
|
| Lymph node
metastasis |
|
|
|
|
|
Negative | 15 | 12 | 3 | 0.019a |
|
Positive | 55 | 24 | 31 |
|
| TNM stage |
|
|
|
|
| I/II | 38 | 31 | 17 | 0.002b |
|
III/IV | 32 | 5 | 17 |
|
| ER status |
|
|
|
|
|
Negative | 37 | 16 | 22 | 0.100 |
|
Positive | 33 | 20 | 12 |
|
| HER2 status |
|
|
|
|
|
Negative | 28 | 17 | 20 | 0.350 |
|
Positive | 42 | 19 | 14 |
|
| PR status |
|
|
|
|
|
Negative |
| 16 | 12 | 0.473 |
|
Positive |
| 20 | 22 |
|
 | Table II.Univariate Cox regression analysis
for the identification of prognostic factors for overall survival
and disease-free survival time in patients with breast cancer. |
Table II.
Univariate Cox regression analysis
for the identification of prognostic factors for overall survival
and disease-free survival time in patients with breast cancer.
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age, years(<45
vs. ≥45) | 1.853
(1.251–2.104) | 0.568 | 1.532
(1.463–1.604) | 0.845 |
| Tumor size, cm
(<2 vs. ≥2) | 1.332
(1.238–1.457) | 0.697 | 1.792
(1.659–1.846) | 0.446 |
| Tumor location
(left vs. right) | 1.643
(1.549–1.784) | 0.297 | 1.406
(1.379–1.523) | 0.684 |
| Differentiation
(moderate/high vs. low) | 2.729
(2.654–2.803) | 0.042 | 2.912
(2.878–3.041) | 0.004 |
| Lymph node
metastasis (negative vs. positive) | 1.478
(1.238–1.739) | 0.007 | 1.649
(1.587–1.882) | 0.015 |
| TNM stage (I/II vs.
III/IV) | 1.193
(1.046–1.254) | 0.026 | 1.264
(1.206–1.321) | 0.043 |
| ER status (negative
vs. positive) | 1.774
(1.536–1.862) | 0.922 | 2.012
(1.897–2.145) | 0.238 |
| HER2 status
(negative vs. positive) | 1.547
(1.502–1.621) | 0.218 | 1.708
(1.654–1.783) | 0.465 |
| PR status (negative
vs. positive) | 2.745
(2.133–3.027) | 0.589 | 1.965
(1.457–2.541) | 0.628 |
| miR-455-5p relative
expression (<7.02 vs. ≥7.02) | 2.612
(2.544–2.764) | 0.024 | 2.832
(2.478–3.007) | 0.017 |
 | Table III.Multivariate Cox regression analysis
for the identification of prognostic factors for overall survival
and disease-free survival time in patients with breast cancer. |
Table III.
Multivariate Cox regression analysis
for the identification of prognostic factors for overall survival
and disease-free survival time in patients with breast cancer.
|
| Overall
survival | Disease-free
survival |
|---|
|
|
|
|
|---|
| Variables | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Differentiation
(moderate/high vs. low) | 1.894
(1.761–2.327) | 0.918 | 1.664
(1.498–1.863) | 0.089 |
| Lymph node
metastasis (negative vs. positive) | 2.003
(1.847–2.214) | 0.028 | 2.784
(2.516–2.943) | 0.003 |
| TNM stage (negative
vs. positive) | 1.339
(1.235–1.456) | 0.026 | 1.649
(1.468–1.892) | 0.011 |
| miR-455-5p relative
expression (<7.02 vs. ≥7.02) | 2.334
(2.178–2.619) | 0.003 | 2.568
(2.511–2.597) | 0.019 |
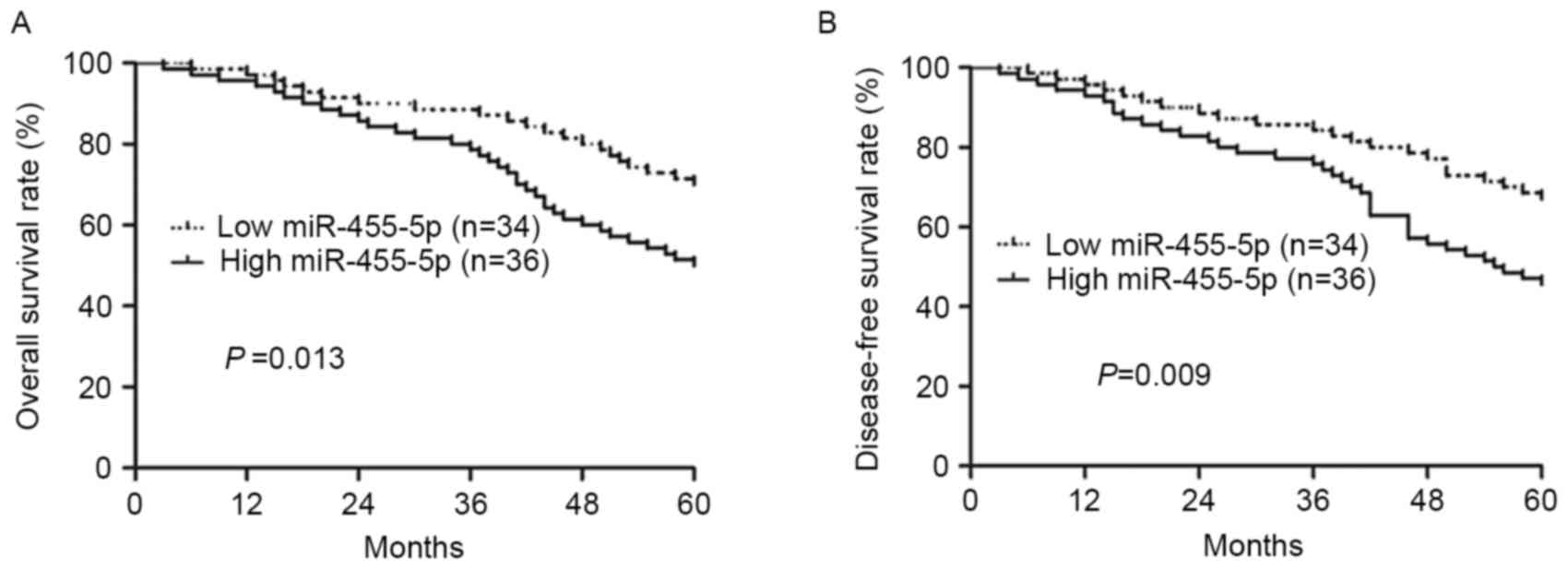
Elevated miR-455-5p is associated with
poor prognosis of breast cancer
The present study compared the survival rate of two
groups of patients using Kaplan-Meier survival analysis, and it was
revealed that patients with increased expression of miR-455-5p
exhibited a poorer OS and DFS rate compared with lower miR-455-5p
expression (P=0.013, P=0.009, respectively; Fig. 2A and B). Furthermore, univariate
and multivariate Cox proportional hazard regression analysis were
conducted to explore whether miR-455-5p acts as an independent
prognostic factor in breast cancer. The data demonstrated that
miR-455-5p expression, TNM stage and lymph node metastasis were
independent prognostic factors of breast cancer patients, which may
be used to predict their prognosis (P<0.05; Table II). The results demonstrated that
miR-455-5p may act as an independent prognostic factor for breast
cancer patients.
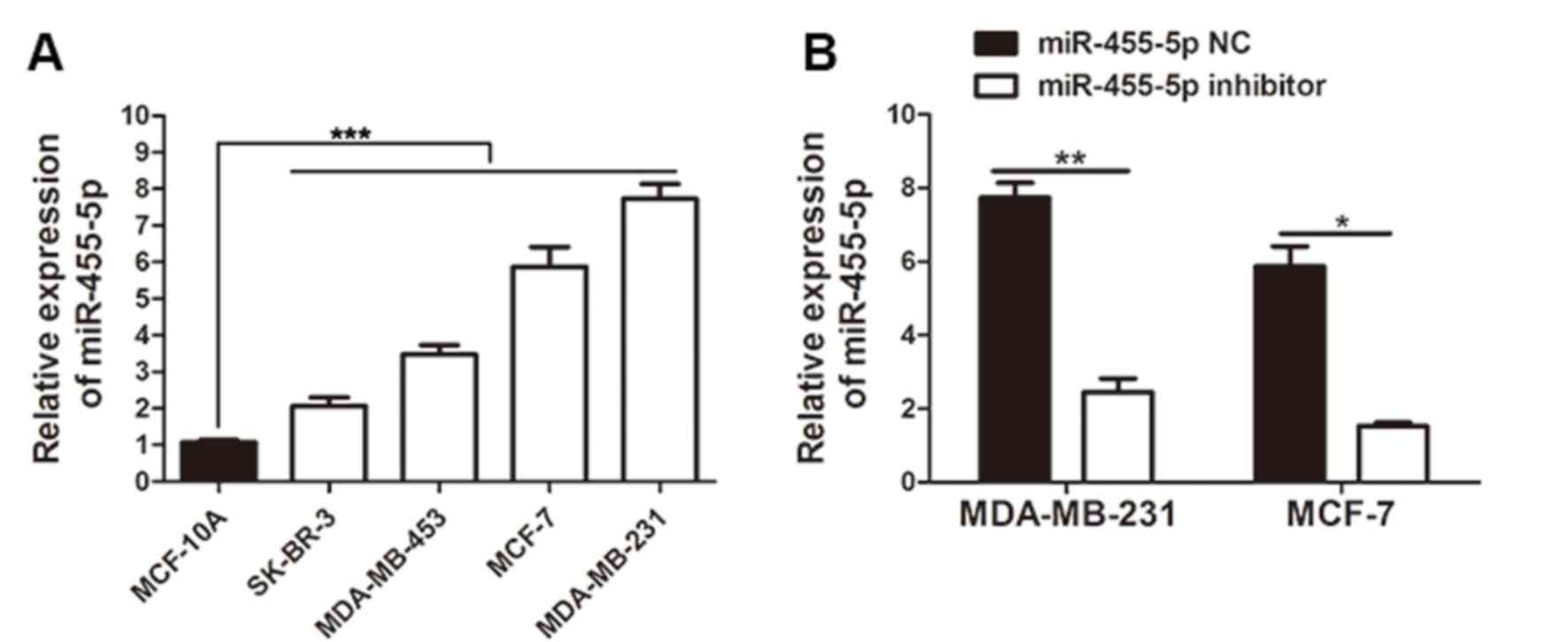
MiR-455-5p is overexpressed in breast
cancer cell lines
RT-qPCR was used to measure the expression of
miR-455-5p in breast cancer cell lines, including MDA-MB-453,
MCF-7, SK-BR-3 and MDA-MB-231, in addition to the normal cell line
MCF-10A. Consistent with the clinical results, the experiment
revealed that when compared with MCF-10A cells, miR-455-5p
expression in all breast cancer cell lines was markedly upregulated
(P<0.001; Fig. 3A). MDA-MB-231
and MCF-7 cells exhibited a relatively increased expression of
miR-455-5p, and the two cell types were therefore selected to be
used in further experiments. To explore the function of miR-455-5p
in breast cancer progression, the miR-455-5p expression in MCF-7
cells and MDA-MB-231 cells was manipulated via transfection with
miR-455-5p inhibitor or NC into cells. The results of the RT-qPCR
demonstrated that the miR-455-5p inhibitor markedly repressed
miR-455-5p expression in these cell lines (Fig. 3B; P<0.05).
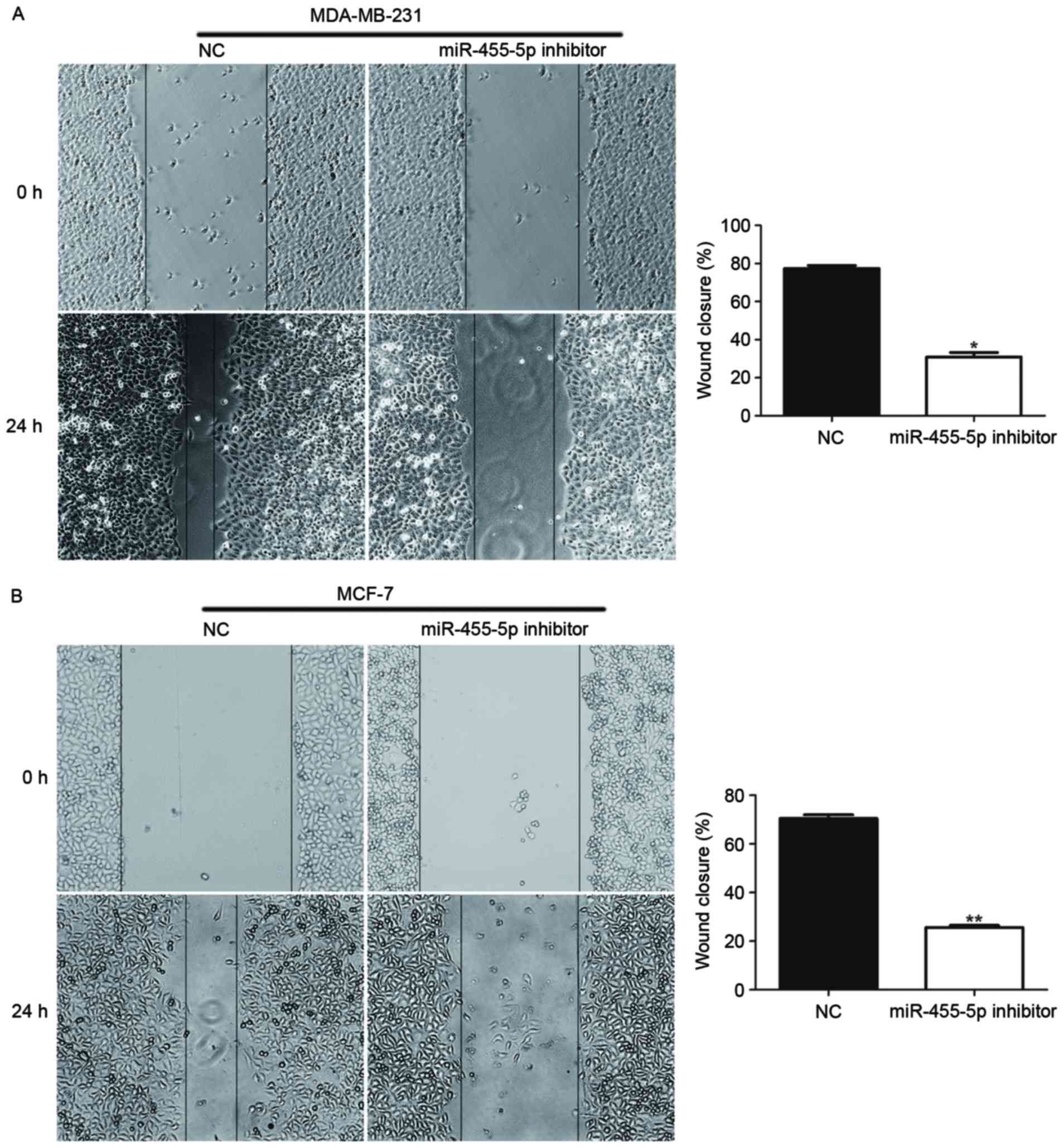
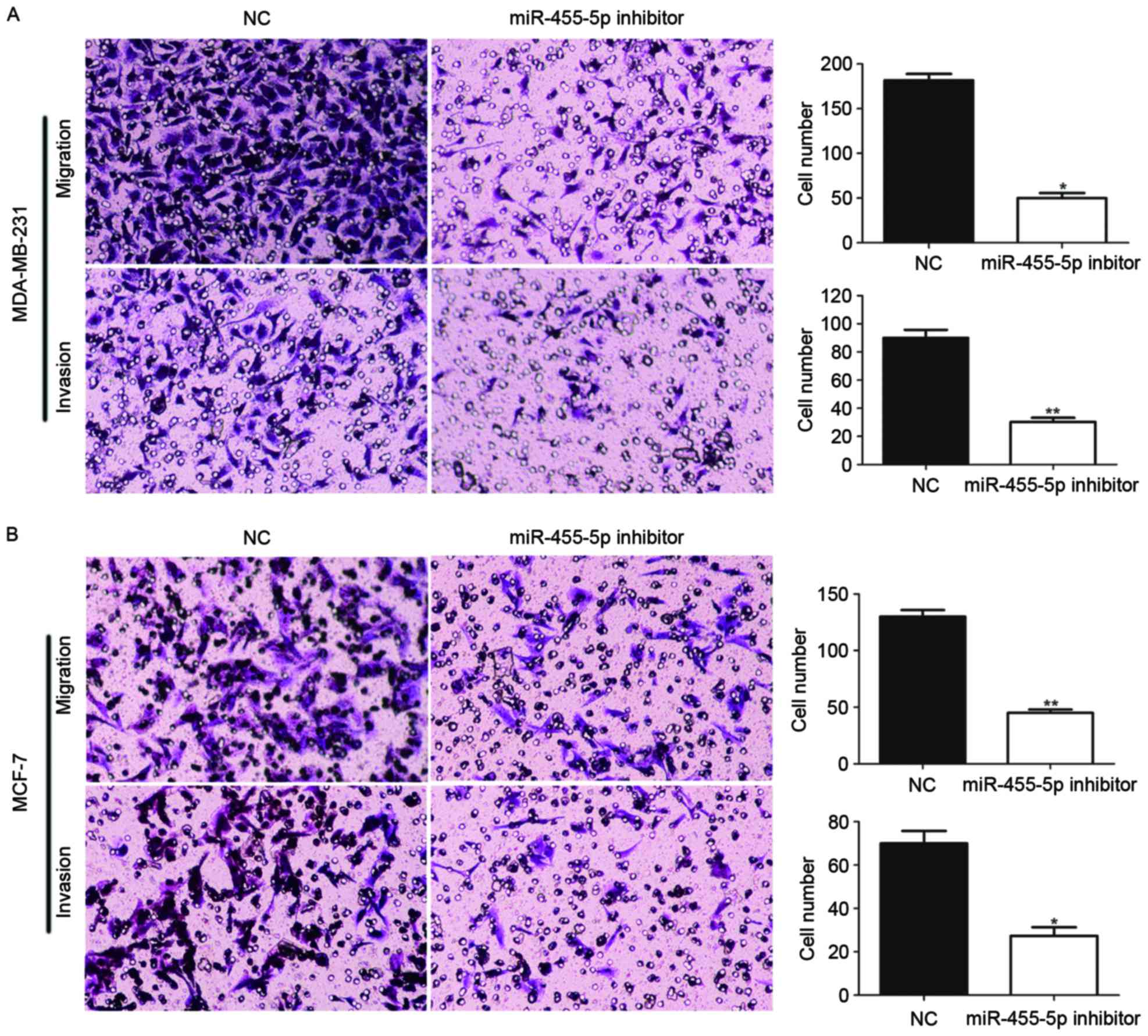
MiR-455-5p promotes cell invasion and
migration
Furthermore, the present study investigated whether
miR-711 promotes invasion and migration abilities of breast cancer
cells, using wound healing and Transwell assays. The results
indicated that compared with the NC, the groups with downregulated
miR-455-5p via miR-455-5p inhibitor transfection, exhibited
suppressed invasion and migration abilities (Figs. 4 and 5; P<0.05). Therefore, the findings
demonstrated that miR-455-5p facilitated breast cancer cell
migratory and invasive abilities.
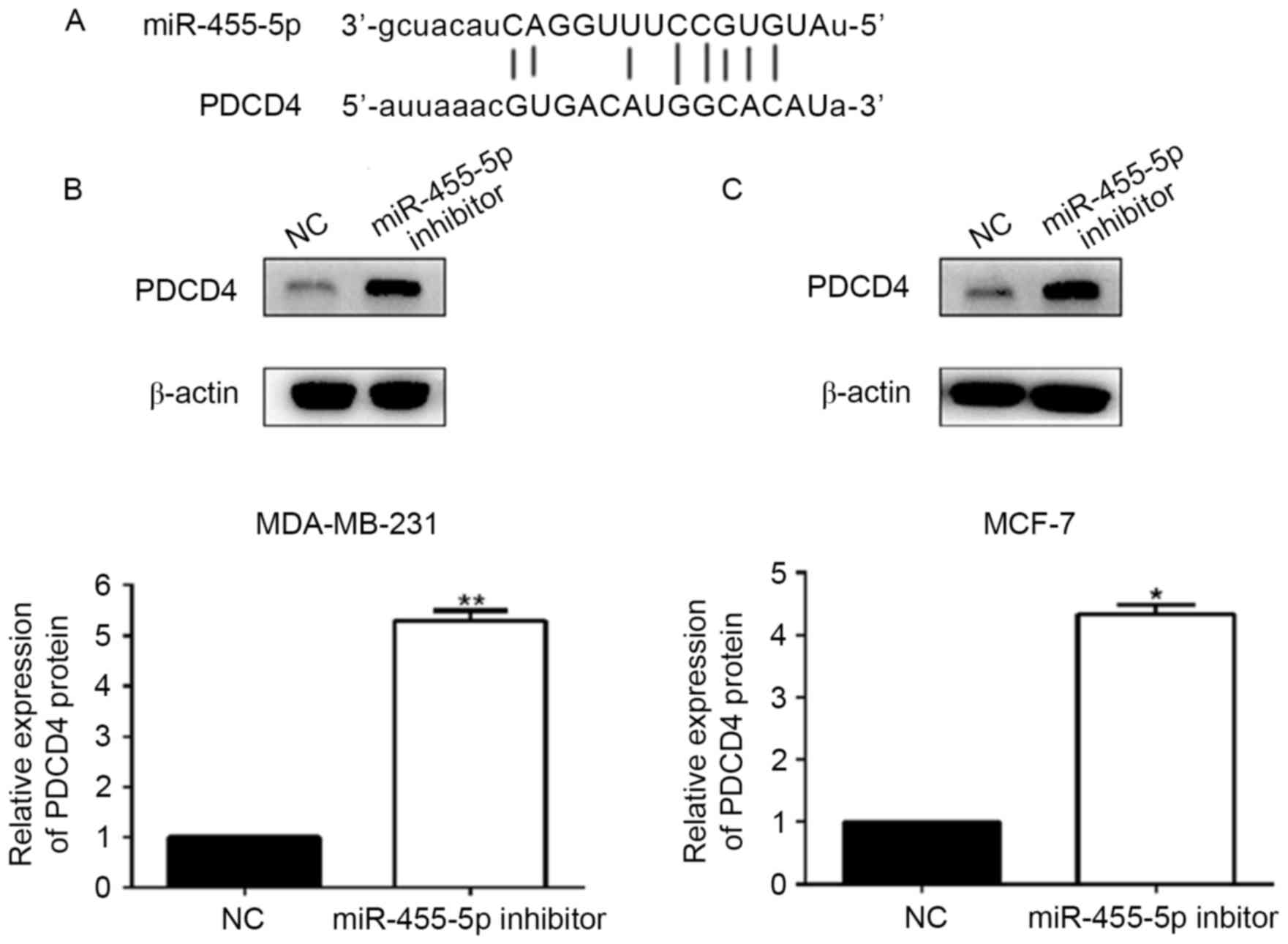
PDCD4 is a downstream target of
miR-455-5p in breast cancer cells
In order to uncover the functional mechanism of
miR-455-5p in breast cancer, the present study explored the
potential targets of miR-455-5p using bioinformatic tools
(TargetScan, miRNA.org and miRbase). The results
suggested that PDCD4 acted as a downstream target of miR-455-5p
(Fig. 6A). Following this, the
expression levels of PDCD4 were detected by western blotting in
MCF-7 and MDA-MB-231 cells, which had been transfected with the
miR-455-5p inhibitor. The results demonstrated that the expression
of PDCD4 was significantly increased in these two breast cancer
cell lines transfected with the miR-455-5p inhibitor, compared with
the NC group (P<0.05; Fig. 6B and
C). Therefore, PDCD4 may be negatively regulated by miR-455-5p
in breast cancer cells. Overall, the findings demonstrated that
PDCD4 may act as a downstream target of miR-455-5p in breast cancer
cells.
Discussion
The diagnosis and treatment strategies regarding
breast cancer have markedly improved over previous years, however
patients currently still exhibit a poor prognosis (15). There are >1 million newly
diagnosed cases of male breast cancer each year, and breast cancer
results in ~520,000 mortalities each year worldwide (16). Therefore, development of novel
treatment strategies for breast cancer is of primary concern.
miRNAs have been extensively investigated as
effective prognostic biomarkers for cancer, including breast cancer
(17,18). It has previously been reported that
miR-145 exhibits a suppressive role in the regulation of breast
cancer cell migration through specifically inhibiting the
expression of fascin and epithelial to mesenchymal transition
progression (19). Zhang et
al (20) demonstrated that low
expression of miR-124-3p promotes breast cancer cell development,
primarily by increasing beclin-1 expression. Wu et al
(21) suggested that in breast
cancer, miR-613 negatively regulates vascular endothelial growth
factor expression and restrains the cell proliferation and
invasion. Notably, miR-455-5p has been identified as an oncogene or
anti-oncogene in previous studies (11,13,22).
In gastric cancer, miR-455-5p has been identified as a tumor
suppressor, which decreases the expression of RAB18 (13). Conversely, TGF-β-induced miR-455-5p
is overexpressed, which results in low expression of UBE2B and
promotes cancer progression (11).
In the present study, the data indicated that in breast cancer
tissues and cell lines, miR-455-5p expression levels were
significantly upregulated, which suggested that miR-455-5p may act
as an oncogene in breast cancer.
Malignant tumors with poor tumor differentiation
tend to present more lymph node metastasis and an advanced TNM
stage, which results in an increased tendency of tumor metastasis
and invasion (23,24). The present study demonstrated that
miR-455-5p was closely correlated with lymph node metastasis, TNM
stage and tumor differentiation. Further analysis indicated that
upregulated miR-455-5p was additionally associated with poorer OS
and DFS survival rates. The results additionally revealed that TNM
stage, lymph node metastasis, tumor differentiation and miR-255-5p
acted as prognostic factors for patients with breast cancer. It was
therefore hypothesized that miR-455-5p acts as an oncogene to
promote migration and invasion of breast cancer.
Next, cytological experiments indicated that
silencing miR-455-5p had a suppressive effect in terms of the
invasive and migratory abilities of breast cancer cells. This
indicated that miR-455-5p may promote breast cancer progression by
accelerating cell migration and invasion.
MiRNAs exert various functions in cancer cells via
interaction with specific targets (11,12).
In oral squamous cell carcinoma, UBE2B has been identified as a
target of miR-455-5p (11). In
gastric cancer, miR-455-5p may directly target RAB18 (13). Therefore, in order to identify a
target of miR-455-5p in breast cancer, the present study applied
bioinformatics tools to analyze potential targets. PDCD4 was
subsequently identified as a potential target, and has previously
been verified to act as a tumor suppressor in multiple cancers,
including breast cancer (25–27).
PDCD4 is regulated by various miRNAs in breast cancer, including
miR-183-5p and miR-21 (28,29).
In order to verify PDCD4 as a target of miR-455-5p, the protein
expression of PDCD4 was detected in breast cancer cells, where the
expression of miR-455-5p had been decreased. It was revealed that
miR-455-5p inversely regulated PDCD4 expression levels in breast
cancer cells.
In conclusion, the results of the present study
identified PDCD4 as a downstream target of miR-455-5p. However,
further studies are required in order to validate this finding, and
explore the underlying molecular mechanism of the role of
miR-455-5p in breast cancer. The findings demonstrated that
miR-455-5p was highly expressed in breast cancer, and therefore may
facilitate cancer development, acting as an oncogene and biomarker
for the prognosis of breast cancer patients.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Rangarajan B, Shet T, Wadasadawala T, Nair
NS, Sairam RM, Hingmire SS and Bajpai J: Breast cancer: An overview
of published Indian data. South Asian J Cancer. 5:86–92. 2016.
View Article : Google Scholar :
|
|
3
|
Onyido EK, Sweeney E and Nateri AS:
Wnt-signalling pathways and microRNAs network in carcinogenesis:
Experimental and bioinformatics approaches. Mol Cancer. 15:562016.
View Article : Google Scholar :
|
|
4
|
Cheng W, Yan K, Xie LY, Chen F, Yu HC,
Huang YX and Dang CX: MiR-143-3p controls TGF-β1-induced cell
proliferation and extracellular matrix production in airway smooth
muscle via negative regulation of the nuclear factor of activated T
cells 1. Mol Immunol. 78:133–139. 2016. View Article : Google Scholar
|
|
5
|
Sun X, Li Y, Zheng M, Zuo W and Zheng W:
MicroRNA-223 increases the sensitivity of triple-negative breast
cancer stem cells to TRAIL-induced apoptosis by targeting HAX-1.
PLoS One. 11:e01627542016. View Article : Google Scholar :
|
|
6
|
Ma L: MicroRNA and metastasis. Adv Cancer
Res. 132:165–207. 2016. View Article : Google Scholar
|
|
7
|
Zhang Z, Zhang M, Chen Q and Zhang Q:
Downregulation of microRNA-145 promotes epithelial-mesenchymal
transition via regulating Snail in osteosarcoma. Cancer Gene Ther.
24:83–88. 2017. View Article : Google Scholar
|
|
8
|
Del Rosario RC, Damasco JR and Aguda BD:
MicroRNA inhibition fine-tunes and provides robustness to the
restriction point switch of the cell cycle. Sci Rep. 6:328232016.
View Article : Google Scholar :
|
|
9
|
Chiu LY, Kishnani PS, Chuang TP, Tang CY,
Liu CY, Bali D, Koeberl D, Austin S, Boyette K, Weinstein DA, et
al: Identification of differentially expressed microRNAs in human
hepatocellular adenoma associated with type I glycogen storage
disease: A potential utility as biomarkers. J Gastroenterol.
49:1274–1284. 2014. View Article : Google Scholar
|
|
10
|
Wong N, Khwaja SS, Baker CM, Gay HA,
Thorstad WL, Daly MD, Lewis JS Jr and Wang X: Prognostic microRNA
signatures derived from The cancer genome atlas for head and neck
squamous cell carcinomas. Cancer Med. 5:1619–1628. 2016. View Article : Google Scholar :
|
|
11
|
Cheng CM, Shiah SG, Huang CC, Hsiao JR and
Chang JY: Up-regulation of miR-455-5p by the TGF-β-SMAD signalling
axis promotes the proliferation of oral squamous cancer cells by
targeting UBE2B. J Pathol. 240:38–49. 2016. View Article : Google Scholar
|
|
12
|
Dong Y, He D, Peng Z, Peng W, Shi W, Wang
J, Li B, Zhang C and Duan C: Circular RNAs in cancer: An emerging
key player. J Hematol Oncol. 10:22017. View Article : Google Scholar :
|
|
13
|
Liu J, Zhang J, Li Y, Wang L, Sui B and
Dai D: MiR-455-5p acts as a novel tumor suppressor in gastric
cancer by down-regulating RAB18. Gene. 592:308–315. 2016.
View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
15
|
Lei L, Wang X and Chen Z: PET/CT imaging
for monitoring recurrence and evaluating response to treatment in
breast cancer. Adv Clin Exp Med. 25:377–382. 2016. View Article : Google Scholar
|
|
16
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
17
|
Huan L, Liang LH and He XH: Role of
microRNAs in inflammation-associated liver cancer. Cancer Biol Med.
13:407–425. 2016. View Article : Google Scholar :
|
|
18
|
Nagini S: Breast cancer: Current molecular
therapeutic targets and new players. Anticancer Agents Med Chem.
17:152–163. 2017. View Article : Google Scholar
|
|
19
|
Zhao H, Kang X, Xia X, Wo L, Gu X, Hu Y,
Xie X, Chang H, Lou L and Shen X: miR-145 suppresses breast cancer
cell migration by targeting FSCN-1 and inhibiting
epithelial-mesenchymal transition. Am J Transl Res. 8:3106–3114.
2016.
|
|
20
|
Zhang F, Wang B, Long H, Yu J, Li F, Hou H
and Yang Q: Decreased miR-124-3p expression prompted breast cancer
cell progression mainly by targeting beclin-1. Clin Lab.
62:1139–1145. 2016. View Article : Google Scholar
|
|
21
|
Wu J, Yuan P, Mao Q, Lu P, Xie T, Yang H
and Wang C: miR-613 inhibits proliferation and invasion of breast
cancer cell via VEGFA. Biochem Biophys Res Commun. 478:274–278.
2016. View Article : Google Scholar
|
|
22
|
Shoshan E, Mobley AK, Braeuer RR, Kamiya
T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al:
Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma
growth and metastasis. Nat Cell Biol. 17:311–321. 2015. View Article : Google Scholar :
|
|
23
|
Zhang DH, Yang ZL, Zhou EX, Miao XY, Zou
Q, Li JH, Liang LF, Zeng GX and Chen SL: Overexpression of Thy1 and
ITGA6 is associated with invasion, metastasis and poor prognosis in
human gallbladder carcinoma. Oncol Lett. 12:5136–5144. 2016.
|
|
24
|
Zhang XF, Liu T, Li Y and Li S:
Overexpression of long non-coding RNA CCAT1 is a novel biomarker of
poor prognosis in patients with breast cancer. Int J Clin Exp
Pathol. 8:9440–9445. 2015.
|
|
25
|
Li C, Deng L, Zhi Q, Meng Q, Qian A, Sang
H, Li X and Xia J: MicroRNA-183 functions as an oncogene by
regulating PDCD4 in gastric cancer. Anticancer Agents Med Chem.
16:447–455. 2016. View Article : Google Scholar
|
|
26
|
Li JZ, Gao W, Lei WB, Zhao J, Chan JY, Wei
WI, Ho WK and Wong TS: MicroRNA 744-3p promotes MMP-9-mediated
metastasis by simultaneously suppressing PDCD4 and PTEN in
laryngeal squamous cell carcinoma. Oncotarget. 7:58218–58233. 2016.
View Article : Google Scholar :
|
|
27
|
Li Y, Jiang D, Zhang Q, Liu X and Cai Z:
Ubiquitin-specific protease 4 inhibits breast cancer cell growth
through the upregulation of PDCD4. Int J Mol Med. 38:803–811. 2016.
View Article : Google Scholar :
|
|
28
|
Cheng Y, Xiang G, Meng Y and Dong R:
MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in
human breast cancer by targeting the PDCD4. Reprod Biol.
16:225–233. 2016. View Article : Google Scholar
|
|
29
|
Venturutti L, Romero LV, Urtreger AJ,
Chervo MF, Russo Cordo RI, Mercogliano MF, Inurrigarro G, Pereyra
MG, Proietti CJ, Izzo F, et al: Stat3 regulates ErbB-2 expression
and co-opts ErbB-2 nuclear function to induce miR-21 expression,
PDCD4 downregulation and breast cancer metastasis. Oncogene.
35:2208–2222. 2016. View Article : Google Scholar
|