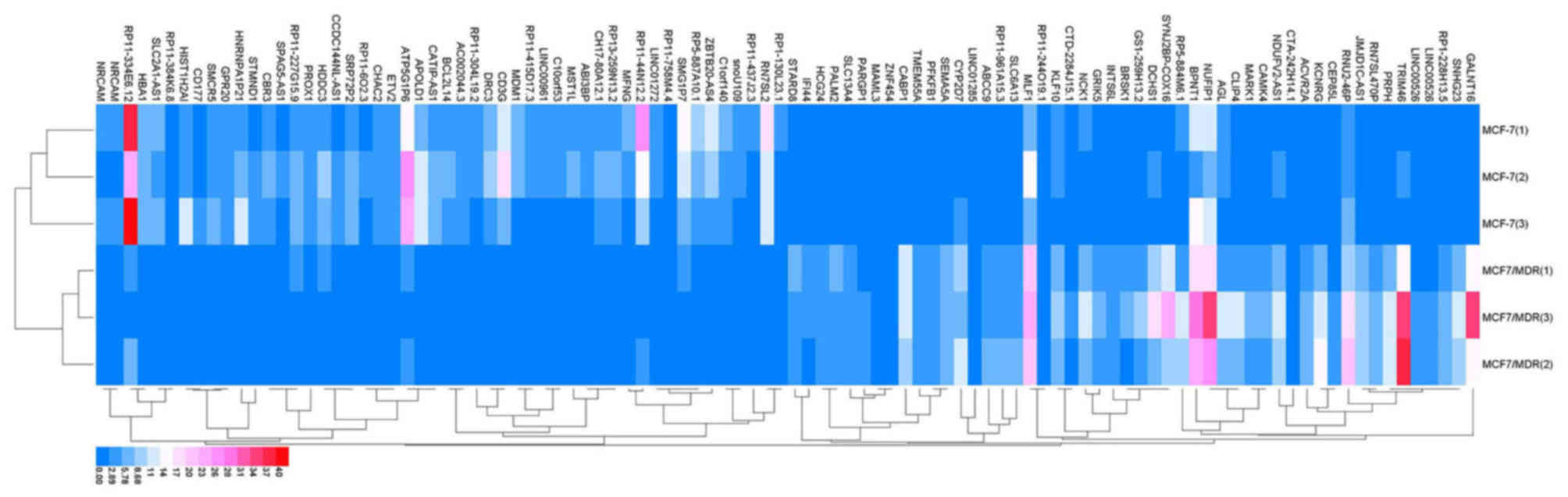
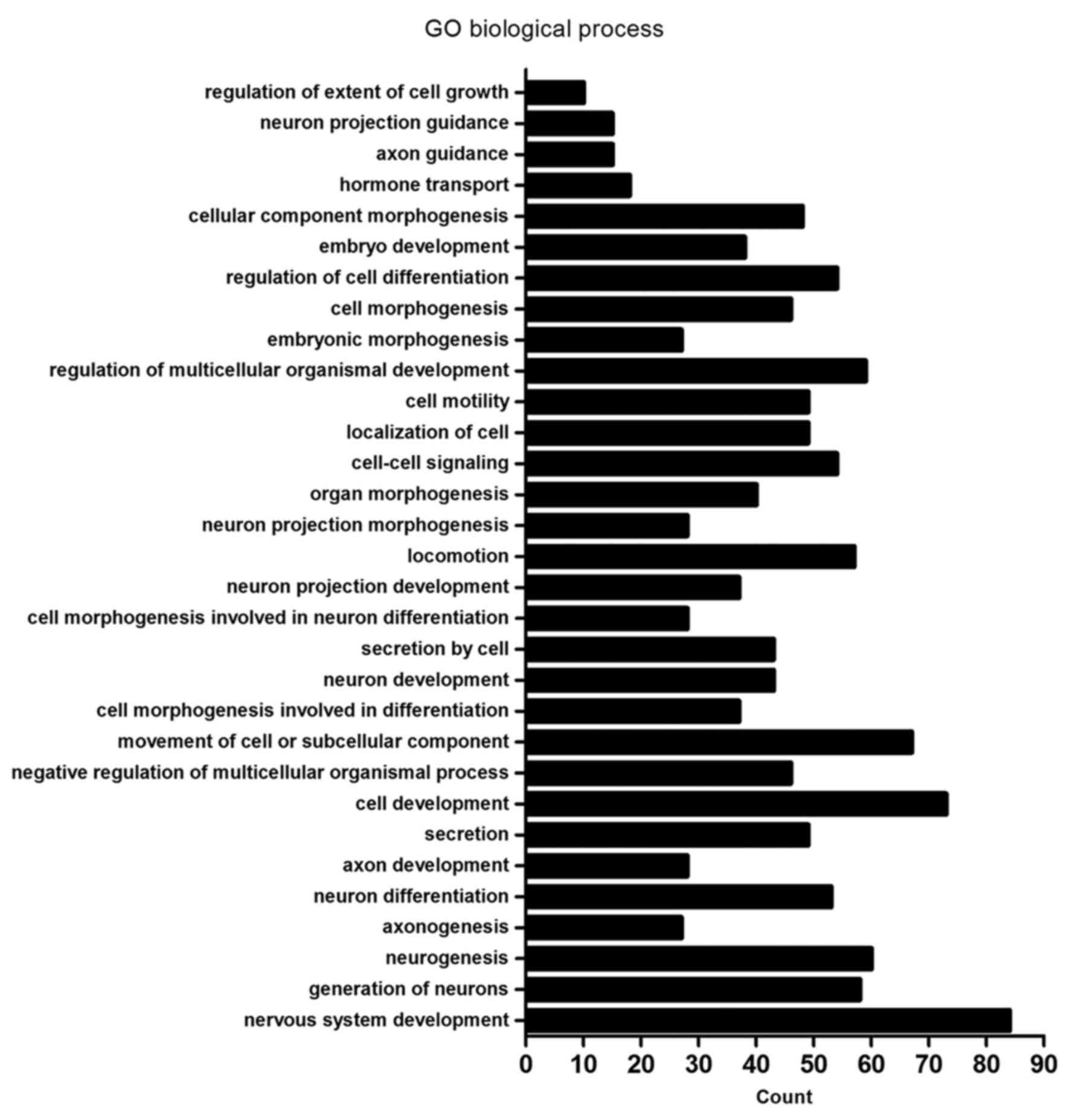
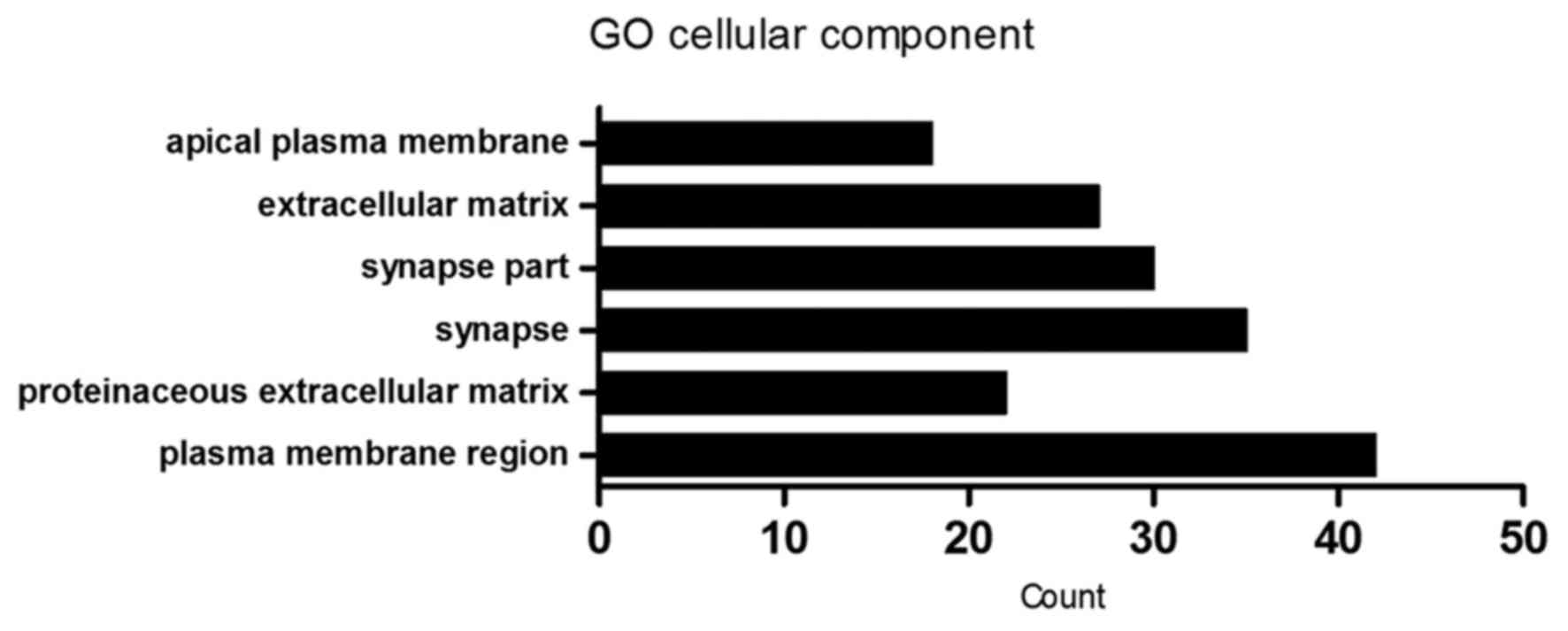
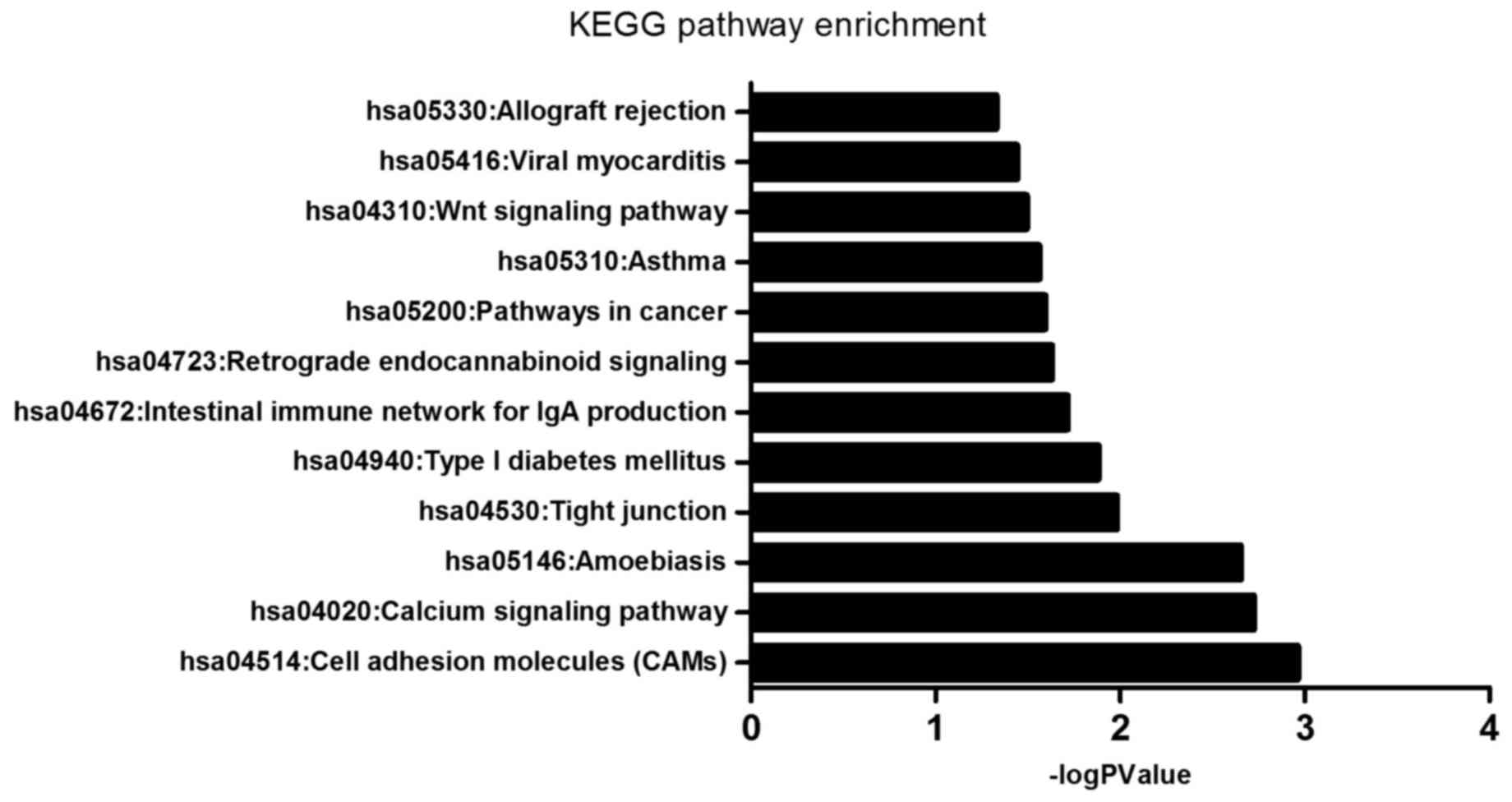
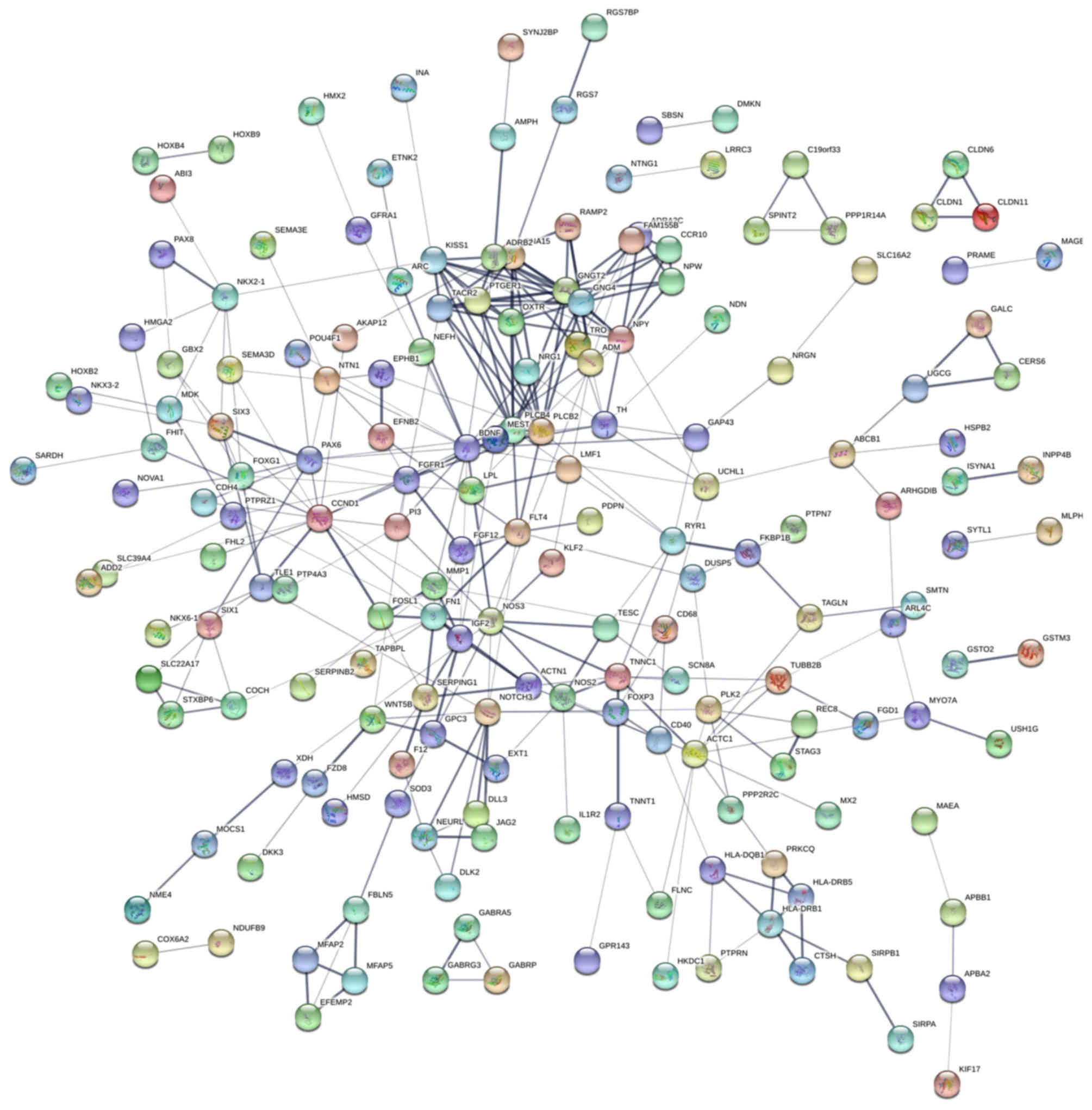
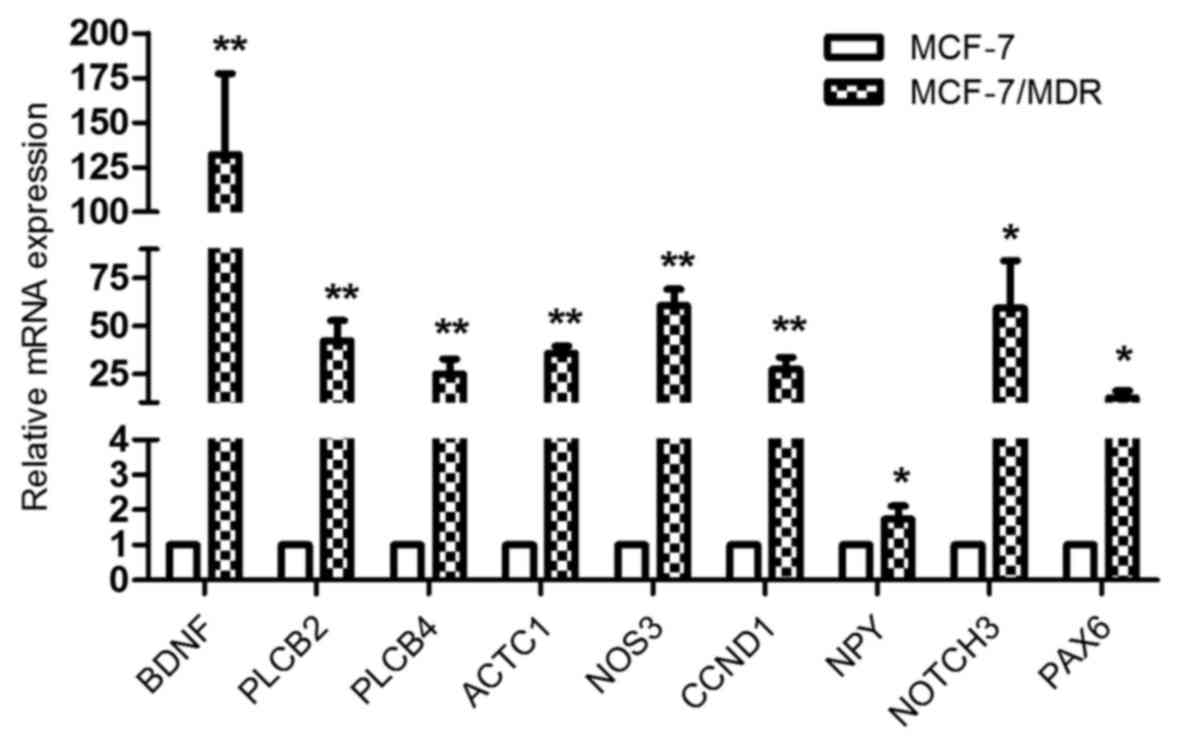
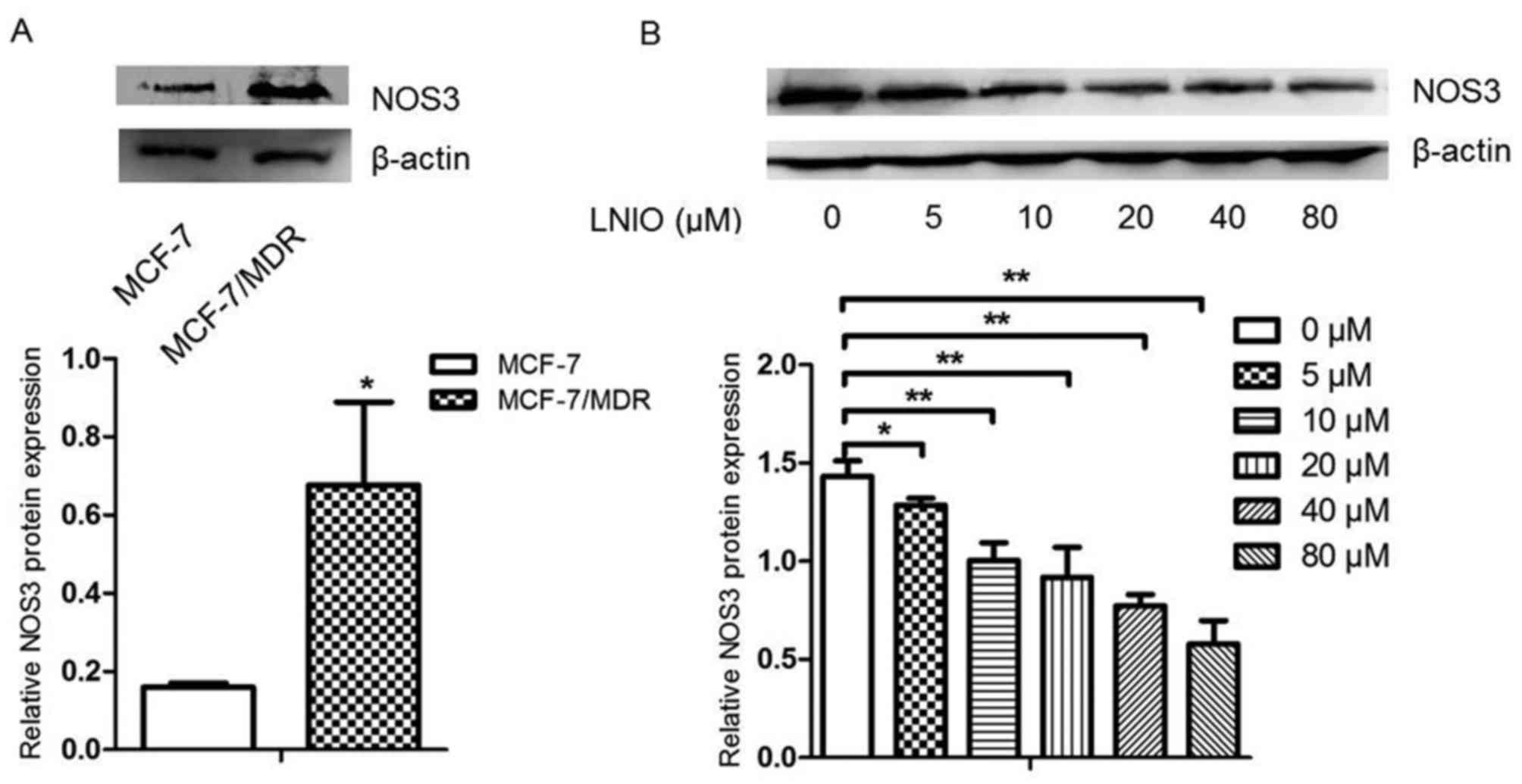
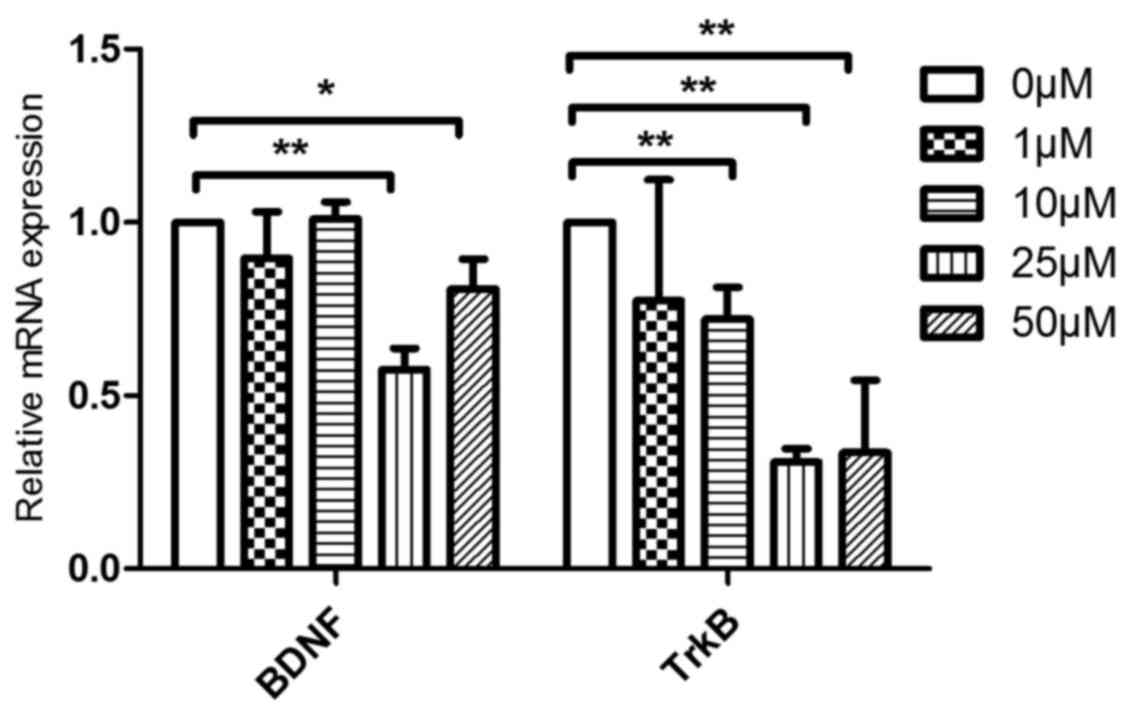
|
1
|
Leonessa F and Clarke R: ATP binding
cassette transporters and drug resistance in breast cancer. Endocr
Relat Cancer. 10:43–73. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Doyle LA, Yang W, Abruzzo LV, Krogmann T,
Gao Y, Rishi AK and Ross DD: A multidrug resistance transporter
from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA.
95:pp. 15665–15670. 1998; View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mechetner E, Kyshtoobayeva A, Zonis S, Kim
H, Stroup R, Garcia R, Parker RJ and Fruehauf JP: Levels of
multidrug resistance (MDR1) P-glycoprotein expression by human
breast cancer correlate with in vitro resistance to taxol and
doxorubicin. Clin Cancer Res. 4:389–398. 1998.PubMed/NCBI
|
|
4
|
Batist G, Tulpule A, Sinha BK, Katki AG,
Myers CE and Cowan KH: Overexpression of a novel anionic
glutathione transferase in multidrug-resistant human breast cancer
cells. J Biol Chem. 261:15544–15549. 1986.PubMed/NCBI
|
|
5
|
Lima RT, Martins LM, Guimarães JE, Sambade
C and Vasconcelos MH: Specific downregulation of bcl-2 and xIAP by
RNAi enhances the effects of chemotherapeutic agents in MCF-7 human
breast cancer cells. Cancer Gene Ther. 11:309–316. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wagener C, Bargou RC, Daniel PT, Bommert
K, Mapara MY, Royer HD and Dörken B: Induction of the
death-promoting gene bax-alpha sensitizes cultured breast-cancer
cells to drug-induced apoptosis. Int J Cancer. 67:138–141. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Devarajan E, Sahin AA, Chen JS,
Krishnamurthy RR, Aggarwal N, Brun AM, Sapino A, Zhang F, Sharma D,
Yang XH, et al: Down-regulation of caspase 3 in breast cancer: A
possible mechanism for chemoresistance. Oncogene. 21:8843–8851.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang XH, Sladek TL, Liu X, Butler BR,
Froelich CJ and Thor AD: Reconstitution of caspase 3 sensitizes
MCF-7 breast cancer cells to doxorubicin- and etoposide-induced
apoptosis. Cancer Res. 61:348–354. 2001.PubMed/NCBI
|
|
9
|
Geisler S, Lønning PE, Aas T, Johnsen H,
Fluge O, Haugen DF, Lillehaug JR, Akslen LA and Børresen-Dale AL:
Influence of TP53 gene alterations and c-erbB-2 expression on the
response to treatment with doxorubicin in locally advanced breast
cancer. Cancer Res. 61:2505–2512. 2001.PubMed/NCBI
|
|
10
|
Davis JM, Navolanic PM,
Weinstein-Oppenheimer CR, Steelman LS, Hu W, Konopleva M,
Blagosklonny MV and McCubrey JA: Raf-1 and Bcl-2 induce distinct
and common pathways that contribute to breast cancer drug
resistance. Clin Cancer Res. 9:1161–1170. 2003.PubMed/NCBI
|
|
11
|
Yu DD, Wu Y, Zhang XH, Lv MM, Chen WX,
Chen X, Yang SJ, Shen H, Zhong SL, Tang JH and Zhao JH: Exosomes
from adriamycin-resistant breast cancer cells transmit drug
resistance partly by delivering miR-222. Tumour Biol. 37:3227–3235.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Conze D, Weiss L, Regen PS, Bhushan A,
Weaver D, Johnson P and Rincón M: Autocrine production of
interleukin 6 causes multidrug resistance in breast cancer cells.
Cancer Res. 61:8851–8858. 2001.PubMed/NCBI
|
|
13
|
Teixeira C, Reed JC and Pratt MA: Estrogen
promotes chemotherapeutic drug resistance by a mechanism involving
Bcl-2 proto-oncogene expression in human breast cancer cells.
Cancer Res. 55:3902–3907. 1995.PubMed/NCBI
|
|
14
|
Li J, Xu LZ, He KL, Guo WJ, Zheng YH, Xia
P and Chen Y: Reversal effects of nomegestrol acetate on multidrug
resistance in adriamycin-resistant MCF7 breast cancer cell line.
Breast Cancer Res. 3:253–263. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang Z, Wu H, Xia J, Li Y, Zhang Y, Huang
K, Wagar N, Yoon Y, Cho HT, Scala S and Shim H: Involvement of
miR-326 in chemotherapy resistance of breast cancer through
modulating expression of multidrug resistance-associated protein 1.
Biochem Pharmacol. 79:817–824. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting p27Kip1.
J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi
Y, Xiong W, Li G, Lu J, Fodstad O, et al: MicroRNA-125b confers the
resistance of breast cancer cells to paclitaxel through suppression
of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J
Biol Chem. 285:21496–21507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kastl L, Brown I and Schofield AC:
miRNA-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Knuefermann C, Lu Y, Liu B, Jin W, Liang
K, Wu L, Schmidt M, Mills GB, Mendelsohn J and Fan Z:
HER2/PI-3K/Akt activation leads to a multidrug resistance in human
breast adenocarcinoma cells. Oncogene. 22:3205–3212. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
deGraffenried LA, Friedrichs WE, Russell
DH, Donzis EJ, Middleton AK, Silva JM, Roth RA and Hidalgo M:
Inhibition of mTOR activity restores tamoxifen response in breast
cancer cells with aberrant Akt activity. Clin Cancer Res.
10:8059–8067. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gritsko T, Williams A, Turkson J, Kaneko
S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, et al:
Persistent activation of stat3 signaling induces survivin gene
expression and confers resistance to apoptosis in human breast
cancer cells. Clin Cancer Res. 12:11–19. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qu C, Zhang W, Zheng G, Zhang Z, Yin J and
He Z: Metformin reverses multidrug resistance and
epithelial-mesenchymal transition (EMT) via activating
AMP-activated protein kinase (AMPK) in human breast cancer cells.
Mol Cell Biochem. 386:63–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mallini P, Lennard T, Kirby J and Meeson
A: Epithelial-to-mesenchymal transition: What is the impact on
breast cancer stem cells and drug resistance. Cancer Treat Rev.
40:341–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Z, Gerstein M and Snyder M: RNA-Seq:
A revolutionary tool for transcriptomics. Nat Rev Genet. 10:57–63.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wacker SA, Houghtaling BR, Elemento O and
Kapoor TM: Using transcriptome sequencing to identify mechanisms of
drug action and resistance. Nat Chem Biol. 8:235–237. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang HJ, Kim N, Seong KM, Youn H and Youn
B: Investigation of radiation-induced transcriptome profile of
radioresistant non-small cell lung cancer A549 cells using RNA-seq.
PLoS One. 8:e593192013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim KY, Kim SH, Yu SN, Park SK, Choi HD,
Yu HS, Ji JH, Seo YK and Ahn SC: Salinomycin enhances
doxorubicin-induced cytotoxicity in multidrug resistant MCF-7/MDR
human breast cancer cells via decreased efflux of doxorubicin. Mol
Med Rep. 12:1898–1904. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Assanhou AG, Li W, Zhang L, Xue L, Kong L,
Sun H, Mo R and Zhang C: Reversal of multidrug resistance by
co-delivery of paclitaxel and lonidamine using a TPGS and
hyaluronic acid dual-functionalized liposome for cancer treatment.
Biomaterials. 73:284–295. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Denoyer D, Perek N, Le Jeune N, Frère D
and Dubois F: The multidrug resistance of in vitro tumor cell lines
derived from human breast carcinoma MCF-7 does not influence
pentavalent technetium-99m-dimercaptosuccinic acid uptake. Cancer
Biother Radiopharm. 18:791–801. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou Y, Li HR, Huang J, Jin G and Fu XD:
Multiplex analysis of polyA-linked sequences (MAPS): An RNA-seq
strategy to profile poly(A+) RNA. Methods Mol Biol. 1125:169–178.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fox-Walsh K, Davis-Turak J, Zhou Y, Li H
and Fu XD: A multiplex RNA-seq strategy to profile poly(A+) RNA:
Application to analysis of transcription response and 3′ end
formation. Genomics. 98:266–271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li B and Dewey CN: RSEM: Accurate
transcript quantification from RNA-Seq data with or without a
reference genome. BMC Bioinformatics. 12:3232011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki R: DAVID: Database for annotation,
visualization and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Leek JT, Monsen E, Dabney AR and Storey
JD: EDGE: Extraction and analysis of differential gene expression.
Bioinformatics. 22:507–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Marioni JC, Mason CE, Mane SM, Stephens M
and Gilad Y: RNA-seq: An assessment of technical reproducibility
and comparison with gene expression arrays. Genome Res.
18:1509–1517. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Crivellari D, Pagani O, Veronesi A,
Lombardi D, Nolè F, Thürlimann B, Hess D, Borner M, Bauer J,
Martinelli G, et al: High incidence of central nervous system
involvement in patients with metastatic or locally advanced breast
cancer treated with epirubicin and docetaxel. Ann Oncol.
12:353–356. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Buttery RC, Rintoul RC and Sethi T: Small
cell lung cancer: The importance of the extracellular matrix. Int J
Biochem Cell Biol. 36:1154–1160. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Liu WH, Chen MT, Wang ML, Lee YY, Chiou
GY, Chien CS, Huang PI, Chen YW, Huang MC, Chiou SH, et al:
Cisplatin-selected resistance is associated with increased motility
and stem-like properties via activation of STAT3/Snail axis in
atypical teratoid/rhabdoid tumor cells. Oncotarget. 6:1750–1768.
2015.PubMed/NCBI
|
|
43
|
Kohmo S, Kijima T, Otani Y, Mori M, Minami
T, Takahashi R, Nagatomo I, Takeda Y, Kida H, Goya S, et al: Cell
surface tetraspanin CD9 mediates chemoresistance in small cell lung
cancer. Cancer Res. 70:8025–8035. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Toole BP and Slomiany MG: Hyaluronan, CD44
and Emmprin: Partners in cancer cell chemoresistance. Drug Resist
Updat. 11:110–121. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hodkinson PS, Mackinnon AC and Sethi T:
Extracellular matrix regulation of drug resistance in small-cell
lung cancer. Int J Radiat Biol. 83:733–741. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Doberstein K, Wieland A, Lee SB, Blaheta
RA, Wedel S, Moch H, Schraml P, Pfeilschifter J, Kristiansen G and
Gutwein P: L1-CAM expression in ccRCC correlates with shorter
patients survival times and confers chemoresistance in renal cell
carcinoma cells. Carcinogenesis. 32:262–270. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Simpson WG: The calcium channel blocker
verapamil and cancer chemotherapy. Cell Calcium. 6:449–467. 1985.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: Long non-coding RNA UCA1 increases chemoresistance of
bladder cancer cells by regulating Wnt signaling. FEBS J.
281:1750–1758. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mohammed MK, Shao C, Wang J, Wei Q, Wang
X, Collier Z, Tang S, Liu H, Zhang F, Huang J, et al: Wnt/β-catenin
signaling plays an ever-expanding role in stem cell self-renewal,
tumorigenesis and cancer chemoresistance. Genes Dis. 3:11–40. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Deng YH, Pu XX, Huang MJ, Xiao J, Zhou JM,
Lin TY and Lin EH: 5-Fluorouracil upregulates the activity of Wnt
signaling pathway in CD133-positive colon cancer stem-like cells.
Chin J Cancer. 29:810–815. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Su HY, Lai HC, Lin YW, Liu CY, Chen CK,
Chou YC, Lin SP, Lin WC, Lee HY and Yu MH: Epigenetic silencing of
SFRP5 is related to malignant phenotype and chemoresistance of
ovarian cancer through Wnt signaling pathway. Int J Cancer.
127:555–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Song JH, Kim SH, Cho D, Lee IK, Kim HJ and
Kim TS: Enhanced invasiveness of drug-resistant acute myeloid
leukemia cells through increased expression of matrix
metalloproteinase-2. Int J Cancer. 125:1074–1081. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Almendro V, Ametller E, Garcia-Recio S,
Collazo O, Casas I, Augé JM, Maurel J and Gascón P: The role of
MMP7 and its cross-talk with the FAS/FASL system during the
acquisition of chemoresistance to oxaliplatin. PLoS One.
4:e47282009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fukuyama R, Ng KP, Cicek M, Kelleher C,
Niculaita R, Casey G and Sizemore N: Role of IKK and oscillatory
NFkappaB kinetics in MMP-9 gene expression and chemoresistance to
5-fluorouracil in RKO colorectal cancer cells. Mol Carcinog.
46:402–413. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou B, Blanchard A, Wang N, Ma X, Han J,
Schroedter I, Leygue E and Myal Y: Claudin 1 promotes migration and
increases sensitivity to tamoxifen and anticancer drugs in
luminal-like human breast cancer cells MCF7. Cancer Invest.
33:429–439. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Kwon MJ: Emerging roles of claudins in
human cancer. Int J Mol Sci. 14:18148–18180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yoshida H, Sumi T, Zhi X, Yasui T, Honda K
and Ishiko O: Claudin-4: A potential therapeutic target in
chemotherapy-resistant ovarian cancer. Anticancer Res.
31:1271–1277. 2011.PubMed/NCBI
|
|
58
|
Hoggard J, Fan J, Lu Z, Lu Q, Sutton L and
Chen YH: Claudin-7 increases chemosensitivity to cisplatin through
the upregulation of caspase pathway in human NCI-H522 lung cancer
cells. Cancer Sci. 104:611–618. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang M, Li Y, Shen X, Ruan Y, Lu Y, Jin X,
Song P, Guo Y, Zhang X, Qu H, et al: CLDN6 promotes chemoresistance
through GSTP1 in human breast cancer. J Exp Clin Cancer Res.
36:1572017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Noel EE, Yeste-Velasco M, Mao X, Perry J,
Kudahetti SC, Li NF, Sharp S, Chaplin T, Xue L, McIntyre A, et al:
The association of CCND1 overexpression and cisplatin resistance in
testicular germ cell tumors and other cancers. Am J Pathol.
176:2607–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Leung EL, Fraser M, Fiscus RR and Tsang
BK: Cisplatin alters nitric oxide synthase levels in human ovarian
cancer cells: Involvement in p53 regulation and cisplatin
resistance. Br J Cancer. 98:1803–1809. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gupta N, Xu Z, El-Sehemy A, Steed H and Fu
Y: Notch3 induces epithelial-mesenchymal transition and attenuates
carboplatin-induced apoptosis in ovarian cancer cells. Gynecol
Oncol. 130:200–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Yao J and Qian C: Inhibition of Notch3
enhances sensitivity to gemcitabine in pancreatic cancer through an
inactivation of PI3K/Akt-dependent pathway. Med Oncol.
27:1017–1022. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kang H, Jeong JY, Song JY, Kim TH, Kim G,
Huh JH, Kwon AY, Jung SG and An HJ: Notch3-specific inhibition
using siRNA knockdown or GSI sensitizes paclitaxel-resistant
ovarian cancer cells. Mol Carcinog. 55:1196–1209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Gu X, Lu C, He D, Lu Y, Jin J, Liu D and
Ma X: Notch3 negatively regulates chemoresistance in breast
cancers. Tumour Biol. Oct 14–2016.(Epub ahead of print). View Article : Google Scholar :
|
|
66
|
Middlemas DS, Kihl BK and Moody NM: Brain
derived neurotrophic factor protects human neuroblastoma cells from
DNA damaging agents. J Neurooncol. 45:27–36. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Lee J, Jiffar T and Kupferman ME: A novel
role for BDNF-TrkB in the regulation of chemotherapy resistance in
head and neck squamous cell carcinoma. PLoS One. 7:e302462012.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Huang BS, Luo QZ, Han Y, Huang D, Tang QP
and Wu LX: MiR-223/PAX6 axis regulates glioblastoma stem cell
proliferation and the chemo resistance to TMZ via regulating
PI3K/Akt pathway. J Cell Biochem. 118:3452–3461. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Elifio-Esposito S, Mahajan A, Fonseca AS,
Galli S, Noronha L, Poncio LC, Figueiredo BC, Kitlinska JB and
Cavalli LR: Increase in protein expression and copy number drives
the activation of NPY/Y5R pro-survival loop in chemotherapy-treated
neuroblastoma. Cancer Res. 77:58222017. View Article : Google Scholar
|
|
70
|
Li Y, Rong G and Kang H: Taxotere-induced
elevated expression of IL8 in carcinoma-associated fibroblasts of
breast invasive ductal cancer. Oncol Lett. 13:1856–1860. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Kraemer B, Crittenden S, Gallegos M,
Moulder G, Barstead R, Kimble J and Wickens M: NANOS-3 and FBF
proteins physically interact to control the sperm-oocyte switch in
Caenorhabditis elegans. Curr Biol. 9:1009–1018. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Fischer KR, Durrans A, Lee S, Sheng J, Li
F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, et al:
Epithelial-to-mesenchymal transition is not required for lung
metastasis but contributes to chemoresistance. Nature. 527:472–476.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Goldstein JL and Brown MS: Regulation of
the mevalonate pathway. Nature. 343:425–430. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wang SJ and Bourguignon LY: Role of
hyaluronan-mediated CD44 signaling in head and neck squamous cell
carcinoma progression and chemoresistance. Am J Pathol.
178:956–963. 2011. View Article : Google Scholar : PubMed/NCBI
|