Introduction
A keloid is defined as an overgrowth of pathological
scar tissue that grows beyond the original boundaries of the wound,
does not regress spontaneously and tends to recur despite treatment
(1,2). It can also spread to the surrounding
skin by invasion (3). The clinical
feature of keloid is a raised growth, usually accompanied by pain
and pruritus (4). Since the
mechanism and etiopathogenesis of keloids remain unknown, keloid
therapy is very limited.
Tumor necrosis factor-stimulated gene-6 (TSG-6) was
identified in differential screening of a cDNA library collected
from tumor necrosis factor-treated human diploid FS-4 fibroblasts
(5). Multiple functions of TSG-6
have been proposed, including interacting with matrix-associated
molecules or acting as an inflammatory factor (6,7).
However, current knowledge on the physiological function of TSG-6
remains limited. It has been reported to be critical in maintaining
the physiological architecture of skin (8). Furthermore, TSG-6 has potent
anti-inflammatory properties, which have been investigated in
several disease models, such as rheumatoid arthritis (9,10).
In our previous rabbit ear model of hypertrophic scarring,
injection of TSG-6 protein into the wound resulted in reduced
hypertrophic scar formation and anti-inflammatory effects during
wound healing (11). However, the
role of TSG-6 in keloid remains unclear.
In the current study, a TSG-6 expression vector was
constructed and stably transfected into human keloid fibroblasts
(KFs). The effects of overexpression of TSG-6 on proliferation and
apoptosis in KFs were determined. In addition, the potential
mechanisms of the signaling pathway regulated via TSG-6 in KFs were
investigated.
Materials and methods
Ethics
The current study was approved by the institutional
review board of Anhui Medical University (Hefei, China), and
written informed consent was obtained from all patients prior to
inclusion. The study was performed according to the principles of
the Declaration of Helsinki.
Keloid fibroblasts and cell
culture
Ten patients were recruited from the First
Affiliated Hospital of Anhui Medical University. The
characteristics of participants with keloid in this study are
listed in Table I. Patients had
received no previous treatment for keloid. KFs were isolated and
cultured as previously described (12). Cells were cultured in Dulbeccos
modified Eagles medium (DMEM), L-glutamine, 100 µg/ml streptomycin,
100 U/ml penicillin and fetal calf serum (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). No morphological and
biochemical differences were found with the passage.
 | Table I.Patient information. |
Table I.
Patient information.
| Patient number | Sex | Age | Position | Cause of scar |
|---|
| 01 | Male | 03 | Anterior neck | Scald |
| 02 | Female | 05 | Chest | Scald |
| 03 | Female | 06 | Shoulder | Burn |
| 04 | Female | 09 | Back | Surgery |
| 05 | Male | 14 | Chest | Burn |
| 06 | Female | 18 | Earlobe | Puncture |
| 07 | Female | 19 | Earlobe | Puncture |
| 08 | Male | 23 | Upper Arm | Surgery |
| 09 | Female | 25 | Abdomen | Surgery |
| 10 | Female | 27 | Earlobe | Puncture |
Lentiviral vector construction and
packaging
Total RNA was extracted from KFs using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturers instructions. cDNA synthesis was conducted
according to the RNA polymerase chain reaction (PCR) kit protocol
(Takara Bio, Inc., Otsu, Japan). The TSG-6 cDNA (GenBank no.
AJ421518) was amplified by PCR (Takara Bio, Inc.) from KF cDNA
using the following oligonucleotide primers: Forward,
5′-GGAATTCATGATCATCTTAATTTACT-3; and reverse,
5′-CGGGATCCTAAGTGGCTAAATCTTCC-3. The cycling for PCR amplifying
reaction was: Reverse transcription (RT) 50°C, 5 min for 1 cycle;
RT inaction 95°C, 20 sec for 1 cycle; denaturing 95°C, 15 sec for
40 cycles; annealing 60°C, 60 sec for 40 cycles. Sequencing results
were analyzed using Basic Local Alignment Search Tool (BLAST) from
National Center for Biotechnology Information (Bethesda, MD, USA).
The amplified DNA was digested by the enzymes EcoRI and
BamHI and ligated into lentiviral plasmid pLVX-Puro
(Clontech Laboratories, Inc., Mountain View, CA, USA). Following
transduction of DH5α-competent E. coli, the positive clones
were selected for PCR identification and DNA sequencing. Clones
containing the target plasmids were selected and named
pLVX-Puro-TSG-6.
pLVX-Puro-TSG-6 was highly purified and extracted
with endotoxin-free solution. pLVX-Puro-TSG-6 was then
co-transfected into 293T cells (Clontech Laboratories, Inc.) with
pHelper 1.0 (Biovector Science Lab, Inc., Beijing, China) and
pHelper 2.0 (Biovector Science Lab, Inc.) and lentiviruses using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.),
according to the manufacturers instructions. The 293T cells were
cultured using Minimum Essential Media (MEM; Gibco; Thermo Fisher
Scientific, Inc.). Cells were cultured at 37°C in 5%
CO2. Cells were seeded at a concentration of
5×105 cells/well in 6 well plates. The plasmids were
mixed in a 1.5 ml Eppendorf tube with 1.5 µg Lentiviruses plus 1.5
µg packing vector (pLVX-Puro-TSG-6: pHelper 1.0: pHelper 2.0=5:3:2)
and 125 µl MEM medium, then 5 µl Lipo in 125 µl MEM added. The Lipo
and plasmids mixture was mixed and incubated for 20 min at 25°C.
During the incubation of the Lipo and plasmids mixture, the culture
medium of the 293T cells was replaced with fresh MEM medium. Then
the transfection mixture was added to the well, and it was moved to
an incubator at 37°C and 5% CO2. At 8 h following
transfection, the medium was replaced with fresh growth medium.
After culture for 48 h, the supernatant containing lentivirus
particles was collected and centrifuged at 1,258 × g for 5 min, and
cell debris was discarded. In order to obtain a high concentration
of lentivirus, the supernatant was further filtered to remove cells
and debris. The virus titer in 293T cells was determined by
end-point dilution assay.
Infection with recombinant
lentiviruses
KFs in logarithmic growth phase were divided into
three groups: pLVX-Puro-TSG-6 group, pLVX-Puro group and KFs group.
In each group, 8×104 cells were seeded into a 6-well
plate and cultured in an incubator with 5% CO2 at 37°C
for 24 h. Following adhesion, the supernatant was discarded, and 2
ml DMEM containing 10% fetal calf serum (Gibco; Thermo Fisher
Scientific, Inc.) was added. Once the cells reached 30% confluence,
the medium was removed. Subsequently, pLVX-Puro-TSG-6 or pLVX-Puro
were added into the wells at multiplicity of infection of 10.
Following the addition of 50 µl polybrene, DMEM containing 10%
fetal calf serum was added to reach a total of 2 ml. For the KF
group (2 wells), only 2 ml DMEM containing 10% fetal calf serum was
added. Stable transfectants of pLVX-Puro-TSG-6 and pLVX-Puro were
continuously selected using 2.5 µg/ml puromycin. Stable
transfection was further confirmed by reverse transcription-PCR
(RT-PCR) and western blot analysis.
RNA isolation and RT-PCR
Levels of specific mRNAs in each group were assessed
by RT-PCR. Total RNA was extracted using the Trizol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Purified RNA (1 µg)
was reverse transcribed using the RNA PCR Kit (Takara Biotechnology
Co., Ltd., Dalian, China) according to the manufacturers protocol.
PCR primers are listed in Table
II. cDNA was amplified and quantified using the RT-PCR Kit and
the following conditions: 94°C for 5 min, then 40 cycles consisting
of 30 sec at 94°C, 30 sec at 54°C and 45 sec at 72°C, followed by
incubation at 72°C for 10 min.
 | Table II.Oligonucleotides used in
reverse-polymerase chain reaction analysis. |
Table II.
Oligonucleotides used in
reverse-polymerase chain reaction analysis.
| Gene | Forward (5–3) | Reverse (5–3) |
|---|
| p21 |
CGTGAGCGATGGAACTTCGA |
CTCTTGGAGAAGATCAGCCG |
| Cyclin D1 |
CCCTCGGTGTCCTACTTCA |
CTCCTCGCACTTCTGTTCCT |
| Bax |
GACGGCCTCCTCTCCTACTT |
CTCAGCCCATCTTCTTCCAG |
| Bcl-2 |
GAACTGGGGGAGGATTGTGG |
CCGTACAGTTCCACAAAGGC |
| Caspase-3 |
TGGAATTGATGCGTGATGTT |
GTCGGCATACTGTTTCAGCA |
| NF-κB |
AAGATCAATGGCTACACAGG |
CCTCAATGTCCTCTTTCTGC |
| β-actin |
CTCCATCCTGGCCTCGCTGT |
GCTGTCACCTTCACCGTTCC |
PCR products with UltraPure Ethidium bromide (EB;
Invitrogen, Thermo Fisher Scientific, Inc.) were electrophoresed on
a 2% agarose gel. An image of the gel was captured and the
intensity of the bands was analyzed using Labworks software (UVP;
LLC, Phoenix, AZ, USA). β-actin was used as the internal loading
control.
Western blot analysis
Total protein was prepared using RIPA Lysis and
Extraction Buffer (Thermo Fisher Scientific, Inc.) at freezing
temperature (Cell Signaling Technology, Inc., Danvers, MA, USA).
Protein concentration was measured using a bicinchoninic acid assay
protein concentration kit (Pierce; Thermo Fisher Scientific, Inc.).
Equal amounts of cell lysate (50 µg of protein) were boiled for 5
min and resolved by SDS-PAGE (12%), then electrophoretically
transferred onto a nitro-cellulose membrane. The membrane was
blocked with 5% non-fat dry milk in Tris-buffered saline with Tween
(TBST) overnight at 4°C, then incubated with appropriate primary
antibodies: TSG-6 (cat. no. ab128266; Abcam, Cambridge, MA, USA;
1:1,000); p21 (cat. no. 2947; Cell Signaling Technology, Inc.;
1:1,000); cyclin D1 (cat. no. 2978; Cell Signaling Technology,
Inc.; 1:1,000); apoptosis regulator Bcl-2-associated X protein
(Bax; cat. no. 5023; Cell Signaling Technology, Inc.; 1:1,000);
apoptosis regulator B-cell lymphoma-2 (Bcl-2; cat. no. 2872; Cell
Signaling Technology, Inc.; 1:1,000); caspase-3 (cat. no. 9662;
Cell Signaling Technology, Inc.; 1:1,000); nuclear factor-κB
(NF-κB; cat. no. 8242; Cell Signaling Technology, Inc.; 1:1,000);
GAPDH (cat. no. 5174; Cell Signaling Technology, Inc.; 1:1,000);
β-actin (cat. no. 4970; Cell Signaling Technology, Inc.; 1:1,000)
in blocking buffer for 2 h at room temperature or 8–12 h at 4°C.
Following washing with TBST, the membranes were then incubated with
appropriate secondary antibody (goat anti-rabbit; cat. no. 7074;
Cell Signaling Technology, Inc.; 1:10,000) for 3 h at room
temperature. After washing with TBST, proteins were visualized
using an enhanced chemiluminescence assay kit (Thermo Fisher
Scientific, Inc.). Grey scale scanning was used to analyze relative
expression by using Quantity One 1D Analysis software (version 4.4,
Bio-Rad Laboratories, Inc., Hercules, CA, USA).
MTT assay
An MTT assay was performed to evaluate cell
proliferation of pLVX-Puro-TSG-6 transfected cells, pLVX-Puro
transfected cells and KFs. Single-cell suspensions
(1×105 cells/ml) were prepared for each group, and 100
µl cell suspension was seeded into 96-well plates and incubated at
37°C with 5% CO2 to form a monolayer. At 24 h, once
cells had adhered, 10 µl MTT stock solution (Promega Corporation,
Madison, WI, USA) was added to each well, and the plate was then
incubated at 37°C for 4 h. The MTT stock solution was discarded and
10 µl MTT solvent was added to each well and incubated for 4 h at
37°C to allow the crystals to be fully dissolved. The OD values at
570 nm were determined and the experiment was repeated three times.
Assay was also performed at 48, 72, 96 and 120 h after plating.
Flow cytometry assay
Flow cytometry was performed to evaluate apoptosis
and cell cycle arrest of the pLVX-Puro-TSG-6 transfected cells,
pLVX-Puro transfected cells and KFs. Cells in each group were
collected, digested with trypsin, washed with PBS three times, and
centrifuged at 4°C for 5 min at 560 × g, and the supernatant was
discarded. The cells were fixed with 70% ethanol at 4°C for ≥24 h
and then washed with PBS twice, following removal of ethanol by
centrifugation. Cell apoptosis was detected using Annexin V-FITC
Apoptosis Staining/Detection kit (Abcam). Cells (1×105)
were collected by centrifugation 4°C for 5 min at 560 × g and
re-suspended in 500 µl 1X binding buffer. Following incubation with
5 µl of Annexin V-fluorescein isothiocyanate (FITC) and 5 µl of
propidium iodide (PI), the cells were maintained at room
temperature for 5 min in the dark. Apoptosis was detected using a
flow cytometer and analyzed with FlowJo 10.0.6 (FlowJo LLC,
Ashland, OR, USA).
For cell cycle analysis, cells from each group were
fixed in 70% ethanol for 1–3 h at 4°C. After washing with PBS,
cells were collected with RNase A and stained with PI for 15 min at
37°C. Cell cycle was analyzed with the flow cytometer, and the
distribution of all cell-cycle phases was determining with Modfit
Software (BD Biosciences, Franklin Lakes, NJ, USA).
Statistical analysis
All data are presented as the mean ± standard
deviation. Statistical analysis was performed using SPSS 17.0
software (SPSS, Inc., Chicago, IL, USA). Tests for homogeneity of
variance (Levenes test) and one-way analysis of variance procedures
were performed, and followed by Turkeys multiple comparison post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Transfection efficiency
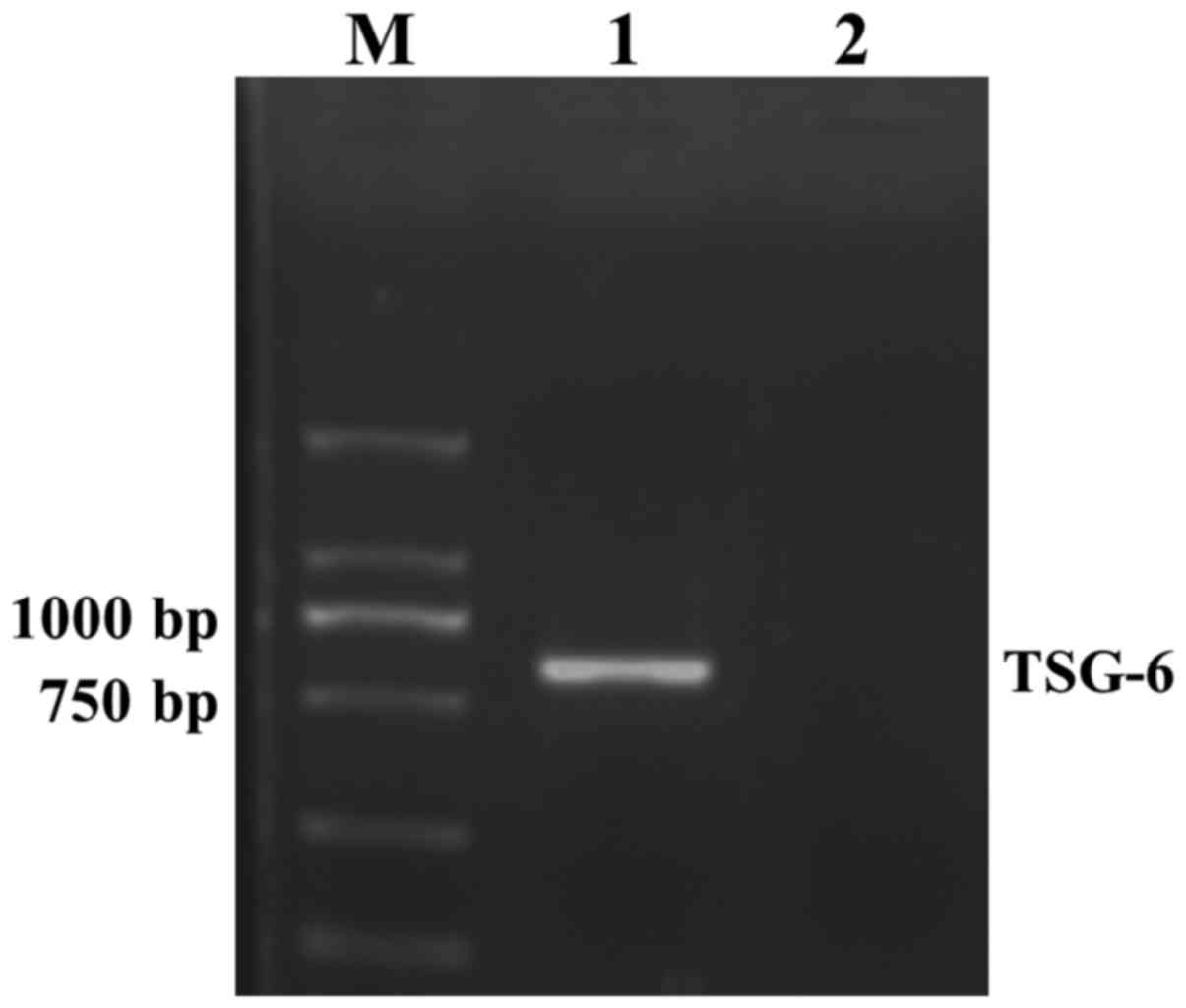
An 828 bp target gene fragment was obtained from
agarose gel electrophoresis of a PCR product, the size of which was
in accordance with TSG-6. The results suggested that TSG-6 gene was
successfully cloned into the pLVX-Puro vector (Fig. 1). Agarose gel electrophoresis
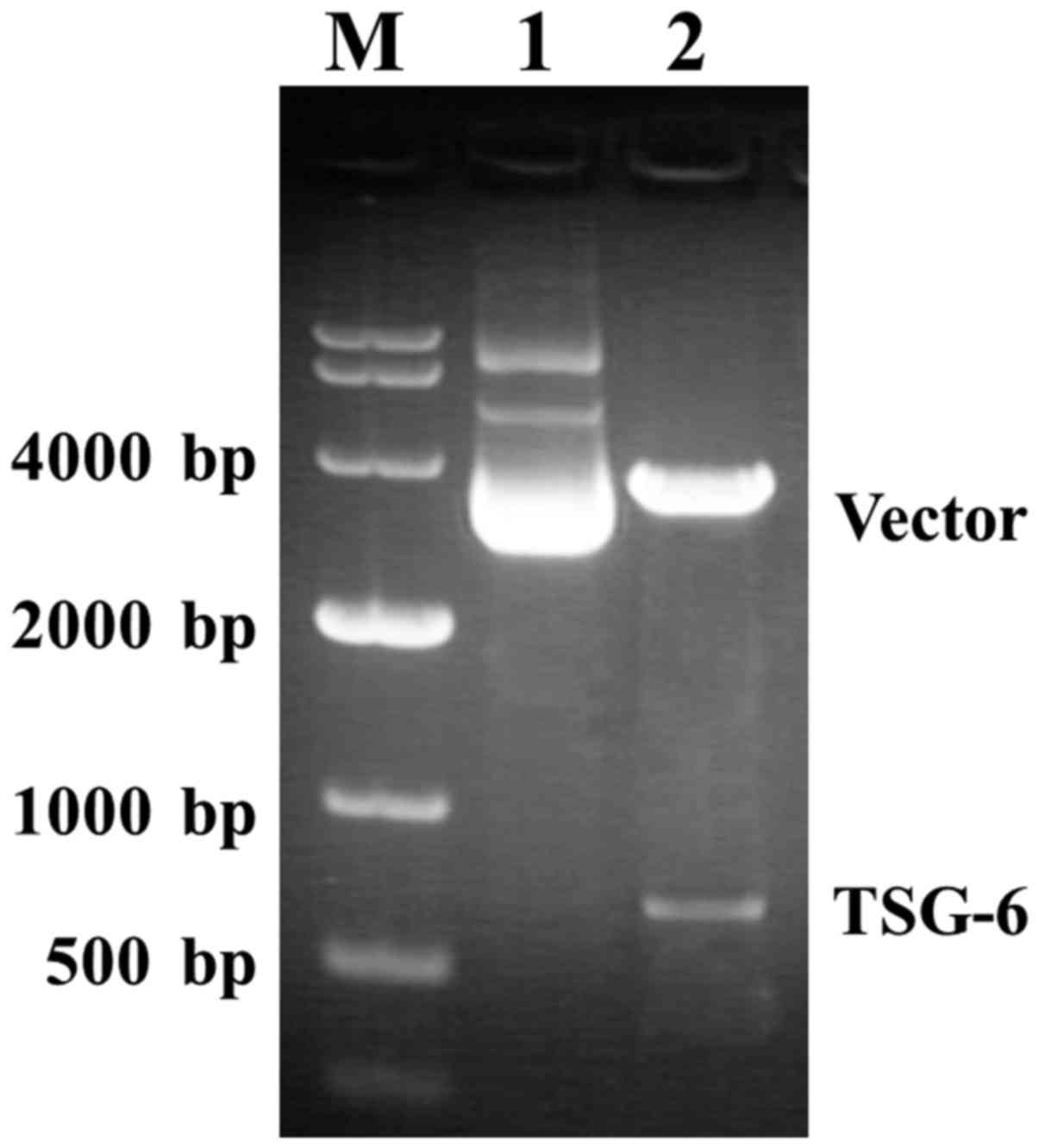
revealed the positive clones after infection, with 1,179 bp and 198
bp size bands for positive and negative transformants, respectively
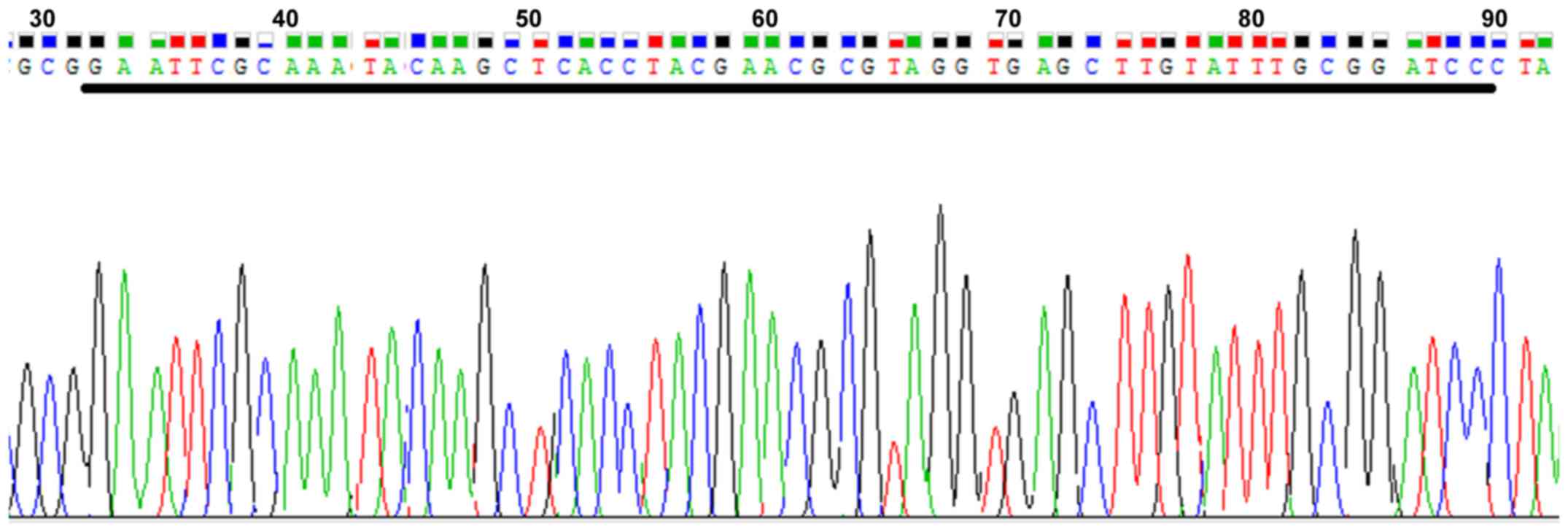
(Fig. 2). After BLAST detection,
the sequencing results of the clones revealed that the sequence
identity was 828/828 (100%) and the target gene lentiviral vector
had been successfully constructed (Fig. 3).
TSG-6 expression in KFs following
infection
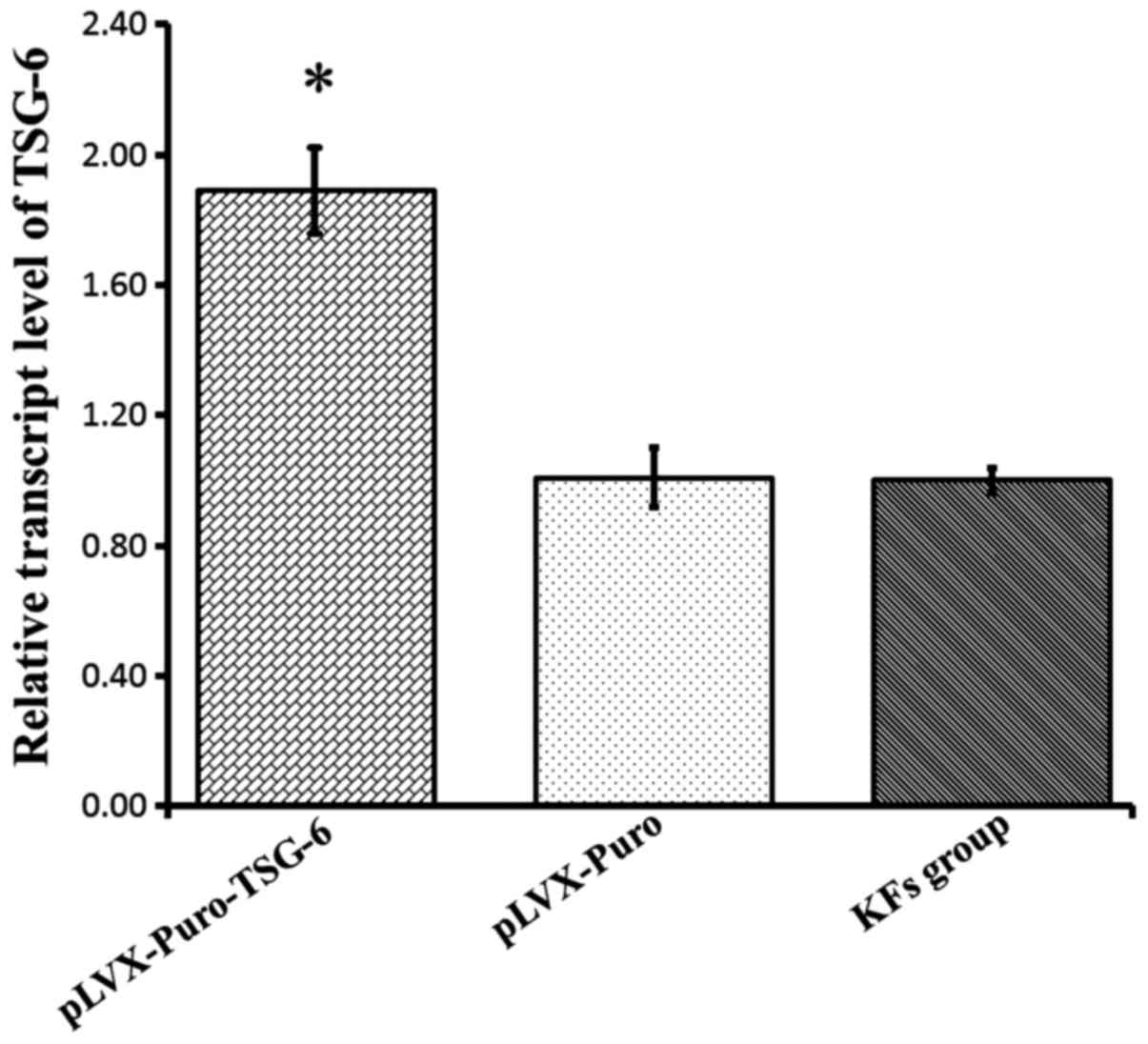
To determine the efficiency of transfection in KFs,
total RNA was extracted and RT-PCR was performed. The data
indicated that a high level of TSG-6 mRNA expression was detected
in the pLVX-Puro-TSG-6 group compared with the KF and Plvx-Puro
groups (P<0.05), while there was no significant difference
between the pLVX-Puro group and the KF group (P>0.05). This
suggested that the exogenous TSG-6 gene was transduced successfully
(Fig. 4). To further verify this
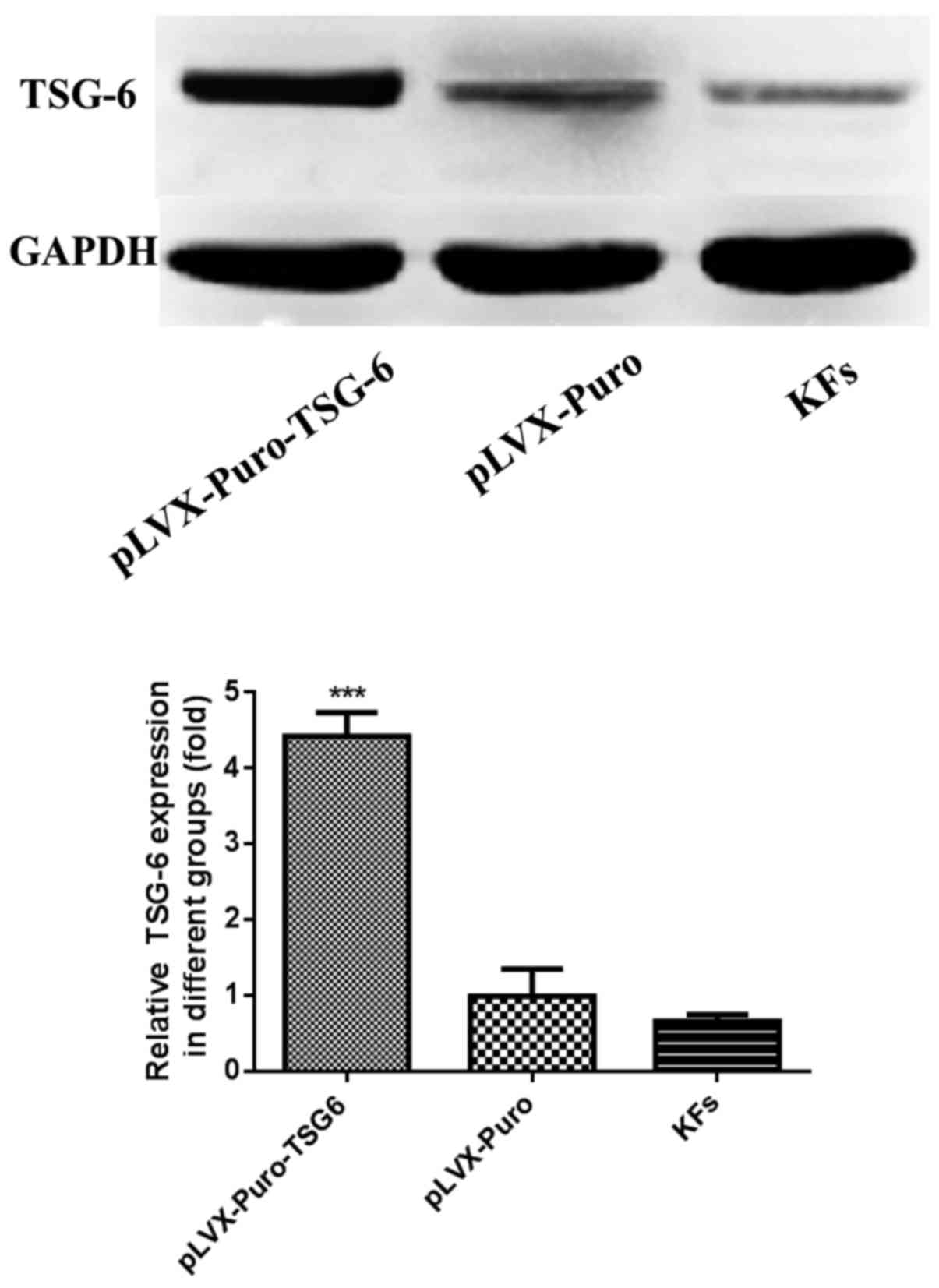
result, TSG-6 protein expression was determined using western blot
analysis. The results suggested that the expression of TSG-6
protein was increased significantly in the pLVX-Puro-TSG-6 group
compared with the pLVX-Puro group and the KF group (P<0.05),
while there was no significant difference between the pLVX-Puro
group and the KF group (P>0.05; Fig. 5).
TSG-6 overexpression inhibited cell
proliferation
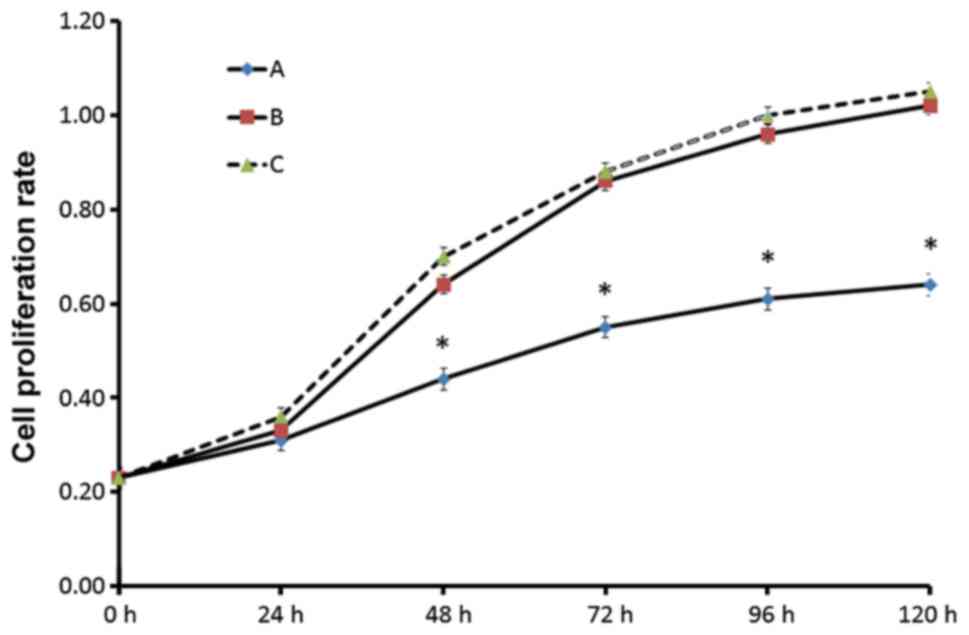
Cell viability was determined using an MTT assay.
Compared with the control and pLVX-Puro group, the proliferation of
the TSG-6 transfected cells was significantly decreased. However,
there was no significant difference between the pLVX-Puro group and
the KF group, which demonstrated that the vector itself had no
effect on the viability of KFs (Fig.
6).
Overexpression of TSG-6 induced
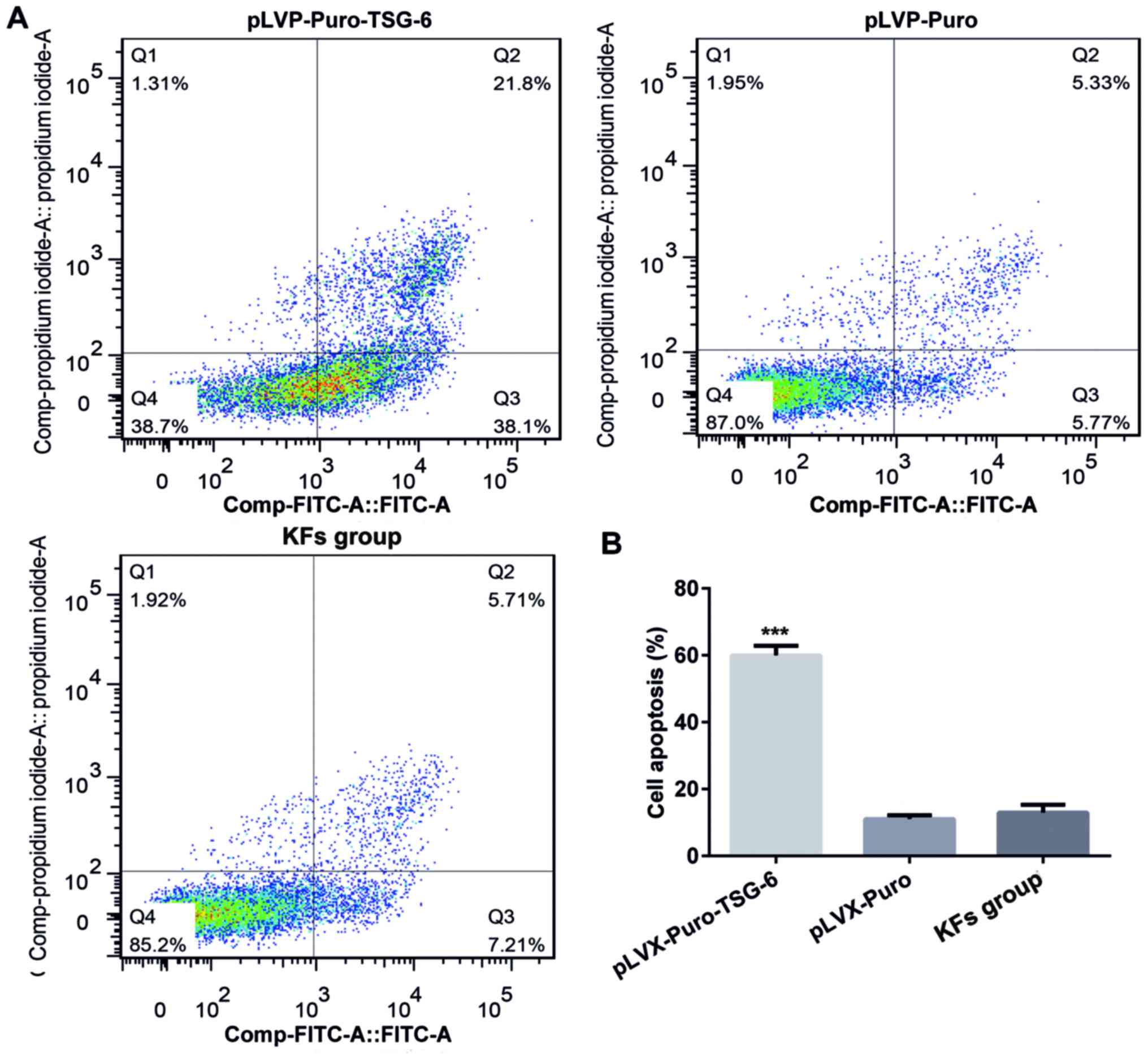
apoptosis of KFs
Flow cytometry was performed to determine the rate
of cell apoptosis. Apoptosis was analyzed by double-staining with
Annexin V-FITC and PI. The apoptosis rate in the pLVX-Puro-TSG-6
group was 51.92%, which was significantly higher than that in
pLVX-Puro group (19.00%) and the KF group (3.59%; P<0.05). There
was no significant difference in apoptosis rate between the
pLVX-Puro group and the KF cell group (Fig. 7).
TSG-6 regulated expression of genes
and proteins involved in proliferation, cell cycle and
apoptosis
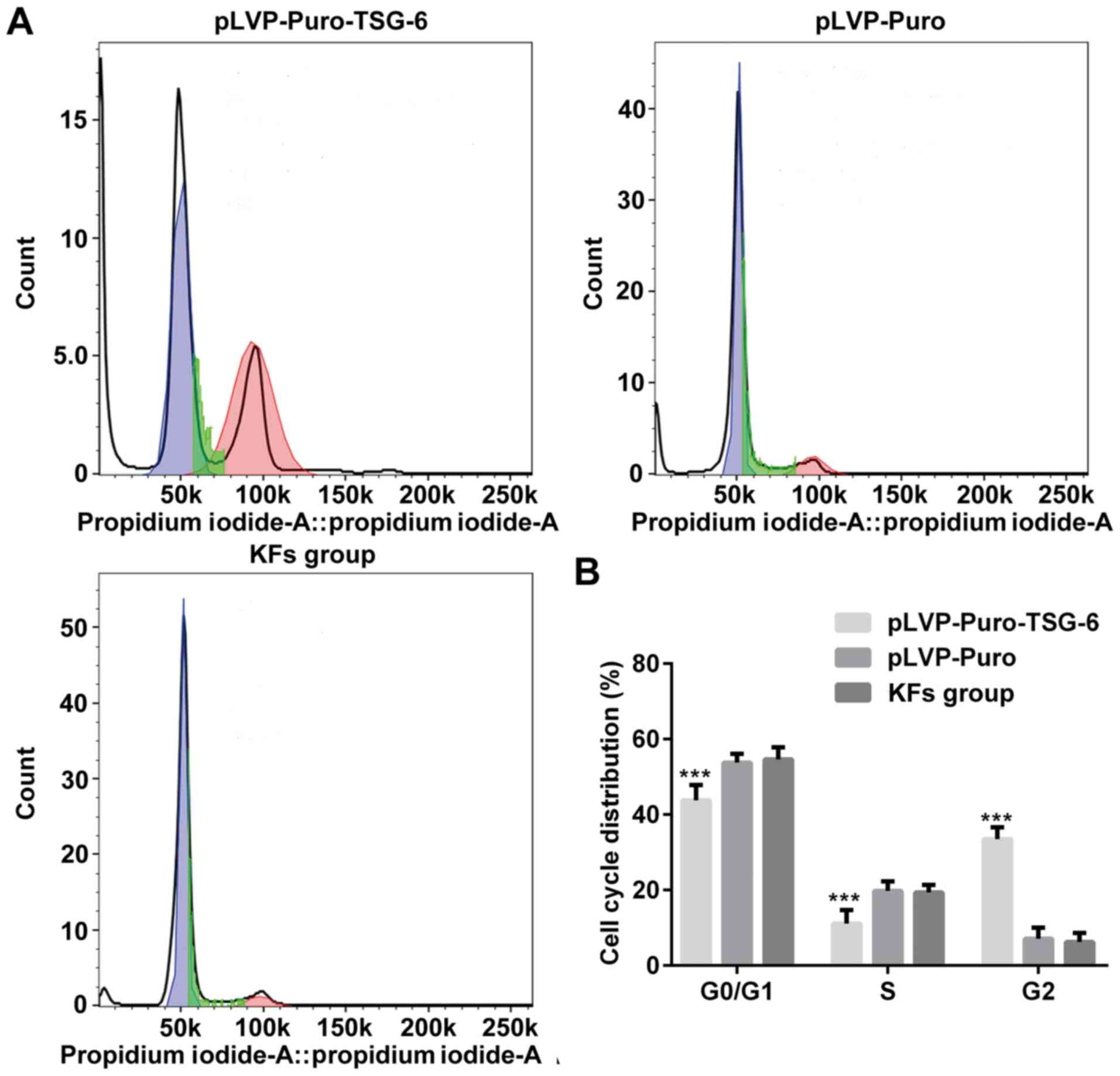
Whether the regulation of TSG-6 affects proteins
involved in proliferation, apoptosis and cell cycle in KFs was
investigated. Total cellular mRNA or protein was isolated from
cells transfected with pLVX-Puro-TSG-6 or pLVX-Puro, and
untransfected cells. Flow cytometry analysis suggested that cells
were arrested in G2/M phase by TSG-6 overexpression (Fig. 8). Levels of p21, cyclin D1, Bax,
Bcl-2, caspase-3 and NF-κB were determined by RT-PCR and western
blot. The RT-PCR results indicated that transfection of TSG-6 led
to an increase in the expression of p21, Bax and caspase-3.
Additionally, overexpression of TSG-6 decreased the mRNA levels of
cyclin D1 and NF-κB (Fig. 9). The
ratio of Bcl-2/Bax was also decreased significantly in the
TSG-6-transfected cells compared with the control group. Similar
results were confirmed by western blot; overexpressed TSG-6
markedly increased the protein levels of p21, Bax and caspase-3
(Fig. 10).
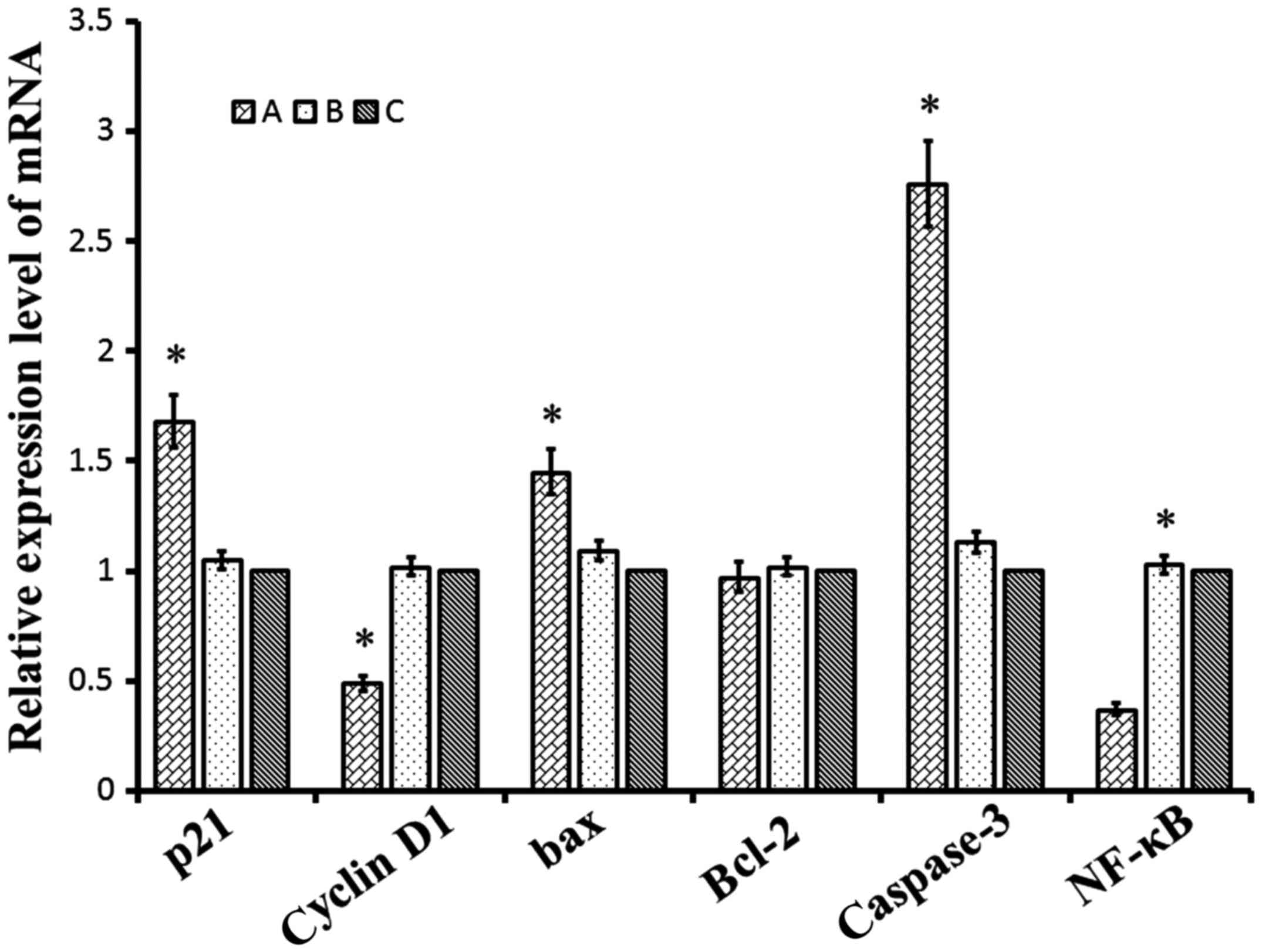 | Figure 9.Relative mRNA expression levels of
p21, cyclin D1, Bax, Bcl-2, caspase-3 and NF-κB. A, pLVX-Puro-TSG-6
transfected cells; B, pLVX-Puro transfected cells; C, keloid
fibroblasts. Data were presented by mean ± SD. *P<0.05 vs. KFs.
TSG-6, tumor necrosis factor-stimulated gene-6; Bax, apoptosis
regulator Bcl-2-associated X protein; Bcl-2, apoptosis regulator
B-cell lymphoma-2; NF-κB, nuclear factor-κB. |
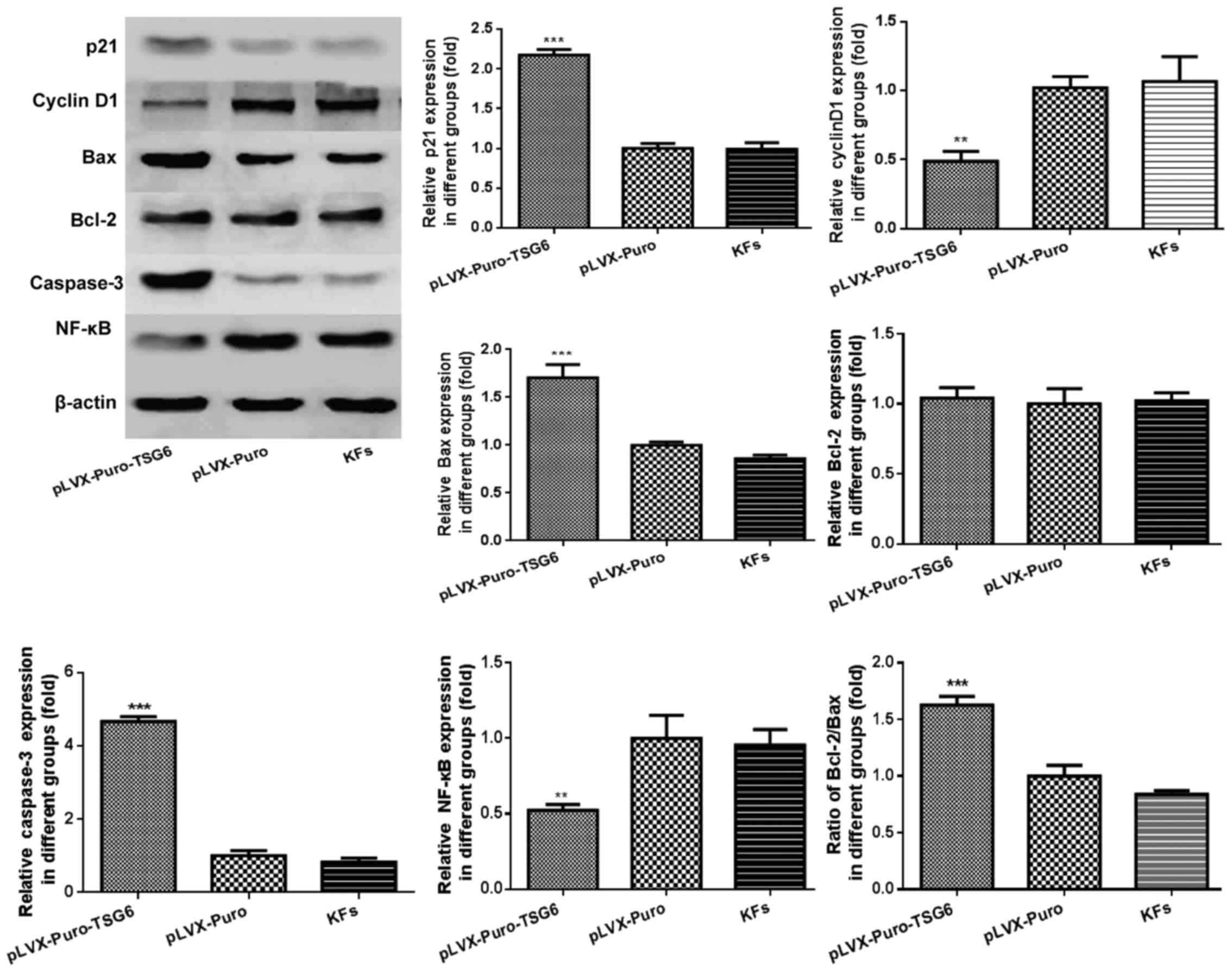 | Figure 10.Protein expression of p21, cyclin D1,
Bax, Bcl-2, caspase-3 and NF-κB. Data are presented as mean ±
standard deviation. **P<0.05, ***P<0.05 vs. KFs. Bax,
apoptosis regulator Bcl-2-associated X protein; Bcl-2, apoptosis
regulator B-cell lymphoma-2; NF-κB, nuclear factor-κB; TSG-6, tumor
necrosis factor-stimulated gene-6; KFs, keloid fibroblasts. |
Discussion
Keloid formation occurs when the normal
wound-healing process is disordered and the evolving scar remains
in the proliferative phase of healing. The scar may grow beyond the
boundaries of the original wound (13). It has been identified that
apoptosis may be a mechanism by which granulation tissue develops,
thus decreasing cellularity and scar formation (14). The mRNA expression of TSG-6 in
keloid was previously compared with normal skin and it was
demonstrated that low expression of TSG-6 may participate in the
formation of keloid. The previous investigations indicated that the
expression level of TSG-6 in primary keloid fibroblasts derived
from individual patients was significantly higher than in normal
skin (15), which is supported by
the results and conclusions of the present study.
TSG-6 is a hyaluronic acid (HA)-binding protein
composed of a single-strand module of two α-helices and two
triple-strand β-sheets arranged around a large hydrophobic core
(16). TSG-6 is involved in the
regulation of leukocyte migration and the pattern of expression
indicates that it may be associated with extracellular matrix
remodeling (17) Increased TSG-6
protein expression has been detected in the synovial fluid of
patients with arthritis, and recombinant TSG-6 protein has been
identified to have potent anti-inflammatory effects in vivo
(18). It has been indicated that
TSG-6 is part of a cytokine-initiated feedback loop that reduces
the inflammatory response. TSG-6 was observed to form a stable,
possibly covalent, 120 kDa complex with a serine protease inhibitor
and inter-α inhibitor (IαI) (19).
Furthermore, anti-plasmin activity was significantly increased by
this complex compared with IαI alone (20). In conclusion, TSG-6 may be a key
factor involved in regulating leukocyte migration and matrix
remodeling, caused by break down of latent metalloproteinases in
the extracellular matrix via enhanced activity of plasmin.
In the current study, a lentiviral vector carrying
the TSG-6 gene was transfected into KFs, and increased the
expression of TSG-6 mRNA confirmed that the transduction was
effective. In addition, western blot results indicated that the
TSG-6 protein in the pLVX-Puro-TSG-6 group was primarily encoded by
the exogenous TSG-6 gene. To determine whether TSG-6 has selective
cytotoxic activity in KFs, MTT and apoptosis assays were performed,
and the mRNA and protein levels of key molecules were determined.
According to the MTT results, TSG-6 inhibited KF proliferation
selectively compared with the negative control. Flow cytometry
analysis suggested that TSG-6 selectively induced KF cell death and
accumulation of cells in G2/M phase by preventing KFs from entering
M phase. RT-PCR and western blot results also supported these
conclusions.
Recent studies have revealed that keloids have
tumor-like biological functions, and the roles of therapeutic
strategies such as microRNAs, bone morphogenetic proteins, activin
membrane bound inhibitor and heat shock protein 70 in keloids have
been investigated (21–23). It has been confirmed that TSG-6 is
lowly expressed in prostate cancer and can be a potential marker
for prostate cancer diagnosis (24). A previous study identified that
TSG-6 combined with Yeast Cytosine Deaminase and 5-Fluorocytosine
exerts anti-tumor effects (25).
Pathogenic mechanisms of keloid have not been elucidated in detail;
however, controlling fibroblast proliferation and apoptosis via
certain targeted genes may be an effective therapeutic strategy for
treating keloids. In the current study, TSG-6 overexpression
increased apoptosis and decreased cell proliferation, which could
offer a protective effect against keloid development.
In summary, KFs transfected with a lentiviral vector
carrying TSG-6 expressed a high level of exogenous TSG-6.
Transduction of the pLVX-Puro-TSG-6 selectively suppressed
proliferation and induced apoptosis in KFs, which confirmed the
role of TSG-6 in KFs. As a result, TSG-6 has potential as a gene
therapy candidate for the treatment of keloid, and may result in
novel and effective therapeutic approaches.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (no. 81272107).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
ZC conceived, designed, coordinated and performed
all the experiments, conducted the statistical analysis and drafted
the manuscript. XYL collected all samples and achieved particle
analysis. HW and XJL assisted in study design and manuscript
editing. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The current study was approved by the institutional
review board of Anhui Medical University (Hefei, China), and
written informed consent was obtained from all patients prior to
inclusion. The study was performed according to the principles of
the Declaration of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gupta J, Gantyala SP, Kashyap S and Tandon
R: Diagnosis, management, and histopathological characteristics of
corneal keloid: A case series and literature review. Asia Pac J
Ophthalmol (Phila). 5:354–359. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeon YR, Ahn HM, Choi IK, Yun CO, Rah DK,
Lew DH and Lee WJ: Hepatocyte growth factor-expressing adenovirus
upregulates matrix metalloproteinase-1 expression in keloid
fibroblasts. Int J Dermatol. 55:356–361. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Syed F, Singh S and Bayat A: Superior
effect of combination vs. single steroid therapy in keloid disease:
A comparative in vitro analysis of glucocorticoids. Wound Repair
Regen. 21:88–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Har-Shai Y and Zouboulis CC: Intralesional
cryotherapy for the treatment of keloid scars: A prospective study.
Plast Reconstr Surg. 136:397e–398e. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee MJ, Kim DH, Ryu JS, Ko AY, Ko JH, Kim
MK, Wee WR, Khwarg SI and Oh JY: Topical TSG-6 administration
protects the ocular surface in two mouse models of
inflammation-related dry eye. Invest Ophthalmol Vis Sci.
56:5175–5181. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim JA, Ko JH, Ko AY, Lee HJ, Kim MK, Wee
WR, Lee RH, Fulcher SF and Oh JY: TSG-6 protects corneal
endothelium from transcorneal cryoinjury in rabbits. Invest
Ophthalmol Vis Sci. 55:4905–4912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baranova NS, Nilebäck E, Haller FM, Briggs
DC, Svedhem S, Day AJ and Richter RP: The inflammation-associated
protein TSG-6 cross-links hyaluronan via hyaluronan-induced TSG-6
oligomers. J Biol Chem. 286:25675–25686. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tan KT, McGrouther DA, Day AJ, Milner CM
and Bayat A: Characterization of hyaluronan and TSG-6 in skin
scarring: Differential distribution in keloid scars, normal scars
and unscarred skin. J Eur Acad Dermatol Venereol. 25:317–327. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mahoney DJ, Swales C, Athanasou NA,
Bombardieri M, Pitzalis C, Kliskey K, Sharif M, Day AJ, Milner CM
and Sabokbar A: TSG-6 inhibits osteoclast activity via an autocrine
mechanism and is functionally synergistic with osteoprotegerin.
Arthritis Rheum. 63:1034–1043. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kehlen A, Pachnio A, Thiele K and Langner
J: Gene expression induced by interleukin-17 in fibroblast-like
synoviocytes of patients with rheumatoid arthritis: Upregulation of
hyaluronan-binding protein TSG-6. Arthritis Res Ther. 5:R186–R192.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang H, Chen Z, Li XJ, Ma L and Tang YL:
Anti-inflammatory cytokine TSG-6 inhibits hypertrophic scar
formation in a rabbit ear model. Eur J Pharmacol. 751:42–49. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang GY, Yi CG, Li X, Zheng Y, Niu ZG,
Xia W, Meng Z, Meng CY and Guo SZ: Inhibition of vascular
endothelial growth factor expression in keloid fibroblasts by
vector-mediated vascular endothelial growth factor shRNA: A
therapeutic potential strategy for keloid. Arch Dermatol Res.
300:177–184. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hochman B, Isoldi FC, Furtado F and
Ferreira LM: New approach to the understanding of keloid:
Psychoneuroimmune-endocrine aspects. Clin Cosmet Investig Dermatol.
8:67–73. 2015.PubMed/NCBI
|
|
14
|
Parikh DA, Ridgway JM and Ge NN: Keloid
banding using suture ligature: A novel technique and review of
literature. Laryngoscope. 118:1960–1965. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hong X, Li X and Ning J: Expression of
TSG-6 and its significance in pathological scar. Acta Univ Med
Anhui. 48:685–687. 2013.(In Chinese).
|
|
16
|
Milner CM, Higman VA and Day AJ: TSG-6: A
pluripotent inflammatory mediator? Biochem Soc Trans. 34:446–450.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park Y, Jowitt TA, Day AJ and Prestegard
JH: Nuclear magnetic resonance insight into the multiple
glycosaminoglycan binding modes of the link module from human
TSG-6. Biochemistry. 55:262–276. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nagyeri G, Radacs M, Ghassemi-Nejad S,
Tryniszewska B, Olasz K, Hutas G, Gyorfy Z, Hascall VC, Glant TT
and Mikecz K: TSG-6 protein, a negative regulator of inflammatory
arthritis, forms a ternary complex with murine mast cell tryptases
and heparin. J Biol Chem. 286:23559–23569. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baranova NS, Foulcer SJ, Briggs DC,
Tilakaratna V, Enghild JJ, Milner CM, Day AJ and Richter RP:
Inter-a-inhibitor impairs TSG-6-induced hyaluronan cross-linking. J
Biol Chem. 288:29642–29653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang S, He H, Day AJ and Tseng SC:
Constitutive expression of inter-a-inhibitor (IaI) family proteins
and tumor necrosis factor-stimulated gene-6 (TSG-6) by human
amniotic membrane epithelial and stromal cells supporting formation
of the heavy chain-hyaluronan (HC-HA) complex. J Biol Chem.
287:12433–12444. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang GY, Wu LC, Liao T, Chen GC, Chen YH,
Zhao YX, Chen SY, Wang AY, Lin K, Lin DM, et al: A novel regulatory
function for miR-29a in keloid fibrogenesis. Clin Exp Dermatol.
41:341–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin L, Wang Y, Liu W and Huang Y: BAMBI
inhibits skin fibrosis in keloid through suppressing TGF-b1-induced
hypernomic fibroblast cell proliferation and excessive accumulation
of collagen I. Int J Clin Exp Med. 8:13227–13234. 2015.PubMed/NCBI
|
|
23
|
Shin JU, Lee WJ, Tran TN, Jung I and Lee
JH: Hsp70 knockdown by siRNA decreased collagen production in
keloid fibroblasts. Yonsei Med J. 56:1619–1626. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garcia GE, Wisniewski HG, Lucia MS,
Arevalo N, Slaga TJ, Kraft SL, Strange R and Kumar AP:
2-Methoxyestradiol inhibits prostate tumor development in
transgenic adenocarcinoma of mouse prostate: Role of tumor necrosis
factor-alpha-stimulated gene 6. Clin Cancer Res. 12:980–988. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park JI, Cao L, Platt VM, Huang Z, Stull
RA, Dy EE, Sperinde JJ, Yokoyama JS and Szoka FC: Antitumor therapy
mediated by 5-fluorocytosine and a recombinant fusion protein
containing TSG-6 hyaluronan binding domain and yeast cytosine
deaminase. Mol Pharm. 6:801–812. 2009. View Article : Google Scholar : PubMed/NCBI
|