Introduction
Diabetic cardiomyopathy (DCM) is a type
cardiomyopathy caused by diabetes mellitus; the main pathological
alterations are cardiac hypertrophy and cardiac dysfunction
(1). Clinical studies have
reported that DCM is the main reason underlying the high incidence
and high mortality rate of heart failure (HF) in patients with
diabetes (2,3). However, the mechanism underlying DCM
is not entirely clear; therefore, there is no effective targeted
therapy (4). Research has
increasingly focused on identifying the mechanism underlying DCM
and exploring potential effective treatments.
Non-coding RNAs (ncRNAs) are functional RNA
molecules in the transcriptome that do not encode proteins,
including microRNAs (miRNAs/miRs) and long ncRNAs (lncRNAs)
(5). miRNAs are single-stranded,
non-coding small RNAs that are highly conserved in evolution and
have post-transcriptional regulatory activity (6), whereas lncRNAs are mRNA-like
transcripts >200 nucleotides long that have no or little
protein-coding function and serve important roles in numerous
biological processes (7). lncRNAs
have been reported to be important in the governing of fundamental
biological processes (8), and
their aberrant expression may be associated with the pathogenesis
of various diseases, including cancer, neurodegenerative diseases,
and cardiovascular diseases (9).
Previous studies have demonstrated that lncRNAs can
act as competitive endogenous RNAs (ceRNAs) to compete with target
genes for miRNA response elements and attenuate the inhibitory
effect of miRNAs on target genes (10,11).
Therefore, they can indirectly regulate the expression of target
genes and affect the occurrence and development of diseases
(12), particularly cardiovascular
disease (13). Zhang et al
(14), reported that the lncRNA
metastasis-associated lung adenocarcinoma transcript 1 serves a
role in the pathogenesis of DCM, and Zhou et al (15) demonstrated that lncRNA myocardial
infarction-associated transcript is a ceRNA that upregulates
death-associated protein kinase 2 by inhibiting miR-22-3p in DCM.
However, the association between ceRNAs and DCM is unclear.
Therefore, the present study aimed to elucidate the pathogenesis of
DCM from the perspective of ceRNA using bioinformatics analysis.
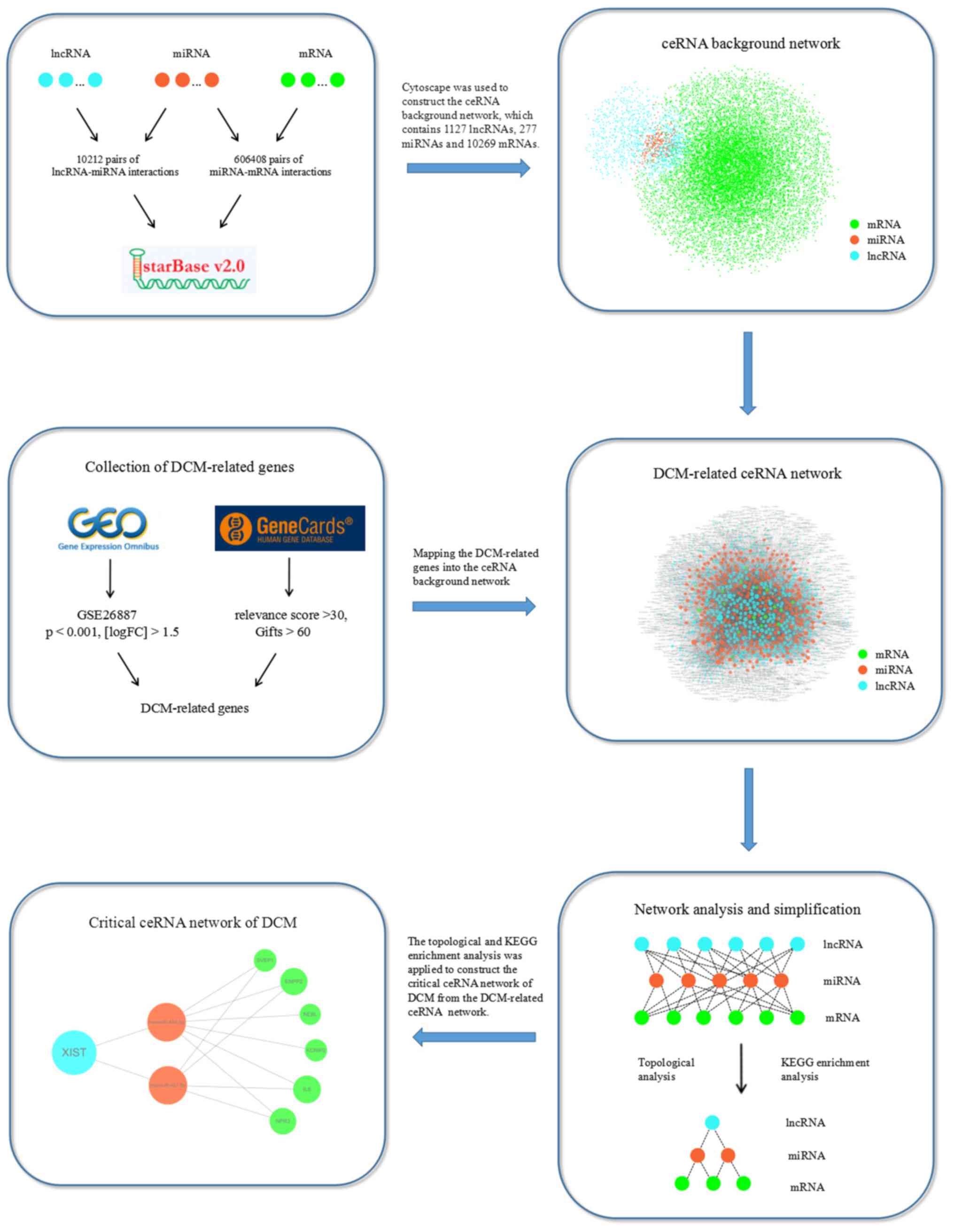
The pipeline of construction and analysis of the lncRNA-miRNA-mRNA
network based on ceRNA in DCM is shown in Fig. 1.
Materials and methods
Identification of DCM-related
genes
DCM-related genes were collected from two main
sources: Gene Expression Omnibus (GEO) and GeneCards. The GEO
(https://www.ncbi.nlm.nih.gov/gds/) is
a public functional genomics data repository supporting minimum
information about a microarray experiment-compliant data
submissions. For the present study, the GSE26887 dataset was
obtained, which includes DCM and normal myocardial tissue gene
expression profiles (16). The
GSE26887 dataset includes microarray data from seven patients with
type 2 diabetes mellitus (T2DM) and HF, and five control
individuals. Subsequently, GeoDiver (https://www.geodiver.co.uk/) (17) was used to develop overview
boxplots, a volcano plot, and a heat map to show the distribution
of differentially expressed genes (DEGs). P<0.01 and [log fold
change (FC)]>1.5 were considered statistically significant.
GeneCards is a database for searchable human gene
annotations (http://www.genecards.org/). Its gene-centric data are
automatically mined from ~125 web sources, including genomic,
transcriptomic, proteomic, genetic, clinical and functional
information. After entering a keyword of ‘diabetic cardiomyopathy’
into GeneCards, relevance score >30 and GeneCards Inferred
Functionality Scores (18) >60
were used as cut-off criteria to collect disease-related genes and
merge them with DEGs.
miRNA-mRNA and lncRNA-miRNA
interactions
The ceRNA theory dictates that the construction of a
ceRNA background network (CBGN) requires large numbers of lncRNAs,
miRNAs, mRNAs and their interactions. Firstly, miRNA-mRNA and
lncRNA-miRNA interactions were obtained from the starBase V2.0
database (http://starbase.sysu.edu.cn/). StarBase was designed
to decode miRNA-mRNA, miRNA-ceRNA, miRNA-lncRNA, miRNA-circRNA,
miRNA-pseudogene and protein-RNA interaction networks from
cross-linking immunoprecipitation-sequencing data.
Construction of the ceRNA background
network
Cytoscape software v3.5.1 (http://www.cytoscape.org/) was used to integrate and
combine miRNA-mRNA and lncRNA-miRNA interactions to construct the
lncRNA-miRNA-mRNA (ceRNA) background network. Cytoscape is an
open-source software platform used to visualize complex networks
and integrate these with any type of attribute data. In Cytoscape,
lncRNAs, miRNAs and mRNAs are expressed in different nodes, which
are connected by lines to indicate an interaction between them.
Finally, an intricate network diagram consisting of lncRNAs, miRNAs
and mRNAs was obtained.
Signaling pathway enrichment
analysis
Signaling pathway enrichment of DCM-related genes
was conducted using the Cytoscape plugin ClueGO (http://apps.cytoscape.org/apps/cluego)
(19). ClueGO is a Cytoscape
plugin that visualizes non-redundant biological terms for large
clusters of genes in a functionally grouped network; it is created
with kappa statistics and reflects the relationships between the
terms based on the similarity of their associated genes. For this
study, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis
(20) was selected in ClueGO to
obtain information about the signaling pathways regulated by
DCM-related genes.
Construction of a DCM-related ceRNA
network (DCMCN)
The DCMCN was extracted from CBGN using the
network-merge tool in Cytoscape. DCM-related genes and CBGN were
imported into Cytoscape together to construct two separate
networks. The two networks were then merged using the network merge
operation (21) of Cytoscape to
build a new network. The new network comprised DCM-related genes
and relevant miRNAs and lncRNAs; this network is known as the
DCMCN.
Analysis of the DCMCN
Hub nodes serve critical roles in gene networks
(22). Therefore, all node degrees
of the lncRNA-miRNA-mRNA network were calculated. Specifically, the
network-analyzer tool (23) in
Cytoscape was used to obtain the topological parameters of the
network, mainly the degree value. Degree is the most direct measure
of centrality in network analysis. Generally, the higher the degree
of a node, the more critical it is to the network (24). Finally, the lncRNA with the largest
degree from the network was selected, which likely serves a key
role in regulating DCM. Subsequently, the lncRNA and its downstream
miRNAs and mRNAs were selected in Cytoscape and isolated from the
DCMCN to form a new network. The miRNAs bound by the lncRNA were
screened synthetically for target sites in starBase, and degree and
number of target genes in Cytoscape. The major miRNAs and their
target genes regulated by the lncRNA were obtained and plotted into
a small ceRNA regulatory network using Cytoscape. Finally, the
ClueGO plugin in Cytoscape was used to analyze the gene function of
each regulatory network to determine whether it was consistent with
the regulatory function of the DCMCN.
Results
Construction of the ceRNA background
network
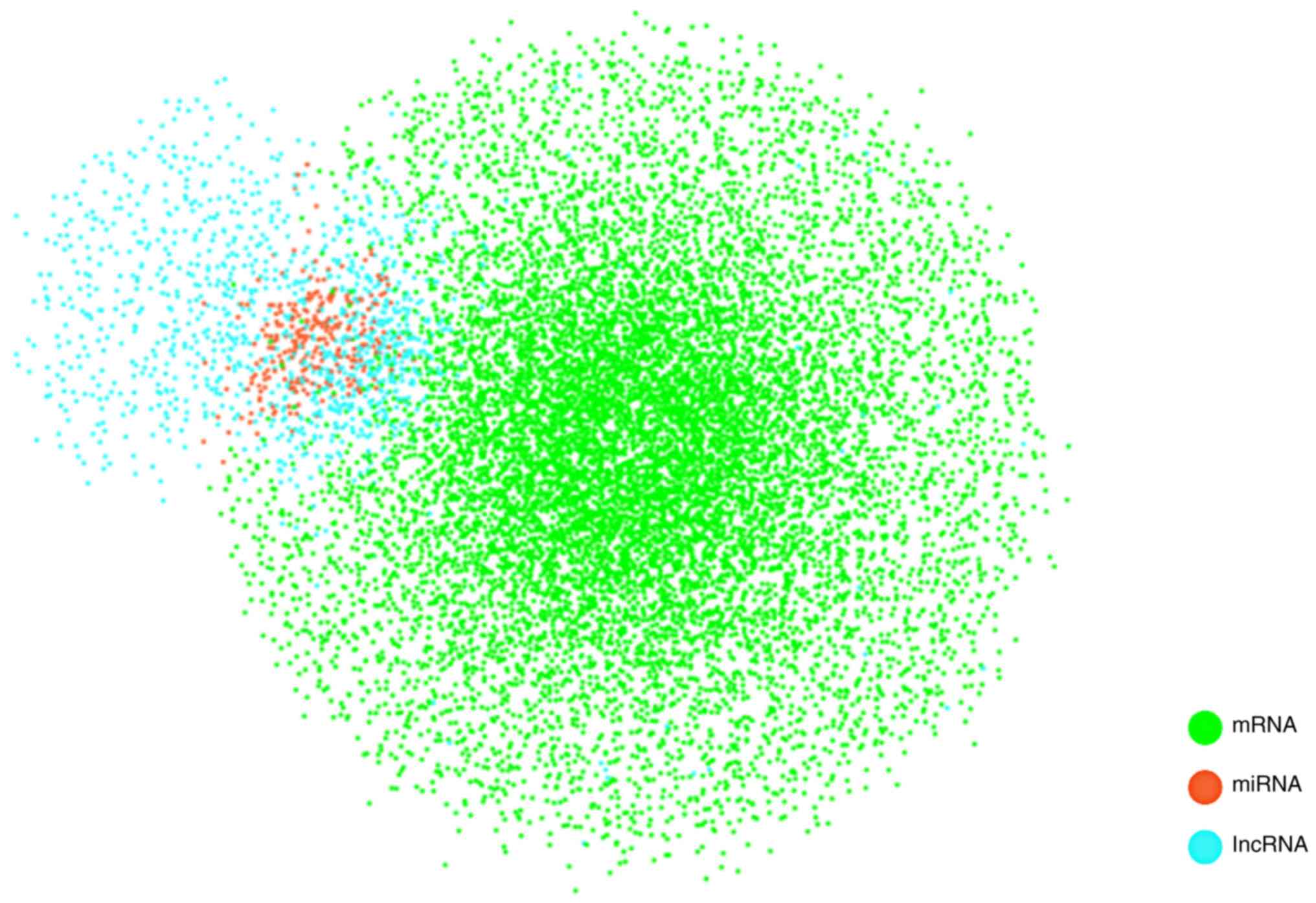
To construct the ceRNA background network, the
miRNA-mRNA and lncRNA-miRNA interactions were downloaded from
starBase v2.0. The data in starBase v2.0 are mainly collected from
TargetScan (http://www.targetscan.org/vert_71/) (25), RNA22 (https://cm.jefferson.edu/rna22/) (26), picTar (http://pictar.mdc-berlin.de/) (27), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html)
and miRanda (http://www.microrna.org/microrna/home.do?) (28). A total of 606,408 pairs of
miRNA-mRNA interactions and 10,212 pairs of lncRNA-miRNA
interactions were obtained and Cytoscape software v3.5.1 was then
used to integrate the miRNA-mRNA and lncRNA-miRNA information to
construct the lncRNA-miRNA-mRNA (ceRNA) background network
(Fig. 2).
Identification of DCM-related
genes
DCM-related genes were collected from the GEO and
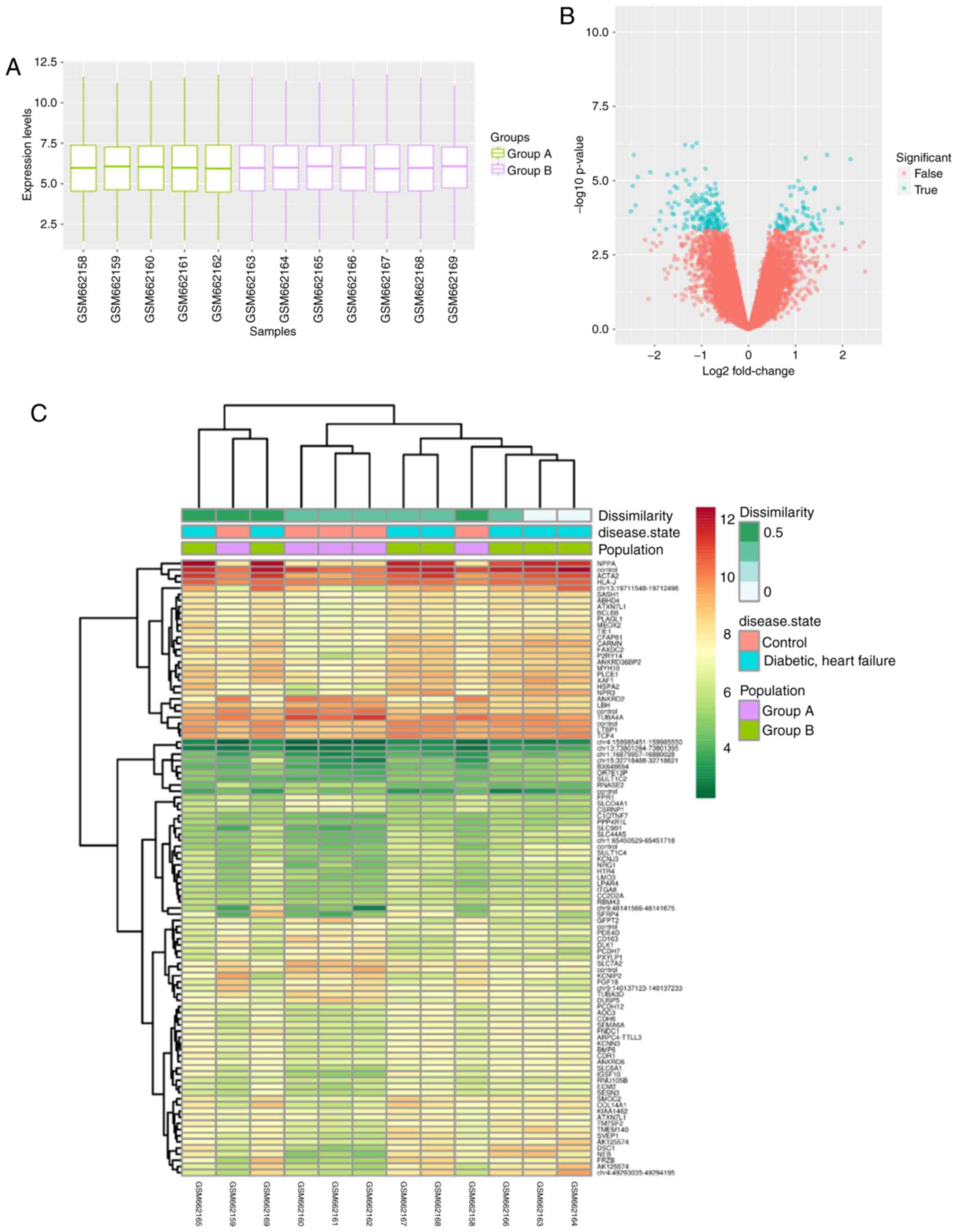
GeneCards. Firstly, the open gene expression profile GSE26887 was
downloaded the from GEO database. Subsequently, GeoDiver was used
to analyze the DEGs between controls and patients with T2DM and HF
in the GSE26887 dataset. An overview boxplot, volcano plot and
heatmap were generated using GeoDiver (Fig. 3A-C), and DEGs were identified.
As shown in Table
I, 23 DEGs were identified from the GEO repository microarray
data using cutoff criteria of P<0.01 and [logFC]>1.5.
Subsequently, GeneCards databases were searched for disease-related
genes (Table II). After removing
redundancy, 53 genes were identified as DCM-related genes,
including NPPA, SFRP4, DSC1, NEB, FRZB, AK125574, ENPP2, PRELP,
NPR3, IGSF10, HSPA2, COL14A1, P2RY14, SLC9B1, FMOD, SVEP1, FAXDC2,
CNN1, GFPT2, CD163, KCNIP2, S100A8, ANKRD2, ACE, F2, IL6, NOS3,
TNF, AGTR1, MTHFR, REN, APOB, APOE, LMNA, TNNI3, TGFB1, NPPB, INS,
APOA1, FBN1, CRP, F5, THBD, CST3, EDN1, SCN5A, GP1BA, PON1, ACTC1,
DES, PTPN11, CCL2, TNNT2.
 | Table I.Differentially expressed genes
identified from the GSE26887 dataset using GeoDiver. |
Table I.
Differentially expressed genes
identified from the GSE26887 dataset using GeoDiver.
| Gene symbol | logFC | P-value |
|---|
| NPPA | −3.53 |
1.36×10−10 |
| SFRP4 | −2.71 |
2.96×10−5 |
| DSC1 | −2.44 |
1.35×10−6 |
| NEB | −2.40 |
6.83×10−5 |
| FRZB | −2.36 |
7.56×10−6 |
| AK125574 | −2.09 |
5.16×10−6 |
| ENPP2 | −2.01 |
4.50×10−5 |
| PRELP | −1.90 |
2.18×10−5 |
| NPR3 | −1.87 |
1.12×10−5 |
| IGSF10 | −1.72 |
6.26×10−6 |
| HSPA2 | −1.71 |
5.91×10−5 |
| COL14A1 | −1.67 |
2.44×10−5 |
| P2RY14 | −1.62 |
1.27×10−5 |
| SLC9B1 | −1.57 |
1.56×10−5 |
| FMOD | −1.56 |
3.42×10−5 |
| SVEP1 | −1.53 |
5.54×10−6 |
| FAXDC2 | −1.52 |
1.77×10−5 |
| CNN1 | 1.57 |
4.55×10−5 |
| GFPT2 | 1.61 |
9.96×10−5 |
| CD163 | 1.67 |
1.35×10−6 |
| KCNIP2 | 1.90 |
8.40×10−5 |
| S100A8 | 1.98 |
2.68×10−5 |
| ANKRD2 | 2.16 |
1.89×10−6 |
 | Table II.Disease-related genes collected from
GeneCards. |
Table II.
Disease-related genes collected from
GeneCards.
| Gene symbol | Gene name | GIFtS | Relevance
score |
|---|
| ACE | Angiotensin I
converting enzyme | 70 | 82.65 |
| F2 | Coagulation factor
II, thrombin | 70 | 55.55 |
| IL6 | Interleukin 6 | 70 | 54.18 |
| NOS3 | Nitric oxide
synthase 3 | 72 | 52.43 |
| TNF | Tumor necrosis
factor | 77 | 51.78 |
| AGTR1 | Angiotensin II
receptor type 1 | 71 | 51.43 |
| MTHFR |
Methylenetetrahydrofolate reductase | 64 | 50.99 |
| REN | Renin | 68 | 49.63 |
| APOB | Apolipoprotein
B | 65 | 47.05 |
| APOE | Apolipoprotein
E | 71 | 46.92 |
| LMNA | Lamin A/C | 66 | 42.33 |
| TNNI3 | Troponin I3,
cardiac type | 69 | 42.21 |
| TGFB1 | Transforming growth
factor β1 | 75 | 42.09 |
| NPPB | Natriuretic peptide
B | 63 | 41.2 |
| INS | Insulin | 67 | 39.05 |
| NPPA | Natriuretic peptide
A | 65 | 36.24 |
| APOA1 | Apolipoprotein
A1 | 70 | 35.38 |
| FBN1 | Fibrillin 1 | 62 | 34.86 |
| CRP | C-reactive
protein | 68 | 34.74 |
| F5 | Coagulation factor
V | 63 | 34.44 |
| THBD | Thrombomodulin | 62 | 33.6 |
| CST3 | Cystatin C | 64 | 33.38 |
| EDN1 | Endothelin 1 | 67 | 32.87 |
| SCN5A | Sodium
voltage-gated channel α subunit 5 | 70 | 32.8 |
| GP1BA | Glycoprotein Ib
platelet alpha Subunit | 63 | 31.03 |
| PON1 | Paraoxonase 1 | 65 | 30.99 |
| ACTC1 | Actin, α, cardiac
muscle 1 | 60 | 30.96 |
| DES | Desmin | 68 | 30.88 |
| PTPN11 | Protein tyrosine
phosphatase, non-receptor type 11 | 71 | 30.71 |
| CCL2 | C-C motif chemokine
ligand 2 | 70 | 30.67 |
| TNNT2 | Troponin T2,
cardiac type | 67 | 30.2 |
Signaling pathway enrichment analysis
for DCM-related genes
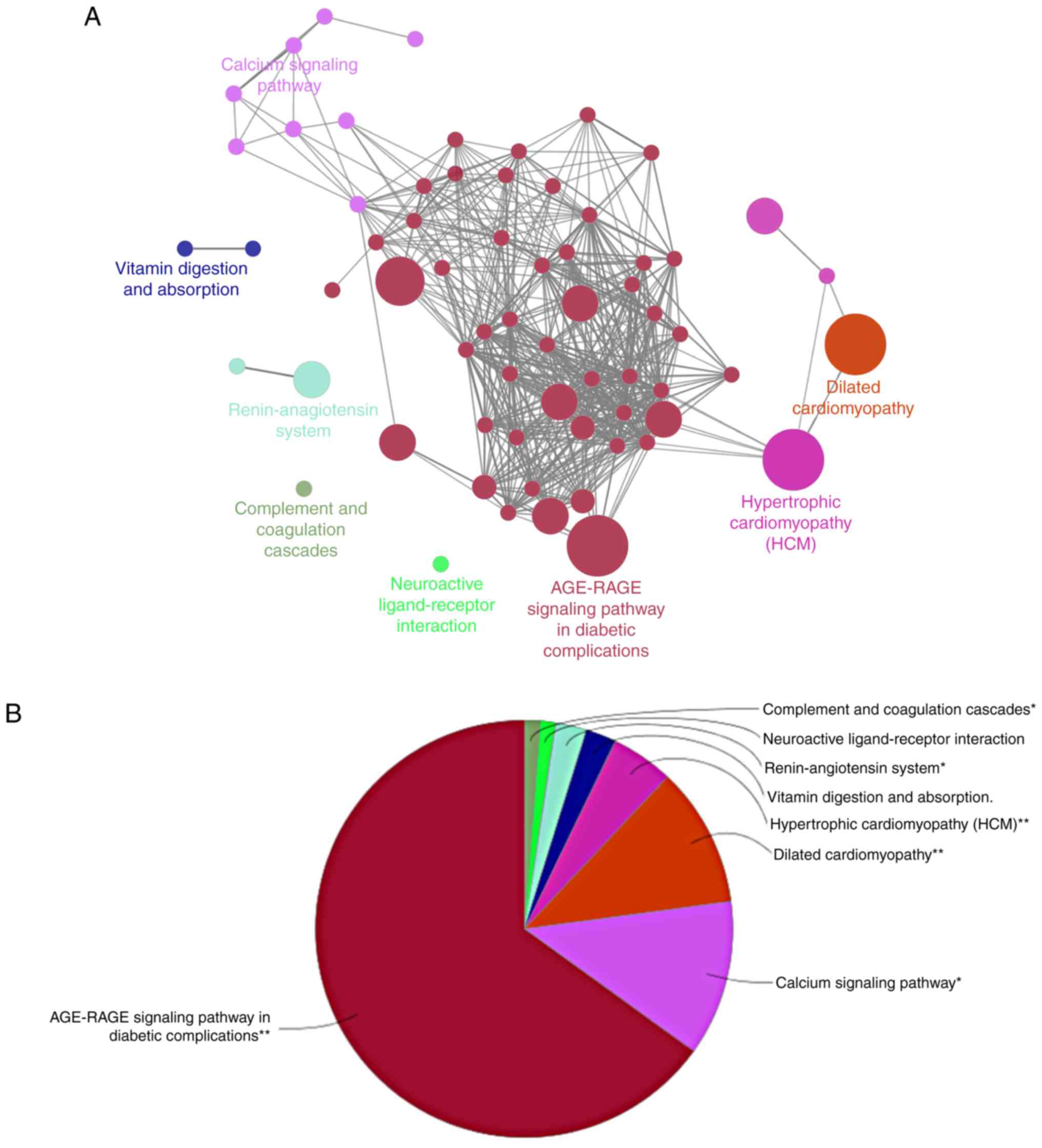
DCM-related genes were examined through
ClueGO-mediated enrichment analysis by employing KEGG terms for the
annotation of gene function. As shown in Fig. 4A and B, DCM-related genes were
mainly associated with the ‘AGE-RAGE signaling pathway in diabetic
complications’.
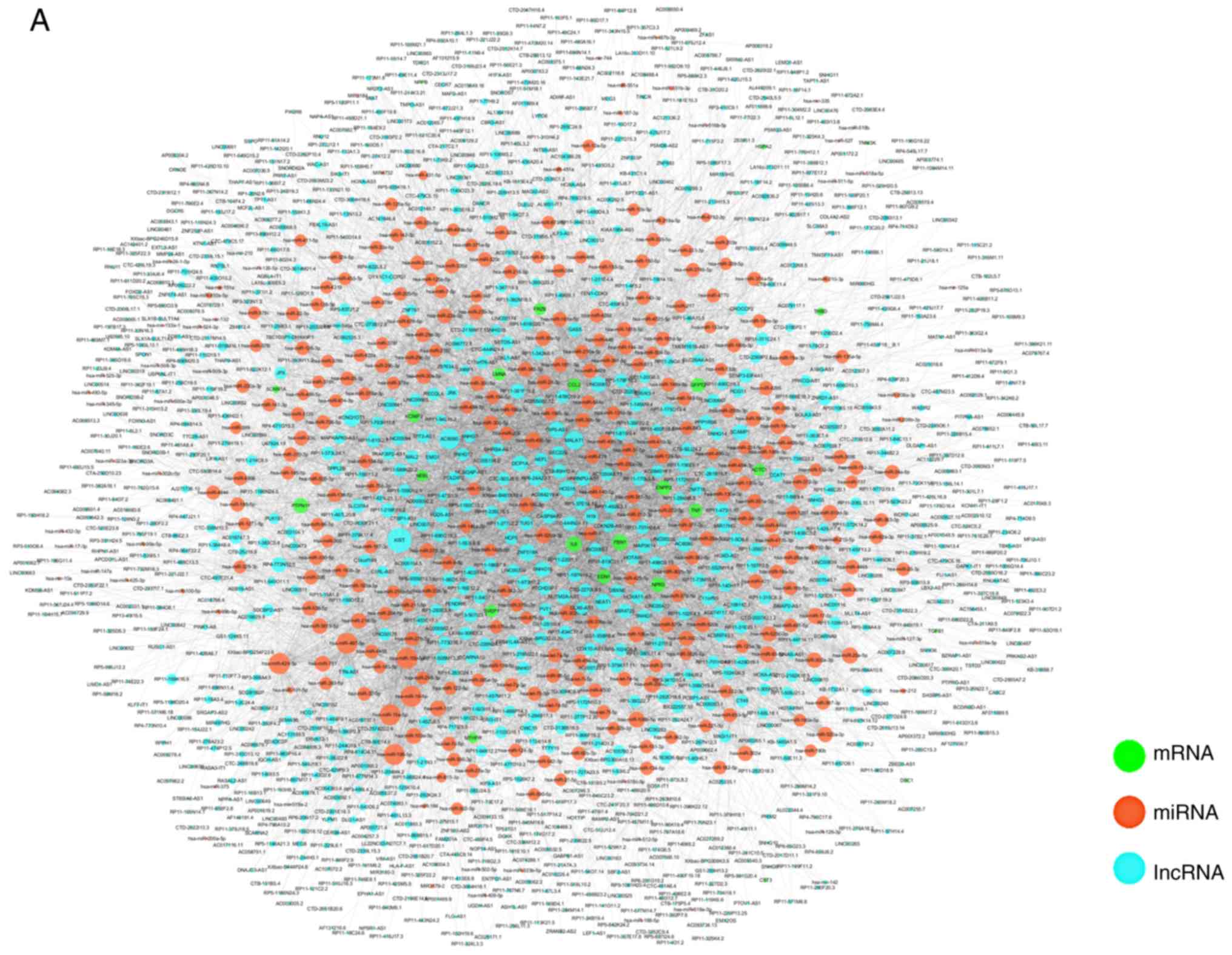
Construction and analysis of the DCMCN
from CBGN
To observe regulation of ceRNAs in DCM and identify
DCM-related lncRNAs, 53 DCM-related genes (mRNAs) were mapped into
the ceRNA background network. As shown in Fig. 5A, the DCMCN was extracted from the
ceRNA background network using the network merge tool in Cytoscape.
The blue, orange and green nodes represent lncRNAs, miRNAs and
DCM-related genes, respectively. In addition, the edges represent
the interactions between lncRNAs, miRNAs and mRNAs. The network
analyzer tool in Cytoscape was used to analyze the topological
parameters of the network; the top 10 degrees are shown in Table III. Notably, lncRNA X-inactive
specific transcript (XIST) had the largest degree in the DCMCN,
suggesting that lncRNA-XIST may be a key lncRNA that regulates the
development of DCM.
 | Table III.Degree value of lncRNAs in the
diabetic cardiomyopathy-related ceRNA network. |
Table III.
Degree value of lncRNAs in the
diabetic cardiomyopathy-related ceRNA network.
| lncRNA | Degree |
|---|
| XIST | 175 |
| CTA-204B4.6 | 130 |
| MALAT1 | 99 |
| ZNF518A | 83 |
| KCNQ1OT1 | 75 |
| OIP5-AS1 | 71 |
| NEAT1 | 69 |
| DCP1A | 69 |
| HCG18 | 64 |
| FGD5-AS1 | 62 |
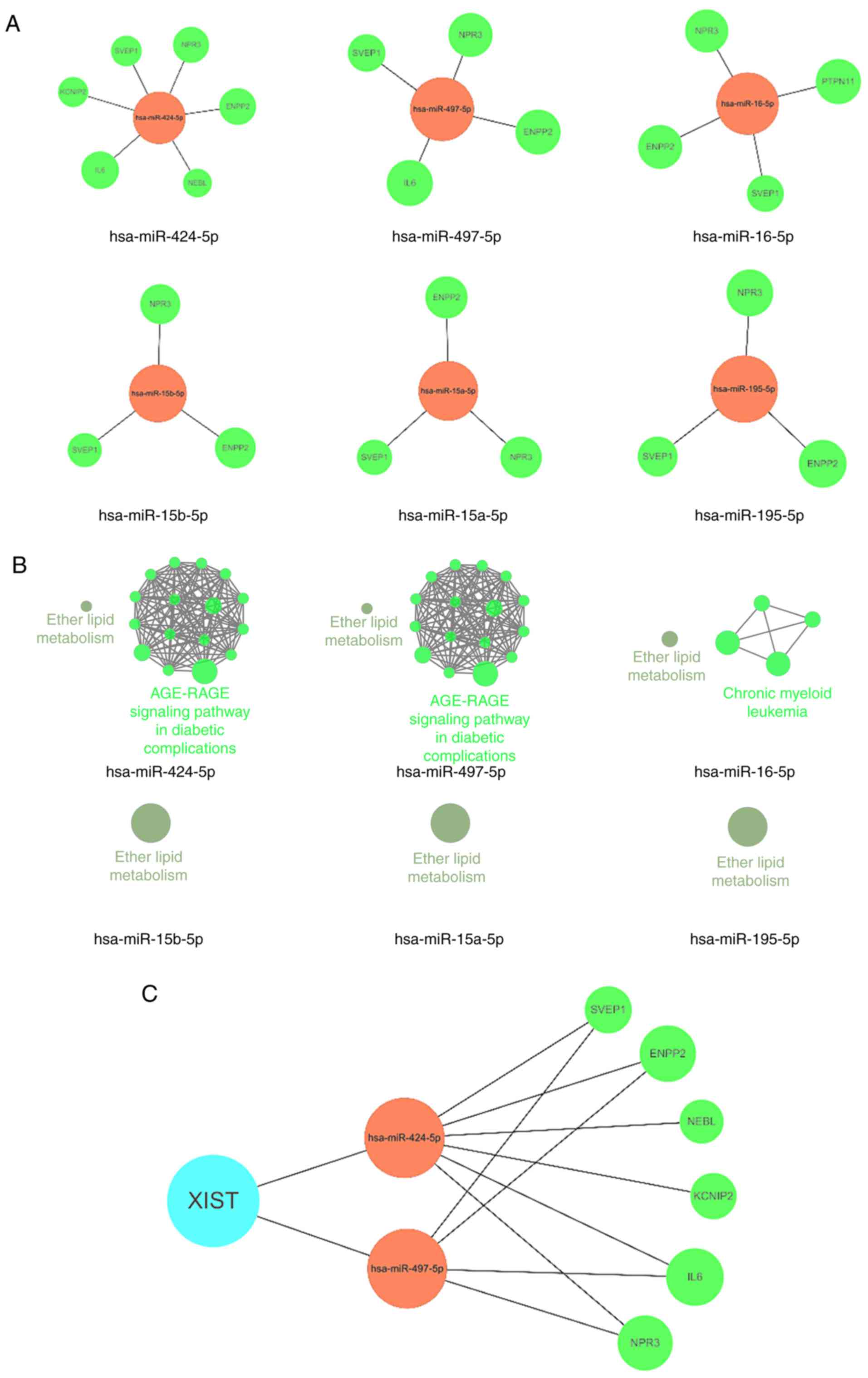
KEGG pathway analysis was performed for lncRNA-XIST.
lncRNA-XIST and its downstream miRNAs and mRNAs were selected in
Cytoscape and isolated from the DCMCN to reconstruct a new ceRNA
network (Fig. 5B). To simplify the
ceRNA XIST network, the number of target sites, degrees, and target
genes of the miRNAs regulated by XIST were further screened
(Table IV). Information regarding
the target sites of the miRNAs was obtained from starBase. After
screening, six miRNAs (hsa-miR-424-5p, hsa-miR-497-5p,
hsa-miR-16-5p, hsa-miR-15b-5p, hsa-miR-15a-5p and hsa-miR-195-5p)
had the maximum parameters of target sites, degrees and target
genes. These six miRNAs and their target genes that were regulated
by the lncRNA were obtained and plotted into a small ceRNA network
using Cytoscape software (Fig.
6A). KEGG enrichment analysis demonstrated that only
hsa-miR-424-5p and hsa-miR-497-5p may act on the ‘AGE-RAGE
signaling pathway in diabetic complications’ (Fig. 6B and C).
 | Table IV.Target sites, degree, and number of
target genes of the miRNAs bound by X-inactive specific
transcript. |
Table IV.
Target sites, degree, and number of
target genes of the miRNAs bound by X-inactive specific
transcript.
| Name | Target sites | Degree | Target genes |
|---|
| hsa-miR-424-5p | 5 | 152 | 6 |
| hsa-miR-497-5p | 5 | 150 | 4 |
| hsa-miR-16-5p | 5 | 150 | 4 |
| hsa-miR-15b-5p | 5 | 150 | 4 |
| hsa-miR-15a-5p | 5 | 149 | 3 |
| hsa-miR-195-5p | 5 | 149 | 3 |
| hsa-miR-485-5p | 1 | 78 | 3 |
| hsa-miR-20a-5p | 4 | 77 | 3 |
| hsa-miR-20b-5p | 4 | 76 | 3 |
| hsa-miR-93-5p | 4 | 77 | 3 |
Discussion
In the present study, a comprehensive bioinformatics
approach was used to examine key lncRNAs involved in DCM and to
elucidate their molecular mechanisms in the development of DCM. In
particular, interaction data from starBase were used to generate a
ceRNA background network based on the theory of ceRNA.
Subsequently, a DCMCN was extracted from the ceRNA background
network by mapping the DCM-related genes. In total, the DCMCN
contained 24 mRNA nodes, 320 miRNA nodes, 1,127 lncRNA nodes, and
10,808 edges. Subsequently, topological properties were assessed
and a cluster analysis was performed on the DCMCN.
The results revealed that the lncRNA XIST could
directly interact with several miRNAs with known relevance to the
development of DCM. Comprehensive analysis revealed that the
downstream targets of XIST, hsa-miR-424-5p and hsa-miR-497-5p, may
be pivotal miRNAs that regulate DCM.
Hsa-miR-424-5p is located at the X chromosome, and
is potentially among the 15% of X-linked genes that escape female
X-chromosome inactivation (XCI), resulting in higher expression in
women (29). A previous study
demonstrated that hsa-miR-424-5p is significantly downregulated in
peripheral blood from patients with HF, thus suggesting that it may
be considered a potential biomarker and contributor to HF (30). In addition, upregulation of
miR-424-5p promotes downstream processes associated with hypoxia,
including angiogenesis and erythropoiesis, to ameliorate myocardial
ischemia, which is considered a cardioprotective factor (31).
Hsa-miR-497-5p is located on chromosome 17 and is
highly conserved in several species (32). This miRNA is a member of the
miR-15/107 group that includes the seed sequence AGCAGC, which is
an important determinant of target recognition (33). It has previously been reported that
miR-497-5p is closely associated with cardiac fibrosis through
activation of latent transforming growth factor (TGF)-β1 anchored
in the extracellular matrix by targeting the 3′-untranslated region
of reversion-inducing cysteine rich protein with Kazal motifs
(34). In addition, a previous
study revealed the negative regulatory effect of hsa-miR-497-5p
against SMAD family member 3 transcripts, which suggests the
possible role of this miRNA in regulation of the TGFβ signaling
pathway (35).
XIST is a lncRNA (17 kb in Homo sapiens)
required for XCI of one of the two X chromosomes in female cells,
thus enabling dosage compensation between XX females and XY males
(36). XCI takes place early in
embryonic development, and is thought to occur in multiple steps:
Counting and choosing the X chromosome to silence, spreading of
XIST over the target X chromosome, and silencing most of its active
genes (37). In recent years,
lncRNA XIST has been reported to serve a role as a regulatory
factor of tumor proliferation. It has been demonstrated that XIST
has an important positive role in pancreatic cancer proliferation
(38), colorectal cancer (39) and other types of cancer by
targeting corresponding miRNAs. Furthermore, lncRNA XIST has been
reported to suppress the proliferation of myocardial cells and
promote apoptosis by targeting miR-130a-3p in myocardial infarction
(40). However, to the best of our
knowledge, there is currently no evidence regarding the regulatory
role of lncRNA XIST in DCM.
To the best of our knowledge, this study is the
first to identify a ceRNA network including lncRNA XIST,
hsa-miR-424-5p, hsa-miR-497-5p and DCM-related genes in DCM. In
addition, KEGG analysis revealed that the ‘AGE-RAGE signaling
pathway in diabetic complications’ may be a major pathway leading
to the development of DCM.
The accumulation of advanced glycation end products
(AGEs) has a crucial role in the onset and progress of diabetic
nephropathy (41). AGEs interact
with the receptor for AGEs (RAGE) on the cell membrane and induce
deleterious effects via activation of nuclear factor κB, ultimately
leading to increased vascular permeability and inflammation
(42). RAGE is formed from the
cleavage of the native membrane receptor mediated by disintegrins
and matrix metalloproteinases (43), and circulates in the blood. It has
previously been reported that AGE-RAGE interaction in diabetes can
negatively affect endothelial cell physiology, resulting in
increased predisposition toward cardiovascular disease (44); this has been reported in clinical
studies wherein patients with diabetes exhibit higher RAGE
expression (45). Animal models of
diabetic atherosclerosis have also demonstrated improved regression
of atherosclerotic plaques following RAGE knockout (46). Therefore, understanding the
AGE-RAGE axis in the development of endothelial dysfunction and its
regulation by lncRNAs may be helpful in designing novel therapies
that target endothelial dysfunction and impair development of
cardiovascular diseases.
In conclusion, the present study constructed a
background network containing mRNAs, miRNAs and lncRNAs, and then a
regulatory network of DCM was screened. Finally, a functional
lncRNA XIST was identified in the network and the main miRNAs
(miR-424-5p and miR-497-5p) that are regulated by XIST were further
screened to obtain the ceRNA regulatory network of DCM. The present
study highlighted the involvement of lncRNA XIST in DCM, and
facilitated the development of lncRNA-directed diagnostic and
therapeutic tools against diabetes mellitus. These findings
improved our knowledge on the mechanism of DCM and may provide
potential therapeutic target in the treatment of DCM in clinic.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant nos. 81072937 and
81473246).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KC, YM, LL and GZ conceived and designed the
experiments. KC and SW performed the experiments. YM and YZ
analyzed the data. KC and XL contributed reagents, materials and
analytical tools and contributed to the analysis and interpretation
of data.. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
DCM
|
diabetic cardiomyopathy
|
|
AGEs
|
advanced glycation end products
|
|
DEGs
|
differentially expressed genes
|
|
GIFtS
|
GeneCards Inferred Functionality
Scores
|
|
HF
|
heart failure
|
References
|
1
|
Jia G, Whaley-Connell A and Sowers JR:
Diabetic cardiomyopathy: A hyperglycaemia-and
insulin-resistance-induced heart disease. Diabetologia. 61:21–28.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seferović PM and Paulus WJ: Clinical
diabetic cardiomyopathy: A two-faced disease with restrictive and
dilated phenotypes. Eur Heart J. 36:1718–1727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lam CS: Diabetic cardiomyopathy: An
expression of stage B heart failure with preserved ejection
fraction. Diab Vasc Dis Res. 12:234–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aneja A, Tang WH, Bansilal S, Garcia MJ
and Farkouh ME: Diabetic cardiomyopathy: Insights into
pathogenesis, diagnostic challenges, and therapeutic options. Am J
Med. 121:748–757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iwakawa H and Tomari Y: The functions of
MicroRNAs: MRNA decay and translational repression. Trends Cell
Biol. 25:651–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jandura A and Krause HM: The New RNA
World: Growing Evidence for long noncoding RNA functionality.
Trends Genet. 33:665–676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li W, Notani D and Rosenfeld MG: Enhancers
as non-coding RNA transcription units: Recent insights and future
perspectives. Nat Rev Genet. 17:207–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thum T and Condorelli G: Long Noncoding
RNAs and MicroRNAs in cardiovascular pathophysiology. Circ Res.
116:751–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA Hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smillie CL, Sirey T and Ponting CP:
Complexities of post-transcriptional regulation and the modeling of
ceRNA crosstalk. Crit Rev Biochem Mol Biol. 53:231–245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong Y, Liu C, Zhao Y, Ponnusamy M, Li P
and Wang K: Role of noncoding RNAs in regulation of cardiac cell
death and cardiovascular diseases. Cell Mol Life Sci. 75:291–300.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Gu H, Chen J and Zhou X:
Involvement of long noncoding RNA MALAT1 in the pathogenesis of
diabetic cardiomyopathy. Int J Cardiol. 202:753–755. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou X, Zhang W, Jin M, Chen J, Xu W and
Kong X: lncRNA MIAT functions as a competing endogenous RNA to
upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy.
Cell Death Dis. 8:e29292017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Greco S, Fasanaro P, Castelvecchio S,
D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC,
Menicanti L and Martelli F: MicroRNA dysregulation in diabetic
ischemic heart failure patients. Diabetes. 61:1633–1641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ismail M, Suresh H, Nazrath N, Anisatu R,
Marian P, Bruno V, Fabrizio S and Conrad B: GeoDiver: Differential
gene expression analysis & gene-set analysis for GEO Datasets.
Queen Mary J Intellectual Property. Apr 15–2017.doi:
https://doi.org/10.1101/127753.
|
|
18
|
Harel A, Inger A, Stelzer G,
Strichman-Almashanu L, Dalah I, Safran M and Lancet D: GIFtS:
Annotation landscape analysis with GeneCards. BMC Bioinformatics.
10:3482009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:1–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han JD, Bertin N, Hao T, Goldberg DS,
Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP and
Vidal M: Evidence for dynamically organized modularity in the yeast
protein-protein interaction network. Nature. 430:88–93. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou B, Wang B and Zhe H: Degree-layer
theory of network topology. Phys Soc. 18–Sep;2014.
|
|
25
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and
Peterson KJ: A Uniform system for the annotation of vertebrate
microRNA genes and the evolution of the human microRNAome. Annu Rev
Genet. 49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nature
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carrel L and Willard HF: X-inactivation
profile reveals extensive variability in X-linked gene expression
in females. Nature. 434:400–404. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Marques FZ, Vizi D, Khammy O, Mariani JA
and Kaye DM: The transcardiac gradient of cardio-microRNAs in the
failing heart. Eur J Heart Fail. 18:1000–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ghosh G, Subramanian IV, Adhikari N, Zhang
X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y and
Ramakrishnan S: Hypoxia-induced microRNA-424 expression in human
endothelial cells regulates HIF-α isoforms and promotes
angiogenesis. J Clin Invest. 120:4141–4154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Flavin RJ, Smyth PC, Laios A, O'Toole SA,
Barrett C, Finn SP, Russell S, Ring M, Denning KM, Li J, et al:
Potentially important microRNA cluster on chromosome 17p13.1 in
primary peritoneal carcinoma. Mod Pathol. 22:197–205. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Finnerty JR, Wang WX, Hébert SS, Wilfred
BR, Mao G and Nelson PT: The miR-15/107 group of microRNA genes:
Evolutionary biology, cellular functions, and roles in human
diseases. J Mol Biol. 402:491–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen X, Shi C, Wang C, Liu W, Chu Y, Xiang
Z, Hu K, Dong P and Han X: The role of miR-497-5p in myofibroblast
differentiation of LR-MSCs and pulmonary fibrogenesis. Sci Rep.
7:409582017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jafarzadeh M, Soltani BM, Dokanehiifard S,
Kay M, Aghdami N and Hosseinkhani S: Experimental evidences for
hsa-miR-497-5p as a negative regulator of SMAD3 gene expression.
Gene. 586:216–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gendrel AV and Heard E: Fifty years of
X-inactivation research. Development. 138:5049–5055. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Payer B and Lee JT: X chromosome dosage
compensation: How mammals keep the balance. Annu Rev Genet.
42:733–772. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei W, Liu Y, Lu Y, Yang B and Tang L:
LncRNA XIST promotes pancreatic cancer proliferation through
miR-133a/EGFR. J Cell Biochem. 118:3349–3358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song H, He P, Shao T, Li Y, Li J and Zhang
Y: Long non-coding RNA XIST functions as an oncogene in human
colorectal cancer by targeting miR-132-3p. J BUON. 22:696–703.
2017.PubMed/NCBI
|
|
40
|
Zhou T, Qin G, Yang L, Xiang D and Li S:
LncRNA XIST regulates myocardial infarction by targeting
miR-130a-3p. J Cell Physiol. 234:8659–8667. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Uribarri J, Cai W, Ramdas M, Goodman S,
Pyzik R, Chen X, Zhu L, Striker GE and Vlassara H: Restriction of
advanced glycation end products improves insulin resistance in
human type 2 diabetes: Potential role of AGER1 and SIRT1. Diabetes
Care. 34:1610–1616. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Prasad K: Low levels of serum soluble
receptors for advanced glycation end products, biomarkers for
disease state: Myth or Reality. Int J Angiol. 23:11–16. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tam XH, Shiu SW, Leng L, Bucala R,
Betteridge DJ and Tan KC: Enhanced expression of receptor for
advanced glycation end-products is associated with low circulating
soluble isoforms of the receptor in Type 2 diabetes. Clin Sci
(Lond). 120:81–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao X, Zhang H, Schmidt AM and Zhang C:
AGE/RAGE produces endothelial dysfunction in coronary arterioles in
type 2 diabetic mice. Am J Physiol Heart Circ Physiol.
295:H491–H498. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Villegas-Rodríguez ME, Uribarri J,
Solorio-Meza SE, Fajardo-Araujo ME, Cai W, Torres-Graciano S,
Rangel-Salazar R, Wrobel K and Garay-Sevilla ME: The AGE-RAGE axis
and its relationship to markers of cardiovascular disease in newly
diagnosed diabetic patients. PLoS One. 11:e01591752016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Soro-Paavonen A, Watson AM, Li J, Paavonen
K, Koitka A, Calkin AC, Barit D, Coughlan MT, Drew BG, Lancaster
GI, et al: Receptor for advanced glycation end products (RAGE)
deficiency attenuates the development of atherosclerosis in
diabetes. Diabetes. 57:2461–2469. 2008. View Article : Google Scholar : PubMed/NCBI
|