Introduction
Preeclampsia (PE) is a serious obstetric
complication, and poses a serious threat to pregnant women and
fetuses (1). The incidence of PE
around the world is 2–8% (2). At
present, PE is one of the main causes of maternal mortality. The
pathogenesis of PE is not fully understood; thus, as an effective
treatment for PE, pregnancies may be terminated. At present, PE is
considered to be associated with dysfunction of the placenta
(3), endothelial dysfunction
(4) and abnormal increases in the
apoptosis of trophoblasts (5). Of
note, increased migration and invasion of trophoblastic cells, and
decreased apoptosis have been hypothesized as potential therapeutic
strategies for the treatment of PE; however, further investigation
is required.
MicroRNAs (miRNAs/miRs) regulate the expression of
their target genes in a negative manner. Numerous studies have
shown that miRNAs are associated with cell migration, apoptosis,
differentiation and proliferation (6–9). It
was reported that the expression profiles of miRNAs in PE were
markedly diverse in umbilical cord blood, maternal serum, placental
samples and mesenchymal stem cells (MSCs) (10–14).
miR-342-3p is one of the most highly expressed miRNAs in placentas
from patients with PE (15), but
its biological mechanism in PE remains unknown. miR-342-3p has been
proposed to inhibit cell migration in cervical cancer (16), and suppress cell invasion and
metastasis in lung cancer (17).
Additionally, miR-342-3p may inhibit the progression of
hepatocellular carcinoma through the nuclear factor-B pathway
(18). These findings suggest that
miR-342-3p may serve vital roles in trophoblastic cell
proliferation, migration and invasion in PE.
Gene expression profiling revealed that the
expression levels of platelet-derived growth factor receptor α
(PDGFRA) were decreased in PE patients compared with the normal
group (19), yet the mechanism of
PDGFRA in PE remains unclear. TargetScan, microcosm and miRanda
analyses identified PDGFRA as one of the putative target genes for
miR-342-3p (20). Further study is
needed to determine the association between miR-342-3p and PDGFRA
in PE.
In the present study, we investigated the roles of
miR-342-3p and PDGFRA in the placental tissues of patients with PE
and in HTR8/SVneo cells. In our research, miR-342-3p was reported
to reduce the migration and invasion of trophoblastic cells by
suppressing PDGFRA. This suggested that miR-342-3p may be
associated with the pathology of PE.
Materials and methods
Tissue collection
A total of 30 placentas were obtained from patients
with PE and healthy controls who underwent cesarean section from
January 2013 to September 2017. The age range of the patients was
23–39 years (mean age, 28.63±2.24 years). The diagnosis of PE was
conducted as previously reported (21). The clinical data of patients was
present in Table I. Patients with
PE were characterized by high blood pressure and a high protein
content in the urine. Tissues were quickly snap-frozen in liquid
nitrogen during surgery and stored at −80°C until use. Written
informed consent was obtained from each patient. The exclusion
criteria for the two groups included: Patients with essential
hypertension or kidney disease, a history of drug or alcohol abuse
half year prior to providing written informed consent. In addition,
the patients with PE were pathologically analyzed by two
specialists.
 | Table I.Clinical information of patients
enrolled in the present study. |
Table I.
Clinical information of patients
enrolled in the present study.
| Variable | Control (n=30) | PE (n=30) | P-value |
|---|
| Age (years) | 28.83±2.42 | 28.63±2.24 | 0.741 |
| Gestational age
(weeks) | 38.67±0.55 | 36.07±0.78 | <0.0001 |
| Systolic pressure
(mmHg) | 113.03±9.02 | 156.67±5.80 | <0.0001 |
| Diastolic pressure
(mmHg) | 67.70±7.26 | 95.80±3.21 | <0.0001 |
| Proteinuria (g/24
h) | N/A | 3.55±0.86 | N/A |
Cell culture
HTR8/SVneo cells were acquired from the American
Type Culture Collection (ATCC). Cells were cultured in RPMI-1640
(Labdel Comércio de Produtos para Laboratório) and 10% fetal bovine
serum (Labdel Comércio de Produtos para Laboratório) and 100
streptomycin mg/ml, 100 penicillin U/ml, in a humidified chamber at
37°C with 5% CO2.
Cell transfection
The cell line was transfected with miR-342-3p mimics
and the negative control (Thermo Fisher Scientific, Inc., cat. no.
4464058.) at a concentration of 100 nmol/l. The cells were
transfected using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). The duration between transfection and subsequent
analysis was 48 h. The sequence of the miR-342-3p mimics was
5′-GAAACUGGGCUCAAGGUGAGGGGUGCUAUCUGUGAUUGAGGGACAUGGUUAAUGGAAUUGUCUCACACAGAAAUCGCACCCGUCACCUUGGCCUACUUA-3′.
Knockdown of PDGFRA in the cell
line
Small interfering RNAs (siRs) against PDGFRA
(siRPDGFRA) and the control (scramble) were purchased from Santa
Cruz Biotechnology, Inc. HTR8/SVneo cells were transfected with 20
µM siRPDGFRA or scramble siRNA utilizing Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from tissues and cells
utilizing TRIzol reagent (Thermo Fisher Scientific, Inc.). For
detecting mRNAs, a PrimeScript™ RT reagent kit (Takara Bio, Inc.,
cat. no. RR047A) was used for RT; the experiment was performed as
follows: Three times at 37°C, 15 min for each time; and
inactivation at 85°C for 5 sec. RT was performed for miRNAs using
an Mir-X™ miRNA First-Strand Synthesis kit (Takara Bio, Inc, cat.
no. 638313) according to the manufacturer's protocols. qPCR was
performed using an SYBR® qRT-PCR kit (Clontech
Laboratories, Inc.) for mRNAs and miRNAs under the following
thermocycling conditions: 94°C for 4 min, followed by 40 cycles of
94°C for 30 sec and 60°C for 60 sec. The sequences of primers
employed were presented in Table
II. Relative miRNA expression levels were standardized to that
of small nucleolar RNA U6, whereas that for relative mRNA
expression was normalized to the expression levels of β-actin using
the 2−ΔΔCq method (22).
 | Table II.Primers for reverse
transcription-quantitative polymerase chain reaction. |
Table II.
Primers for reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence
(5′→3′) |
|---|
| Hsa-miR-342-3p
stem-loop primer |
GCGCGTGAGCAGGCTGGAGAAATTAACCACGCGCACGGGT |
| Hsa-miR-342-3p
forward primer |
TCTCACACAGAAATCGC |
| Hsa-miR-342-3p
reverse primer |
GAGCAGGCTGGAGAA |
| U6 forward
primer |
GCTTCGGCAGCACATATACTAAAT |
| U6 reverse
primer |
CGCTTCACGAATTTGCGTGTCAT |
| PDGFRA forward
primer |
TTGAAGGCAGGCACATTTACA |
| PDGFRA reverse
primer |
GCGACAAGGTATAATGGCAGAAT |
| BCL-2 forward
primer |
CCGTTGGCCCCCGTTGCTTT |
| BCL-2 reverse
primer |
CTGGCGGAGGGTCAGGTGGA |
| Caspase-3 forward
primer |
CTCGGTCTGGTACAGATGTCGATG |
| Caspase-3 reverse
primer |
GGTTAACCCGGGTAAGAATGTGCA |
| β-actin forward
primer |
TGGCACCCAGCACAATGAA |
| β-actin reverse
primer |
CTAAGTCATAGTCCGCCTAGAAGCA |
Western blot analysis
Cells were collected 48 h after transfection and
prepared for lysis in radioimmunoprecipitation assay buffer
(BioVision, Inc.). Protein concentration was determined via a
bicinchoninic acid assay. Protein (50 µg/lane) was separated via 8%
SDS-PAGE. Separated proteins were transferred to polyvinylidene
fluoride membranes (EMD Millipore). The membranes were blocked with
5% fat-free milk powder for 2 h at room temperature, and then
incubated with primary antibodies against PDGFRA (1:1,000; Santa
Cruz Biotechnology, Inc., cat. no. sc-338), anti-BCL-2 (1:1,000;
Abcam, cat. no. ab59348), anti-Caspase-3 (1:500; Abcam, cat. no.
ab13847) and β-actin antibody (1:1,000; Santa Cruz Biotechnology,
Inc., cat. no. sc-47778) at 4°C overnight. Subsequently, the
membranes were incubated with horseradish peroxidase-conjugated
antimouse IgG (H+L) (1:10,000; Invitrogen; Thermo Fisher
Scientific, Inc., cat. no. 62-6520) or anti-rabbit IgG (1:5,000;
Invitrogen; Thermo Fisher Scientific, Inc., cat. no. 65-6122)
antibody for 1 h at room temperature. The membranes were developed
utilizing an ECL kit (Pierce; Thermo Fisher Scientific, Inc.) and
visualized with X-ray film. Protein expression levels were
standardized to those of β-actin. Data was analyzed by Quantity One
version 4.62 software (Bio-Rad Laboratories, Inc.).
Cell proliferation analysis
To analyze cell proliferation, a Cell Counting Kit-8
(CCK-8) assay was conducted using a CCK-8 proliferation assay kit
(Dojindo Molecular Technologies, Inc.) at 0, 12, 24 and 48 h
following transfection, according to the manufacturer's protocols.
Cells were transfected with mimics or negative control (ctrl
group), or treated with Lipofectamine 2000 without miRNA molecules
in the mock group. Cells (1×105) were seeded in 96-well
plates for analysis. Then, 100 µl CCK-8 reagent was added to the
well; at 2 h later, the absorbance was measured at 450 nm with a
microplate reader.
Transwell invasion and migration
assay
Cell invasion was investigated based on the capacity
of the cells invade through the 8 mm pores of polycarbonate
membranes. Briefly, HTR8/SVneo cells (1.0×105
cells/well) transfected with siRPDGFRA or scramble were placed in
the upper chamber, and 600 µl of RPMI1640 medium with 10% fetal
bovine serum was placed in the lower chamber. After incubating for
1 day under standard conditions at 37°C, the cells on the upper
surface that had not migrated were removed with a sterile cotton
swab. Migrated cells were stained with 0.1% crystal violet for 5
min at room temperature and analyzed under a light microscope in 5
random fields (magnification, ×200). The techniques used in the
Transwell invasion assay were based on the cell migration assay;
however, Matrigel was used.
Cell cycle assay
The cell cycle was analyzed via flow cytometry.
After 48 h post-transfection, HTR8/SVneo cells were trypsinized in
chilled PBS, fixed in 70% ethanol for 24 h at −20°C, and then
stained with propidium iodide for 10 min on ice. The samples were
calculated using a flow cytometer and CellQuest Pro version 5.1
software (BD Biosciences).
miRNA target prediction and
dual-luciferase reporter assay
TargetScan (version 6.0; http://www.targetscan.org/vert_60/), microcosm
(version 1.1; http://tools4mirs.org/software/mirna_databases/microcosm-targets/)
and miRanda (version August 2010; http://www.microrna.org/microrna/home.do) were used to
predict miRNAs that could potentially target PDGFRA and identify
possible binding regions. The fragment of the human PDGFRA with
[wild type (wt)] or without [mutant (mut)] the miR-342-3p binding
site at the 3′-untranslated region (3′-UTR) was cloned and inserted
into the pGL3-basic luciferase report plasmid (Promega Corporation)
to generate the luciferase reporter vectors, PDGFRA 3′-UTR-wt and
PDGFRA 3′-UTR-mut. 293T cells (American Type Culture Collection)
were treated in 96-well plates at 5,000 cells per well and
incubated for 24 h at 37°C with 5% CO2 prior to
transfection. Then, miR-342-3p mimics or miR-negative control was
transfected into 293T cells using Lipofectamine 2000 with 100 ng of
PDGFRA 3′-UTR-wt or PDGFRA 3′-UTR-mut, or 10 ng of pRL-TK
Renilla plasmid (Promega Corporation). Following incubation
for 48 h, the luciferase activities were determined with a Dual
Luciferase Reporter System (Promega Corporation); Renilla
luciferase was used for normalization.
Statistical analysis
SPSS 19.0 (IBM Corp.) was employed for statistical
analysis. Data were presented as the mean ± standard deviation, and
an independent samples t-test was conducted for comparisons
between two groups; analysis was performed in a two-tailed manner.
Cell viability was analyzed by one-way analysis of variance with a
Student-Newman-Keuls post hoc test. P<0.05 was considered to
indicate a statistically significant manner.
Results
Analysis of tissue samples
The clinical information of patients was presented
in Table I. Compared with the
control group, patients with PE exhibited significantly higher
systolic pressure and diastolic pressure, notable proteinuria and a
significantly shorter duration of gestation (P<0.0001).
Expression levels of miR-342-3p and
PDGFRA in the two patient groups
The expression of miR-342-3p and PDGFRA was analyzed
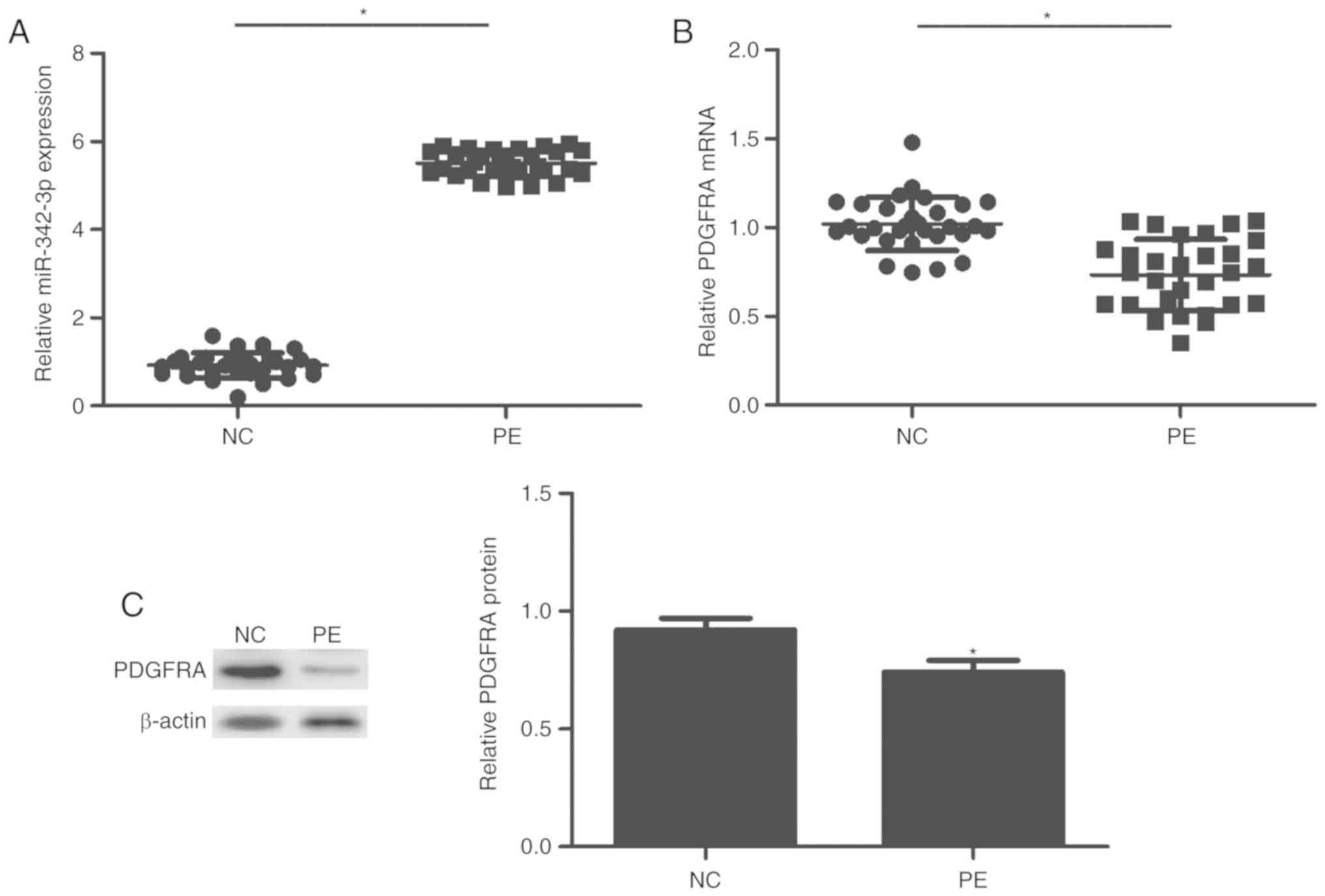
in tissues via RT-qPCR. As presented in Fig. 1, miR-342-3p was significantly
upregulated in the PE group compared with the control group
(P<0.01; Fig. 1A). On the
contrary, the expression of PDGFRA was significantly decreased in
patients with PE compared with the control (P<0.01; Fig. 1B and C).
miR-342-3p may affect cell
proliferation and the cell cycle
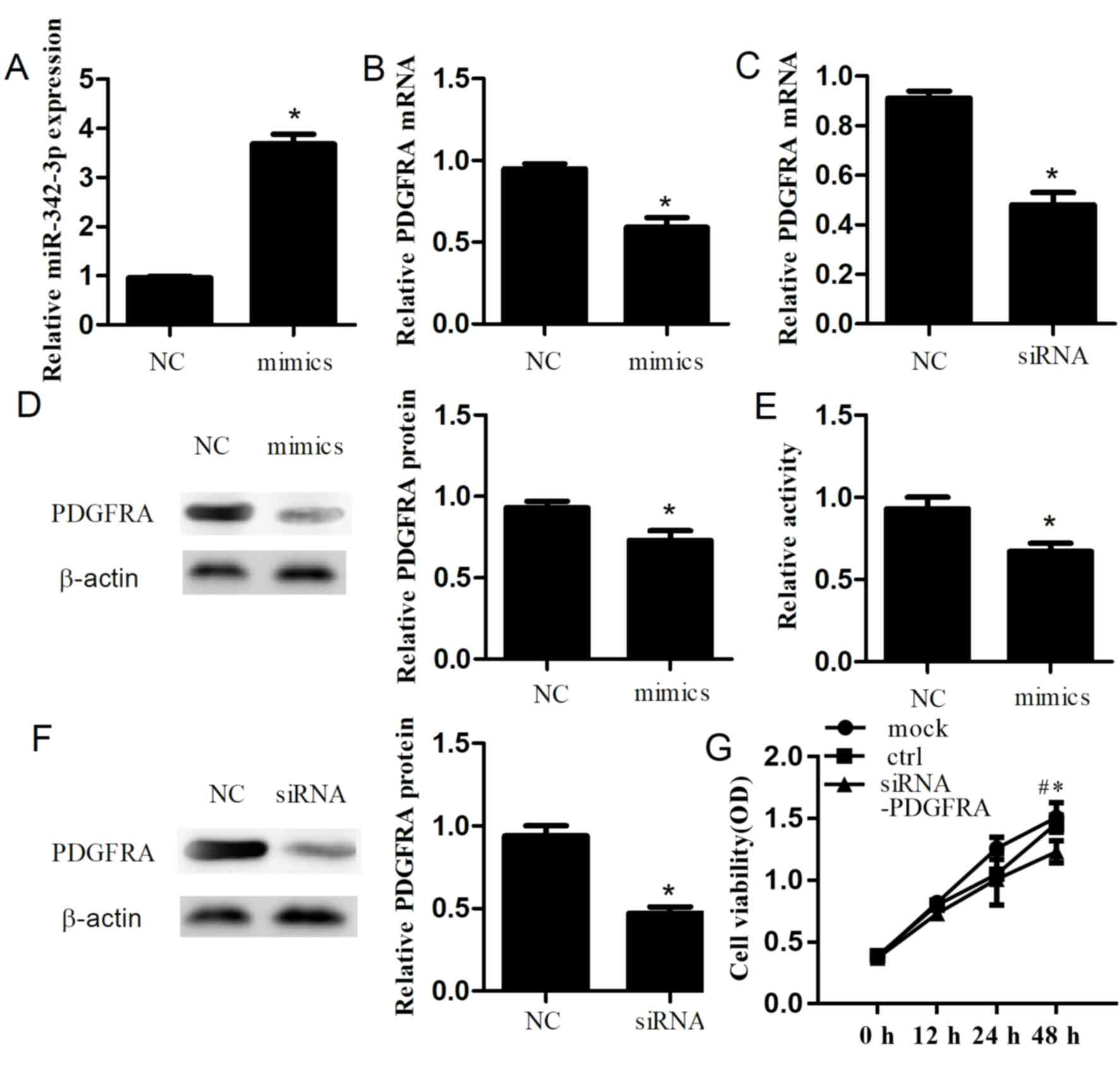
Additionally, to explore the role of miR-342-3p, we
performed a CCK-8 assay to determine the effects of miR-342-3p on
cell proliferation. After 48 h, we confirmed that transfection of
miR-342-3p mimics into cells significantly increased miR-342-3p
expression compared with the control group (P<0.01; Fig. 2A). Furthermore, RT-qPCR and western
blotting were conducted to investigate the effects of miR-342-3p on
PDGFRA in a cell line. Overexpression of miR-342-3p resulted in a
significant decrease in the expression of PDGFRA at the mRNA
(P=0.001; Fig. 2B) and protein
levels (P=0.006; Fig. 2D) compared
with the control. Cell proliferation was significantly inhibited
(P=0.006; Fig. 2E) after
transfection with miR-342-3p mimics compared with the control. In
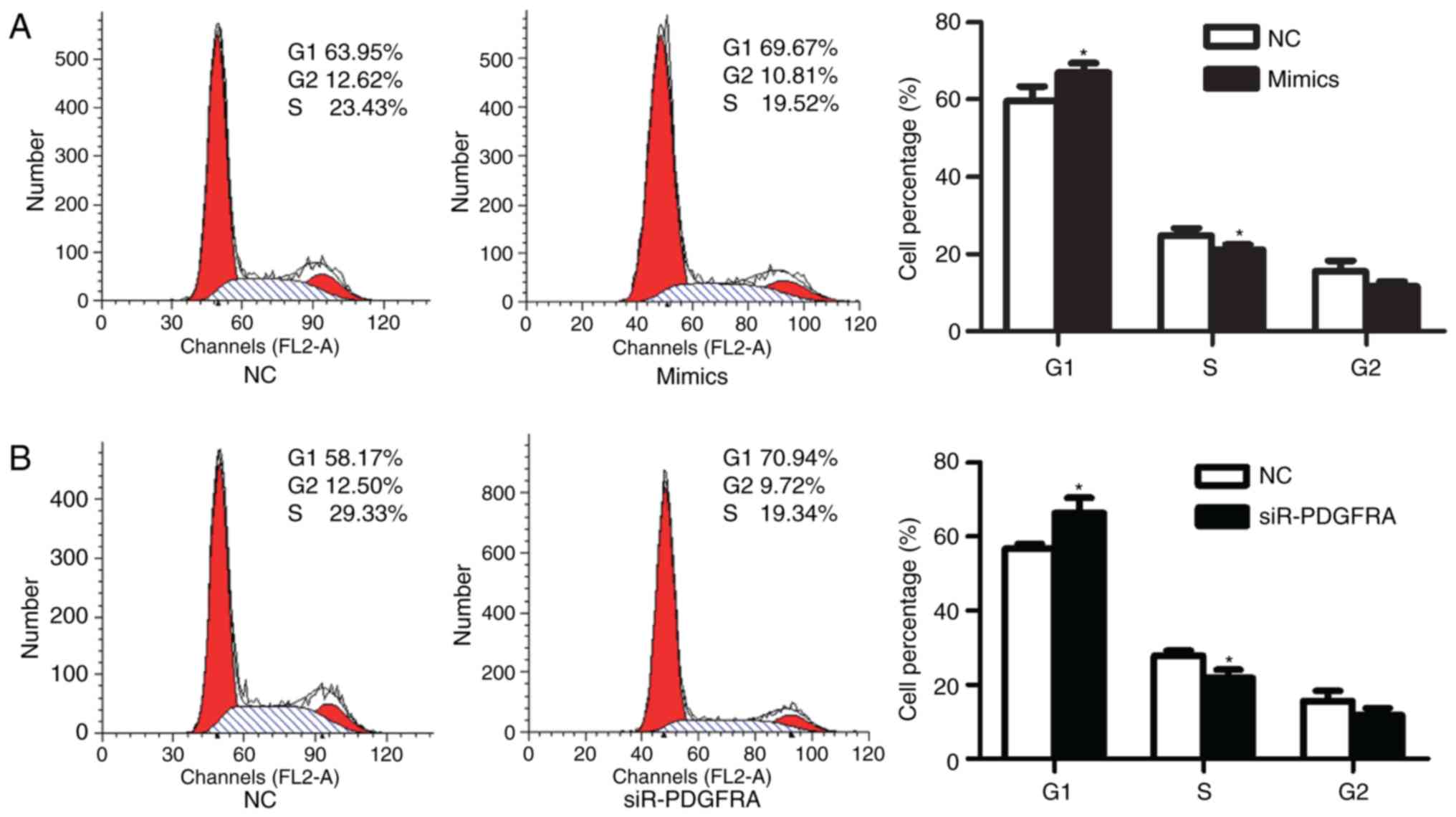
addition, the effects of miR-342-3p on the cell cycle of
HTR-8/SVneo cells were analyzed via flow cytometry. The results
demonstrated that the percentage of HTR-8/SVneo cells in G1 phase
significantly increased from 59.66±3.75% prior transfection to
66.94±2.39% at 48 h post-transfection (P=0.047; Fig. 3A). The percentage of cells in S
phase was significantly reduced from 24.79±1.87% prior transfection
to 21.07±1.36% following transfection (P=0.049; Fig. 3A).
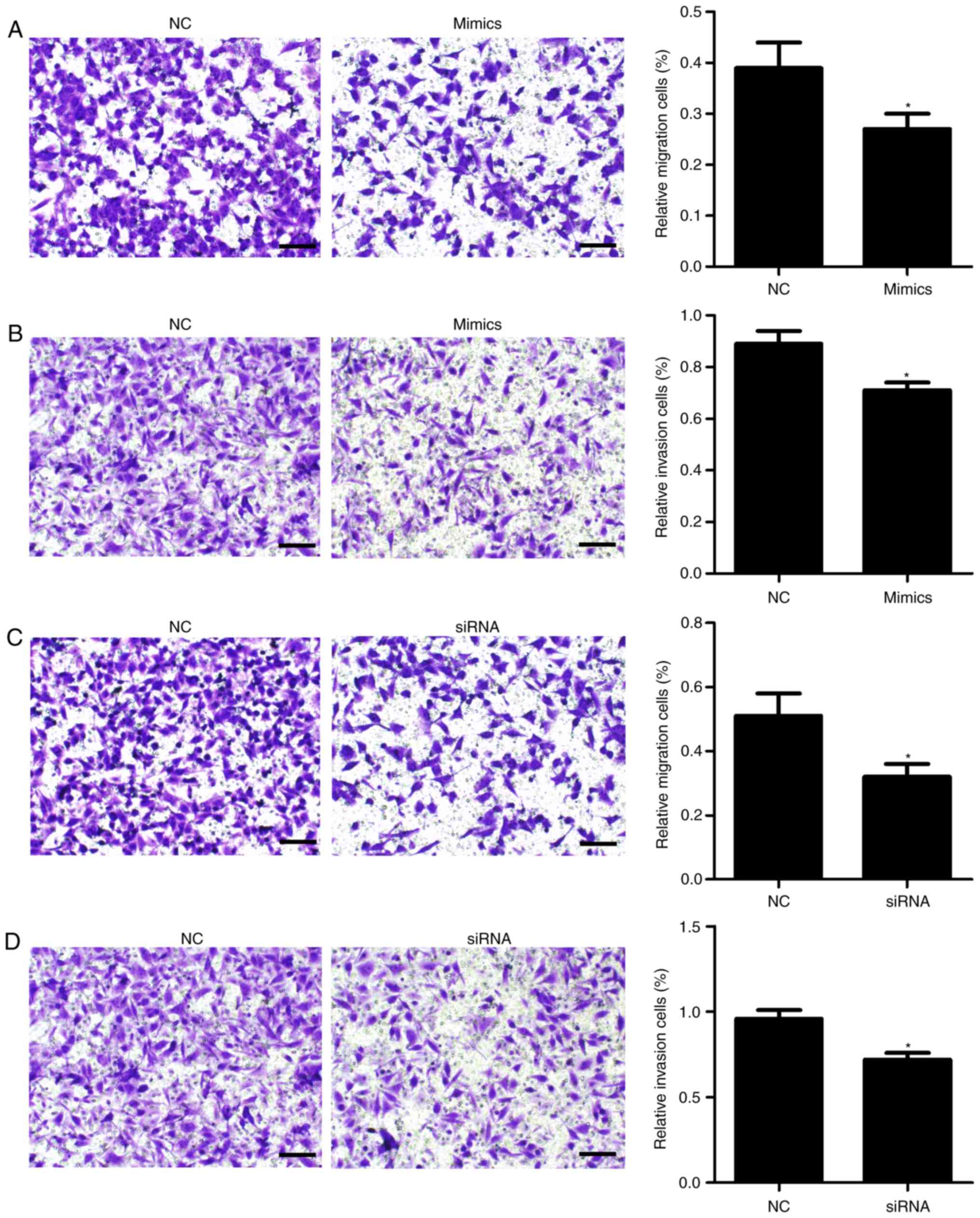
miR-342-3p suppresses cell migration
and invasion
Additionally, Transwell migration and invasion
assays were conducted. As presented in Fig. 4, transfection with miR-342-3p
mimics significantly inhibited cell migration (P=0.027; Fig. 4A) and invasion (P=0.022; Fig. 4B) compared with the control
group.
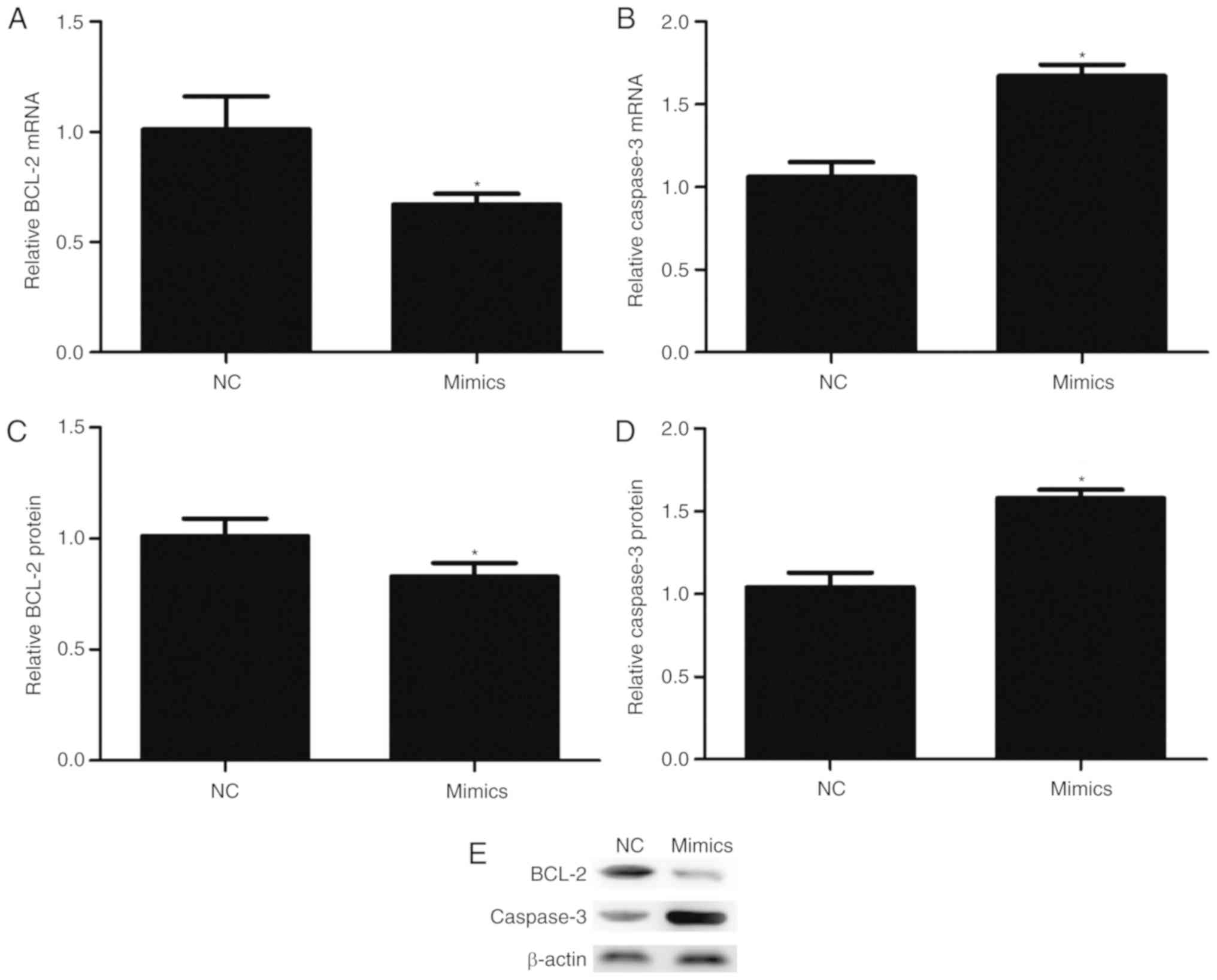
miR-342-3p may affect the expression
of BCL-2 and Caspase-3 in cells
Transfection with miR-342-3p mimics significantly
reduced the mRNA (P=0.017; Fig.
5A) and protein expression (P=0.035; Fig. 5C and E) levels of BCL-2. On the
contrary, overexpression of miR-342-3p significantly increased the
mRNA (P=0.001; Fig. 5B) and
protein expression (P=0.001; Fig. 5D
and E) levels of Caspase-3.
siR-PDGFRA exhibits similar effects to
miR-342-3p mimics
PDGFRA knockdown was performed in cells to its
effects in PDGFRA in HTR-8/SVneo cells. After 48 h
post-transfection, the mRNA and protein expression levels of PDGFRA
were significantly decreased by 43.7±3.2% (P=1.56×10−4;
Fig. 2C) and 47.0±5.0%
(P=3.08×10−4; Fig. 2F),
respectively, compared with the control. Additionally, PDGFRA
knockdown significantly increased the percentage of HTR-8/SVneo
cells in G1 phase from 56.63±1.40% prior to knockdown to
66.30±4.07% at 48 h following siR-PDGFRA transfection (P=0.018;
Fig. 3B) compared with the
control. Furthermore, cell proliferation (P=0.023; Fig. 2G), migration (P=0.01; Fig. 4C) and invasion (P=0.003; Fig. 4D) were significantly inhibited
following PDGFRA knockdown compared with the control.
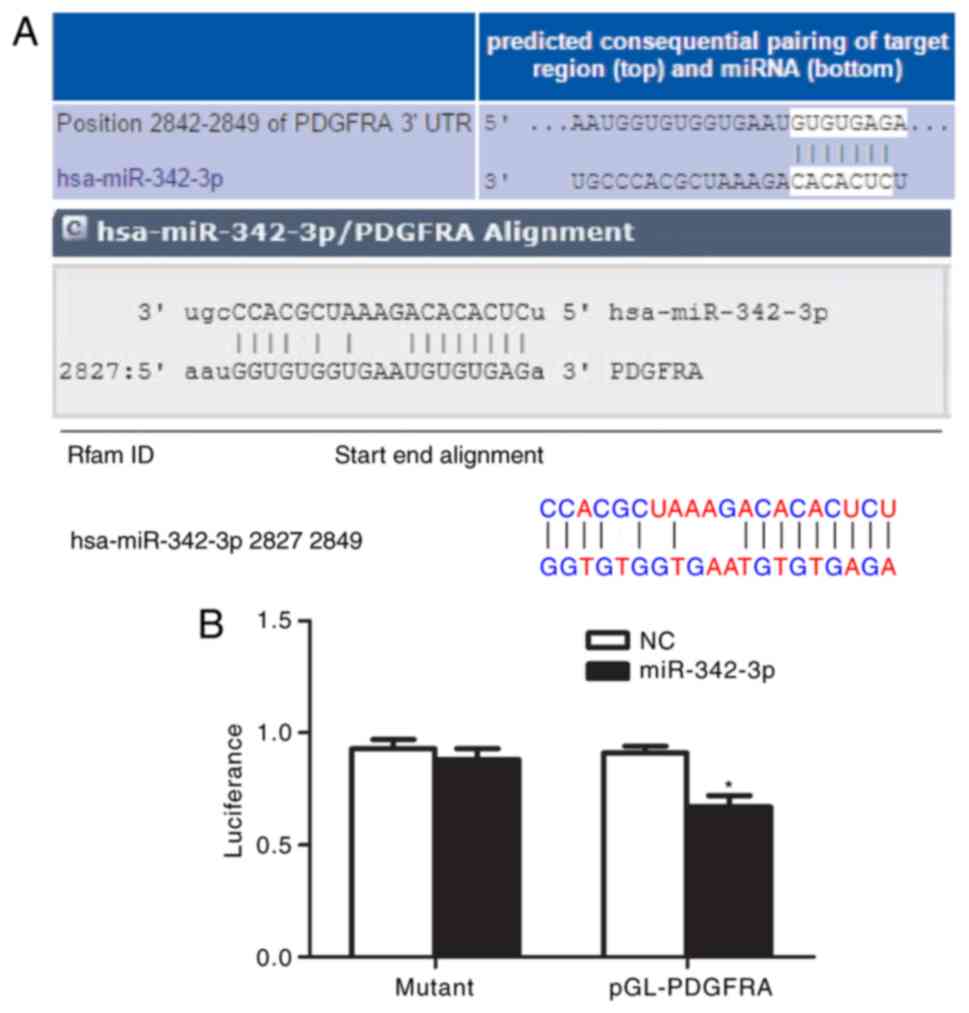
PDGFRA is a direct target of
miR-342-3p
As presented in Fig.
6A, bioinformatics analysis indicated that the 3′-UTR of PDGFRA
has a target sequence for miR-342-3p. Transfection with miR-342-3p
mimics significantly decreased the luciferase activity of cells
possessing PDGFRA 3′-UTR-wt compared with the control (P<0.01),
while transfection with miR-342-3p mimics had no significant
effects on the luciferase activity of cells possessing PDGFRA
3′-UTR-mut (Fig. 6B). These
findings indicated that PDGFRA is likely to be a target of
miR-342-3p.
Discussion
In the first trimester of pregnancy, extravillous
trophoblasts (EVTs) with an invasive phenotype serve an important
role in the formation of the maternal-fetal interface (23). At present, the importance of
incomplete EVT invasion and subsequent abnormal remodeling of the
spiral arteries was highlighted in the etiology of PE; these
processes are modulated by a variety of inflammatory and immune
cells (23). The migration ability
of trophoblasts is important in deep placentation for the normal
and healthy development of the fetus (23). Incomplete trophoblast migration and
invasion could lead to insufficient deep placentation, which has
been associated with PE (23).
Reduced trophoblast proliferation has been reported to serve an
important role in the development of PE (24).
The importance of miRNAs in PE has been reported.
The transient upregulation of miR-136 could induce the apoptosis of
MSCs, and affect the formation and development of capillaries
(25). miR-125b may be associated
with the occurrence of PE by targeting sphingosine-1-phosphate
lyase 1 (26). In addition,
miR-128a could be involved in the pathogenesis of PE by means of
initiating apoptosis (27).
miR-195 was associated with the pathogenesis of PE by targeting the
activin receptor type-2B (28). In
the present study, the expression of miR-342-3p was upregulated in
the placentas of patients with PE; the transient overexpression of
miR-342-3p decreased cell proliferation, migration and invasion.
Overexpression of miR-342-3p also inhibited G1/S phase transition;
the expression of BCL-2 was decreased while that of Caspase-3 had
increased, which suggested miR-342-3p may serve a role in the
proliferation, migration, invasion and the cell cycle of
trophoblastic cells. miR-342-3p may play a key role in the
pathogenesis of PE.
In the field of obstetrics, miR-342-3p was
downregulated in intrauterine growth restriction pregnancies if the
gestational weeks was <34 weeks (29). In addition, miR-342-3p was reported
to be increased in the plasma in pregnancies with severe PE
(30), which was in line with our
outcomes. miR-342-3p has been suggested to target certain genes,
including Forkhead box protein M1, RAP2B, member of RAS oncogene
family, IκB kinase, transforming growth factor-β activated kinase 1
binding protein (TAB)2, TAB3, anterior gradient 2, astrocyte
elevated gene-1, T-lymphoma invasion and metastasis-inducing
protein 1, E2F transcription factor 1 and C-terminal binding
protein 2 (16–18,31–36).
We revealed PDGFRA to be a novel direct target of miR-342-3p in
trophoblastic cells. In our study, PDGFRA expression in the
placental tissues of patients with PE was decreased, whereas that
of miR-342-3p was increased. In addition, the expression of PDGFRA
decreased following transfection with miR-342-3p mimics in cells;
siR-PDGFRA transfection appeared to exhibit similar effects to
those of miR-342-3p mimics. Furthermore, the results of the
dual-luciferase assay indicated a direct connection between
miR-342-3p and PDGFRA. Collectively, these findings suggest PDGFRA
as a direct target of miR-342-3p in PE.
PDGFRA is a type III receptor tyrosine kinase that
regulates cell proliferation, differentiation, adhesion and
survival. Ligand binding activates the kinase, stimulating cellular
signaling proteins, including mitogen-activated protein kinases
(MAPKs), signal transducers and activators of transcription and
phosphatidylinositol-3 kinases (37). High expression of PDGFRA has been
detected in various cancers, such as breast cancer (38), ovarian cancer (37), gastrointestinal stromal tumors
(39) and melanoma (40). These findings indicate that PDGFRA
is associated with various physiological and pathological
processes, such as cell survival, migration and wound healing. In
our study, PDGFRA was determined to promote cell proliferation,
migration and invasion, and affected the cell cycle in
trophoblastic cells. Increases in PDGFRA expression could induce
cell proliferation, and activate the AKT and MAPK signal pathways.
On the contrary, these pathways were suppressed in response to
reductions in PDGFRA expression (41); further investigation into the
mechanism underlying the roles of PDGFRA in PE is required.
Our study has certain limitations. Only 30 cases of
PE placentas and 30 normal placentas were employed for histological
analysis. In the future, a larger sample size is required to
validate our findings. In addition, in vitro experiments
were performed; however, further insight from investigations may be
obtained by conducted in vivo research in rats or mice as
models. The present study aimed to determine the mechanism
underlying incomplete trophoblastic invasion in PE. However,
endothelial dysfunction is also one of the major causes of PE
(4), yet this was not explored in
our study this paper; thus, this should be investigated in the
future.
In conclusion, our research demonstrated that
miR-342-3p was upregulated and PDGFRA expression was decreased in
PE. Functional experiments showed that miR-342-3p could
affect the proliferation, migration, invasion and cell cycle of
trophoblastic cells by targeting PDGFRA. The present study proposed
that miR-342-3p may be a novel clinical indicator or prognostic
marker for PE.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author on reasonable request.
Authors' contributions
XY analyzed and interpreted the patient data, and
was a major contributor in writing the manuscript. FG performed the
statistical analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Ethics Committee in the First Hospital of China
Medical University agreed our research. Written informed consent
for participation in the study or use of their tissues was obtained
from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PE
|
preeclampsia
|
|
miRNA
|
microRNA
|
|
MSCs
|
mesenchymal stem cells
|
|
PDGFRA
|
platelet-derived growth factor
receptorα
|
|
EVT
|
extravillous trophoblast
|
|
MAPK
|
mitogen-activated protein kinase
|
References
|
1
|
Cheung HC, Leung KY and Choi CH:
Diagnostic accuracy of spot urine protein-to-creatinine ratio for
proteinuria and its association with adverse pregnancy outcomes in
Chinese pregnant patients with pre-eclampsia. Hong Kong Med J.
22:249–255. 2016.PubMed/NCBI
|
|
2
|
Askie LM, Duley L, Henderson-Smart DJ and
Stewart LA; PARIS Collaborative Group, : Antiplatelet agents for
prevention of pre-eclampsia: A meta-analysis of individual patient
data. Lancet. 369:1791–1798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Turner RJ, Bloemenkamp KW, Bruijn JA and
Baelde HJ: Loss of thrombomodulin in placental dysfunction in
preeclampsia. Arterioscler Thromb Vasc Biol. 36:728–735. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hastie R, Brownfoot FC, Pritchard N,
Hannan NJ, Cannon P, Nguyen V, Palmer K, Beard S, Tong S and
Kaitu'u-Lino TJ: EGFR (epidermal growth factor receptor) signaling
and the mitochondria regulate sFlt-1 (soluble FMS-like tyrosine
kinase-1) secretion. Hypertension. 73:659–670. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang X and Meng T: MicroRNA-431 affects
trophoblast migration and invasion by targeting ZEB1 in
preeclampsia. Gene. 683:225–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liang T, Hu XY, Li YH, Tian BQ, Li ZW and
Fu Q: MicroRNA-21 regulates the proliferation, differentiation, and
apoptosis of human renal cell carcinoma cells by the mTOR-STAT3
signaling pathway. Oncol Res. 24:371–380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu M, Chen Y, Song G, Chen B, Wang L, Li
X, Kong X, Shen Y and Qian L: MicroRNA-29c overexpression inhibits
proliferation and promotes apoptosis and differentiation in P19
embryonal carcinoma cells. Gene. 576:304–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou N, Fei D, Zong S, Zhang M and Yue Y:
MicroRNA-138 inhibits proliferation, migration and invasion through
targeting hTERT in cervical cancer. Oncol Lett. 12:3633–3639. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang X, Lei P, Huang Y, Zhang Z and Zhang
Y: MicroRNA-133b inhibits the migration and invasion of non-small
cell lung cancer cells via targeting FSCN1. Oncol Lett.
12:3619–3625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao G, Zhou X, Chen S, Miao H, Fan H,
Wang Z, Hu Y and Hou Y: Differential expression of microRNAs in
decidua-derived mesenchymal stem cells from patients with
pre-eclampsia. J Biomed Sci. 21:812014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Biró O, Nagy B and Rigó J Jr: Identifying
miRNA regulatory mechanisms in preeclampsia by systems biology
approaches. Hypertens Pregnancy. 36:90–99. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang S, Li H, Ge Q, Guo L and Chen F:
Deregulated microRNA species in the plasma and placenta of patients
with preeclampsia. Mol Med Rep. 12:527–534. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li H, Ge Q, Guo L and Lu Z: Maternal
plasma miRNAs expression in preeclamptic pregnancies. Biomed Res
Int. 2013:9702652013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hromadnikova I, Kotlabova K, Ivankova K,
Vedmetskaya Y and Krofta L: Profiling of cardiovascular and
cerebrovascular disease associated microRNA expression in umbilical
cord blood in gestational hypertension, preeclampsia and fetal
growth restriction. Int J Cardiol. 249:402–409. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choi SY, Yun J, Lee OJ, Han HS, Yeo MK,
Lee MA and Suh KS: MicroRNA expression profiles in placenta with
severe preeclampsia using a PNA-based microarray. Placenta.
34:799–804. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li XR, Chu HJ, Lv T, Wang L, Kong SF and
Dai SZ: miR-342-3p suppresses proliferation, migration and invasion
by targeting FOXM1 in human cervical cancer. FEBS Lett.
588:3298–3307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xie X, Liu H, Wang M, Ding F, Xiao H, Hu
F, Hu R and Mei J: miR-342-3p targets RAP2B to suppress
proliferation and invasion of non-small cell lung cancer cells.
Tumour Biol. 36:5031–5038. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao L and Zhang Y: miR-342-3p affects
hepatocellular carcinoma cell proliferation via regulating NF-κB
pathway. Biochem Biophys Res Commun. 457:370–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jarvenpaa J, Vuoristo JT, Savolainen ER,
Ukkola O, Vaskivuo T and Ryynanen M: Altered expression of
angiogenesis-related placental genes in pre-eclampsia associated
with intrauterine growth restriction. Gynecol Endocrinol.
23:351–355. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Saito T and Saetrom P: MicroRNAs-targeting
and target prediction. N Biotechnol. 27:243–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang C, Li Q, Ren N, Li C, Wang X, Xie M,
Gao Z, Pan Z, Zhao C, Ren C and Yang W: Placental miR-106a~363
cluster is dysregulated in preeclamptic placenta. Placenta.
36:250–252. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schatz F, Guzeloglu-Kayisli O, Arlier S,
Kayisli UA and Lockwood CJ: The role of decidual cells in uterine
hemostasis, menstruation, inflammation, adverse pregnancyoutcomes
and abnormal uterine bleeding. Hum Reprod Update. 22:497–515. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Redline RW and Patterson P: Pre-eclampsia
is associated with an excess of proliferative immature intermediate
trophoblast. Hum Pathol. 26:594–600. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ji L, Zhang L, Li Y, Guo L, Cao N, Bai Z,
Song Y, Xu Z, Zhang J, Liu C and Ma X: MiR-136 contributes to
pre-eclampsia through its effects on apoptosis and angiogenesis of
mesenchymal stem cells. Placenta. 50:102–109. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang W, Wang A, Zhao C, Li Q, Pan Z, Han
X, Zhang C, Wang G, Ji C, Wang G, et al: miR-125b enhances IL-8
production in earlyonset severe preeclampsia by targeting
sphingosine-1-phosphate lyase 1. PLoS One. 11:e01669402016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ding GC, Chen M, Wang YX, Rui C, Xu W,
Ding HJ and Shi ZH: MicroRNA-128a-induced apoptosis in HTR-8/SVneo
trophoblast cells contributes to pre-eclampsia. Biomed
Pharmacother. 81:63–70. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu H, Wang H, Liu M, Bai Y, Li YX, Ji L,
Peng C, Yu Y and Wang YL: MiR-195 participates in the placental
disorder of preeclampsia via targeting activin receptortype-2B in
trophoblastic cell. J Hypertens. 34:1371–1379. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hromadnikova I, Kotlabova K, Hympanova L
and Krofta L: Cardiovascular and cerebrovascular disease associated
microRNAs are dysregulated in placental tissues affected with
gestational hypertension, preeclampsia and intrauterine growth
restriction. PLoS One. 10:e01383832015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu L, Zhou H, Lin H, Qi J, Zhu C, Gao Z
and Wang H: Circulating microRNAs are elevated in plasma from
severe preeclamptic pregnancies. Reproduction. 143:389–397. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401.
2014.PubMed/NCBI
|
|
32
|
Xue X, Fei X, Hou W, Zhang Y, Liu L and Hu
R: miR-342-3p suppresses cell proliferation and migration by
targeting AGR2 in non-small cell lung cancer. Cancer Lett.
412:170–178. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang S, Liu L, Lv Z, Li Q, Gong W and Wu
H: MicroRNA-342-3p inhibits the proliferation, migration, and
invasion of osteosarcoma cells by targeting astrocyte-elevated
gene-1 (AEG-1). Oncol Res. 25:1505–1515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang H, Fan L, Zhan R, Wu S and Niu W:
Expression of microRNA-10a, microRNA-342-3p and their predicted
target gene TIAM1 in extranodal NK/T-cell lymphoma, nasal type.
Oncol Lett. 11:345–351. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tai MC, Kajino T, Nakatochi M, Arima C,
Shimada Y, Suzuki M, Miyoshi H, Yatabe Y, Yanagisawa K and
Takahashi T: miR-342-3p regulates MYC transcriptional activity via
direct repression of E2F1 in human lung cancer. Carcinogenesis.
36:1464–1473. 2015.PubMed/NCBI
|
|
36
|
Wang L, Xu L, Xu M, Liu G, Xing J, Sun C
and Ding H: Obesity-Associated miR-342-3p promotes adipogenesis of
mesenchymal stem cells by suppressing CtBP2 and releasing C/EBPα
from CtBP2 binding. Cell Physiol Biochem. 35:2285–2298. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lan H, Chen W, He G and Yang S: miR-140-5p
inhibits ovarian cancer growth partially by repression of PDGFRA.
Biomed Pharmacother. 75:117–122. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu H, Han Y, Lou J, Zhang H, Zhao Y,
Győrffy B and Li R: PDGFRA, HSD17B4 and HMGB2 are potential
therapeutic targets in polycystic ovarian syndrome and breast
cancer. Oncotarget. 8:69520–69526. 2017.PubMed/NCBI
|
|
39
|
Kalfusová A and Kodet R: Molecular
mechanisms of primary and secondary resistance, molecular-genetic
features and characteristics of KIT/PDGFRA non-mutated GISTs. Cesk
Patol Winter. 53:167–173. 2017.(In Czech).
|
|
40
|
Bai X, Kong Y, Chi Z, Sheng X, Cui C, Wang
X, Mao L, Tang B, Li S, Lian B, et al: MAPK pathway and TERT
promoter gene mutation pattern and its prognostic value in melanoma
patients: A retrospective study of 2,793 cases. Clin Cancer Res.
23:6120–6127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen W, Kuang Y, Qiu HB, Cao Z, Tu Y,
Sheng Q, Eilers G, He Q, Li HL, Zhu M, et al: Dual targeting of
insulin receptor and KIT in imatinib-resistant gastrointestinal
stromal tumors. Cancer Res. 77:5107–5117. 2017. View Article : Google Scholar : PubMed/NCBI
|