Introduction
Globally, colorectal cancer ranks third in cancer
incidence and fourth in cancer-associated mortality. It is
recognized as one of the most severe malignant tumors worldwide,
exhibiting high incidence and mortality (1–3), and
it has become a major global health problem. At present,
traditional treatment methods for colon cancer include surgery,
radiotherapy and chemotherapy (1).
Although traditional therapy improves the survival rate of patients
with colon cancer, its invasiveness and biological toxicity
considerably affect patient quality of life (4). Therefore, it is necessary to develop
new and more effective methods for treating colon cancer.
In recent years, with advancements in tumor biology
and immunology, cell-based cancer immunotherapy has become a
potential method of tumor treatment (5–8).
Typical immunotherapies include the use of tumor-infiltrating
lymphocytes, T cell receptor-engineered T cells and chimeric
antigen receptor (CAR)-modified T cells (9–11).
CARs are fusion molecules that couple antibody molecules that
recognize tumor antigens with T cell activation signal (12). CARs are composed of the
extracellular antigen recognition region through the transmembrane
region, including the hinge region and the intracellular signal
region (5). The precise targeting
specificity of monoclonal antibodies allied with the strong
toxicity and persistence of cytotoxic CAR-modified T cells allow
these cells to specifically recognize tumor-associated antigens
without relying on major histocompatibility complex (MHC)
restriction, thereby efficiently and permanently killing tumor
cells (13). This immunotherapy
technology has opened new avenues for the treatment of colon
cancer.
Epithelial cell adhesion molecule (EpCAM), which
promotes the proliferation and metastasis of tumor cells, is one of
the strongest and most ubiquitous tumor surface antigens, and has
potential as a target for tumor immunotherapy (14). Since the 1990s, EpCAM-specific
monoclonal antibodies (mAbs) have been used in the treatment of
human colon cancer, increasing the 5-year survival rate of patients
by 30% and reducing the recurrence rate by 27% within 7 years of
treatment (15). It was recently
reported that a new treatment for colon cancer involving
single-chain fragment variable (scFv) antibody-truncated
protamine-small interfering RNA, which recognizes and binds to
colon cancer cells through EpCAM antigen activity (16). This RNA specifically inhibits
Wnt/β-catenin signaling, effectively interrupting the functional
cycle between EpCAM and Wnt/β-catenin signaling, thus providing a
new strategy for the effective treatment of colon cancer (16).
In the present study, EpCAM-targeting CAR-T cells
were constructed and their apoptotic effect on EpCAM+ colon cancer
cells was evaluated. EpCAM-CAR-T cells were transfected with a
recombinant lentivirus carrying the EpCAM-CAR gene expression
cassette and tested for their killing efficacy against colon cancer
cells in vitro. The results indicated that EpCAM-CAR-T cells
may be able to induce EpCAM+ colon cancer cell apoptosis, and this
ability may be dependent on the expression of EpCAM on the surface
of colon cancer cells and on the number of T cells. In summary, the
EpCAM-CAR-T cells developed in this study exhibited antitumor
potential and may serve as a basis for further research and
development of colon cancer treatment.
Materials and methods
Cell culture
All cell lines (SW620, SW480, HCT116, LoVo, HT-29
and 293T; preserved by the Department of Digestive Tumor
Microenvironment of the First Affiliated Hospital, Sichuan, China)
were cultured in RPMI 1640 medium supplemented with 10% fetal calf
serum (FCS; Gibco; Thermo Fisher Scientific, Inc.).
Short tandem repeat (STR)
profiling
In total, 20 STR loci, plus the gender determining
locus amelogenin, were amplified using the commercially available
PowerPlex® 21 System from Promega Corporation. The
amplified products were processed using the Applied Biosystems
3730×l DNA Analyzer and data were analyzed using GeneMapper 5.0
software (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Appropriate positive and negative controls were run and confirmed
for each sample submitted.
Blood donor samples
For all experiments, blood samples were collected
with informed consent from healthy volunteers using a protocol
approved by the Ethics Committee of the First Affiliated Hospital
of Chengdu Medical College (Chengdu, China). Between March and May
2017, peripheral blood samples were collected from 3 healthy
volunteers (2 males and 1 female), aged 20–35 years. The lymphocyte
density gradient centrifugation kit (GE Healthcare Life Sciences)
was used to isolate peripheral blood mononuclear cells (PBMCs) from
blood. Briefly, peripheral blood was slowly added to the upper
layer of the equal volume lymphocyte liquid and centrifuged at 400
× g for 30 min at room temperature. Then, the mononuclear cell
layer was collected and washed with PBS buffer.
Construction of EpCAM-CAR
The EpCAM scFv was cloned from the vector
pET-26b-EpCAM (Novagen, Inc.), which contained the sequence for the
scFv antibody for EpCAM. This vector was established in our
laboratory as previously described (17). Then, the EpCAM-scFv was linked the
CD8 α hinge-transmembrane region with 4-1BB co-stimulatory domain
and CD3 ζ chain, and the DNA sequence encoding this cassette was
digested by HindIII and XhoI, and cloned into the lentiviral
backbone pCLK-EF-1 (Invitrogen; Thermo Fisher Scientific, Inc.)
vector as previously described (18). The plasmids pCLK-EpCAM-CAR, psPAX-2
(Invitrogen; Thermo Fisher Scientific, Inc.) and pMD2.G (Addgene,
Inc.) were transfected into Escherichia coli HD5a (cat. no.
CD201-01; Beijing Transgen Biotech Co., Ltd.); 1 µl plasmids (1
µg/µl) were incubated with E. coli DH5a on ice for 30 min, heated
for 90 sec in a water bath at 42°C and incubated on ice for 2 min.
DH5a were cultured in 900 µl LB liquid medium (cat. no. 12795027;
Invitrogen; Thermo Fisher Scientific, Inc.) on 37°C for 1 h at 200
RPM. Then, the mixture was coated on the surface of LB solid medium
(cat. no. 22700025; Invitrogen; Thermo Fisher Scientific, Inc.)
plates and static-cultured at 37°C for 16 h. Mono-bacteria were
selected and cultured for plasmid extraction. The plasmid
extraction kit (cat. no. 12381; Qiagen, Inc.) was used to extracted
these three plasmids for detection and lentiviral packaging. Then,
a 0.5% agarose gel was used to detect the size of plasmid (1 µl
DNA/lane). Goldview was used as the visualization reagent.
Packaging and concentration of the
lentivirus
A lentiviral supernatant was generated from 293T
cells transfected with PCLK-EF-1-CAR, pMD2.G and psPAX-2. 293T
cells were cultured and used for packaging lentivirus at 70–80%
confluence. The vectors (9 µg PCLK-EF-1-CAR; 9 µg pMD2.G; 4.5 µg
psPAX-2) were transfected into 293T cells using the calcium
phosphate method. Following transfection, 293T cells were cultured
at 37°C with 5% CO2 for 72 h. The lentivirus suspension was
collected and filtered with a 0.22-µm filter. Then, the lentivirus
suspension was ultracentrifuged at 70,000 × g at 20°C for 2 h to
concentrate the virus. Reverse transcription-quantitative PCR
(RT-qPCR) was used to determine the titer of concentrated
virus.
Transduction and expansion of T
cells
Human PBMCs were cultured in RPMI 1640 medium with
10% FCS, and activated with CD3 antibodies (cat. no. MA1-10175; 50
ng/ml; Invitrogen; Thermo Fisher Scientific, Inc.) and interleukin
(IL)-2 (cat. no. 0208AF12; 300 U/ml; PeproTech, Inc.) for 24 h
(19). Then, T cells were
transduced with the concentrated lentiviral at a multiplicity of
infection of 4 on RetroNectin-coated plates (Takara Bio, Inc.).
Transduced cells were cultured with IL-2 (300 U/ml) for 14 days
before subsequent analysis. Non-transduced T cells were used as
negative controls and were cultured under the same conditions. In
all trials, the functions of transduced and non-transduced T cells
obtained from the same donor were compared.
Flow cytometry
In order to detect the expression of EpCAM on the
cell surface, the colon cancer cell lines SW620, SW480, HCT116 and
LoVo, and the colorectal cancer cell line HT-29, were incubated
with EpCAM antibody (1:500; cat. no. 2929; Cell Signaling
Technology, Inc.) at 37°C for 30 min. Then, cells were washed and
incubated with a Cy3-conjugated fluorescent secondary antibody
(1:100; cat. no. SA00009-1; ProteinTech Group, Inc.) at 37°C for 30
min in the dark. In order to detect the expression of CAR on the
surface of T cells, T cells were incubated with an antigen-binding
fragment 2 [F(ab)2] antibody (1:200; cat. no. NBP1-51900; Novus
Biologicals, Ltd.) at 37°C for 30 min. Cells were then washed, and
incubated with a Cy3-conjugated fluorescent secondary antibody
(1:100; cat. no. SA00009-4; ProteinTech Group, Inc.) at 37°C for 30
min in the dark. Cells were washed with PBS and detected using a
flow cytometer. BD Accuri™ C6 software was used for data analysis
(BD Biosciences).
Western blotting
Total proteins were obtained using RIPA buffer
(Beyotime Institute of Biotechnology) and quantified using a
bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). Proteins (30 µg/lane) were then transferred to PVDF
membranes (Beyotime Institute of Biotechnology) after separation by
10% SDS-PAGE. Membranes were blocked using 5% nonfat milk for 1 h
at 26°C. In order to detect the expression of EpCAM, membrane-bound
proteins from colon cancer cells were incubated with an anti-EpCAM
primary antibody (1:1,000; cat. no. 2929; Cell Signaling
Technology, Inc.) at 4°C overnight. Membranes were reacted with
secondary horseradish peroxidase-conjugated antibody (1:2,000; cat.
no. 7076; Cell Signaling Technology, Inc.) for 2 h at 37°C.
Membranes containing T cell-derived proteins were instead incubated
with an anti-F(ab)2 antibody primary (1:5,000; cat. no. NBP1-51900;
Novus Biologicals, Ltd.) at 4°C overnight, and then with secondary
horseradish peroxidase-conjugated antibody (1:1,000; cat. no.
HAF109; R&D Systems, Inc.) for 2 h at 37°C. After washing,
protein bands were measured using an enhanced chemiluminescence
assay kit (EMD Millipore) and imaged with a chemiluminescence
detection system (Bio-Rad Laboratories, Inc.). The relative
expression of a target protein was determined as the ratio of the
grayscale value of the target protein to that of β-actin (1:1,000;
cat. no. 58169; Cell Signaling Technology, Inc.) by Image Lab
(version 4.0; Bio-Rad Laboratories, Inc.). All experiments were
repeated three times.
Gene expression analysis by
RT-qPCR
Total RNA was isolated from lentiviruses or
EpCAM-CAR-T cells and untransfected T cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The primers used for viral titer determination were:
Lentiviral Rev response element forward,
5′-TTTGTTCCTTGGGTTCTTGGG-3′ and reverse,
5′-GATTCTTGCCTGGAGCTGCTT-3′. For CAR mRNA expression analysis, the
primers were: Forward, 5′-CAAGATTACACTCAGGAGTCCC-3′ and reverse,
5′-GTGGGTATTACTGGATGGTGGG-3′. The primers used to measure GAPDH
were: Forward, 5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGCCAAA-3′. RNA was reverse transcribed using a
PrimeScript™ RT reagent kit (cat. no. RR047A; Takara Bio, Inc.) to
obtain cDNA as follows: 37°C for 15 min, 85°C for 5 sec and 4°C for
1 h. qPCR was performed using a TB Green® Premix Ex Taq™
II kit (cat. no. RR820A; Takara Bio, Inc.). The thermocycling
conditions were as follows: 95°C for 30 sec, then 39 cycles of 95°C
for 5 sec and 60°C for 30 sec, followed by 95°C for 10 sec, then
65°C for 5 sec, and finally 95°C for 0.5 sec. The Cq values of the
target genes were normalized to that of GAPDH (20).
Cytotoxicity assay
The antitumor effect of EpCAM-CAR-T cells on colon
cancer cells was measured using a LDH-Glo™ Cytotoxicity Assay
(J2380, Promega). Briefly, EpCAM-CAR-T cells and untransfected T
cells were added as effector cells to each well, followed by the
addition of the target colon cancer cells (105; SW620, SW480,
HCT116, LoVo or HT-29). The final Effector: Target (E:T) ratios
were 0.5:1, 1:1, 2:1, 4:1, 8:1 or 16:1. The cell mixtures were
incubated at 37°C under 5% CO2 for 4 h. Collecting 50 µl culture
supernatant, mixture with 50 µl LDH Detection Reagent, then
transferred to fresh 96-well flat-bottom plates. Record
luminescence after incubate for 60 min at room temperature. The
percentage of cell lysis was calculated as: Specific lysis
(%)=(Effector spontaneous release-Target spontaneous
release)/(Target maximum release-Target spontaneous release) ×100.
Each assay was performed in triplicate.
Cytokine production analysis
To measure cytokine production in vitro,
colon and colorectal cancer cells were co-cultured with EpCAM-CAR-T
cells or untransfected T cells at the ratios of 0.5:1, 1:1, 2:1,
4:1, 8:1 or 16:1 at 37°C under 5% CO2. After 24 h, the supernatant
was collected, and the levels of IL-2 (cat. no. EK0397), IL-6 (cat.
no. EK0410) and IFN-γ (cat. no. EK0373) were analyzed by ELISA (all
Boster Biological Technology).
Statistical analysis
Data are expressed as the mean ± standard deviation
from at least three independent experiments. Differences between
different treatment groups were analyzed using one-way ANOVA
analysis. P<0.05 was considered to indicate a statistically
significant difference. All statistical analyses were performed
using SPSS 16.0 (SPSS, Inc.).
Results
Construction and identification of
EpCAM-CAR plasmid
Firstly, the primary plasmid EpCAM-CAR for
lentiviral packaging, as well as the helper plasmids psPAX-2 and
pMD2.G, were constructed. The structure of the main plasmid
EpCAM-CAR is shown in Fig. 1A and
B. Then, the plasmids were identified by agarose gel
electrophoresis. As shown in Fig.
1C, the sizes of the plasmids EpCAM-CAR, psPAX-2, and pMD2.G
were 10.8, 11.0 and 5.8 kb, respectively.
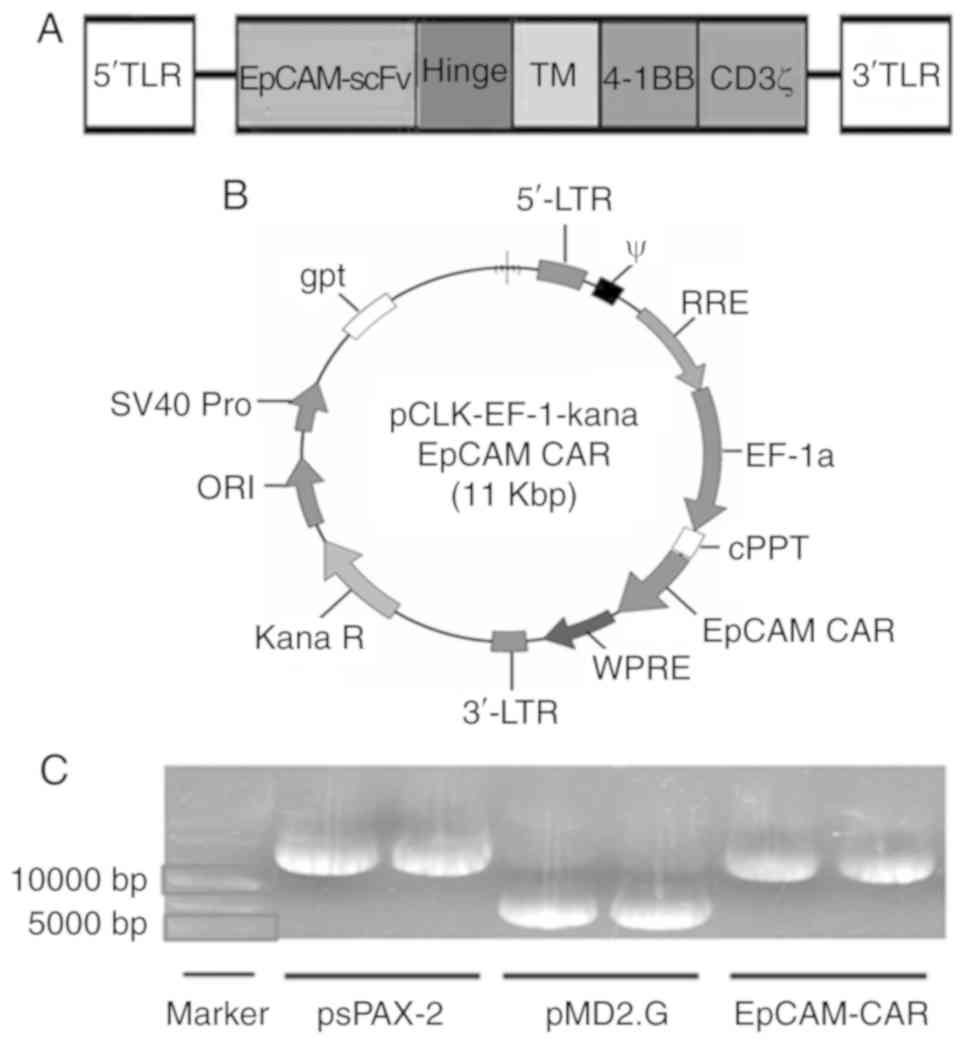 | Figure 1.Plasmid carrying the EpCAM-CAR gene.
(A) The EpCAM gene consists of EpCAM-scFv, along with a hinge
region, a TM region and a signal region. Its signal variable region
includes the 4-1BB costimulatory molecule and CD3ζ activation
domain. (B) EpCAM-CAR is a pCLK-EF-EpCAM-CAR plasmid construct
carrying the EpCAM-CAR gene, obtained by cloning the EpCAM fragment
into the pCLK-EF-1 vector containing the lentiviral packaging
component. (C) 0.5% agarose gel was used to detect the size of
plasmid (1 µl DNA/lane). The sizes of the plasmids EpCAM-CAR,
psPAX-2 and pMD2.G were 11.0, 11.0 and 5.8 kb, respectively, which
matched the expected molecular weights. EpCAM, epithelial cell
adhesion molecule; CAR, chimeric antigen receptor; scFv,
single-chain fragment variable; 4-1BB, TNF superfamily member 9;
TM, transmembrane; LTR, long terminal repeat; RRE, Rev response
element; WPRE, woodchuck hepatitis virus post-transcriptional
response element; cPPT, central polypurine tract. |
Packaging and concentration of
lentivirus
The viral-packaged master plasmid (EpCAM-CAR) and
the helper plasmids (psPAX-2 and pMD2.G) were transfected into 293T
cells using the calcium phosphate method to obtain a recombinant
plasmid carrying the EpCAM-CAR gene expression cassette. Cells were
also transfected with a construct whereby the main plasmid was
replaced with a fluorescent plasmid with the same fragment size and
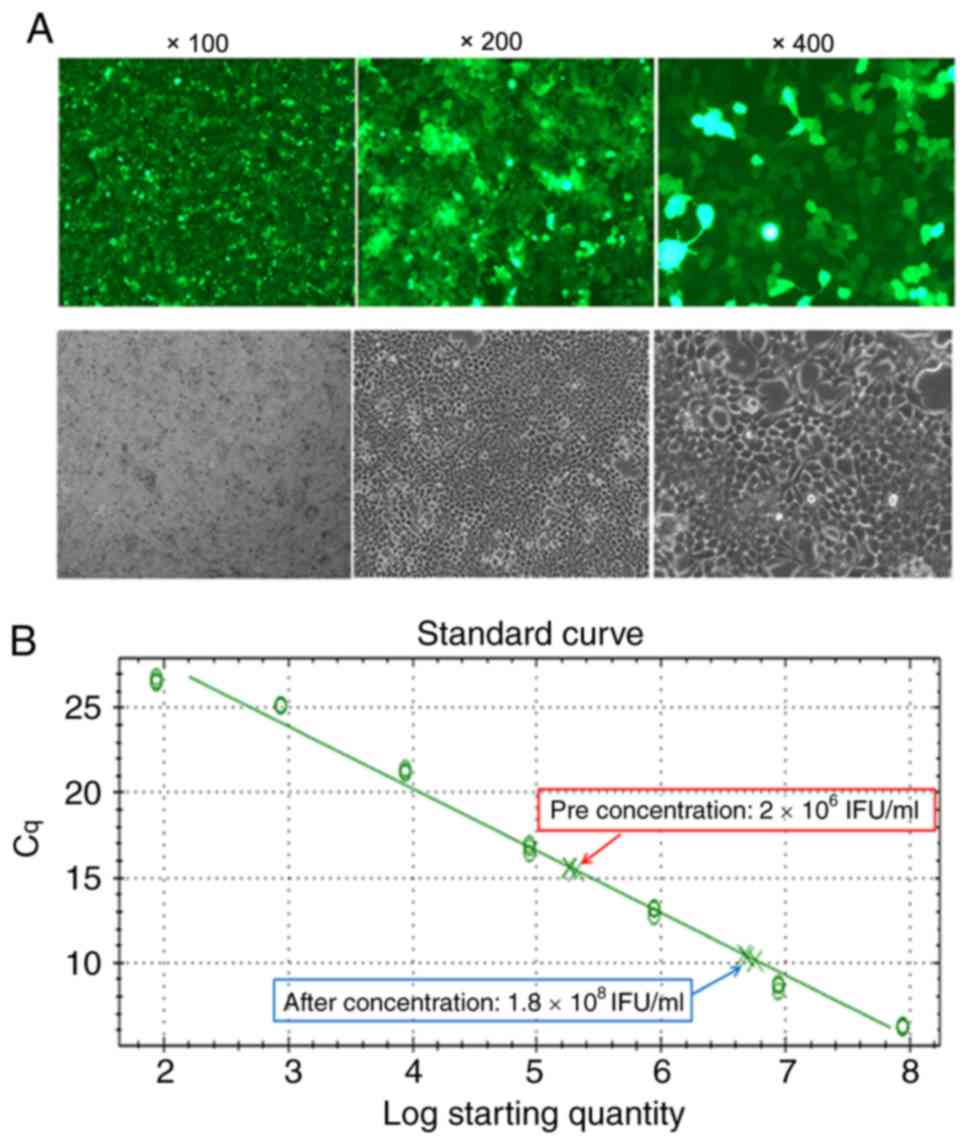
skeleton for use as a positive control. After 48 h, the
fluorescence of the positive control 293T cells was ~90% (Fig. 2A), indicating that the lentivirus
was successfully packaged. Then, virus supernatants were
concentrated by ultracentrifugation. The concentrated lentiviral
titer was assessed by RT-qPCR, and a standard curve was drawn
according to the Cq value and the copy number. The lentiviral
titers before and after concentration were 4.3×106 infection
function units (IFU)/ml and 1.8×108 IFU/ml, respectively (Fig. 2B). Based on these results, the
recombinant lentivirus carrying the EpCAM-CAR gene was considered
to have been successfully packaged and concentrated.
Expression of EpCAM in cancer cell
lines
The present study used the colon cancer cell lines
SW620, SW480, HCT116 and LoVo, and the colorectal cancer cell line
HT-29, as target cells to detect the antitumoral effects of
EpCAM-CAR-T cells. All cell lines were authenticated using STR
profiling, and all cells had a matching degree >95%. Western
blotting and flow cytometry were used to detect EpCAM expression in
these cancer cell lines. The results of the western blotting
indicated that EpCAM protein expression was observed in all five
cancer cell lines, and the molecular weight of the protein was 40
kDa. Among the lines, SW620 exhibited the highest expression, while
HT-29 exhibited the lowest expression of EpCAM (Fig. 3A and B). The results of the flow
cytometry showed that the expression rates of EpCAM on the surfaces
of HCT116, HT-29, LoVo, SW620 and SW480 cells were 78.3, 67.3,
75.4, 97.5 and 85.4%, respectively (Fig. 3C).
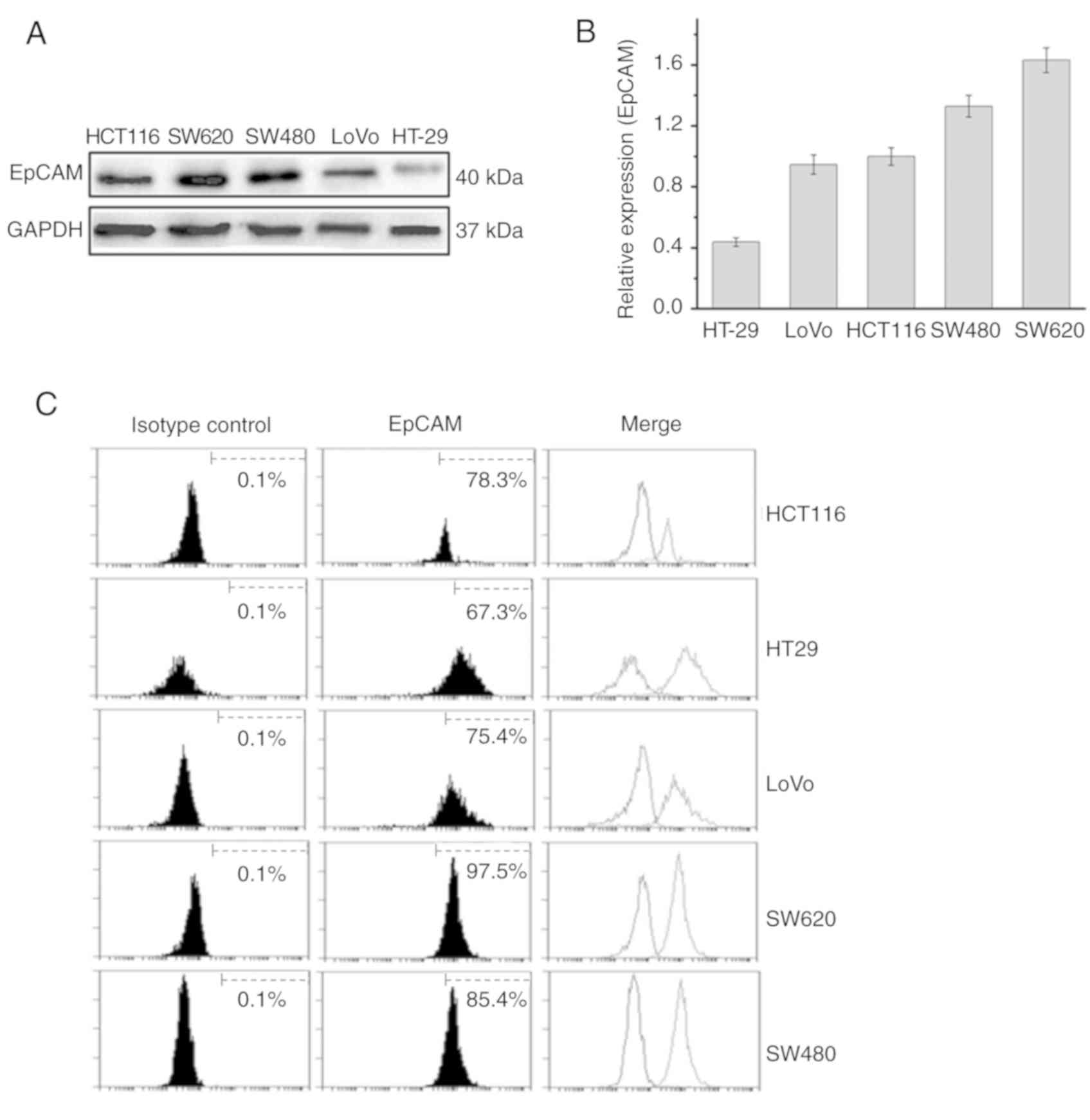 | Figure 3.Expression of EpCAM in cancer cell
lines. (A) Western blot analysis of EpCAM in HCT116, SW620, SW480,
LoVo and HT-29 cells. (B) Relative grayscale levels of EpCAM
protein in HCT116, SW620, SW480, LoVo and HT-29 cells (n=3/line).
(C) Flow cytometry was used to detect the expression of EpCAM on
HCT116, HT-29, LoVo, SW620 and SW480 cells. The expression rates
were 78.3, 67.3, 75.4, 97.5 and 85.4%, respectively. EpCAM,
epithelial cell adhesion molecule; CAR, chimeric antigen
receptor. |
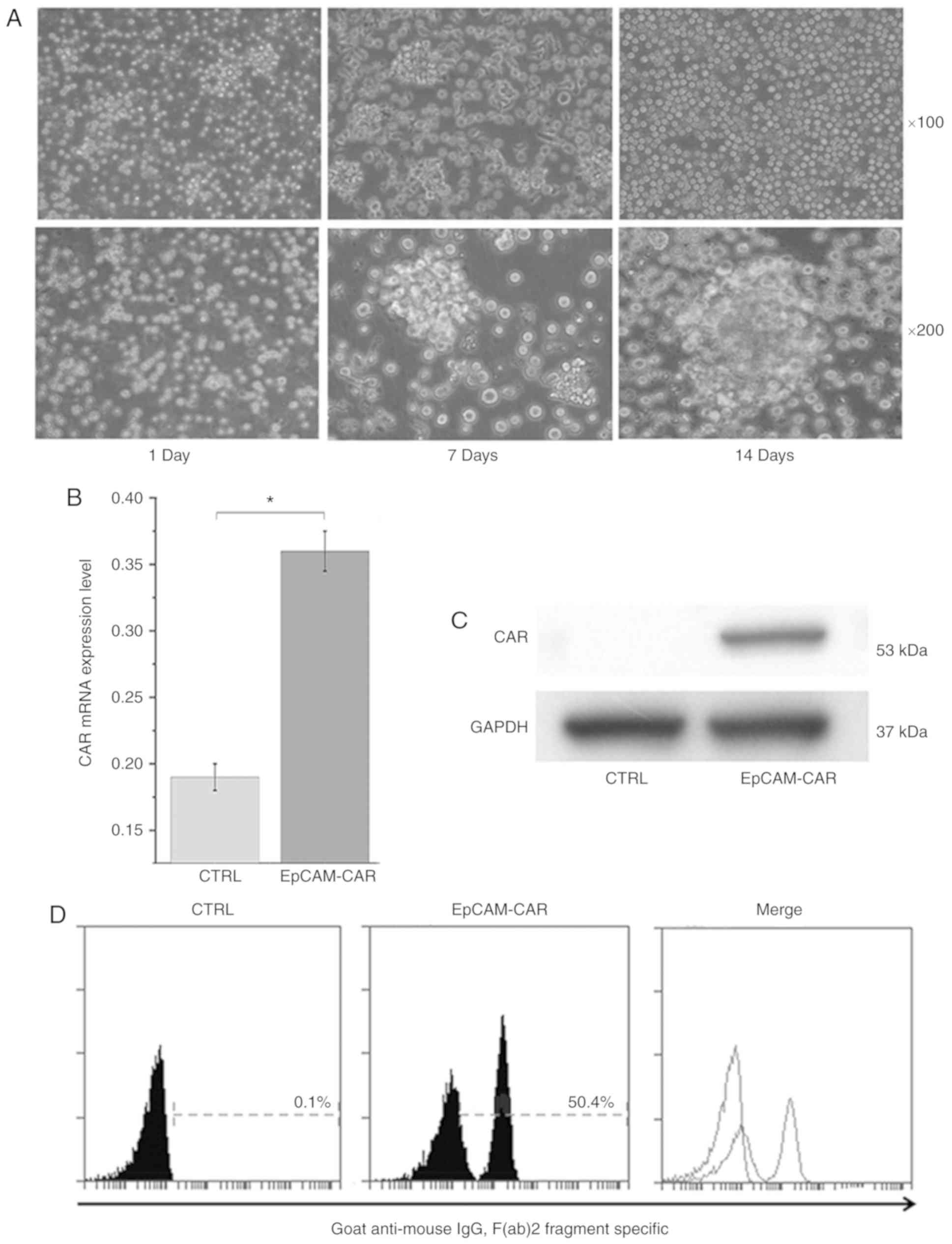
Construction of T cells stably
expressing EpCAM-CAR
To construct T cells that stably expressed
EpCAM-CAR, T cells were transfected with recombinant lentivirus.
The co-stimulatory domain 9/4-1BB and CD3 chain in the CAR can
activate T cells and promote the proliferation of T cells following
transfection of CAR into T cells. The results indicated that the
number of cells increased over time, and T cells tended to become
activated (Fig. 4A). On the 14th
day following viral transfection, the expression of CAR mRNA in T
cells was detected by RT-qPCR, confirming that CAR expression
levels were higher in transfected cells than in control cells
(Fig. 4B). In addition, the
expression of CAR in T cells evaluated by protein immunoblotting
confirmed that CAR was only expressed in transfected T cells, and
its molecular weight was 53 kDa (Fig.
4C), which was consistent with the theoretical molecular weight
(21). The expression of CAR on
the surface of T cells facilitates recognition of and binding to
antigens on the surface of tumor cells. Therefore, the expression
of CAR on the surface of T cells was evaluated by flow cytometry.
The results showed that CAR was expressed on the surface of 50.4%
of transfected T cells (Fig. 4D),
indicating that the transfected T cells stably expressed
EpCAM-CAR.
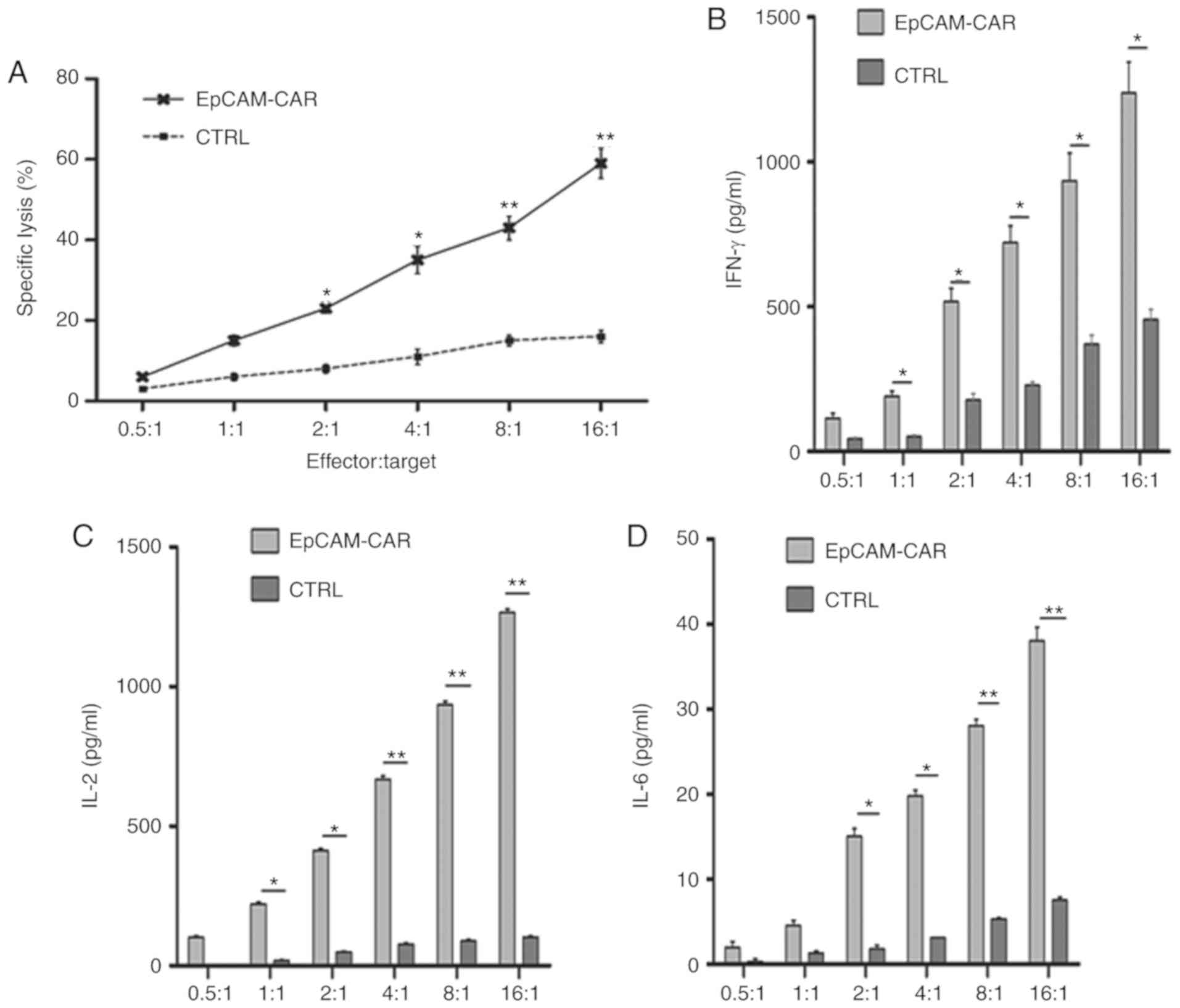
In vitro cytotoxicity effect of
EpCAM-CAR-T on cancer cells
In order to evaluate the antitumoral effect of
EpCAM-CAR-T cells on cancer cells, SW620, SW480, HCT116, LoVo and
the colorectal cancer cell line HT-29, all of which exhibit,
different EpCAM expression rates, were used as target cells for
EpCAM-CAR-T cells and untransfected T cells (effector cells).
First, T cells were co-cultured with colon cancer SW620 cells,
which exhibited the highest EpCAM expression, at E:T ratios of
0.5:1, 1:1, 2:1, 4:1, 8:1, and 16:1 for 4 h. The supernatant was
then extracted for analysis of the cytotoxic effects of T cells at
different effector ratios using an LDH release assay kit. The
results showed that as the E:T ratio increased, the LDH release of
the EpCAM-CAR-T cell group also increased, indicating that the
antitumor effect of EpCAM-CAR-T cells was dependent on the number
of EpCAM-CAR-T cells (Fig. 5A).
Studies have shown that IL-2, IFN-γ and IL-6 have antitumor roles
in tumor immunotherapy by regulating immune responses (22–24).
Therefore, levels of the inflammatory cytokines IL-2, IL-6 and
IFN-γ in the co-culture supernatants of EpCAM-CAR-T cells and SW620
cells were evaluated by ELISA. The results showed that the levels
of these inflammatory cytokines released by EpCAM-CAR-T cell
co-culture were significantly higher than those in the control
group. In addition, cytokine levels increased as the E:T ratio
increased (Fig. 5B-D).
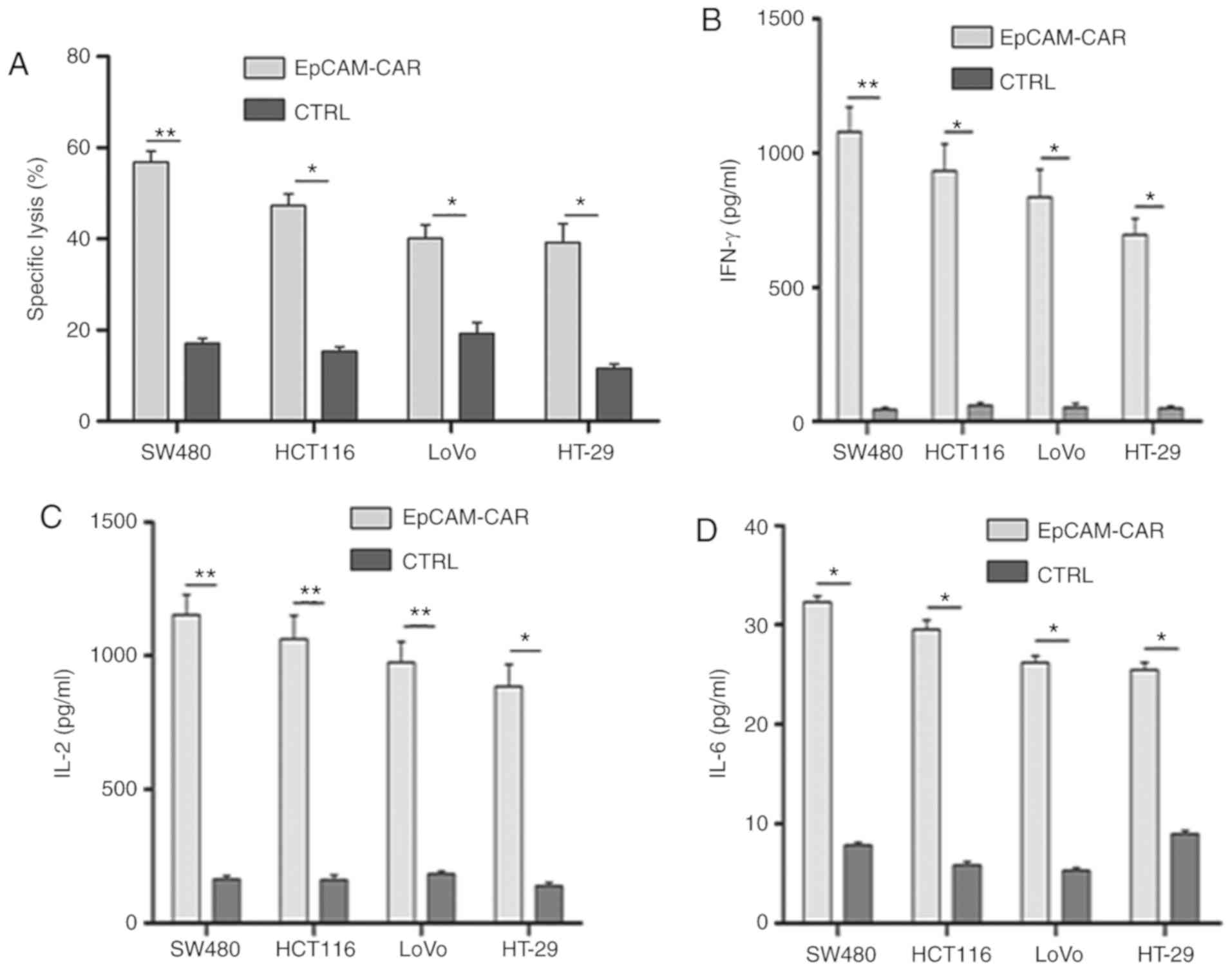
The results indicated that the antitumor effects of
EpCAM-CAR-T cells were strongest, and the release of cytokines was
highest at an E:T ratio of 16:1 when T cells were co-cultured with
colon cancer SW620 cells. Therefore, the remaining cell lines
(SW480, HCT116, LoVo and HT-29) were co-cultured with T cells at an
E:T ratio of 16:1 for 4 h. Compared with the control T cells,
EpCAM-CAR-T cells exhibited a stronger antitumor effect on the four
cancer cell lines (Fig. 6A).
Furthermore, the levels of IL-2, IL-6 and IFN-γ released by
EpCAM-CAR-T cells were also significantly higher than those
released by control T cells (Fig.
6B-D). Moreover, the antitumor effect and inflammatory cytokine
release may also be associated with the expression of EpCAM on the
surface of cancer cells, as cell lines with higher expression rates
of EpCAM appeared to exhibit more potent antitumor effects and to
release higher levels of inflammatory cytokines (Fig. 6).
Discussion
Tumor immunotherapy is a method for treating
malignant tumors by regulating the immune status of the body
(25). CAR-T cell therapy is also
known for its role in the treatment of B cell hematological
malignancies (26). As a promising
treatment modality, CAR-T cell therapy offers the following
advantages: i) Binding to tumor surface antigens in a
non-MHC-restricted manner; ii) simultaneous recognition of multiple
antigens; and iii) large-scale ex vivo acquisition of CAR-T cells
(4). Studies have shown that
immune cell therapy has achieved successful results in acute
lymphoblastic leukemia (ALL), chronic lymphocytic leukemia and
non-Hodgkin's lymphoma (26–29).
In 2017, the US Food and Drug Administration approved a CAR-T cell
therapy, which has been used in B-cell ALL (30), inspiring further development of
various CAR-T cells for immunotherapy in the future.
Studies have shown that EpCAM may play a role in
cell proliferation, migration, differentiation and morphogenesis
(31). Overexpression of EpCAM is
associated with progression and poor prognosis in gastric cancer
(32) and pancreatic cancer
(33). In a large retrospective
study, truncated EpCAM was observed to be associated with several
factors related to cancer stem cell formation and
epithelial-mesenchymal transition, such as poor differentiation,
vascular and limb invasion and lymph node metastasis (34). In addition, EpCAM is considered to
be a hallmark of numerous cancer stem cells (35). Tumor stem cells are characterized
by high tumorigenicity and high drug resistance (35). Traditional treatment of tumors is
unable to remove tumor stem cells, resulting in limited antitumor
efficacy and recurrence (36). In
this study, the expression of EpCAM was assessed in four colon
cancer cell lines and one colorectal cancer cell line using western
blotting analysis and flow cytometry. EpCAM was selected as both a
marker of cancer cells and a target for CAR-T cells. EpCAM-CAR-T
cells can specifically recognize cancer cells expressing EpCAM,
thereby achieving greater efficacy in killing cancer cells.
After successful construction of EpCAM-CAR-T cells,
their lysis efficiency was initially evaluated on the co-cultured
colon cancer cell line SW620, which had the highest expression of
EpCAM in vitro. This was tested at different E:T ratios
(ranging between 0.5:1 and 16:1) for 4 h. The LDH release assay
results showed that EpCAM-CAR-T cells were capable of causing SW620
cell lysis, and this effect was dependent on the number of
EpCAM-CAR-T cells. The lysis efficiency of EpCAM-CAR-T cells was
also evaluated in the other four cancer cell lines (SW480, HCT116,
LoVo and HT-29), which exhibit different expression levels of
EpCAM, at an E:T ratio of 16:1. The results showed that the killing
efficiency of EpCAM-CAR-T cells may have been dependent on the
levels of EpCAM expressed by the different cell lines, with higher
levels of LDH being detected in cell lines that express higher
levels of EpCAM.
Studies have shown that IL-2, IFN-γ and IL-6 play an
antitumor role in tumor immunotherapy by regulating immune
responses (37–39). Therefore, the secreted levels of
IFN-γ, IL-2 and IL-6 were examined in the co-culture supernatants
of EpCAM-CAR-T cells and EpCAM+ tumor cells. The secretion of these
cytokines was found to be increased with larger E:T ratios and with
higher expression of EpCAM on the surface of target cancer cells.
These results indicated that the antitumoral effect and
inflammatory cytokine release by EpCAM-CAR-T cells may be
associated with the levels of EpCAM on the surface of cancer cells,
with higher expression rates of EpCAM leading to more potent
antitumor effects and more inflammatory cytokine release.
Studies have found that the main side effect of
CAR-T cell therapy is cytokine release syndrome (CRS), which
primarily manifests as fatigue, high fever, hypotension and
hypoxia, and may even cause cardiopulmonary dysfunction, organ
failure and mortality in some cases (40). The secretion of a large number of
pro-inflammatory cytokines by activated T cells may lead to this
adverse reaction. It has been reported that the use of tropizumab,
an IL-6 receptor antagonist, may help control severe CRS without
impairing T cell efficacy (41).
Beyond cytokine use, certain studies have reported that steroids
may be used to control CRS (42,43).
In addition, it has also been reported that the severity of CRS may
be associated with the tumor load when CAR-T is injected.
Therefore, early reinfusion of CAR-T cells or drug pretreatment to
reduce the tumor burden can significantly reduce the occurrence of
severe CRS (42).
In summary, the present study described the
successful construction of an EpCAM-CAR plasmid and the subsequent
establishment of EpCAM-CAR-T cells that target EpCAM expressed on
the surface of colon and colorectal cancer cells. EpCAM-CAR-T cells
effectively killed cancer cells, and significantly promoted
cytokine release in vitro, indicating that CAR-T cells
targeting EpCAM may have the potential to treat colon or colorectal
cancer. Future studies should aim to conduct experiments to test
the antitumor effects of EpCAM-CAR-T cells in vivo. Overall, these
findings indicate a new avenue for the treatment of cancer by
immunotherapy.
Acknowledgements
The authors would like to thank Professor Wang Wei
(State Key Laboratory of Biological Therapy, Sichuan University,
Chengdu, China) for providing technical support and donating the
293T cells.
Funding
The present study was supported by the Natural
Science Foundation of Science and Technology (grant no.
2016JY0090), the Science and Technology Project of The Health
Planning Committee of Sichuan (grant no. 17ZD012), the National
Natural Science Foundation of China (grant no. 81302170), the
Natural Science Foundation of Education Department of Sichuan
Province (grant no. 16ZA0280) and the Innovative Group Foundation
of Education Department of Sichuan Province (grant no.
16TD0028).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YZ and XAL designed the study. PW, MML and YQL
performed the experiments. YZ and PW analyzed and interpreted the
data, and drafted the manuscript. All authors critically revised
the manuscript, and read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of the First Affiliated Hospital of Chengdu
Medical College (Chengdu, China). All participants provided written
informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD, Fedewa SA, Ahnen DJ,
Meester RGS, Barzi A and Jemal A: Colorectal cancer statistics,
2017. CA Cancer J Clin. 67:177–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pang Y, Hou X, Yang C, Liu Y and Jiang G:
Advances on chimeric antigen receptor-modified T-cell therapy for
oncotherapy. Mol Cancer. 17:912018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Couzin-Frankel J: Breakthrough of the year
2013. Cancer immunotherapy. Science. 342:1432–1433. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mellman I, Coukos G and Dranoff G: Cancer
immunotherapy comes of age. Nature. 480:480–490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rosenberg SA and Restifo NP: Adoptive cell
transfer as personalized immunotherapy for human cancer. Science.
348:62–68. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tran E, Robbins PF and Rosenberg SA:
‘Final common pathway’ of human cancer immunotherapy: Targeting
random somatic mutations. Nat Immunol. 18:255–262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Crompton JG, Klemen N and Kammula US:
Metastasectomy for tumor-infiltrating lymphocytes: An emerging
operative indication in surgical oncology. Ann Surg Oncol.
25:565–572. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kunert A, Obenaus M, Lamers CHJ,
Blankenstein T and Debets R: T-cell receptors for clinical therapy:
In vitro assessment of toxicity risk. Clin Cancer Res.
23:6012–6020. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hay KA and Turtle CJ: Chimeric antigen
receptor (CAR) T cells: Lessons learned from targeting of CD19 in
B-cell malignancies. Drugs. 77:237–245. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dominguez G: The CART gene: Structure and
regulation. Peptides. 27:1913–1918. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang C, Liu J, Zhong JF and Zhang X:
Engineering CAR-T cells. Biomark Res. 5:222017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martowicz A, Seeber A and Untergasser G:
The role of EpCAM in physiology and pathology of the epithelium.
Histol Histopathol. 31:349–355. 2016.PubMed/NCBI
|
|
15
|
Jin Z, Maiti S, Huls H, Singh H, Olivares
S, Mátés L, Izsvák Z, Ivics Z, Lee DA, Champlin RE and Cooper LJ:
The hyperactive sleeping beauty transposase SB100X improves the
genetic modification of T cells to express a chimeric antigen
receptor. Gene Therapy. 18:849–856. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hao H, Zhen Y, Wang Z, Chen F and Xie X: A
novel therapeutic drug for colon cancer: EpCAM scFv-truncated
protamine (tp)-siRNA. Cell Biol Int. 37:860–864. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mala J, Puthong S, Maekawa H, Kaneko Y,
Palaga T, Komolpis K and Sooksai S: Construction and sequencing
analysis of scFv antibody fragment derived from monoclonal antibody
against norfloxacin (Nor155). J Genet Eng Biotechnol. 15:69–76.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Carpenito C, Milone MC, Hassan R, Simonet
JC, Lakhal M, Suhoski MM, Varela-Rohena A, Haines KM, Heitjan DF,
Albelda SM, et al: Control of large, established tumor xenografts
with genetically retargeted human T cells containing CD28 and CD137
domains. Proc Natl Acad Sci USA. 106:3360–3365. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Savoldo B, Ramos CA, Liu E, Mims MP,
Keating MJ, Carrum G, Kamble RT, Bollard CM, Gee AP, Mei Z, et al:
CD28 costimulation improves expansion and persistence of chimeric
antigen receptor-modified T cells in lymphoma patients. J Clin
Invest. 121:1822–1826. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ang WX, Li Z, Chi Z, Du SH, Chen C, Tay
JC, Toh HC, Connolly JE, Xu XH and Wang S: Intraperitoneal
immunotherapy with T cells stably and transiently expressing
anti-EpCAM CAR in xenograft models of peritoneal carcinomatosis.
Oncotarget. 8:13545–13559. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fisher DT, Appenheimer MM and Evans SS:
The two faces of IL-6 in the tumor microenvironment. Semin Immunol.
26:38–47. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nakajima C, Uekusa Y, Iwasaki M, Yamaguchi
N, Mukai T, Gao P, Tomura M, Ono S, Tsujimura T, Fujiwara H and
Hamaoka T: A role of interferon-gamma (IFN-gamma) in tumor
immunity: T cells with the capacity to reject tumor cells are
generated but fail to migrate to tumor sites in IFN-gamma-deficient
mice. Cancer Res. 61:3399–3405. 2001.PubMed/NCBI
|
|
24
|
Rosenberg SA: IL-2: The first effective
immunotherapy for human cancer. J Immunol. 192:5451–5458. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lichty BD, Breitbach CJ, Stojdl DF and
Bell JC: Going viral with cancer immunotherapy. Nat Rev Cancer.
14:559–567. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rodgers DT, Mazagova M, Hampton EN, Cao Y,
Ramadoss NS, Hardy IR, Schulman A, Du J, Wang F, Singer O, et al:
Switch-mediated activation and retargeting of CAR-T cells for
B-cell malignancies. Proc Natl Acad Sci USA. 113:E459–E468. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grupp SA, Kalos M, Barrett D, Aplenc R,
Porter DL, Rheingold SR, Teachey DT, Chew A, Hauck B, Wright JF, et
al: Chimeric antigen receptor-modified T cells for acute lymphoid
leukemia. N Engl J Med. 368:1509–1518. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Porter DL, Levine BL, Kalos M, Bagg A and
June CH: Chimeric antigen receptor-modified T cells in chronic
lymphoid leukemia. N Engl J Med. 365:725–733. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schuster SJ, Svoboda J, Chong EA, Nasta
SD, Mato AR, Anak Ö, Brogdon JL, Pruteanu-Malinici I, Bhoj V,
Landsburg D, et al: Chimeric antigen receptor T cells in refractory
B-cell lymphomas. N Engl J Med. 377:2545–2554. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Singh N, Shi J, June CH and Ruella M:
Genome-editing technologies in adoptive T cell immunotherapy for
cancer. Curr Hematol Malig Rep. 12:522–529. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schmelzer E and Reid LM: EpCAM expression
in normal, non-pathological tissues. Front Biosci. 13:3096–3100.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dai M, Yuan F, Fu C and Shen G, Hu S and
Shen G: Relationship between epithelial cell adhesion molecule
(EpCAM) overexpression and gastric cancer patients: A systematic
review and meta-analysis. PLoS One. 12:e01753572017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fong D, Moser P, Kasal A, Seeber A, Gastl
G, Martowicz A, Wurm M, Mian C, Obrist P, Mazzoleni G and Spizzo G:
Loss of membranous expression of the intracellular domain of EpCAM
is a frequent event and predicts poor survival in patients with
pancreatic cancer. Histopathology. 64:683–692. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Seeber A, Untergasser G, Spizzo G,
Terracciano L, Lugli A, Kasal A, Kocher F, Steiner N, Mazzoleni G,
Gastl G and Fong D: Predominant expression of truncated EpCAM is
associated with a more aggressive phenotype and predicts poor
overall survival in colorectal cancer. Int J Cancer. 139:657–663.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yahyazadeh Mashhadi SM, Kazemimanesh M,
Arashkia A, Azadmanesh K, Meshkat Z, Golichenari B and Sahebkar A:
Shedding light on the EpCAM: An overview. J Cell Physiol.
234:12569–12580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cooper LJ, Topp MS, Serrano LM, Gonzalez
S, Chang WC, Naranjo A, Wright C, Popplewell L, Raubitschek A,
Forman SJ and Jensen MC: T-cell clones can be rendered specific for
CD19: Toward the selective augmentation of the
graft-versus-B-lineage leukemia effect. Blood. 101:1637–1644. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Davila ML, Riviere I, Wang X, Bartido S,
Park J, Curran K, Chung SS, Stefanski J, Borquez-Ojeda O, Olszewska
M, et al: Efficacy and toxicity management of 19-28z CAR T cell
therapy in B cell acute lymphoblastic leukemia. Sci Transl Med.
6:224ra252014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Zhang WY, Han QW, Liu Y, Dai HR,
Guo YL, Bo J, Fan H, Zhang Y, Zhang YJ, et al: Effective response
and delayed toxicities of refractory advanced diffuse large B-cell
lymphoma treated by CD20-directed chimeric antigen
receptor-modified T cells. Clin Immunol. 155:160–175. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Till BG, Jensen MC, Wang J, Qian X, Gopal
AK, Maloney DG, Lindgren CG, Lin Y, Pagel JM, Budde LE, et al:
CD20-specific adoptive immunotherapy for lymphoma using a chimeric
antigen receptor with both CD28 and 4-1BB domains: Pilot clinical
trial results. Blood. 119:3940–3950. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dholaria BR, Bachmeier CA and Locke F:
Mechanisms and management of chimeric antigen receptor T-cell
therapy-related toxicities. BioDrugs. 33:45–60. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee DW, Gardner R, Porter DL, Louis CU,
Ahmed N, Jensen M, Grupp SA and Mackall CL: Current concepts in the
diagnosis and management of cytokine release syndrome. Blood.
2:188–195. 2014. View Article : Google Scholar
|
|
42
|
Lee DW, Kochenderfer JN, Stetler-Stevenson
M, Cui YK, Delbrook C, Feldman SA, Fry TJ, Orentas R, Sabatino M,
Shah NN, et al: T-cells expressing CD19 chimeric antigen receptors
for acute lymphoblastic leukaemia in children and young adults: A
phase 1 dose-escalation trial. Lancet. 385:517–528. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Oluwole OO and Davila ML: At the bedside:
Clinical review of chimeric antigen receptor (CAR) T cell therapy
for B cell malignancies. J Leukoc Biol. 100:1265–1272. 2016.
View Article : Google Scholar : PubMed/NCBI
|