Introduction
Thoracic aortic diseases, including aneurysms and
dissections of the thoracic aorta, are a major cause of morbidity
and mortality in developed countries (1). The incidence of aortic diseases, such
as thoracic aortic aneurysm (TAA) and thoracic aortic dissection
(TAD), has been estimated at 6 per 100,000 individuals/year
(2). TAAs are localized
dilatations of the supra-diaphragmatic aorta that result from the
weakening and expansion of the arterial wall (3). TAD is characterized by the separation
of the thoracic aortic wall layers by extraluminal blood that
typically enters the vessel wall via an intimal tear (4). The primary cause of aortic diseases
is the degeneration of the aortic media layer due to hereditary and
environmental factors (5). The
process of the degeneration of the media layer, consisting of
elastin and smooth muscle cells (SMC) involves changes to the SMC
phenotype, loss of elastin layers and the increased production of
matrix metalloproteinases and proteoglycans (6).
MicroRNAs (miRNAs/miRs) are small non-coding
single-stranded RNAs (~21 nucleotides in length) that negatively
regulate or repress target gene expression. miRNAs regulate gene
expression in numerous physiological mechanisms, in addition to
pathological conditions within the cells (7). A number of miRNAs have been
identified that are associated with TAAs and TADs (8). Hsa-miR-143-3p is one of the most
important miRNAs expressed by vascular SMCs (VSMC) and interacts
with Kirsten rat sarcoma viral oncogene homolog (KRAS) and
mitogen-activated protein kinases (MAPKs), associated with the
pathogenesis of aortic diseases (9,10).
This miRNA regulates the MAPK pathways that induce TAA formation
via non-canonical transforming growth factor β signaling (11). The KRAS gene transcription product
is the K-RAS protein, which is the part of a signaling pathway
known as RAS type GTPase family/MAPK pathway (12). Another miRNA, hsa-miR-22-3p, has
been associated with MAPK14, which is implicated in VSMC
differentiation, proliferation, migration and fibrosis (13). In addition to these, SMCs from
thoracic aneurysms are characterized by the decreased expression of
SMC contractile proteins, including transgelin (TAGLN) (14).
In the present study, hsa-miR-143-3p and
hsa-miR-22-3p expression profiles were compared between patients
with TAA and TAD, and healthy volunteers. The present study also
investigated the potential target genes of these miRNAs. Selected
KRAS, MAPK7, MAPK14 and TAGLN genes expression analyses were
performed. In addition, the present study performed pathway
analysis for each miRNA-mRNA pair.
Materials and methods
Study design and subjects
The present study included a total of 28 serum
samples obtained from 9 patients with TAA, 9 patients with TAD and
10 healthy individuals. Patients who had been operated on for TAA
(n=9) and TAD (n=9) between 2017 and 2018 at the Cardiovascular
Surgery Clinic of Kartal Kosuyolu Heart Research Hospital
(Istanbul, Turkey) were enrolled in the present study. A total of 9
patients were diagnosed with acute Stanford type A aortic
dissection and received surgery on an emergency basis, while 9
patients were diagnosed with an ascending aortic aneurysm, and
received surgery electively. The experimental group refers to both
groups of patients. Patient data were collected from clinical
records of the hospital prospectively, following the recieval of
ethical approval of the study by the Ethics Committee of Istanbul
University. All patients provided written informed consent to
participate in the study. All patients with acute aortic dissection
were diagnosed via a computed tomography (CT) scan and were further
evaluated with transthoracic echocardiography prior to surgery.
Patients with asending aortic aneurysms were diagnosed using
routine echocardiographic evaluation and were followed-up by serial
CT scans until a surgical decision was made by the treating
physician. A further 10 individuals were admitted to the hospital
for routine examinations and were identified as healthy without any
aortic pathology, and these individuals agreed to be included in
the present study as the control group. The control group also
received a transthoracic echocardiograhic evaluation in order to
demonstrate the absence of any aortic and valvular pathology.
Sample demographics
The demographic characteristics of patients with TAA
and TAD constituting the experimental groups, and the healthy
individuals constituting the control group, are presented in
Table I. In all groups
(experimental and healthy), individuals with Marfan syndrome,
Loeys-Dietz syndrome, aneurysm osteoarthritis syndrome, arterial
gangrene syndrome and Ehlers Danlos syndrome were excluded from the
present study. Patients with syndromic thoracic aortic aneurysms,
known or suspected connective tissue disorders, and patients with
bicuspid aortic valves were not included in the present study.
Patients with inflammatory, autoimmune diseases and traumatic
etiology were not included in the present study. The present study
did not include patients >65 years old in the TAA and control
groups. All patients were male, with a mean age of 47±11 years
(range, 22–63 years) in the acute dissection group; mean 58±4 years
(range, 45–65 years) in the asending aortic aneurysm group; and
mean 53±1 years (range, 42–63 years) in the control group.
 | Table I.Patients and healthy controls
demographics. |
Table I.
Patients and healthy controls
demographics.
|
| Study groups |
|
|---|
|
|
|
|
|---|
| Demographic
characteristics | TAA | TAD | Control group |
|---|
| Number of
patients/individuals | 9 | 9 | 10 |
| Mean age,
years | 58.4 | 47.1 | 53.1 |
| Mean height,
cm | 169 | 173 | 173 |
| Mean weight,
kg | 89.8 | 85 | 80 |
| Hypertension, n
(%) | 9 (100) | 7 (77.7) | – |
| Diabetes, n
(%) | – | 1 (11.1) | – |
| COPD, n (%) | – | 1 (11.1) | – |
| Smoking, n (%) | 2 (22.2) | 6 (66.6) | – |
| CVD, n (%) | – | – | – |
| Obesity, n (%) | 1 (11.1) | 1 (11.1) | – |
Bioinformatics analysis
Bioinformatic tools were used in the present studyin
order to not only determine candidate miRNAs and their subsequent
sequences, but also to determine the target genes associated with
these miRNAs and sequences. The present study aimed to decrease the
rate of false positivity in the bioinformatic tools used by making
use of features including seed matching, conservation, free energy
and region accessibility to predict miRNA-mRNA interaction. To
identfy putative target mRNAs, the miRGator bioinformatic tool
(http://mirgator.kobic.re.kr/) was used
which shows the comparative results of other bioinformatics tools
such as microRNA.org (http://www.microrna.org/microrna/home.do), miRBase
(http://www.mirbase.org/), PITA (https://www.mybiosoftware.com/pita-6-microrna-prediction-tool.html)
and PicTar (https://pictar.mdc-berlin.de/). Also, TargetScan
(http://www.targetscan.org/vert_72/),
mirDB (http://www.mirdb.org/) and RNAHybrid
(https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid)
bioinformatic tools were used to confirmation target mRNAs found by
miRGator. In addition, Pathway Studio® and ENSEMBL were
used for the pathway analysis of candidate miRNAs and their target
mRNA (details nad URLs are presented in Table II).
 | Table II.Table of bioinformatic tools utilized
in the present study. |
Table II.
Table of bioinformatic tools utilized
in the present study.
Biological samples and total RNA
extraction
Peripheral blood samples were collected from healthy
individuals and patients. Blood samples were transferred to the
Istanbul University Molecular Biology and Genetics Department from
Cardiovascular Surgery Clinic at Kartal Kosuyolu Training and
Research Hospital within 36 h at the latest using the cold chain
method and stored at 4°C until the genetic analyses were
performed.
The peripheral blood samples were centrifuged at 937
× g for 20 min at 4°C and the supernatant was transferred into
microcentrifuge tubes. Serum was then aliquoted and stored at −80°C
for long-term storage. Total RNA, including miRNA, was extracted
from the serum using an mirVana PARIS kit (Ambion; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol. The concentrations of the RNA were determined on the
basis of the absorbance at 260 nm, and the purity was assessed on
the basis of the absorbance ratio at 260/280 nm using a NanoDrop
spectrophotometer. The samples were preserved as total RNA at −80°C
until use for the reverse transcription-quantitative polymerase
chain reaction (RT-qPCR).
miRNA and mRNA analyis via
RT-qPCR
The present study used RT-qPCR in the analyis of the
expression levels of the 2 miRNAs and mRNA of 4 genes. Each serum
sample was analyzed for the following miRNAs and mRNAs using
gene-specific TaqMan primer/probe sets (Applied Biosystems; Thermo
Fisher Sceintific, Inc.): Hsa-miR-143-3p (assay ID: 002249),
hsa-miR-22-3p (assay ID: 000398), KRAS (assay ID: Hs00364284_g1),
MAPK7 (assay ID: Hs0061114_g1), MAPK14 (assay ID: Hs01051152_m1)
and TAGLN (assay ID: Hs01038777_g1). Synthetic cel-miR-39 and GAPDH
served as the internal controls for the normalization of the miRNA
and mRNA expressions analyses. Negative PCR controls were run to
verify the absence of genomic DNA contamination.
For the miRNA expression analysis, selected miRNAs
underwent RT to form complementary DNA (cDNA) using a TaqMan
MicroRNA Reverse Transcription kit (Ambion; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The RT
product was amplified using gene-specific TaqMan primer/probe sets
and the TaqMan Universal PCR Master Mix II (Ambion; Thermo Fisher
Scientific, Inc.) with uracil-N glycosylase by the following a
2-step procedure: Initial denaturation for 10 min at 95°C, followed
by 40 cycles of 15 sec at 95°C, and 60 sec at 60°C.
First, for the mRNA expression analysis, cDNA was
synthesized from total RNA using a High Capacity cDNA Reverse
Transcription kit and random primers (Agilent SureCycler 8800;
Agilent Technologies, Inc., Santa Clara, CA, USA). cDNA was used to
analyze specific gene expression with specific Taqman Gene
Expression Assays on a CFX96 Touch™ Real-Time PCR detection system.
The thermal cycling protocol was performed as follows: 2 min at
95°C, 2 min at 95°C, followed by 40 cycles of 95°C for 15 sec, and
60°C for 1 min.
Data analysis
All statistical analyses were performed using
GraphPad Prism (version 7.04; GraphPad Software, Inc., La Jolla,
CA, USA). The Cq values of the samples were automatically
determined on a CFX96 Touch™ Real-Time PCR. The expression level of
each miRNA and mRNA was calculated using the 2−∆∆Cq
method (15). Data were analyzed
for outliers. D'Agostino & Pearson's normality test was
performed in order to determine whether the data were parametric or
non-parametric. All data were nonparametric and comparisons between
different groups were performed using Kruskal-Wallis tests. Dunn's
Multiple Comparison test was performed to determine differences
between specific groups. Results are presented as the mean ±
standard error of the mean. Receiver operating characteristic
curves were generated, and the area under the curve (AUC),
sensitivity and specificity were calculated to evaluate the
diagnostic values of candidate miRNAs and mRNA of targeted genes.
Correlation analysis was performed using a Pearson correlation
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
miRNA expression analysis
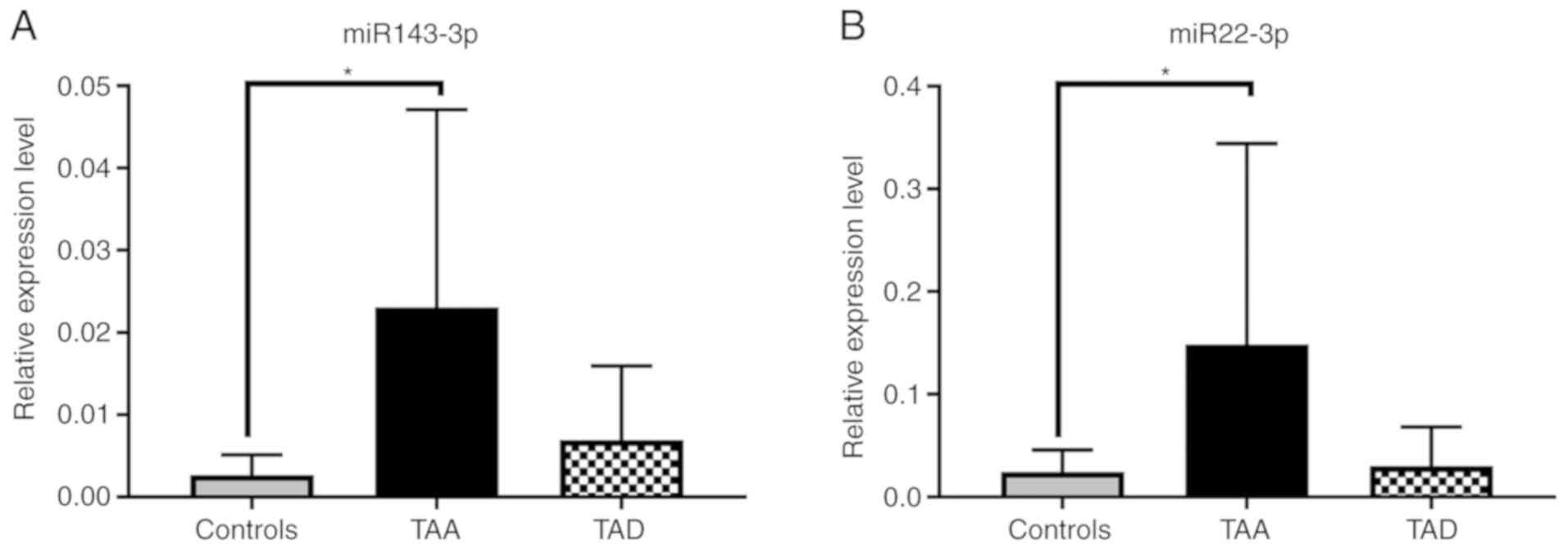
The expression level of hsa-miR-143-3p increased in
the TAA and TAD groups compared with the control group. While this
increase was statistically significant in the TAA group when
compared with the control group (P<0.05), there was no
significant difference observed in the TAD group compared with the
control group. The expression level of hsa-miR-22-3p increased in
the two experimental groups when compared with the control group.
While this increase was statistically significant in TAA group when
compared with the control group (P<0.05), no significant
difference was observed in the TAD group when compared with the
control group (Fig. 1).
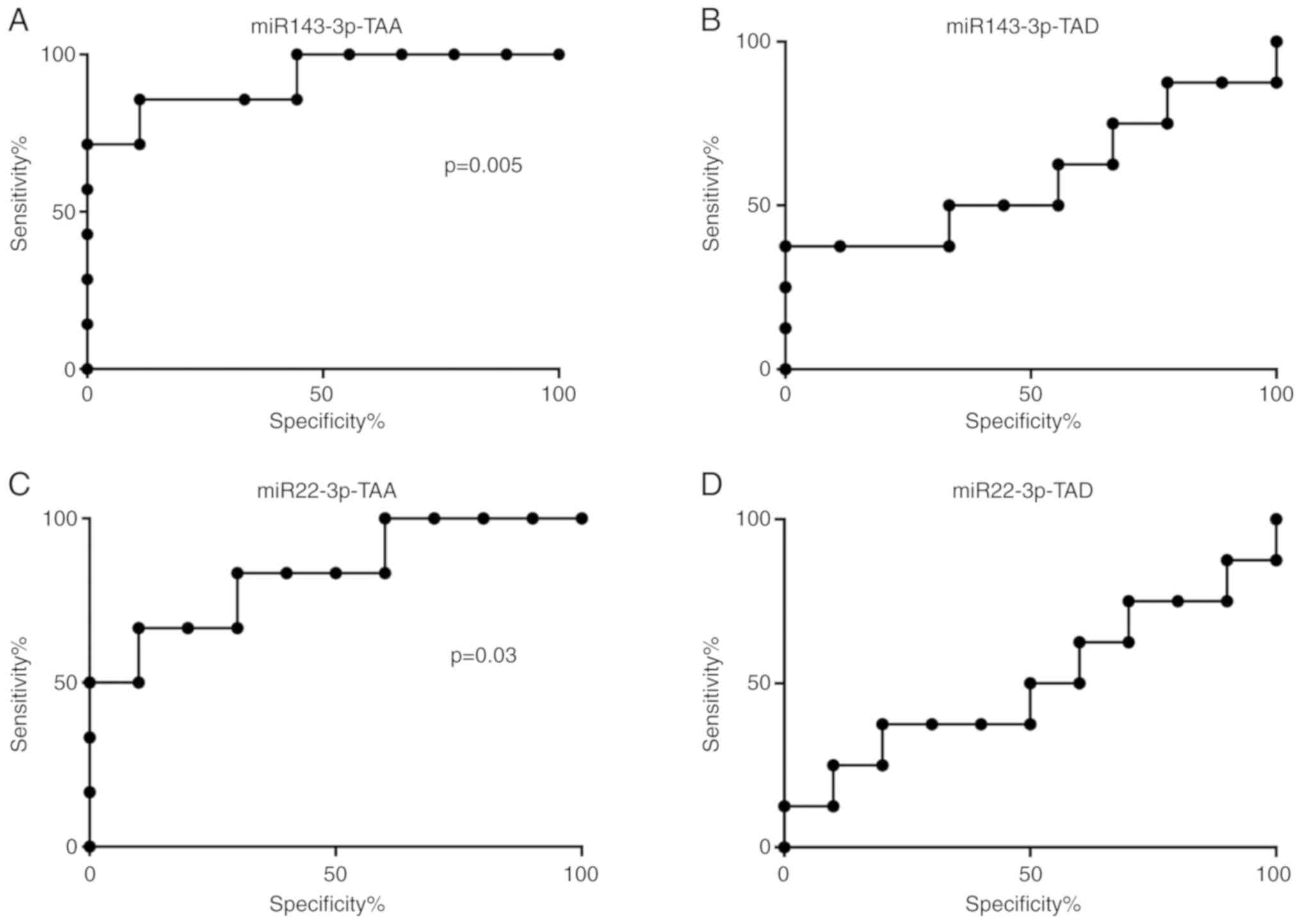
Hsa-miR-143-p and hsa-miR-22-3p exhibited a
diagnostic value in differentiating patients with TAA from healthy
volunteers, with AUC values of 0.92 and 0.80, respectively.
Hsa-miR-143-p with AUC values of 0.60 exhibited diagnostic value
when differentiating patients with TAD from healthy volunteers. The
present study also demonstrated that the AUC values of
hsa-miR-22-2p were not useful as a diagnostic tool when
differentiating patients with TAD from healthy volunteers. The
receiver operating characteristic (ROC) curves analysis of this
miRNA revealed that the AUC was 0.50 (Fig. 2).
Determination of target genes
Tarbase, MirTarbase, TargetScan, MiRNAorg, PITA,
PicTar and MirDB were selected as target prediction tools. These
tools demonstrated that hsa-miR-143-3p and hsa-miR-22 may target
certain associated genes, including KRAS, MAPK7, MAPK14 and TAGLN.
As a result of comparing the different computer programs and
databases, MAPK7, MAPK14 and TAGLN were predicted by at least five
of the seven algorithms, and KRAS was predicted by six of the
algorithms (data not shown).
mRNA expression analysis
The expression levels of KRAS and MAPK7 target mRNAs
were decreased in the TAA and TAD groups, but this was not
statistically significant (P>0.05). The expression of MAPK14
mRNA was increased in the TAA group, but decreased in the TAD
groups, however, this increase and decrease was not statistically
significant. The level of expression of TAGLN was increased in each
of the patient groups, but was not significant. However, the
expression levels of TAGLN were 40-fold higher in the TAD group
compared with the control group (Fig.
3).
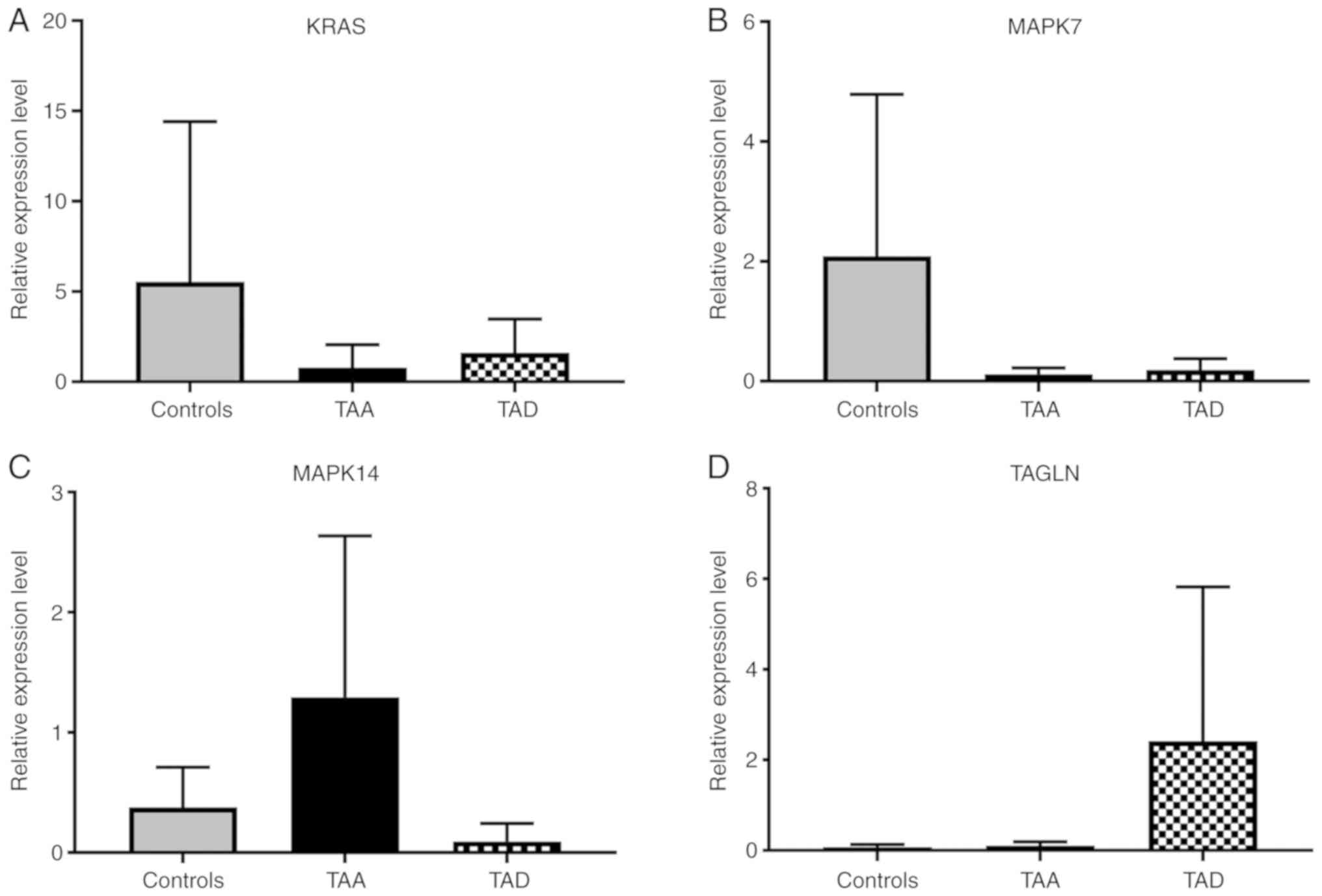 | Figure 3.Expression levels of genes targeted by
hsa-miR-143-3p and hsa-miR-22 in TAA, TAD and control cells. (A)
Expression levels of KRAS, (B) MAPK7, (C) MAPK14 and (D) TAGLN.
Values are presented as the mean ± standard error of the mean. ‘n’
values 10 (controls), 9 (TAA), 9 (TAD). TAA, thoracic aortic
aneurysm; TAD, thoracic aortic dissection; KRAS, Kirsten rat
sarcoma viral oncogene homolog; MAPK, mitogen-activated protein
kinase; TAGLN, transgelin. |
The ROC curves analysis of these mRNAs demonstrated
(Fig. 4) that MAPK14 exhibited
diagnostic value in differentiating each of the patient groups from
the healthy volunteers, with AUC values of 0.7 and 0.8 in the TAA
and TAD groups, respectively. TAGLN with AUC values of 0.8
exhibited diagnostic value when differentiating patients with TAD
from healthy volunteers.
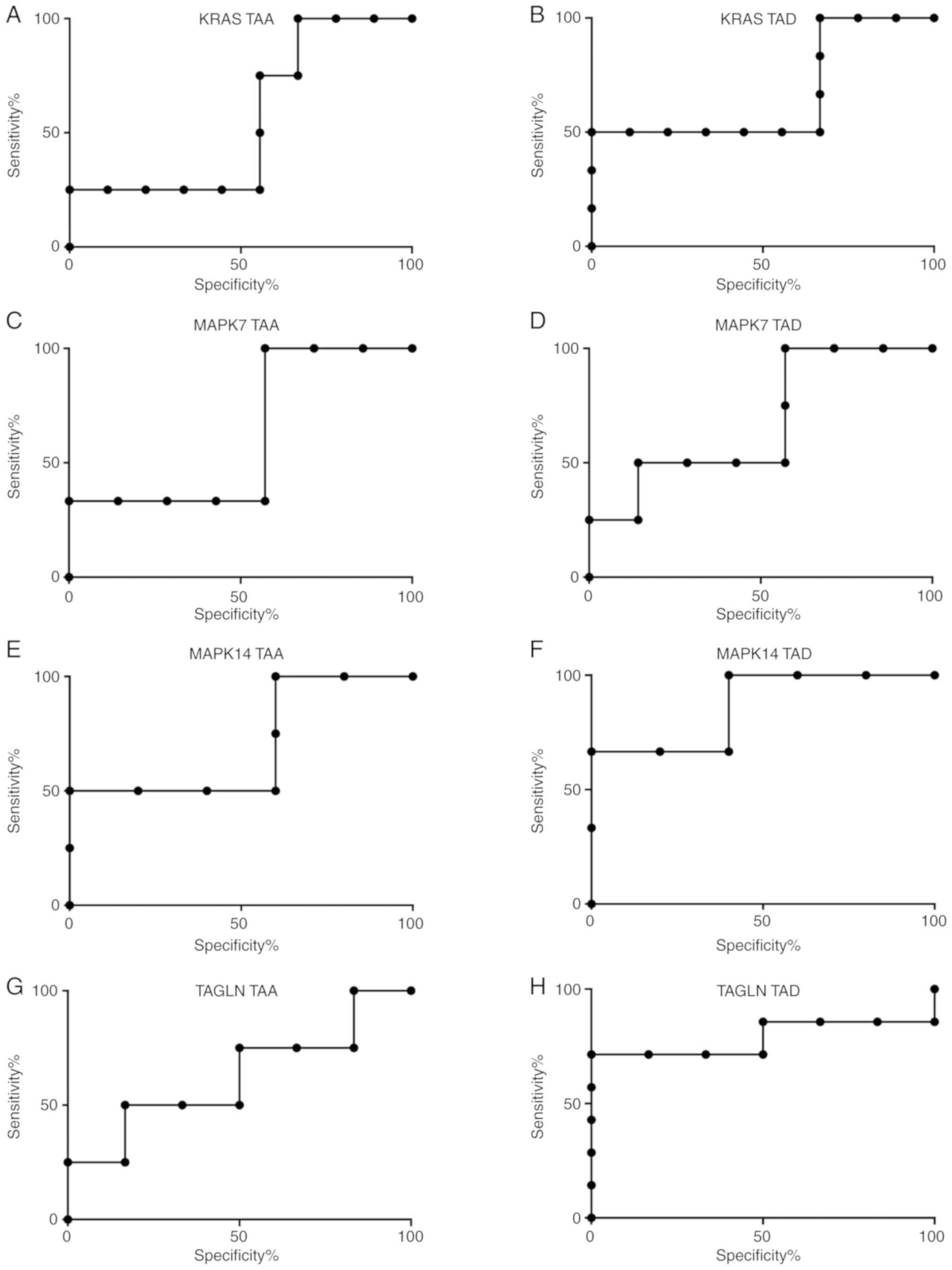 | Figure 4.ROC analysis of the target genes. ROC
analysis of (A) KRAS in the TAA group, (B) KRAS in the TAD group,
(C) MAPK7 in the TAA group, (D) MAPK7 in the TAD group, (E) MAPK14
in the TAA group, (F) MAPK14 in the TAD group, (G) TAGLN in the TAA
group and (H) TAGLN in the TAD group. TAA, thoracic aortic
aneurysm; TAD, thoracic aortic dissection; ROC, receiver operating
characteristic; KRAS, Kirsten rat sarcoma viral oncogene homolog;
MAPK, mitogen-activated protein kinase; TAGLN, transgelin. |
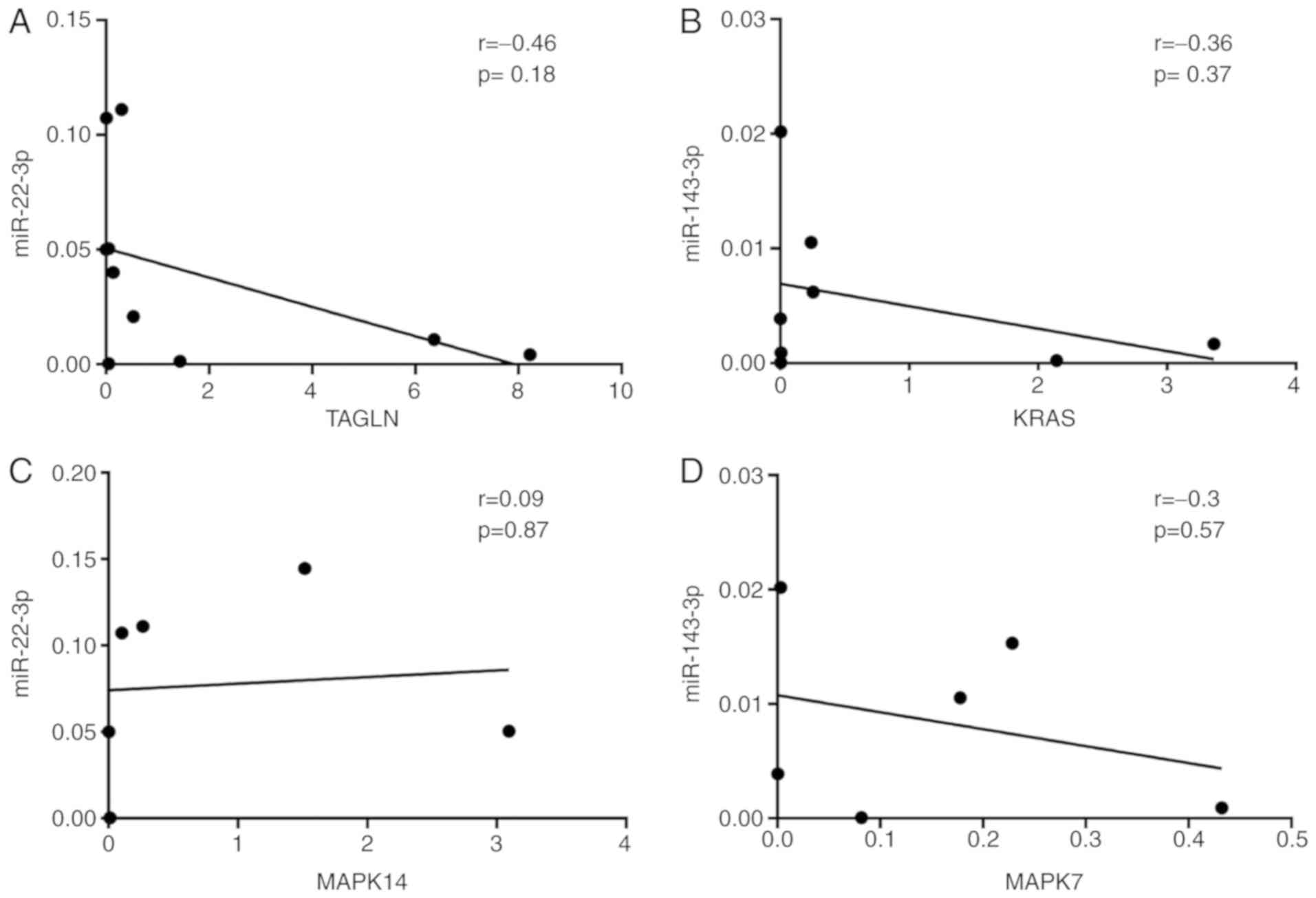
Correlation analysis between miRNA and
target genes
The present study investigated whether miR levels
were correlated with their respective mRNA targets. The positive
and negative correlations are summarized in Fig. 5. A negative correlation between hsa
miR-22-3p and TAGLN (r2=−0.46; P=0.18) and positive
correlation between hsa-miR-22-3p and MAPK14 (r2=0.09;
P=0.087) was observed. A negative correlation was observed between
hsa-miR-143-3p and KRAS (r2=−0.36; P=0.37) and MAPK7
(r2=−0.3; P=0.57). All correlation analysis results were
not statistically significant (P>0.05).
Discussion
TAA and TAD are the most common aortic diseases to
be diagnosed following an extended period of subclinical
development or presentation of an acute complication (16). TAA, which usually progresses in an
asymptomatic manner, is not noticeable and can result in
dissection, rupture or mortality if left untreated (1). According to Kent (17), the majority of patients with aortic
rupture are unlikely to survive, so it is necessary to identify the
aneurysms prior to this stage being reached. The limitations in the
diagnosis and treatment options currently available increase the
necessity of investigating clinically important biomarkers for the
early diagnosis of the disease with a noninvasive approach
(18). TAD is a rare but
life-threatening condition with a lethality rate of 1–2% per hour
following the onset of symptoms in untreated patients. Therefore,
prompt and proper diagnoses are vital in order to increase the
patient's chance of survival and to prevent grievous complications
(19).
miRNAs are small non-coding RNA molecules that are
involved in the regulation of target mRNA molecules and are
detectable in tissue, blood or urine samples. One previous
characterization study confirmed that circulating miRNAs are stable
in plasma and serum (20). The
miRNA expression profile was evaluated in the clinical patient
samples due to their function in the pathogenesis of TAD and
TAA.
The aim of the present study was, first, to
determine the differences in the expression of hsa-miR-143-3p and
hsa-miR-22-3p in serum levels of TAA (n=9) and TAD (n=9) groups
compared with healthy controls (n=10). Secondly, the aim was to
determine those mRNAs targeted by candidate miRNAs with
network-based bioinformatics programs and algorithms. Thirdly, to
reveal the changes in the expression levels of the mRNAs, detected
by qPCR. The validation of hsa-miR-143-3p and hsa-miR-22-3p, which
were selected as candidate miRNA following microarray studies in
TAA and TAD groups (4,21,22),
were performed in the present study.
Hsa-miR-143-3p is a highly expressed miRNA in VSMC.
The gene encoding hsa-miR-143-3p, which is localized in the 5q32
region of the human genome, is highly conserved among species
(23). It has been demonstrated
that they serve important functions in the phenotypic change of
VSMCs and in regulating the pathogenesis of vascular diseases
(24). Its functions include
targeting numerous transcription factors, including Kruppel-like
factor-4l, myocardin and ETS Transcription Factor, and induces the
differentiation of VSMCs and suppresses their proliferation
(21). According to Climent et
al (25), hsa-miR-143-3p
regulates the vessel stabilization properties of endothelial cells
by functioning as a communication molecule between SMCs and
endothelial cells. Hsa-miR-143-3p inhibits cell proliferation by
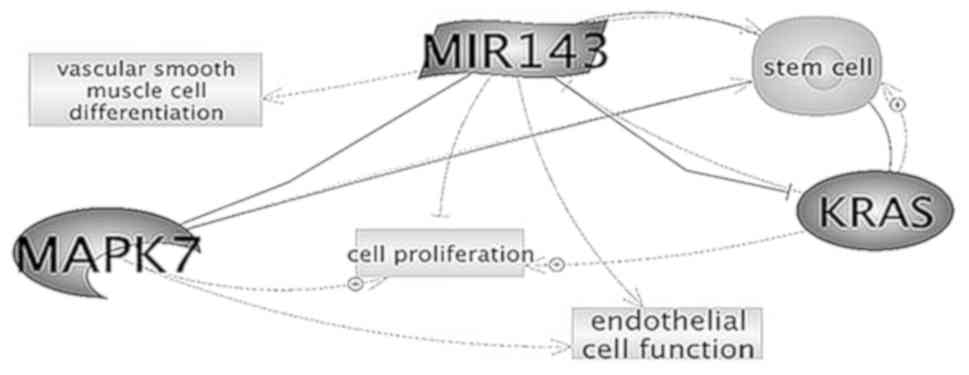
targeting KRAS and extracellular signal regulated kinase (ERK)5
directly (Fig. 6) (26).
In the present study, the expression levels of KRAS
mRNA, which is the target gene of hsa-miR-143-3p, decreased by
7-fold in the TAA group and 3.5-fold in the TAD group when compared
with the control group. Wang et al (27) demonstrated that KRAS is a target of
miR-143. The KRAS/ERK pathway is one of the most important pathways
associated with the proliferation of VSMCs. The accumulation of
VSMCs migrating from media to the intima in the aortic wall and
resulting in proliferation results in vascular remodelling
(28). VSMC proliferation is
necessary for diseases characterized by ballooning in the aortic
structure (29). Hsa-miR-143-3p
regulates VSMC proliferation by inhibiting KRAS. The increased
expression level of hsa-miR-143-3p results in a decrease in KRAS
expression and VSMC proliferation. These results may indicate that
hsa-miR-143-3p is a regulator of VSMC proliferation with KRAS
(28).
It was observed, in the present study, that the
expression level of MAPK7, which was determined and validated as a
target of hsa-mir-143-3p via bioinformatics tools, decreased
17-fold in the TAA group and 11-fold in the TAD group when compared
with the control group. Osaki et al (10) demonstrated that MAPK7 is the target
of hsa-miR-143-3p, while Roberts et al (30) revealed that endothelial cells
increased the survival rate of VEGF-mediated MAPK7 (ERK5).
Decreased MAPK7 expression increased the apoptosis rate of VSMC and
endothelial cells, causing functional changes in the aortic wall
structure. Modifications of endothelial cells may induce
degradation of aortic wall structure by affecting VSMCs (30).
The level of expression of another candidate miRNA,
hsa-miR-22-3p, was observed to be significantly higher compared
with the control group, with a 6-fold increase in the TAA group,
but no statistically significant difference in the TAD group, with
a 1.5-fold increase, compared with the control group. Hsa-miR-22-3p
is localized on the long arm of chromosome 17 in region p13.3 of
the human genome according to the National Center for Biotechnology
Information. According to Liao et al (4), the decrease in the expression levels
of hsa-miR-22-3p were statistically significant in patients with
TAD when investigated via microarray and qPCR processes. Patuzzo
et al (21) demonstrated
that the expression level of hsa-miR-22-3p decreased by 1.5-fold,
but this decrease was not statistically significant. Hsa-miR-22-3p
has been indicated to serve a function as a regulator during stem
cell VSMC differentiation, but little is currently known about
target genes and their associated pathways in mature VSMC phenotype
modification or its clinical effects (31); it has also been reported to
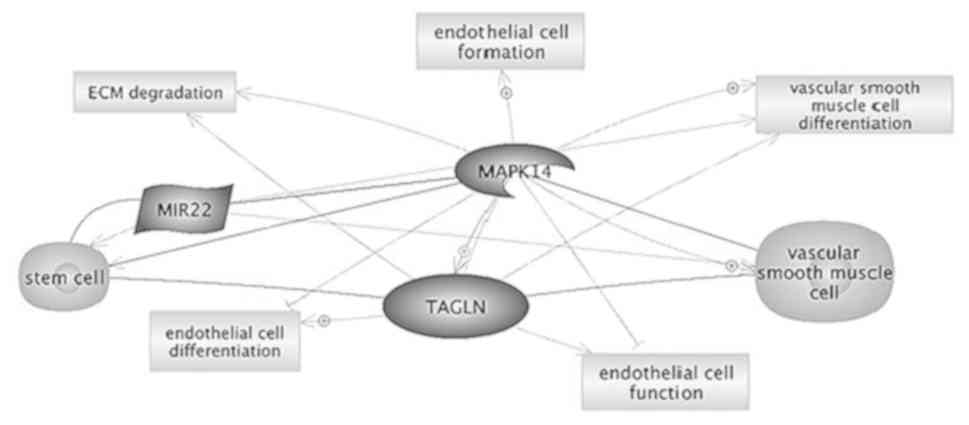
regulate apoptosis in cardiomyocytes by targeting MAPK14 (Fig. 7) (31).
As a result of the relative comparison, MAPK14
expression was increased 3-fold in the TAA group, and decreased
4-fold in the TAD group. The ROC analysis results for the MAPK14
gene revealed AUC values of 0.7 and 0.8 in the TAA and TAD groups,
respectively.
A statistically significant increase in
hsa-miR-22-3p may result in the decreased expression of the MAPK14
gene in the TAA group. As a result of the ROC analysis, high AUC
values obtained in the TAD and TAA groups indicated that
hsa-miR-22-3p and target mRNA MAPK14 may be an important biomarkers
for clinical tests. Decreases in MAPK14 mRNA expression in patients
with TAA may result in aneurysms due to the decreased VSMC
contractile phenotype. However, another study performed by the same
study group revealed that if p38MAPK suppressed the dominant
isoform MAPK14, the markers including VSMCs and TAGLN in the
contractile phenotype increased (32). Increased levels of MAPK14 mRNA
expression in the TAD group may result in a decreased VSMC
contractile phenotype. Considering that the TAD group did not
include patients with aneurysms, MAPK14 may be an important
biomarker in distinguishing between patients with TAA and those
with TAD due to the difference in expression levels of this gene
between these patient groups.
TAGLN gene expression level increased 1.5- and
40.0-fold in the TAA and TAD groups, respectively, in the present
study. As a result of the ROC analysis of the TAGLN target gene,
the AUC values were calculated as 0.6 and 0.8 in the TAA and TAD
groups, respectively.
TAGLN is an early distinguishing marker of smooth
muscle progenitor cell differentiation, and low levels of the TAGLN
protein have been detected in non-differentiated multipotent access
progenitor cells. Increased expression of TAGLN in mesenchymal stem
cells is considered as evidence that mesenchymal stem cells differ
in an SMC phenotype (33). It has
been demonstrated that the overexpression of miR-22 in stem cells
promotes SMC differentiation in vivo (33). All of these studies indicate that
the activation of the injured tissue, and increase in the amount of
TAGLN mRNA through inducing the differentiation of VSMCs in stem
cells, causes the increased expression of hsa-miR-22-3p.
In the scope of the present study, investigating
fresh serum samples obtained from patient groups is necessary in
order to reflect the real situation in terms of identifying
biomarkers that represent the pathophysiology of the diseases in
question. The results from the present study revealed that the
increase in expression levels of hsa-miR-143-3p and hsa-miR-22
candidate miRNAs were statistically significantly higher in the TAA
group compared with the control group. Therefore, it was
hypothesized that extracellular miRNA originating from SMC,
endothelial cells or fibroblasts may result in endothelial cell
dysfunction and/or aberrant vascular remodeling, which may initiate
the intimal or media tear in aortic diseases including TAA. The
expression of KRAS and MAPK7 mRNAs decreased by 7- and 17-fold in
the TAA group, respectively. Therefore, it may be suggested that,
at the molecular level, hsa-miR-143-3p regulates VSMC proliferation
and endothelial cell survival rate by targeting KRAS and MAPK7.
Notably, the expression level of MAPK14 decreased in the TAD group
while it increased in the TAA group. The results of the MAPK14 ROC
analysis demonstrated that MAPK14 may be an important biomarker in
the differentiation between TAA and TAD groups.
In conclusion, the results obtained in the present
study may form a basis for further research into physiological
studies to reflect the associations between hsa-miR-143-3p and
hsa-miR-22-3p in TAA as biomarkers, and to target mRNA-miRNA
associations. The main limitation of the present study was the
small sample size, which was partly as a result of the scarcity of
the diseases in question, and partly due to limited funding. Future
studies should strive to include a large number of TAA and TAD
samples to confirm these results. Another limitation is that the
mean age is not similar in the TAA and TAD groups. One young
patient (aged 22) in the TAD group is the reason for this
difference, but conventional cardiovascular risk factors develop
with advancing age and may attenuate the impact of polymorphism
(34). Therefore, the present
study did not include patients >65 years old in the TAA and
control groups. For patients with TAD, the only exclusion criteria
was presence of connective tissue disorders and syndromic diseases.
Another minor limitation of the present study may be that the
direct association between miRNA and the target genes was not
demonstrated in an in vitro experiment. However, the aim was
to validate the candidate miRNAs and their target genes. An in
vitro study examining the direct association between miRNA and
its target genes may be conducted in future studies. Additionally,
although the result of the correlation analysis were not
statistically significant, in light of this evidence there may
still be an important association between the candidate miRNAs
(hsa-miR-143-3p and hsa-miR-22-2p) and their target mRNAs (KRAS,
MAPK7, MAPK14 and TAGLN). In addition, other experimental
techniques including protein expression analysis by western
blotting and in vivo/in vitro experiments on miRNA mimics
may be used to validate the results of the present study obtained
from gene expression analysis by RT-qPCR. Further research will
strengthen these results and further determine the diagnostic and
prognostic value of miRNAs as biomarkers in aortic diseases, and
should investigate their function as a therapeutic target.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Istanbul
University Scientific Research Projects Department (grant no.
27359).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are not publicly available due to the datasets
containing clinical data, and the authors therefore have an ethical
and legal responsibility to respect the participants' rights to
privacy and to protect their identity, but are available from the
corresponding author upon reasonable request.
Authors' contributions
TS, TG and AA conceived and designed the study. TS
and TG performed the experiments. AA was involved in the clinical
study and preparation of the manuscript. TG and TS reviewed the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Istanbul University (Istanbul, Turkey; project no.
329785), and all patients provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Milewicz DM, Prakash SK and Ramirez F:
Therapeutics targeting drivers of thoracic aortic aneurysms and
acute aortic dissections: Insights from predisposing genes and
mouse models. Annu Rev Med. 68:51–67. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Howard DP, Banerjee A, Fairhead JF,
Perkins J, Silver LE and Rothwell PM; Oxford Vascular Study, :
Population-based study of incidence and outcome of acute aortic
dissection and premorbid risk factor control: 10-year results from
the Oxford Vascular Study. Circulation. 127:2031–2037. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martufi G, Gasser TC, Appoo JJ and Di
Martino ES: Mechano-biology in the thoracic aortic aneurysm: A
review and case study. Biomech Model Mechanobiol. 13:917–928. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liao M, Zou S, Weng J, Hou L, Yang L, Zhao
Z, Bao J and Jing Z: A microRNA profile comparison between thoracic
aortic dissection and normal thoracic aorta indicates the potential
role of microRNAs in contributing to thoracic aortic dissection
pathogenesis. J Vasc Surg. 53:1341–1349.e3. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ikonomidis JS, Ivey CR, Wheeler JB,
Akerman AW, Rice A, Patel RK, Stroud RE, Shah AA, Hughes CG,
Ferrari G, et al: Plasma biomarkers for distinguishing etiologic
subtypes of thoracic aortic aneurysm disease. J Thorac Cardiovasc
Surg. 145:1326–1333. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu R, Lo L, Lay AJ, Zhao Y, Ting KK,
Robertson EN, Sherrah AG, Jarrah S, Li H, Zhou Z, et al: ARHGAP18
protects against thoracic aortic aneurysm formation by mitigating
the synthetic and proinflammatory smooth muscle cell phenotype.
Circ Res. 121:512–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Boon RA, Seeger T, Heydt S, Fischer A,
Hergenreider E, Horrevoets AJ, Vinciguerra M, Rosenthal N, Sciacca
S, Pilato M, et al: MicroRNA-29 in aortic dilation: Implications
for aneurysm formation. Circ Res. 109:1115–1119. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Moushi A, Michailidou K, Soteriou M,
Cariolou M and Bashiardes E: MicroRNAs as possible biomarkers for
screening of aortic aneurysms: A systematic review and validation
study. Biomarkers. 23:253–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Wu B, Dong L, Wang C, Wang X and
Shu X: Circulating matrix metalloproteinase patterns in association
with aortic dilatation in bicuspid aortic valve patients with
isolated severe aortic stenosis. Heart Vessels. 31:189–197. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kim HW and Stansfield BK: Genetic and
epigenetic regulation of aortic aneurysms. Biomed Res Int.
2017:72685212017.PubMed/NCBI
|
|
12
|
Jancík S, Drábek J, Radzioch D and Hajdúch
M: Clinical relevance of KRAS in human cancers. J Biomed
Biotechnol. 2010:1509602010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li Y and Maegdefessel L: Non-coding RNA
contribution to thoracic and abdominal aortic aneurysm disease
development and progression. Front Physiol. 8:4292017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ignatieva E, Kostina D, Irtyuga O,
Uspensky V, Golovkin A, Gavriliuk N, Moiseeva O, Kostareva A and
Malashicheva A: Mechanisms of smooth muscle cell differentiation
are distinctly altered in thoracic aortic aneurysms associated with
bicuspid or tricuspid aortic valves. Front Physiol. 8:5362017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Erbel R, Aboyans V, Boileau C, Bossone E,
Bartolomeo RD, Eggebrecht H, Evangelista A, Falk V, Frank H,
Gaemperli O, et al: 2014 ESC Guidelines on the diagnosis and
treatment of aortic diseases: Document covering acute and chronic
aortic diseases of the thoracic and abdominal aorta of the adult.
The Task Force for the Diagnosis and Treatment of Aortic Diseases
of the European Society of Cardiology (ESC). Eur Heart J.
35:2873–2926. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kent KC: Clinical practice. Abdominal
aortic aneurysms. N Engl J Med. 371:2101–2108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Burillo E, Lindholt JS, Molina-Sánchez P,
Jorge I, Martinez-Pinna R, Blanco-Colio LM, Tarin C, Torres-Fonseca
MM, Esteban M, Laustsen J, et al: ApoA-I/HDL-C levels are inversely
associated with abdominal aortic aneurysm progression. Thromb
Haemost. 113:1335–1346. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gawinecka J, Schönrath F and von
Eckardstein A: Acute aortic dissection: Pathogenesis, risk factors
and diagnosis. Swiss Med Wkly. 147:w144892017.PubMed/NCBI
|
|
20
|
Turchinovich A, Weiz L, Langheinz A and
Burwinkel B: Characterization of extracellular circulating
microRNA. Nucleic Acids Res. 39:7223–7233. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Patuzzo C, Pasquali A, Malerba G, Trabetti
E, Pignatti P, Tessari M and Faggian G: Preliminary microRNA
analysis of non syndromic thoracic aortic aneurysms. Balkan J Med
Genet. 15 (Suppl):S51–S55. 2012. View Article : Google Scholar
|
|
22
|
Licholai S, Blaż M, Kapelak B and Sanak M:
Unbiased profile of MicroRNA expression in ascending aortic
aneurysm tissue appoints molecular pathways contributing to the
pathology. Ann Thorac Surg. 102:1245–1252. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rangrez AY, Massy ZA, Metzinger-Le Meuth V
and Metzinger L: miR-143 and miR-145: Molecular keys to switch the
phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet.
4:197–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Climent M, Quintavalle M, Miragoli M, Chen
J, Condorelli G and Elia L: TGFβ triggers miR-143/145 transfer from
smooth muscle cells to endothelial cells, thereby modulating vessel
stabilization. Circ Res. 116:1753–1764. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Song B and Ju J: Impact of miRNAs in
gastrointestinal cancer diagnosis and prognosis. Expert Rev Mol
Med. 12:e332010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Lu L and Huang K: miR-143 inhibits
osteosarcoma cell proliferation by downregulating K-ras expression.
Int J Clin Exp Pathol. 9:3436–3441. 2016.
|
|
28
|
Yu ML, Wang JF, Wang GK, You XH, Zhao XX,
Jing Q and Qin YW: Vascular smooth muscle cell proliferation is
influenced by let-7d microRNA and its interaction with KRAS. Circ
J. 75:703–709. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Inoue T and Node K: Molecular basis of
restenosis and novel issues of drug-eluting stents. Circ J.
73:615–621. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roberts OL, Holmes K, Müller J, Cross DA
and Cross MJ: ERK5 is required for VEGF-mediated survival and
tubular morphogenesis of primary human microvascular endothelial
cells. J Cell Sci. 123:3189–3200. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang F, Chen Q, He S, Yang M, Maguire EM,
An W, Afzal TA, Luong LA, Zhang L and Xiao Q: miR-22 is a novel
mediator of vascular smooth muscle cell phenotypic modulation and
neointima formation. Circulation. 137:1824–1841. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Long X, Cowan SL and Miano JM:
Mitogen-activated protein kinase 14 is a novel negative regulatory
switch for the vascular smooth muscle cell contractile gene
program. Arterioscler Thromb Vasc Biol. 33:378–386. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao H, Wen G, Huang Y, Yu X, Chen Q,
Afzal TA, Luong le A, Zhu J, Ye S, Zhang L and Xiao Q: MicroRNA-22
regulates smooth muscle cell differentiation from stem cells by
targeting methyl CpG-binding protein 2. Arterioscler Thromb Vasc
Biol. 35:918–929. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dhingra R and Vasan RS: Age as a risk
factor. Med Clin North Am. 96:87–91. 2012. View Article : Google Scholar : PubMed/NCBI
|