Introduction
Rheumatoid arthritis (RA) is a chronic autoimmune
inflammatory response in the synovial tissue of joints, which
affects ~1% of population worldwide (1). However, the exact cause remains
unclear (2). RA is characterized
by a progressive erosion of the articular cartilage and chronic
inflammation of the synovial joint (3). It has been reported that oxidative
stress, inflammation and lipid peroxidation may affect the
progression of RA (4). RA, which
is also a common cause of permanent disability, leads to increased
medical cost, use of social services and a reduction in the quality
of life of patients (5), as the
disease may lead to joint stiffness, swelling, pain and muscle
wasting. A previous study indicated that muscle wasting was an
important cause of RA-associated morbidity and mortality (6). However, the genes regulating the
proliferation of skeletal muscle in RA require additional
study.
MicroRNAs (miRNAs/miRs), are a type of non-coding
RNA (21–24 nucleotides in length) that post-transcriptionally
regulate gene expression (7).
Aberrant expression of miRNAs has been demonstrated to be involved
in several diseases, including various types of cancer, and
rheumatic and other autoimmune diseases (8–10).
Previous studies have reported that imbalances in miR-146a,
miR-155, miR-223 and miR-16 levels existed in the immune cells of
patients with RA (11–14). Lai et al found that
decreased expression of miR-760 was affected by tumor necrosis
factor-a in patients with RA (7).
In addition, it has been reported that miR-760 regulated the
proliferation and apoptosis of a variety of cells, such as human
pulmonary artery smooth muscle cells, and human colon cancer and
ovarian cancer cells, by targeting specific genes (15–17).
However, there are few studies regarding the regulatory mechanisms
via which miR-760 affects skeletal muscle proliferation in patients
with RA, to the best of our knowledge.
Skeletal muscles are primarily composed of different
types of fibers (18). Myoblasts
are precursor cells, which are hypothesized to serve an important
role in injured skeletal muscle (19). Satellite cells mobilize and
proliferate as myoblasts when the muscle needs to be repaired
and/or remodeled (20). Myosin-18b
(Myo18b) is a myosin protein associated with human tumor
progression, and loss-of-function mutations of Myo18b have been
found in certain patients with nemaline myopathy (21). Notably, according to Berger et
al (22), a number of
myopathies are associated with molecular defects in sarcomeres, and
a complete loss of Myo18b function leads to a complete lack of
sarcomeric structure, suggesting that Myo18b may serve an important
role in sarcomere assembly. In the present study, it was
demonstrated that certain miRNAs were differentially expressed in
the tarsus joint of a collagen-induced RA mouse model using the
Gene Expression Omnibus (GEO) database.
Based on these previous results, factors affecting
the proliferation of myoblasts were assessed. Mueller et al
(23) hypothesized that chronic
systemic inflammation may negatively affect myonuclei number and
the regenerative potential of satellite cells; however, this
hypothesis has not been confirmed. Therefore, additional factors
may affect proliferation, differentiation and thus, muscle
strength. In the present study, the targeting effects of miR-760 on
Myo18b were determined, and it was hypothesized that miR-760
regulated skeletal muscle proliferation in RA by targeting
Myo18b.
Materials and methods
Identification of differentially
expressed genes
GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used on
the GEO dataset (24,25), GSE61140 (26), to obtain the gene expression
profile in the tarsus joint of a collagen-induced RA mouse
model.
Cell culture
Mouse C2C12 myoblasts were obtained from the
American Type Culture Collection and cultured in DMEM (Gibco;
Thermo Fisher Scientific, Inc.) containing 1%
penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.) and
10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in an incubator
with 5% CO2 at 37°C. The cells were passaged using 0.25%
trypsin (Gibco; Thermo Fisher Scientific, Inc.) when the confluence
reached 50%. Subsequently, the cells were moved to a
differentiation medium containing DMEM with 2% horse serum (Gibco;
Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin when
the cells reached 75–80% confluence, and then myogenic
differentiation was induced via the addition of differentiation
medium. The media was replaced every 24 h.
Cell transfection
A total of 1×105 C2C12 myoblasts were
cultured in 6-well plates, and divided into 4 groups. The first
group consisted of: Control cells, untreated cells; negative
control (NC) cells, cells transfected with NC siRNA (sense,
CAUGUGGUCUGUCGCAUAAUA and antisense, CGGUACACCAGACAGCGUAUU); and
small interfering (si)Myo18b cells, cells transfected with siMyo18b
(sense, 5′-GAGCCAAAGAACAAAUAAAUU-3′ and antisense,
3′-UUCUCGGUUUCUUGUUUAUUU-5′). The second group consisted of:
Control cells; mock cells, cells transfected with pcDNA3.1 (+)
(GenomeDitech Co., Ltd.) only; and Myo18b cells, cells transfected
with Myo18b-pcDNA3.1 (+) (GenomeDitech Co., Ltd.). The third group
consisted of: NC cells; siMyo18b cells; mock cells and Myo18b
cells. The fourth group consisted: Mimics control cells, cells
transfected with mimics control (forward,
5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′); miR-760 mimics cells, cells
transfected with miR-760 mimics (forward,
5′-CGGCUCUGGGUCUGUGGGGA-3′ and reverse, 5′-UCCCACAGACCCAGAGCCG-3′);
inhibitors control cells, cells transfected with inhibitors control
(5′-CAGUACUUUUGUGUAGUACAA-3′); miR-760 inhibitors cells, cells
transfected with miR-760 inhibitors (5′-TCCCCACAGACCCAGAGCCG). The
siMyo18b, NC siRNA, miR-760 mimics, miR-760 inhibitors, mimic
control and inhibitor control were purchased from Shanghai
GenePharma Co., Ltd.; transfection with siRNAs (50 nM), mimics (50
nM) and inhibitors (50 nM) transfection was performed using
Lipofectamine® 2000 kit (Invitrogen; Thermo Fisher
Scientific, Inc.). The transfected cells were cultured in DMEM at
37°C with 5% CO2 for another 48 h prior to subsequent
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
RT-qPCR analysis was performed to determine the
expression levels of Myo18b, miR-760, cyclin-dependent kinase 2
(CDK2), cyclin D1, matrix metalloproteinase (MMP)-2, MMP-9,
myogenin (MyOG) and myosin heavy chain IId/x (MyH6) in C2C12
myoblasts. To determine the expression of Myo18b during the
myogenic differentiation of C2C12 myoblasts, cells were harvested
after 0, 1, 3, 5 or 7 days. Relative expression levels of Myo18b,
miR-760, CDK2, cyclin D1, MMP-2, MMP-9, MyOG and MyH6 in the C2C12
myoblasts were detected after 48 h of transfection with control,
NC, siMyo18b, mock, Myo18b, mimics control, miR-760 mimics,
inhibitor control and miR-760 inhibitor. Total RNA was extracted
from C2C12 myoblasts using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). The RNAs were reverse transcribed
to cDNAs using 1 µg RNA and a High Capacity cDNA Reverse
Transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.), the reverse transcription reaction conditions were set to
37°C for 30 min, and reverse transcriptase inactivation was
conducted at 85°C for 5 min. Subsequently, the cDNA products were
used for qPCR using a Light Cycler 480 RT-PCR (Roche Diagnostics)
and SYBR Green Master mix (Roche Diagnostics) in a final volume of
20 µl/well. The thermocycling conditions for miR-760 were: 95°C for
15 min, followed by 40 cycles of 94°C for 15 sec, 55°C for 30 sec
and 70°C for 30 sec. Relative expression of miR-760 was normalized
to U6 expression levels. SYBR Green on the 7500 qPCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to
determine the relative expression levels of Myo18b, CDK2, cyclin
D1, MMP-2, MMP-9, MyOG and MyH6. The thermocycling conditions were
as follows: Initial denaturation at 94°C for 5 min, followed by 40
cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec.
GAPDH was used as an internal reference gene, and relative
expression was calculated using the 2−ΔΔCq method
(27). The sequences of the
primers used are listed in Table
I.
 | Table I.Sequences of primers used for
quantitative PCR. |
Table I.
Sequences of primers used for
quantitative PCR.
| Gene | Forward, 5′-3′ | Reverse, 5′-3′ |
|---|
| Myo18b |
GGAAGCAGTTAGCTGTCGC |
TTGACTGGTCGTCCTGAGAGA |
| U6 |
GCTTCGGCAGCACATATACTAA |
AACGCTTCACGAATTTGCGT |
| miR-760 |
GGCTCTGGGTCTGTGGG |
GAACATGTCTGCGTATCTC |
| CDK2 |
TCATGGATGCCTCTGCTCTCAC |
TGAAGGACACGGTGAGAATGGC |
| Cyclin D1 |
TGTTCGTGGCCTCTAAGATGAAG |
GGAAGTGTTCGATGAAATCGTG |
| MMP-2 |
CAAGGATGGACTCCTGGCACAT |
TACTCGCCATCAGCGTTCCCAT |
| MMP-9 |
GCTGACTACGATAAGGACGGCA |
CCCTCAGAGAATCGCCAGTACT |
| MyOG |
CCATCCAGTACATTGAGCGCCT |
CTGTGGGAGTTGCATTCACTGG |
| MyH6 |
GCTGGAAGATGAGTGCTCAGAG |
CCAGCCATCTCCTCTGTTAGGT |
| GAPDH |
CATCACTGCCACCCAGAAGACTG |
ATGCCAGTGAGCTTCCCGTTCAG |
Western blot analysis
Following transfection, western blotting was used to
determine the protein expression levels of Myo18b, MyOG and MyH6 in
C2C12 myoblasts in the control, NC, siMyo18b, mock, Myo18b, mimics
control, miR-760 mimics, inhibitor control and miR-760 inhibitor
groups. PBS was used to wash C2C12 myoblasts twice. Total proteins
were extracted from C2C12 myoblasts using a protein lysis buffer
(pH 7.4, 150 mmol/l NaCl, 20 mmol/l Tris, 1 mmol/l EGTA, 1 mmol/l
EDTA, 1 mmol/l β-glycerolphosphate, 1% Triton X-100, 2.5 mmol/l
sodium pyrophosphate, 1 µg/ml leupeptin, 1 mmol/l
Na3VO4 and 1 mmol/l phenylmethyanesulfonyl
fluoride). Following lysis, the lysates were incubated on ice for
30 min. The supernatant was collected by centrifuged at 10,000 × g
for 30 min at 4°C. A bicinchoninic acid assay kit (Beijing Solarbio
Science & Technology Co., Ltd.) was used to measure the protein
concentration. Equivalent quantities of proteins (30 µg) were
resolved on a 12% gel using SDS-capillary gel electrophoresis and
transferred onto PVDF membranes (Bio-Rad Laboratories, Inc.), which
were probed with the following primary antibodies after blocking in
5% non-fat milk at room temperature for 2 h: Myo18b (cat. no.
36810002; 1:1,000; Novus Biologicals, LLC); GAPDH (cat. no.
ab181602; 1:10,000; Abcam); MyOG (cat. no. ab77232; 1:1,000;
Abcam); or MyH6 (cat. no. ab185967; 1:1,000; Abcam). Membranes were
incubated with primary antibodies overnight at 4°C, and
subsequently with the goat anti-rabbit immunoglobulin G (HRP)
secondary antibody (cat. no. ab205718; 1:3,000; Abcam) at room
temperature for 2 h. An enhanced chemiluminescence kit (Pierce;
Thermo Fisher Scientific, Inc.) was used to visualize the signal,
and Quantity One software (version 4.62; Bio-Rad Laboratories) was
used to perform densitometry analysis.
Cell proliferation assay
Cells were transfected as aforementioned. The C2C12
myoblasts were divided into 4 groups as follows: NC group, siMyo18b
group, mock group and Myo18b group. To determine proliferation,
2×103 C2C12 myoblasts were seeded into 96-well plates
and cultured in DMEM with 5% CO2 at 37°C. Cell Counting
Kit-8 (CCK-8; Beyotime Institute of Biotechnology) reagent (10
µl/well) was added at 24 and 48 h, and the cells were further
incubated for 3 h at 37°C. Subsequently, the absorbance at 450 nm
was detected using a plate reader (Thermo LabSystems; Thermo Fisher
Scientific, Inc.).
Cell cycle assay
In the cell cycle assay, cells were collected and
fixed in 70% ethanol overnight at 4°C. Subsequently, the cells were
stained in PBS containing propidium iodide and RNase for 30 min at
4°C in the dark. Cell cycle distribution was determined using a
FACSCalibur flow cytometer (BD Biosciences) with CellQuest software
(version 5.1; BD Biosciences).
Wound-healing assay
Following transfection, cells were seeded in a
6-well plate with complete medium (DMEM with 10% FBS). Then, cells
were starved in serum-free complete medium for 6–8 h, and a 200 µl
pipette tip was used to create a straight wound in the cell
monolayers when the cells had reached ~100% confluence. The cells
were washed once with PBS. The scratch was observed and imaged
under a fluorescence microscope (magnification, ×100) at 0 and 24
h, and the wound width was measured. All experiments were performed
in triplicate.
Dual-luciferase reporter assay
The putative binding sites of miR-760 in the
Myo18b-3′-untranslated region (3′-UTR) were analyzed using
TargetScan 7.2 ((http://www.targetscan.org/vert_72/). The site of
interaction between miR-760 and target gene Myo18b-3′-UTR was
verified using a dual-luciferase reporter assay. After amplifying
and purifying Myo18b-3′-UTR and Myo18b-3′-UTR mutant (mut), both
PCR products were cloned into the pGL3 vector (Promega Corporation)
to generate Myo18b-3′-UTR plamids and Myo18b-3′-UTR mut plamids.
The C2C12 myoblasts (2×103 cells/ml) were co-transfected
with Myo18b-3′-UTR/Myo18b-3′-UTR mut (0.5 µg) and miR-760
mimics/control (50 nM) using Lipofectamine 2000, according to the
manufacturer's protocol. Relative luciferase activity was measured
using a Dual-Luciferase Reporter assay kit (Promega Corporation) 48
h of transfection on a fluorescence spectrophotometer (Infinite
M200; Tecan Group, Ltd.) according to the manufacturer's protocol.
Firefly luciferase activity was normalized to Renilla
luciferase activity.
Statistical analysis
All data analysis was performed using SPSS version
20.0 (IBM Corp.) or GraphPad Prism version 7 (GraphPad Software,
Inc.). Data are presented as the mean ± standard deviation. The
aforementioned experiments were independently repeated three times.
A one-way ANOVA followed by a post-hoc Tukey's test was used to
analyze differences among the experimental groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
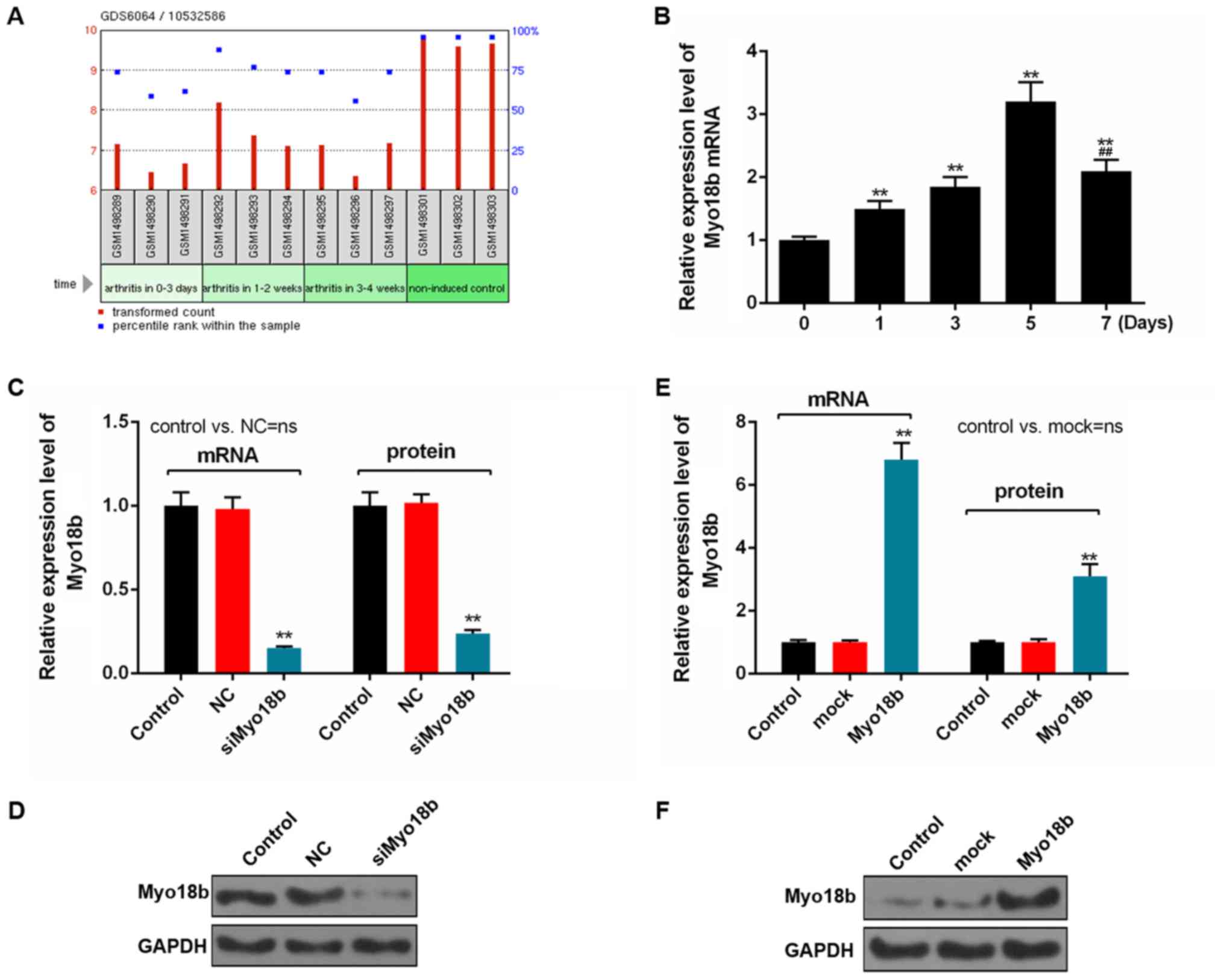
Myo18b expression is downregulated in
the tarsus joint of collagen-induced RA
The gene expression profile in the tarsal joint of a
mouse model of RA was first determined, and the data suggested that
expression of Myo18b was lower in the tarsus joint of mice with
collagen-induced RA, compared with non-induced controls (Fig. 1A).
Myo18b expression increases during the
myogenic differentiation of C2C12 cells
To determine the expression of Myo18b during the
myogenic differentiation of C2C12 myoblasts, cells were harvested
and the expression levels of Myo18b were determined in cells after
0, 1, 3, 5 and 7 days. The RT-qPCR analysis showed that the
expression levels of Myo18b increased significantly on days 1, 3, 5
and 7 compared with 0 days (P<0.01), and Myo18b level decreased
significantly on day 7 compared with 5 days (P<0.01; Fig. 1B). Additionally, the expression of
Myo18b on day 7 was significantly upregulated compared with day 0
(P<0.01; Fig. 1B).
Myo18b regulates proliferation and
cell cycle progression in C2C12 myoblasts
The efficiency of siMyo18b and Myo18b transfection
was determined using RT-qPCR and western blot analyses. The results
showed that the expression of Myo18b in the cells transfected with
siMyo18b was significantly decreased compared with the control or
NC groups (P<0.01; Fig. 1C and
D). However, Myo18b expression in the Myo18b group was
significantly increased compared with the control or mock groups
(P<0.01; Fig. 1E and F).
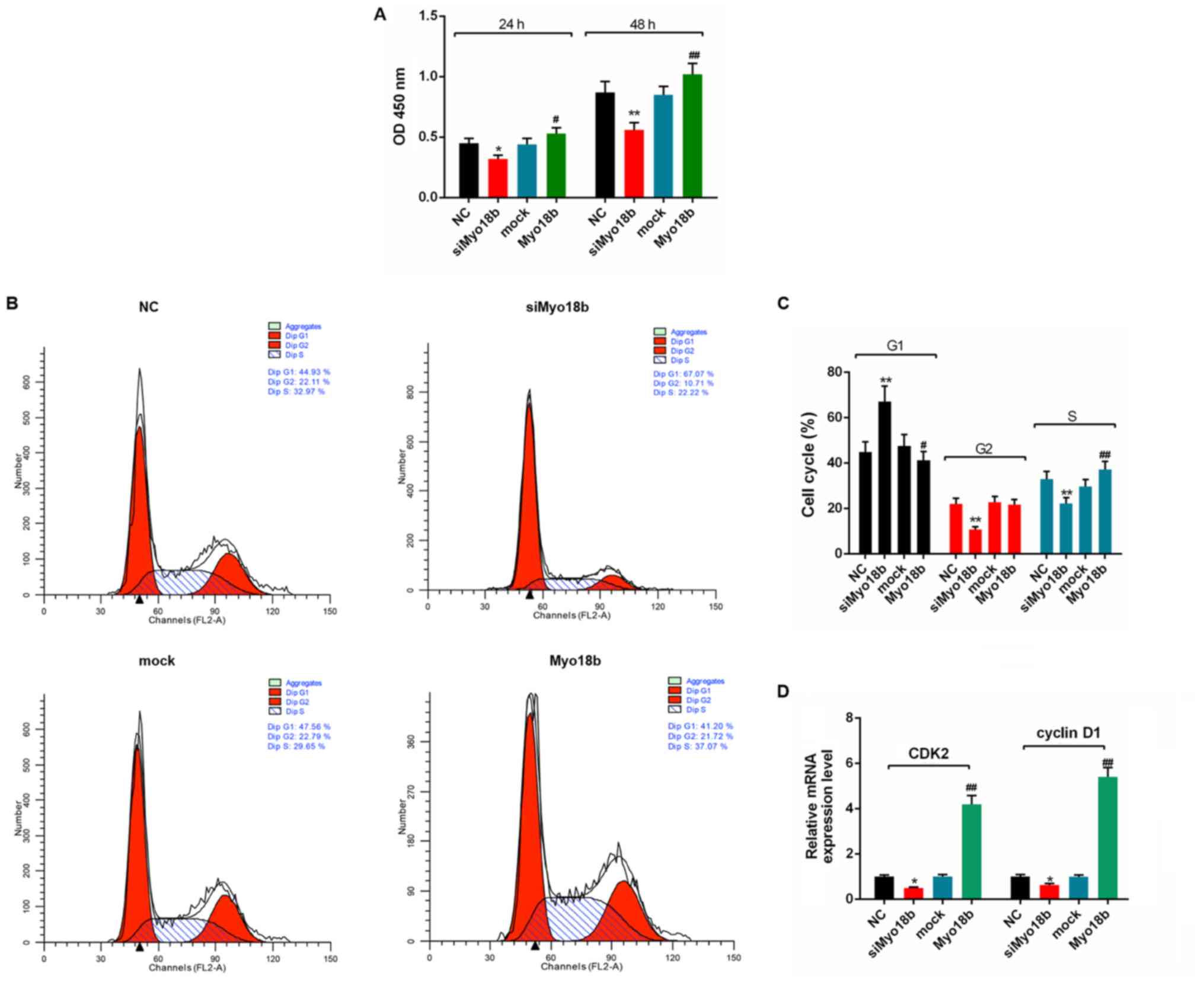
Subsequently, the effects of Myo18b on the growth of
C2C12 myoblasts were assessed. The CCK-8 assay results showed that
the cell viability in the siMyo18b group was significantly reduced
compared with the NC group at 24 (P<0.05) and 48 h (P<0.01)
after transfection with siMyo18b, whereas the cell viability in the
Myo18b group was significantly increased compared with the mock
group at 24 (P<0.05) and 48 h (P<0.01) after transfection
with Myo18b (Fig. 2A).
Furthermore, flow cytometry analysis showed that the number of
C2C12 myoblasts in the G1 phase was significantly increased in the
siMyo18b group (P<0.01), and the number of cells in G2 and S
phase were significantly decreased (P<0.01) compared with the NC
group (Fig. 2B and C). The number
of C2C12 myoblasts in the G1 phase were significantly decreased
when cells were transfected with Myo18b (P<0.05), whereas the
number of cells in the S phase were significantly increased
(P<0.01), compared with the mock group (Fig. 2B and C). RT-qPCR analysis showed
that the expression levels of CDK2 and cyclin D1 in C2C12 myoblasts
transfected with siMyo18b were significantly decreased (P<0.05),
and the expression levels in C2C12 myoblasts transfected with
Myo18b were significantly increased (P<0.01; Fig. 2D), compared with the respective
controls.
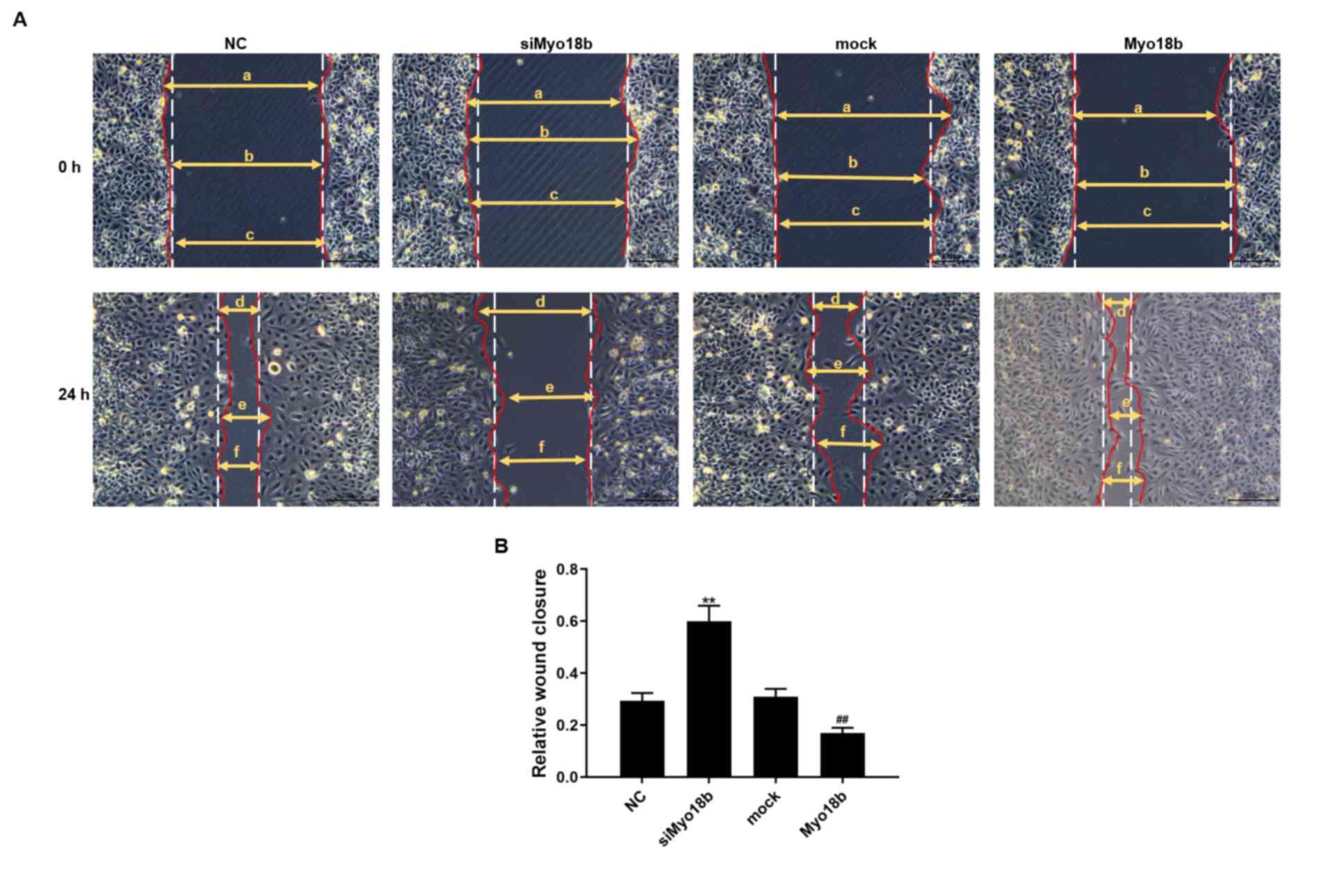
Myo18b regulates the migration of
C2C12 myoblasts
The results of the wound-healing assay showed that
the migratory rate of C2C12 myoblasts transfected with siMyo18b was
significantly decreased compared with the NC group (P<0.01). The
migratory rate of C2C12 myoblasts transfected with Myo18b was
significantly increased compared with the mock cells (P<0.01;
Fig. 3).
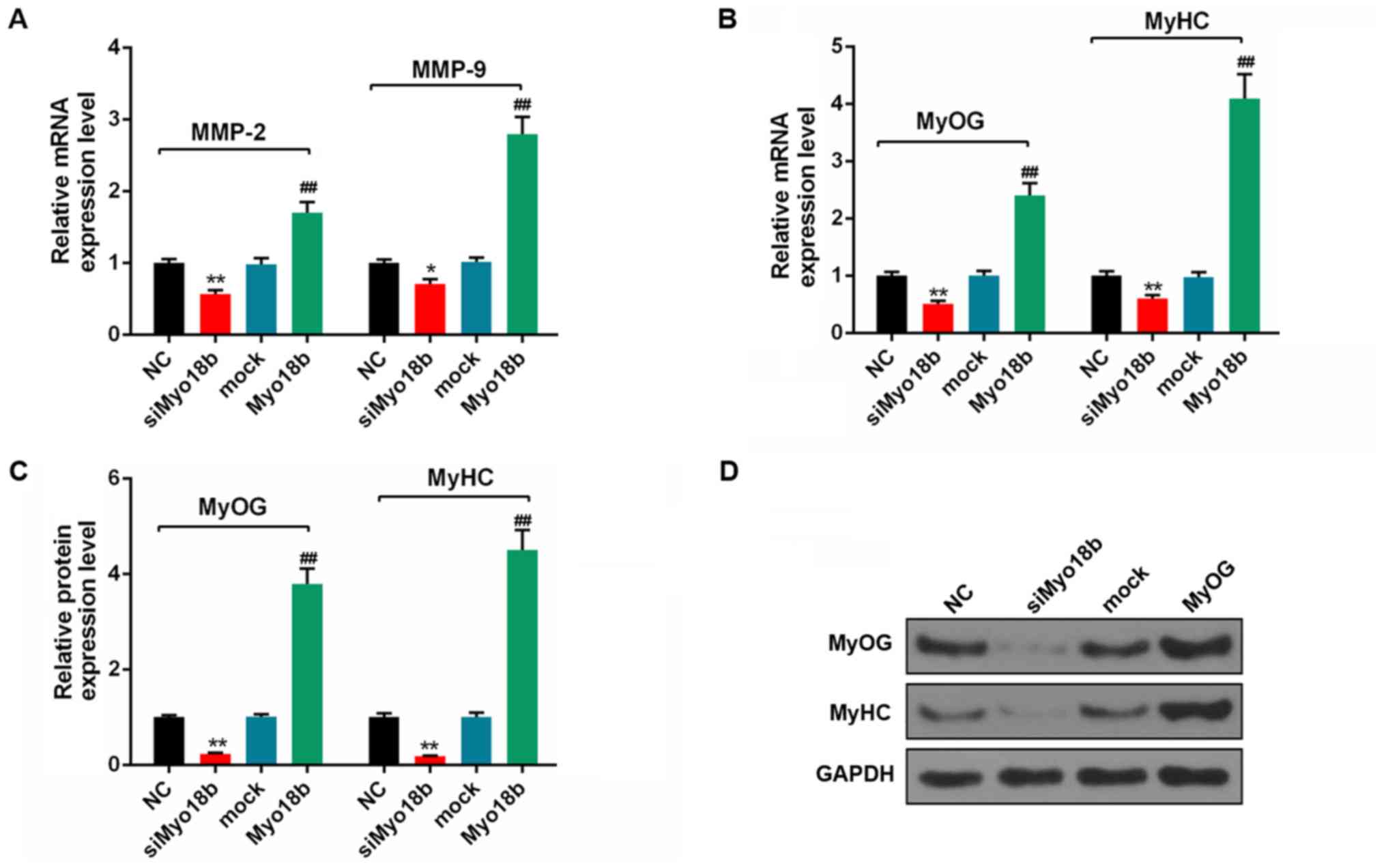
Myo18b regulates differentiation of
C2C12 myoblasts
The results of RT-qPCR and western blotting analysis
results showed that siMyo18b downregulated the mRNA expression
levels of MMP-2 and MMP-9, whereas Myo18b overexpression increased
the expression of these genes in C2C12 myoblasts (Fig. 4A). Furthermore, the mRNA and
protein expression levels of MyOG and MyH6 were downregulated in
the siMyo18b group compared with the NC group, and increased in
cells overexpressing Myo18b compared with the mock group
(P<0.01; Fig. 4B-D). These
results suggested that siMyo18b decreased differentiation in C2C12
myoblasts and Myo18b overexpression induced opposing effects.
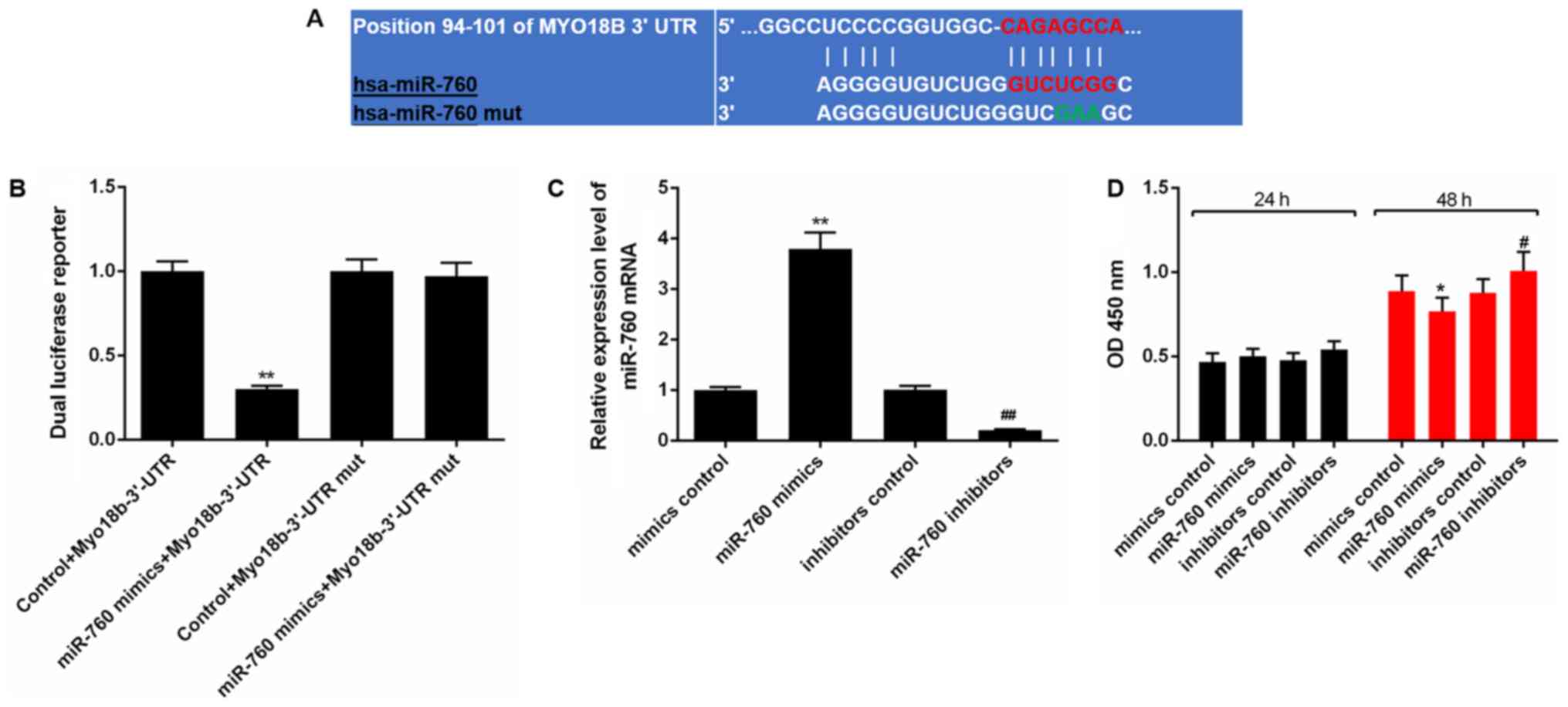
Myo18b is a target gene of miR-760 in
C2C12 myoblast
A binding site between miR-760 and Myo18b-3′-UTR was
identified using TargetScan (Fig.
5A). A dual-luciferase reporter assay revealed that miR-760
mimics significantly decreased the luciferase activity when the
reporter gene contained the Myo18b-3′-UTR in C2C12 myoblasts
(P<0.01). However, no decrease in luciferase activity was
observed when the reporter gene contained the Myo18b-3′-UTR mut
(P>0.05; Fig. 5B).
Upregulation of miR-760 decreases proliferation and
cell cycle progression of C2C12 myoblasts. The expression levels of
miR-760 in C2C12 myoblasts were determined using RT-qPCR analysis
following transfection. The results showed that the expression of
miR-760 in C2C12 myoblasts was significantly increased in cells
transfected with miR-760 mimics compared with cells transfected
with the control mimics (P<0.01). After transfection with
miR-760 inhibitor, the miR-760 expression level was decreased,
compared with cells in the inhibitor control group (P<0.01;
Fig. 5C).
A CCK-8 assay showed there were no significant
differences observed in terms of the cell viability at 24 h among
the groups (P>0.05), and that the cell viability at 48 h was
decreased significantly in the cells transfected with miR-760
mimics, compared with the mimics control group (P<0.05) after 48
h. Cell viability at 48 h was increased significantly in the cells
transfected with miR-760 inhibitor compared with the inhibitors
control group (P<0.05; Fig.
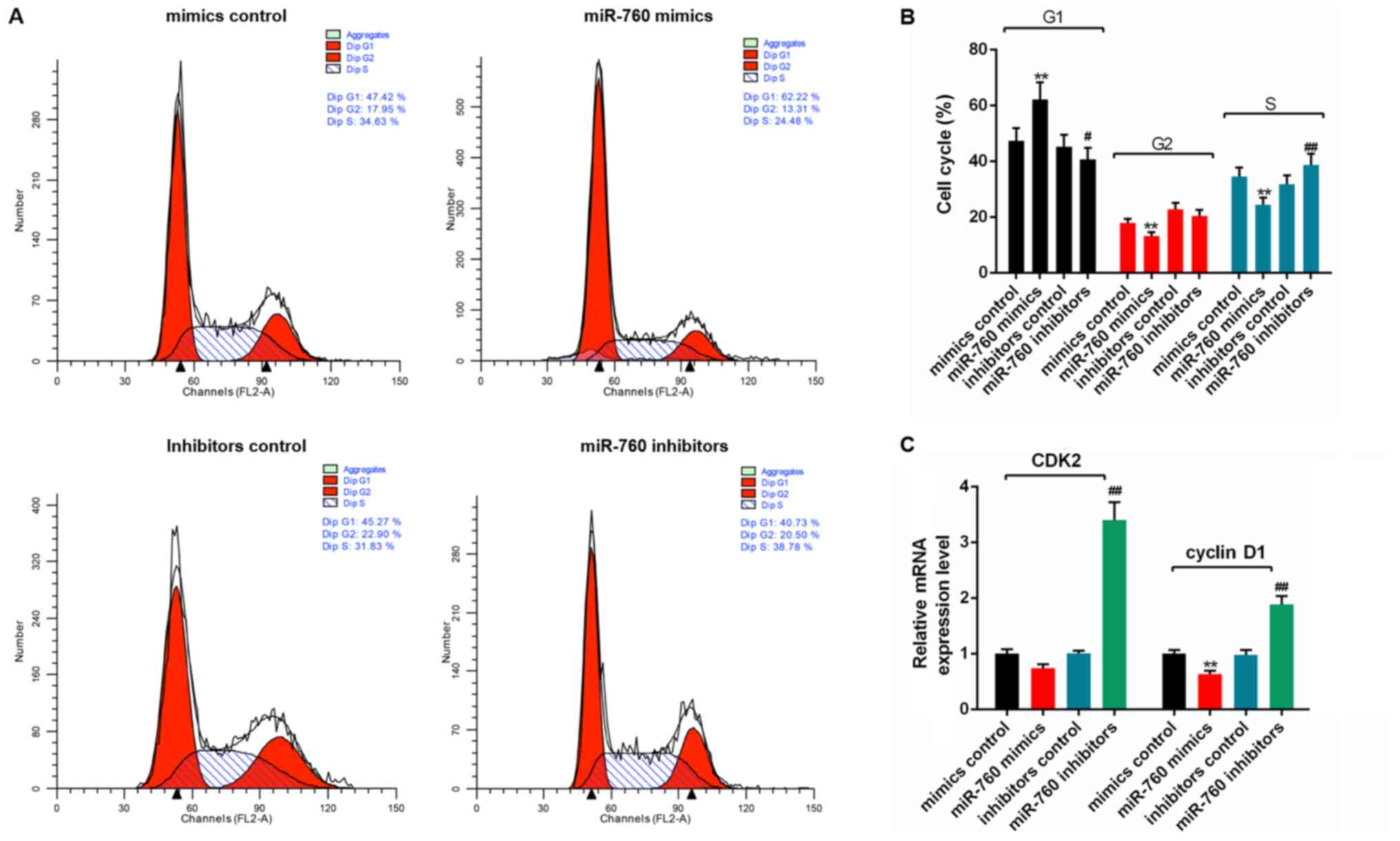
5D). Furthermore, flow cytometry analysis showed that the
percentage of C2C12 myoblasts transfected with miR-760 mimics in G1
was significantly increased (P<0.01), whereas the percentages of
cells in G2 and S were significantly decreased (P<0.01) compared
with the mimics control group (Fig. 6A
and B). The percentage of cells transfected with miR-760
inhibitors in G1 were decreased (P<0.05), and the percentage of
cells in S phase were significantly increased (P<0.01) compared
with the inhibitors control group (Fig. 6A and B). RT-qPCR analysis revealed
that the mRNA expression levels of cyclin D1 were significantly
decreased in the miR-760 mimics group compared with the mimics
control group (P<0.01), while the CDK2 level was slightly
reduced but not significant. The expression levels of CDK2 and
cyclin D1 in miR-760 inhibitor group were significantly higher
compared with the mimics control group (P<0.01; Fig. 6C).
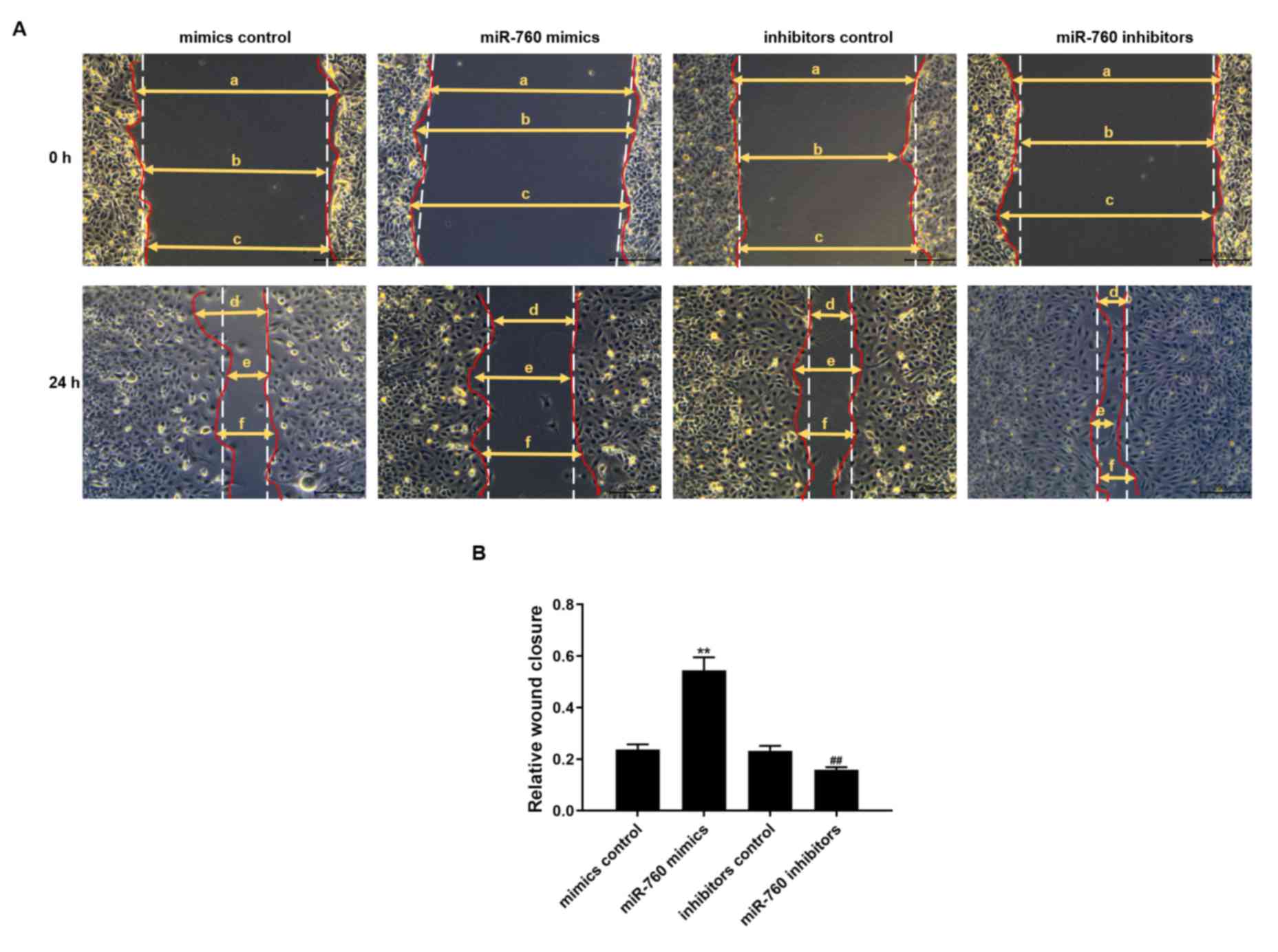
miR-760 decreases the migration of
C2C12 myoblasts
A wound healing assay was used to determine the
effects of miR-760 on the migratory capacity of C2C12 myoblasts
following transfection with a miR-760 mimic or inhibitor. The
results showed that the migratory abilities of C2C12 myoblasts
transfected with miR-760 mimics for 24 h were significantly reduced
compared with the mimics control group (P<0.01). The migratory
capacity in cells transfected with miR-760 inhibitors at 24 h was
significantly increased compared with the inhibitors control group
(P<0.01; Fig. 7).
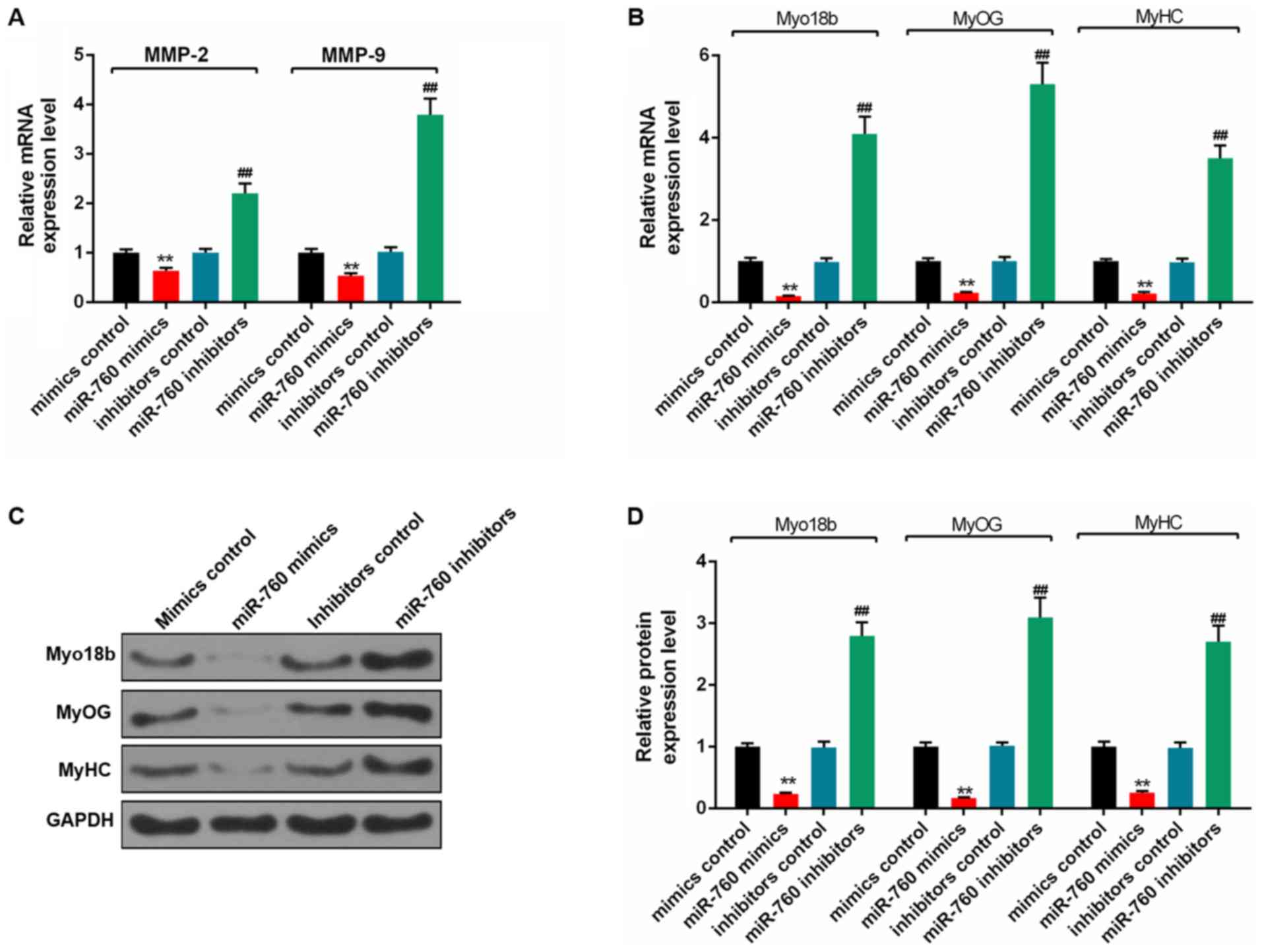
miR-760 decreases differentiation of
C2C12 myoblasts
RT-qPCR and western blot analyses showed that the
expression levels of MMP-2, MMP-9, Myo18b, MyOG and MyH6 in C2C12
myoblasts transfected with miR-760 mimics were significantly
decreased compared with the mimics control group (P<0.01;
Fig. 8A and B). The expression
levels of these genes were significantly increased in cells
transfected with miR-760 inhibitor, compared with the inhibitor
control group (P<0.01; Fig. 8C and
D).
Discussion
RA is a chronic autoimmune disease that causes
progressive articular destruction, functional loss of joints and
related comorbidities of bone, vasculature and metabolism, and thus
physiologically affects patients (28). RA results from complex interactions
between genes and the environment (29). Studies have shown that reducing
inflammation, inhibiting angiogenesis and inducing apoptosis of
fibroblast-like synoviocytes may effectively improve RA (30–32).
Skeletal muscle atrophy has been observed in patients with RA, and
Myo18b is an important component involved in the rapid formation of
skeletal muscle (33,34). miRNAs have been reported as
important regulators of skeletal muscle development and
differentiation (35). Previously,
expression of miR-760 was demonstrated to be altered in several
diseases (36,37). However, the role of miR-760 in
regulating the proliferation of RA skeletal muscle remains unclear.
The aims of the present study were to examine the regulatory
effects of miR-760 during skeletal muscle proliferation in RA, and
to determine the underlying mechanism.
RA is associated with muscle wasting, which impedes
prognosis. Therefore, promoting skeletal muscle proliferation has
significant clinical value for patients with RA (38,39).
Certain genes and multiple proteolytic systems are involved in
muscle atrophy (40). Furthermore,
the release of cytokines during chronic inflammation may promote
satellite cell activation and myogenesis that help promote skeletal
muscle proliferation (41).
According to Malfatti et al (42), Myo18b encodes an unconventional
myosin protein which is primarily expressed in skeletal and cardiac
muscles that are associated with nemaline myopathy and
cardiomyopathy. Similarly, Alazami et al (43) showed that a rare syndrome,
presenting Klippel-Feil anomaly and myopathy, may have been caused
by a mutation in Myo18b. In the present study, it was demonstrated
that Myo18b was downregulated in the tarsus joint of RA; thus, it
was suggested that miR-760 may have regulated skeletal muscle
proliferation in RA by targeting Myo18b. To confirm a link between
miR-760 and Myo18b, a dual-luciferase reporter assay was performed,
and the results showed that Myo18b was a direct target gene of
miR-760.
Myoblast fusion is a crucial step in skeletal muscle
differentiation, and the regulation of genes such as cyclin D1 and
MyOG may affect the proliferation and differentiation of myoblasts
(44,45). CDKs are proline-directed
serine/threonine-protein kinases, which are primarily involved in
cell cycle regulation, and intracellular and extracellular signal
fusion (46). CDKs can bind to
cyclins to form heterodimers and promote the progression of the
cell cycle. As a type of protein kinase, CDK2 is involved in the
regulation of the G1/S phases of the cell cycle in patients with RA
(47). In the present study, it
was demonstrated that Myo18b expression was increased as C2C12
myoblasts differentiated at 1, 3 and 5 days, but decreased on day
7. Based on this result, it may be the case that the rate of
apoptosis increases after day 5. In a previous study, miRNAs were
found to regulate cell proliferation, cell cycle progression and
migration by altering the expressions of various factors, such as
MALAT1 (48). In the present
study, the results showed that miR-760 overexpression and Myo18b
silencing significantly decreased the expression levels of CDK2 and
cyclin D1 in C2C12 myoblasts. Additionally, it was demonstrated
that miR-760 overexpression and Myo18b silencing decreased the
proliferation and migration of C2C12 myoblasts. These results
suggested that miR-760 may inhibit the proliferation and migration
of C2C12 myoblasts by suppressing Myo18b and thus slowing down
skeletal muscle formation. Furthermore, MMP-2 and MMP-9 are the
primary MMPs responsible for the degradation of type IV collagen,
primarily in the basement membrane close to the muscular layer
(49,50). MMPs are associated with increased
motility, differentiation and regeneration of skeletal muscle. Zhai
et al found that salicin decreased the levels of
inflammatory factors and inhibited the expression of MMP-1/-3 in
interleukin-1β-induced RA-fibroblast-like synoviocytes, thereby
improving RA (30). Therefore,
regulation of MMPs may prove to be an effective method of treating
patients with RA.
Skeletal muscles are composed of heterogeneous types
of fibers based on MyHC expression (51). In patients with RA, varying degrees
of muscle mass decline and reduction in cross-sectional area have
been reported, and a slight but significant decrease in MyHC
content has also been observed (52). MyOG, a key transcription factor in
myogenesis, plays an important role in the regulation of myoblast
differentiation and is critical for the terminal differentiation of
differentiated myoblasts (45). In
addition, a previous study suggested that upregulation of MyOG
expression was required for myoblasts to fuse into new or existing
myotubes, and that the knockdown of MTMR7 promoting C2C12 myoblast
early differentiation was associated with the upregulation of MyOG
(53). In the present study, the
results showed that miR-760 inhibited MMP-2, MMP-9, MyOG and MyH6
expression levels in C2C12 myoblasts by downregulating Myo18b.
Based on these results, it was hypothesized that miR-760 may affect
skeletal muscle proliferation and differentiation by regulating
cell cycle and myocyte differentiation, and such a phenomenon may
be associated with the regulation or the expression of Myo18b, MyOG
and MyHC.
In conclusion, it was revealed that Myo18b was a
target gene of miR-760. Overexpression of miR-760 downregulated the
expression levels of CDK2, cyclin D1, MMP-2, MMP-9, MyOG and MyH6.
miR-760 decreased proliferation, cell cycle progression, migration
and differentiation in C2C12 myoblasts by targeting Myo18b. The
results of the present study may improve understanding of the
molecular mechanisms underlying the proliferation of skeletal
muscle in patients with RA. There are certain limitations to the
present study. For example, all experiments were performed in
vitro and thus, whether the results are replicated in
vivo remains to be determined. Additionally, miR-760 targets
other genes involved in the proliferation and differentiation of
C2C12 myoblasts; thus, the effects miR-760 on the various cellular
behaviors may have been mediated via other proteins.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the author on reasonable request.
Authors' contributions
XT conceived and designed the study. JW, SZ, HW, GF,
JZho, JZha, GJ and CX acquired analyzed and interpreted the data.
XT drafted the article and revised for important intellectual
content. All the authors approved publication of the final version
and agree to be held accountable for all aspects of the work.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gibofsky A: Overview of epidemiology,
pathophysiology and diagnosis of rheumatoid arthritis. Am J Manag
Care. 18 (13 Suppl):S295–S302. 2012.PubMed/NCBI
|
|
2
|
Nogueira E, Gomes AC, Preto A and
Cavaco-Paulo A: Folate-targeted nanoparticles for rheumatoid
arthritis therapy. Nanomedicine. 12:1113–1126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Wei T, Gao J, He H, Chang X and
Yan T: Effects of Naringenin on inflammation in complete freund's
adjuvant-induced arthritis by regulating Bax/Bcl-2 balance.
Inflammation. 38:245–251. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shahmohamadnejad S, Vaisi-Raygani A,
Shakiba Y, Kiani A, Rahimi Z, Bahrehmand F, Shakiba E and
Pourmotabbed T: Association between butyrylcholinesterase activity
and phenotypes, paraoxonase192 rs662 gene polymorphism and their
enzymatic activity with severity of rheumatoid arthritis:
Correlation with systemic inflammatory markers and oxidative
stress, preliminary report. Clin Biochem. 48:63–69. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Souza SD, Bansal RK and Galloway J:
Managing patients with rheumatoid arthritis. BDJ Team. 4:170642017.
View Article : Google Scholar
|
|
6
|
Huffman KM, Jessee R, Andonian B, Davis
BN, Narowski R, Huebner JL, Kraus VB, McCracken J, Gilmore BF, Tune
KN, et al: Molecular alterations in skeletal muscle in rheumatoid
arthritis are related to disease activity, physical inactivity, and
disability. Arthritis Res Ther. 19:122017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lai NS, Yu HC, Tung CH, Huang KY, Huang HB
and Lu MC: The role of aberrant expression of T cell miRNAs
affected by TNF-α in the immunopathogenesis of rheumatoid
arthritis. Arthritis Res Ther. 19:2612017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Furer V, Greenberg JD, Attur M, Abramson
SB and Pillinger MH: The role of microRNA in rheumatoid arthritis
and other autoimmune diseases. Clin Immunol. 136:1–15. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Filkova M, Jungel A, Gay RE and Gay S:
MicroRNAs in rheumatoid arthritis: Potential role in diagnosis and
therapy. BioDrugs. 26:131–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stanczyk J, Pedrioli DM, Brentano F,
Sanchez-Pernaute O, Kolling C, Gay RE, Detmar M, Gay S and Kyburz
D: Altered expression of MicroRNA in synovial fibroblasts and
synovial tissue in rheumatoid arthritis. Arthritis Rheum.
58:1001–1009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang
Y, Zhang J, Zhang J, Fu X, Liu H, et al: Altered microRNA
expression profile with miR-146a upregulation in CD4+ T cells from
patients with rheumatoid arthritis. Arthritis Res Ther. 12:R812010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fulci V, Scappucci G, Sebastiani GD,
Giannitti C, Franceschini D, Meloni F, Colombo T, Citarella F,
Barnaba V, Minisola G, et al: miR-223 is overexpressed in
T-lymphocytes of patients affected by rheumatoid arthritis. Hum
Immunol. 71:206–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Filkova M, Aradi B, Senolt L, Ospelt C,
Vettori S, Mann H, Filer A, Raza K, Buckley CD, Snow M, et al:
Association of circulating miR-223 and miR-16 with disease activity
in patients with early rheumatoid arthritis. Ann Rheum Dis.
73:1898–1904. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang YZ, Zhang YF, Yang L, Xu J, Mo XM and
Peng W: miR-760 mediates hypoxia-induced proliferation and
apoptosis of human pulmonary artery smooth muscle cells via
targeting TLR4. Int J Mol Med. 42:2437–2446. 2018.PubMed/NCBI
|
|
16
|
Cao L, Liu Y, Wang D, Huang L, Li F, Liu
J, Zhang C, Shen Z, Gao Q, Yuan W and Zhang Y: MiR-760 suppresses
human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1
signaling. J Exp Clin Cancer Res. 37:832018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liao Y, Deng Y, Liu J, Ye Z, You Z, Yao S
and He S: MiR-760 overexpression promotes proliferation in ovarian
cancer by downregulation of PHLPP2 expression. Gynecol Oncol.
143:655–663. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Goloviznina NA and Kyba M: Twist of fate
for skeletal muscle mesenchymal cells. Nat Cell Biol. 19:153–154.
2017. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ceafalan LC, Fertig TE, Popescu AC,
Popescu BO, Hinescu ME and Gherghiceanu M: Skeletal muscle
regeneration involves macrophage-myoblast bonding. Cell Adh Migr.
12:228–235. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sin J, Andres AM, Taylor DJ, Weston T,
Hiraumi Y, Stotland A, Kim BJ, Huang C, Doran KS and Gottlieb RA:
Mitophagy is required for mitochondrial biogenesis and myogenic
differentiation of C2C12 myoblasts. Autophagy. 12:369–380. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gurung R, Ono Y, Baxendale S, Lee SL,
Moore S, Calvert M and Ingham PW: A Zebrafish model for a human
myopathy associated with mutation of the unconventional myosin
MYO18B. Genetics. 205:725–735. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Berger J, Berger S, Li M and Currie PD:
Myo18b is essential for sarcomere assembly in fast skeletal muscle.
Hum Mol Genet. 26:1146–1156. 2017.PubMed/NCBI
|
|
23
|
Mueller SM, Aguayo D, Aeberli D, Vögelin E
and Toigo M: Myocellular characteristics in rheumatoid arthritis
and osteoarthritis patients. Arthritis Res Ther. 20:512018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41((Database Issue)):
D991–D995. 2013.PubMed/NCBI
|
|
26
|
Denninger KC, Litman T, Marstrand T,
Moller K, Svensson L, Labuda T and Andersson Å: Kinetics of gene
expression and bone remodelling in the clinical phase of
collagen-induced arthritis. Arthritis Res Ther. 17:432015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
McInnes IB and Schett G: Pathogenetic
insights from the treatment of rheumatoid arthritis. Lancet.
389:2328–2337. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hensvold AH, Magnusson PK, Joshua V,
Hansson M, Israelsson L, Ferreira R, Jakobsson PJ, Holmdahl R,
Hammarström L, Malmström V, et al: Environmental and genetic
factors in the development of anticitrullinated protein antibodies
(ACPAs) and ACPA-positive rheumatoid arthritis: An epidemiological
investigation in twins. Ann Rheum Dis. 74:375–380. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhai KF, Duan H, Khan GJ, Xu H, Han FK,
Cao WG, Gao GZ, Shan LL and Wei ZJ: Salicin from Alangium chinense
ameliorates rheumatoid arthritis by modulating the Nrf2-HO-1-ROS
pathways. J Agric Food Chem. 66:6073–6082. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhai KF, Duan H, Chen Y, Khan GJ, Cao WG,
Gao GZ, Shan LL and Wei ZJ: Apoptosis effects of imperatorin on
synoviocytes in rheumatoid arthritis through
mitochondrial/caspase-mediated pathways. Food Funct. 9:2070–2079.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhai KF, Duan H, Cui CY, Cao YY, Si JL,
Yang HJ, Wang YC, Cao WG, Gao GZ and Wei ZJ: Liquiritin from
Glycyrrhiza uralensis attenuating rheumatoid arthritis via reducing
inflammation, suppressing angiogenesis, and inhibiting MAPK
signaling pathway. J Agric Food Chem. 67:2856–2864. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Andrade Fernandes de Mello R, Garcia
Rondina R, Valim V, Santos Belisario S, Burgomeister Lourenço R,
Francisco Batista E and Horst Duque R: Isolated atrophy of the
abductor digiti quinti in patients with rheumatoid arthritis.
Skeletal Radiol. 46:1715–1720. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berger J, Berger S, Li M and Currie PD:
Myo18b is essential for sarcomere assembly in fast skeletal muscle.
Hum Mol Genet. 26:1146–1156. 2017.PubMed/NCBI
|
|
35
|
Moresi V, Marroncelli N, Coletti D and
Adamo S: Regulation of skeletal muscle development and homeostasis
by gene imprinting, histone acetylation and microRNA. Biochim
Biophys Acta. 1849:309–316. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sun D, Lu J, Hu C, Zhang Q, Wang X, Zhang
Z and Hu S: Prognostic role of miR-760 in hepatocellular carcinoma.
Oncol Lett. 16:7239–7244. 2018.PubMed/NCBI
|
|
37
|
Yan C, Zhang W, Shi X, Zheng J, Jin X and
Huo J: MiR-760 suppresses non-small cell lung cancer proliferation
and metastasis by targeting ROS1. Environ Sci Pollut Res Int.
25:18385–18391. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Matschke V, Murphy P, Lemmey AB, Maddison
P and Thom JM: Skeletal muscle properties in rheumatoid arthritis
patients. Med Sci Sports Exerc. 42:2149–2155. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gomez-SanMiguel AB, Gomez-Moreira C,
Nieto-Bona MP, Fernández-Galaz C, Villanúa MÁ, Martín AI and
López-Calderón A: Formoterol decreases muscle wasting as well as
inflammation in the rat model of rheumatoid arthritis. Am J Physiol
Endocrinol Metab. 310:E925–E937. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dutt V, Saini V, Gupta P, Kaur N, Bala M,
Gujar R, Grewal A, Gupta S, Dua A and Mittal A: S-allyl cysteine
inhibits TNFα-induced skeletal muscle wasting through suppressing
proteolysis and expression of inflammatory molecules. Biochim
Biophys Acta Gen Subj. 1862:895–906. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Boutrup RJ, Farup J, Vissing K, Kjaer M
and Mikkelsen UR: Skeletal muscle stem cell characteristics and
myonuclei content in patients with rheumatoid arthritis: A
cross-sectional study. Rheumatol Int. 38:1031–1041. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Malfatti E, Böhm J, Lacène E, Beuvin M,
Romero NB and Laporte J: A premature stop codon in MYO18B is
associated with severe nemaline myopathy with cardiomyopathy. J
Neuromuscul Dis. 2:219–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Alazami AM, Kentab AY, Faqeih E, Mohamed
JY, Alkhalidi H, Hijazi H and Alkuraya FS: A novel syndrome of
Klippel-Feil anomaly, myopathy, and characteristic facies is linked
to a null mutation in MYO18B. J Med Genet. 52:400–404. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Panda AC, Abdelmohsen K, Martindale JL, Di
Germanio C, Yang X, Grammatikakis I, Noh JH, Zhang Y, Lehrmann E,
Dudekula DB, et al: Novel RNA-binding activity of MYF5 enhances
Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids
Res. 44:2393–2408. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Luo W, Li E, Nie Q and Zhang X: Myomaker,
regulated by MYOD, MYOG and miR-140-3p, promotes chicken myoblast
fusion. Int J Mol Sci. 16:26186–26201. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang Y, Qin X, Guo T, Liu P, Wu P and Liu
Z: Up-regulation of CDK16 by multiple mechanisms in hepatocellular
carcinoma promotes tumor progression. J Exp Clin Cancer Res.
36:972017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Okada Y, Raj T and Yamamoto K: Ethnically
shared and heterogeneous impacts of molecular pathways suggested by
the genome-wide meta-analysis of rheumatoid arthritis. Rheumatology
(Oxford). 55:186–189. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hadler-Olsen E, Solli AI, Hafstad A,
Winberg JO and Uhlin-Hansen L: Intracellular MMP-2 activity in
skeletal muscle is associated with type II Fibers. J Cell Physiol.
230:160–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Silva MT, Nascimento TL, Pereira MG,
Siqueira AS, Brum PC, Jaeger RG and Miyabara EH: β2-Adrenoceptor is
involved in connective tissue remodeling in regenerating muscles by
decreasing the activity of MMP-9. Cell Tissue Res. 365:173–186.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Huang L, Chen L, Qiu Y and Li S:
Abnormalities in the fiber composition and contractility in
diabetic skeletal muscles. Int J Clin Exp Med. 11:753–763.
2018.
|
|
52
|
Yamada T, Steinz MM, Kenne E and Lanner
JT: Muscle weakness in rheumatoid arthritis: The role of Ca2+ and
free radical signaling. Ebiomedicine. 23:12–19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yuan Z, Chen Y, Zhang X, Zhou X, Li M,
Chen H, Wu M, Zhang Y and Mo D: Silencing myotubularin related
protein 7 enhances proliferation and early differentiation of C2C12
myoblast. Biochem Biophys Res Commun. 484:592–597. 2017. View Article : Google Scholar : PubMed/NCBI
|