Introduction
PTEN-induced kinase 1 (PINK1) is located in
the mitochondrial membrane, and helps to regulate mitochondrial
morphology and autophagy (1).
Mutations in PINK1 have been recognized to be the second
most common cause of autosomal recessive adolescent Parkinson's
disease (PD) (2). Loss of
Drosophila PINK1 leads to mitochondrial morphological
disorders and mitochondrial complex function damage (3). Furthermore, neurons are dependent on
mitochondria, which act as the major energy producers (4). Therefore, when PINK1 is
mutated, neurotoxicity is increased and this may be associated with
mitochondrial defects, thus contributing to the loss of
dopaminergic (DA) neurons (5).
Oxidative stress is a prominent and common feature of all forms of
PD, and may have a toxic effect that causes neuronal cell death
(6). Moreover, oxidative stress in
PD is closely associated with a series of pathogenic factors,
including mitochondrial dysfunction, DA metabolism and metal ion
dysregulation (7).
The human Wnt gene family, known as the
wingless-type MMTV integration site family, consists of 19 members,
and the interaction between Wnt1 and Wnt5a promotes
the development of DA neurons in the midbrain (8). Moreover, the Wnt2 gene is one
of the 19 Wnt family members that is highly expressed in the
human thalamus, and plays an important role in the late development
of human genes and the brain (9,10). A
previous functional study has shown that Wnt2 promotes the
migration of primitive neurons and increases the number of DA
neurons (11), thus enhancing DA
function in the midbrain, and affecting mRNA and protein expression
levels. The Wnt signaling pathway can be cross-linked with a number
of signaling pathways (12). The
interaction of β-catenin with the forkhead box sub-group O (FOXO)
signaling pathway can inhibit Huntington protein toxicity (13). Furthermore, the Wnt signaling
pathway also regulates mitochondrial energy, metabolism and
oxidative stress (14,15).
The present study selected Drosophila as the
experiment model, as Drosophila genes have been fully
sequenced and annotated, and are highly homologous to human genes
(16,17). Compared with other models, the
Drosophila life cycle is very short; therefore, it is easier
to observe the whole process of disease development (18). Mature genetic systems, abundant
strain resources and powerful genome editing techniques also make
Drosophila one of the primary choices for genetic research
in neurodegenerative diseases (19).
The present study identified that overexpression of
Wnt2 gene had a significant effect on
PINK1B9 transgenic Drosophila. While the
Wnt pathway may have neuroprotective effects, the role of
Wnt2 in neurodegenerative diseases remains unknown.
Therefore, the aims of the present study were to investigate the
function of Wnt2 in PINK1 mutant transgenic
Drosophila, and to identify its association with the
Wnt/β-catenin and PPARG coactivator 1α (PGC-1α)/FOXO/manganese
superoxide dismutase (MnSOD) signaling pathways.
Materials and methods
Drosophila stocks
A total of five fly stocks were used: two stocks of
Drosophila melanogaster (UAS-Wnt2OE and
UAS-Wnt2RNAi) were purchased from the Bloomington
Drosophila Stock Center. In total, three stocks
(UAS-PINK1B9/FM7; MHC-Gal4, W1118 and MHC-GAL4)
were provided by Institute of Life Sciences of Fuzhou University.
W1118 is a wild-type genotype. UAS-Wnt2OE is a stock that
overexpresses the Wnt2 gene. UAS-Wnt2RNAi is a
Drosophila that has lost the function of the Wnt2
gene by using RNA interference technology. MHC-GAL4 is a
Drosophila with an indirect flying muscle promoter.
UAS-PINK1B9/FM7; MHC-Gal4 is a PD
Drosophila model, in which the PINK1 mutation gene
can be specifically expressed in indirect flying muscles. The
classic GAL4/UAS system is divided into two parts: GAL4 and UAS.
The GAL4 stock and UAS stock are two independent stocks (20). The fusion of GAL4 and
tissue-specific promoter can regulate the expression of GAL4
protein in different tissues of Drosophila (21). Furthermore, UAS and target genes
are fused to construct a transgenic line with a UAS-target gene
(22). Only when the two
hybridize, can GAL4 recognize the UAS promoter and induce
expression of UAS downstream genes in specific tissues (23). Drosophila were placed in
Drosophila culture tubes containing corn medium and cultured
at a constant temperature of 25°C and 60% relative humidity.
Drosophila construction
The flies were driven via the muscular driver
MHC-GAL4. MHC-GAL4 virgin flies were crossed with W1118 male flies,
and the F1 generation genes were W1118/+; MHC-GAL4/+, which served
as the control group. In the PINK1B9 disease
group, UAS-PINK1B9/FM7; MHC-GAL4 virgin flies
were crossed with W1118 male flies, which produced the F1
generation with a genotype of UAS-PINK1B9/+;
MHC-GAL4/+. In the overexpression (OE) intervention group,
UAS-PINK1B9/FM7; MHC-GAL4 virgin flies were
hybridized with male flies of UAS-Wnt2OE, and the F1
generation genotype was UAS-PINK1B9/y;
MHC-GAL4/Wnt2 OE, UAS-PINK1B9/y. In the
RNA interference (RNAi) intervention group,
UAS-PINK1B9/ FM7; MHC-GAL4 virgin flies were
crossed with males of UAS-Wnt2 RNAi Drosophila, and
the F1 generation obtained had the genotype
UAS-PINK1B9/y; MHC-GAL4/Wnt2RNAi.
Morphological observation of
Drosophila (24)
Flies carrying the MHC-GAL4/UAS systems were grouped
as follows: Normal control group (W1118), disease control group
(PINK1B9), Wnt2OE intervention group
(PINK1B9; Wnt2OE) and Wnt2 knockdown
intervention group (PINK1B9; Wnt2 RNAi).
On day 5, ~100 male flies were selected from each group. After
being anesthetized by CO2, the 100 flies were divided
into transparent glass tubes with five flies per tube. After the
flies completely woke up (after ~1 h), the shape of their wings and
whether they could fly were observed. The number and the ratio of
abnormal wings and flying were calculated. All assays were
performed in triplicate and independently repeated three times.
Drosophila mRNA expression
detection
The experimental groups were the same as
aforementioned. On day 5, the head and abdomen were removed, and
the chest was kept from 30 male flies from each group.
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to extract the total RNA from the chest, according
to the manufacturer's protocol. The primers were synthesized by
Sangon Biotech Co., Ltd. and are presented in Table I. Subsequently, RNA samples were
reverse transcribed into cDNA using PrimeScript™ reverse
transcription (RT) reagent kit with gDNA Eraser (Takara Bio, Inc.).
Following the manufacturer's instructions, this reaction was a
two-step process. The first step was to remove the genomic DNA,
adding 1 µl gDNA eraser and 2 µl gDNA eraser buffer to the RNA
sample, which was then incubated at 42°C for 2 min. The second step
was to synthesize cDNA by adding the 1 µl enzyme, 1 µl primer, 4 µl
buffer and 4 µl RNase-free H2O to the 10 µl gDNA
eraser-treated sample. The reaction temperature was 37°C for 15 min
and 85°C for 5 sec. Quantitative PCR (qPCR) was conducted using an
Applied Biosystems ABI 7500 system (Thermo Fisher Scientific, Inc.)
using Power SYBR® Green PCR Master mix (Thermo Fisher
Scientific, Inc.). The RT PCR conditions were as follows: Initial
denaturation at 50°C for 20 sec and 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec; annealing at 60°C for 1 min and
extension at 72°C for 1 min. Moreover, 18S served as an endogenous
control for data normalization. The 2−ΔΔCq method
(25) was used to analyze the
relative expression.
 | Table I.Primer sequences used in the reverse
transcription-quantitative PCR assay. |
Table I.
Primer sequences used in the reverse
transcription-quantitative PCR assay.
| Gene | Primer sequence
(5′à3′) |
|---|
| 18S | Forward:
TCTAGCAATATGAGATTGAGCAATAAG |
|
| Reverse:
AATACACGTTGATACTTTCATTGTAGC |
| ND1 | Forward:
GTTATAGTAGCTGGTTGGTCGTC |
|
| Reverse:
AAGGAGTCCGATTAGTTTCAGC |
| ND42 | Forward:
CAAGAAGATGCTCGACTGGC |
|
| Reverse:
TGTCTGCATTGTAGCCAGGA |
| ND75 | Forward:
AAGCTCTTCCTTACCGAACTG |
|
| Reverse:
ATCGATGCTGCTCACCTTAC |
| sdhB | Forward:
CCATCGCCGAGATCAAGAAG |
|
| Reverse:
GGGACGAACTGGGAGTAGA |
| cytb | Forward:
GGATACGTATTACCTTGAGGACAAA |
|
| Reverse:
CAACAGCAAATCCACCTCATAATC |
| COX1 | Forward:
TGGTGGATTTGGAAATTGATTAGTG |
|
| Reverse:
GTAAAGAAAGAGCAGGAGGTAGAA |
| PGC-1α | Forward:
AAGACGTGCCTTCTGTCGTTCATC |
|
| Reverse:
ATTCGGTGCTGGTGCTTCCTTG |
| FOXO | Forward:
CTCATCCAATGCCAGTTCCT |
|
| Reverse:
TCATCGTTGTGTTCTGGTAGTC |
Western blotting for detection of
protein expression
Drosophila grouping was the same as
aforementioned. The the chest tissue of 20 Drosophila was
cut on ice. Fresh tissues were lysed with 200 µl ice-cold RIPA
buffer (Solarbio Science & Technology Co., Ltd.) containing 1
mM phenylmethylsulfonyl fluoride (Beijing Solarbio Science &
Technology Co., Ltd.). Subsequently, tissues were ground to a
homogenate, followed by centrifugation at 14,000 × g for 15 min at
4°C. Then, 120 µl supernatant liquid was collected and added to 40
µl 4Xloading buffer (Beijing Solarbio Science & Technology Co.,
Ltd.), mixed and boiled at 100°C for 10 min. The samples were
stored at −20°C prior to further experiments.
For analytical SDS-PAGE, 10 µl protein was loaded
per lane to 5% concentrated gel (30% acrylamide, 10% SDS; 10% APS;
1 M Tris-HCl pH 6.8; TEMED) and dissociated in 10% separation gel
(30% acrylamide; 10% SDS; 10% APS; 1.5 M Tris-HCl pH 8.8; TEMED).
The proteins were then transferred to 0.2-µm PVDF membranes
(Beijing Solarbio Science and Technology Co., Ltd.), blocked in 5%
milk for 2 h at room temperature and washed in TBS-T (TBS, 3 M
NaCl, 200M Tris; pH 7.5, containing 0.1% Tween-20). Subsequently,
overnight incubation at 4°C was performed with the following
primary antibodies: Rabbit anti-MnSOD (1:1,000; cat. no. ab13534;
Abcam; polyclonal), rabbit anti-α tubulin (1:1,000; cat. no.
ab52866; Abcam; monoclonal) and mouse anti-β-catenin (1:1,000; cat.
no. AB_528089; Developmental Studies Hybridoma Bank; monoclonal).
All the primary antibodies were Drosophila-specific.
Following washing with TBS-T, the membrane was incubated with the
corresponding secondary antibody, horseradish peroxidase-AffiniPure
Goat Anti-Rabbit/Mouse IgG (H+L; 1:5,000; cat. nos. EM35111–01 and
EM35110-01; Emarbio Science and Technology Co., Ltd.), for 1 h at
room temperature. All the resulting immune complexes were
visualized with chemiluminescence reagent (Thermo Fisher
Scientific, Inc.,), followed by imaging using Image Lab 5.1
(National Institutes of Health). The target protein bands were
quantified by scanning densitometry using ImageJ software (v.1.49v;
National Institutes of Health).
ATP, malondialdehyde (MDA) and
reactive oxygen species (ROS)
In total, ten Drosophila thoraxes were cut on
ice from each group, and 1,000 µl lysate was added to homogenize
the chest tissue on ice. Samples were then heated at 100°C for 2
min and centrifuged at 12,000 × g for 5 min at 4°C. The thoracic
ATP level was measured using a luciferase-based bioluminescence
assay (cat. no. S0027; Beyotime Institute of Biotechnology). Then,
40 fly chests from each group were ground by adding 500 µl 1X PBS
and centrifuge at 1,425 × g for 10 min at 4°C. The supernatant was
collected and the MDA content was measured by the thiobarbituric
acid method (26). Analyses were
performed according to the instructions of the reagent kit for MDA
(cat. no. A003-1-2; Nanjing Jiancheng Bioengineering Institute).
ROS were measured using the CellROX Orange reagent (cat. no.
BB-470512; Shanghai Bio-Tech Co., Ltd.). The thoraxes of 30
Drosophila were obtained, homogenized, centrifuged at 1,000
× g for 10 min at 4°C and the supernatant was collected. The
supernatant and 20 µM CellROX Orange Reagent were mixed and
incubated at 37°C in the dark for 30 min, and the fluorescence
intensity at 510 and 610 nm (maximum excitation light and maximum
emission wavelength) was measured using a multi-function microplate
reader.
Transmission electron microscopy
analysis
Drosophila was grouped as aforementioned, and
10 male flies from each group F1 generation were randomly selected
on day 5. Flies were anesthetized by CO2, and the chest
was cut carefully so as not to damage the muscle tissue. Thoraxes
were fixed overnight at 4°C in 2.5% glutaraldehyde, washed several
times with 0.1 mol/l phosphate buffer and post-fixed in 1%
osmiumtetroxide in distilled water for 2 h at room temperature. The
samples were dehydrated in a 50, 70 and 90% graded ethanol series,
and embedded in Epoxy resin for 48 h at room temperature. The
polymerization conditions in the polymerization tank were 36°C for
24 h, 45°C for 12 h and 65°C for 48 h.
The embedded polymer samples were cut into 1 µm
sections using a Leica UC7 ultrathin slicer, stained with 1%
toluidine blue for 30 sec at room temperature and observed under an
optical microscope (magnification, ×100). The samples were then cut
into ultra-thin sheets (70 nm), stained with 3% uranyl acetate for
15 min at room temperature and 3% lead citrate for 15 min at room
temperature. Sections were observed with a Hitachi H-7650
transmission electron microscope (magnification, ×30,000).
Statistical analysis
Statistical analysis of data was performed using
SPSS 16.0 (IBM Corp.). Normally distribution continuous variable
data were compared by one-way ANOVA followed by Bonferroni's post
hoc test. Data are presented as the mean ± SD. P<0.05 was
considered to indicate a statistically significant difference.
Results
Wnt2 overexpression rescues the
abnormal phenotype caused by PINK1B9 mutation
The Wnt2 gene was specifically expressed in
Drosophila flight muscles using the MHC-GAL4 promoter. The
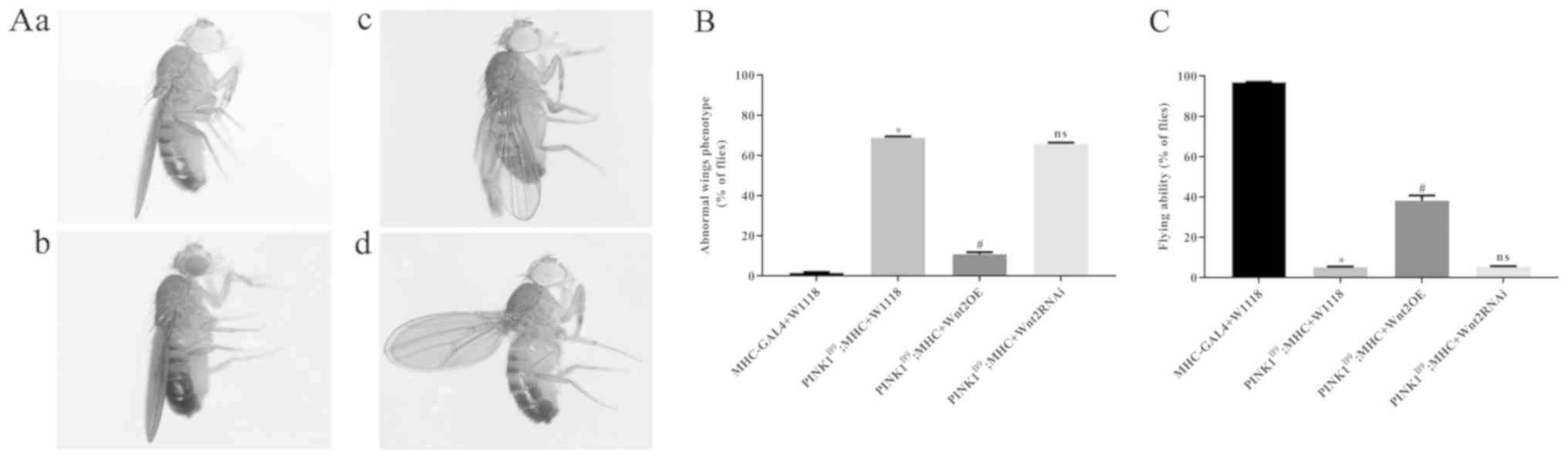
phenotypic changes of Drosophila were demonstrated by
changes in wing morphology and flight ability. In the normal
control group, most of the wings completely overlapped and were
parallel to the body (Fig. 1A).
However, in the transgenic disease model of
PINK1B9 the wings had bifurcations, erections and
sagged. Furthermore, compared with the low rate of abnormal wings
in the control group, the disease group had a significantly higher
rate of abnormal wings (Fig. 1B)
and a significant decrease in flight ability (Fig. 1C). In the Wnt2OE
intervention group, the incidence of wing anomalies was
significantly reduced and the flight capabilities were improved,
compared with the disease group. Moreover, there were no
significant differences between the Wnt2 RNAi intervention
group and the PD disease model group. Therefore, the present
results suggested that Wnt2OE may have a protective effect
on the phenotype of PINK1B9 transgenic
Drosophila.
Wnt2 gene overexpression enhances
mitochondrial function in PINK1B9 transgenic
Drosophila
RT-qPCR results demonstrated that in
PINK1B9 disease model group, the mRNA expression
levels of the mitochondrial complex subunit-related genes, Complex
I [NADH-ubiquinone oxidoreductase chain 1 (ND1), ND42
and ND75], Complex II (succinate dehydrogenase complex
subunits B), Complex III (Cytochrome b) and Complex IV
(Cyclooxygenase 1), decreased significantly. While Wnt2OE
intervention in PINK1B9 transgenic
Drosophila increased the mRNA expression levels of these
related genes (P<0.05), in the Wnt2 RNAi intervention
group there was no significant difference compared with the disease
model group (Fig. 2A). The results
of the ATP assay demonstrated that ATP production in the PD model
group was decreased. Furthermore, the amount of ATP produced by
mitochondria in the Wnt2OE intervention group was ~1.5 times
higher compared with the disease model group (Fig. 2B). Ultrastructural transmission
electron microscopy analysis identified that mitochondria were
disrupted in PINK1B9 transgenic
Drosophila, and mitochondrial morphology was not
recognizable. Moreover, Wnt2OE could rescue mitochondrial
defects in PINK1B9 flies (Fig. 2C). Therefore, the present results
suggested that overexpression of Wnt2 can improve the
mitochondrial function of PINK1B9 transgenic
Drosophila.
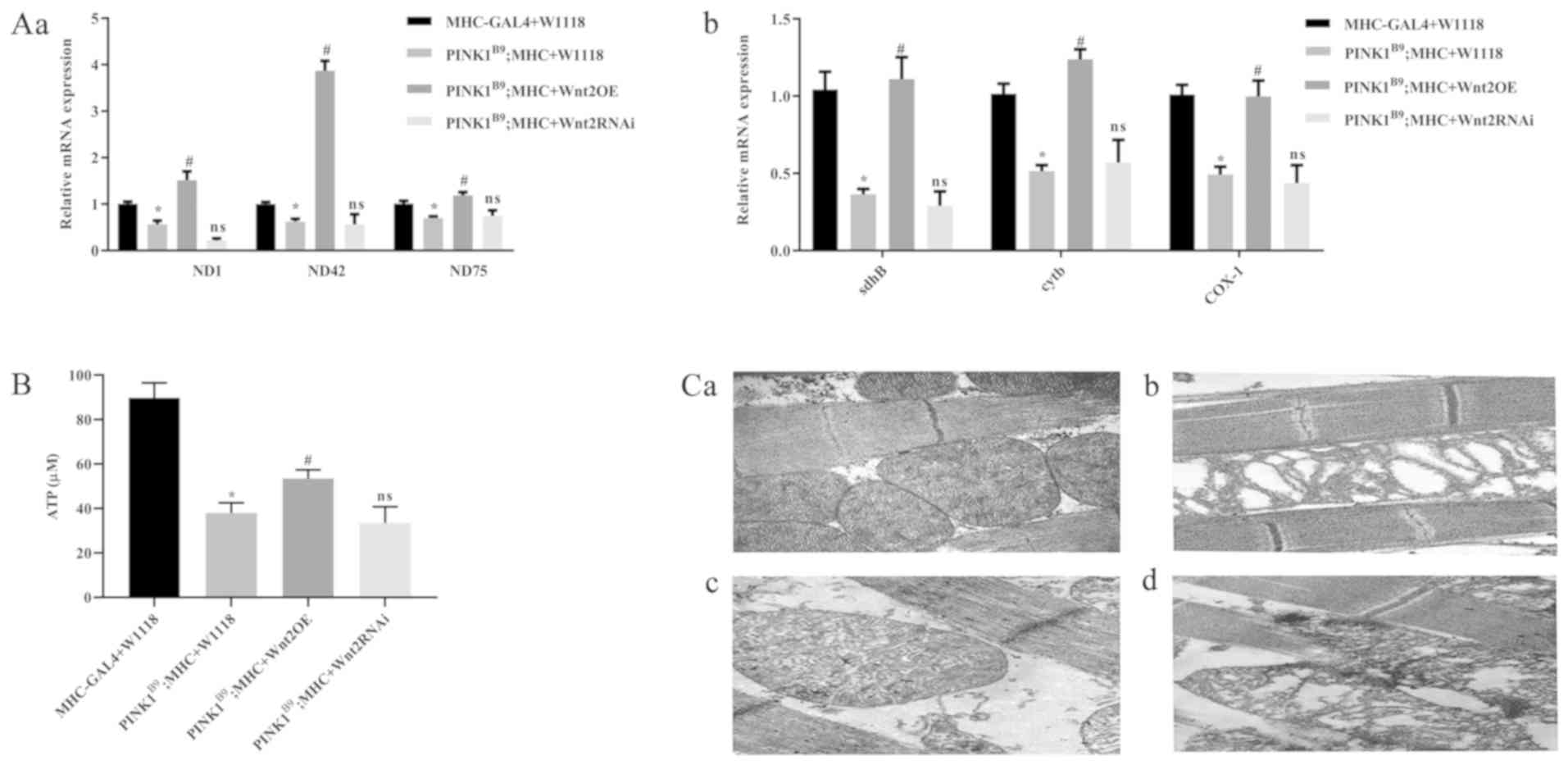 | Figure 2.Overexpression of Wnt2 rescues
mitochondrial function. (A) Overexpression of Wnt2 increased
mitochondrial complex subunit-related genes. (Aa) mRNA level of
mitochondrial complex subunit ND1, ND42 and ND75, (Ab) mRNA level
of mitochondrial complex subunit sdhB, cytb and COX-1. (B) ATP
levels in flies. (C) Abnormal mitochondrial morphology of
PINK1B9 flies was restored by Wnt2
overexpression. (Ca) Control flies, (Cb) PINK1B9 flies, (Cc)
Wnt2OE flies and (Cd) Wnt2RNAi flies. Magnification,
×30,000. Scale bar, 1 µm. n=30, 5-day-old males. *P<0.05 vs. the
control flies. #P<0.05 vs. the
PINK1B9 flies. ns vs. the
PINK1B9 flies. PINK1, PTEN induced putative
kinase 1; Wnt2OE, Wnt2 overexpression;
Wnt2RNAi, Wnt2 RNA interference; ns, not significant;
ND, NADH-ubiquinone oxidoreductase chain 1; sdhB, succinate
dehydrogenase complex subunits B; cytb, Cytochrome b; COX1,
Cyclooxygenase 1. |
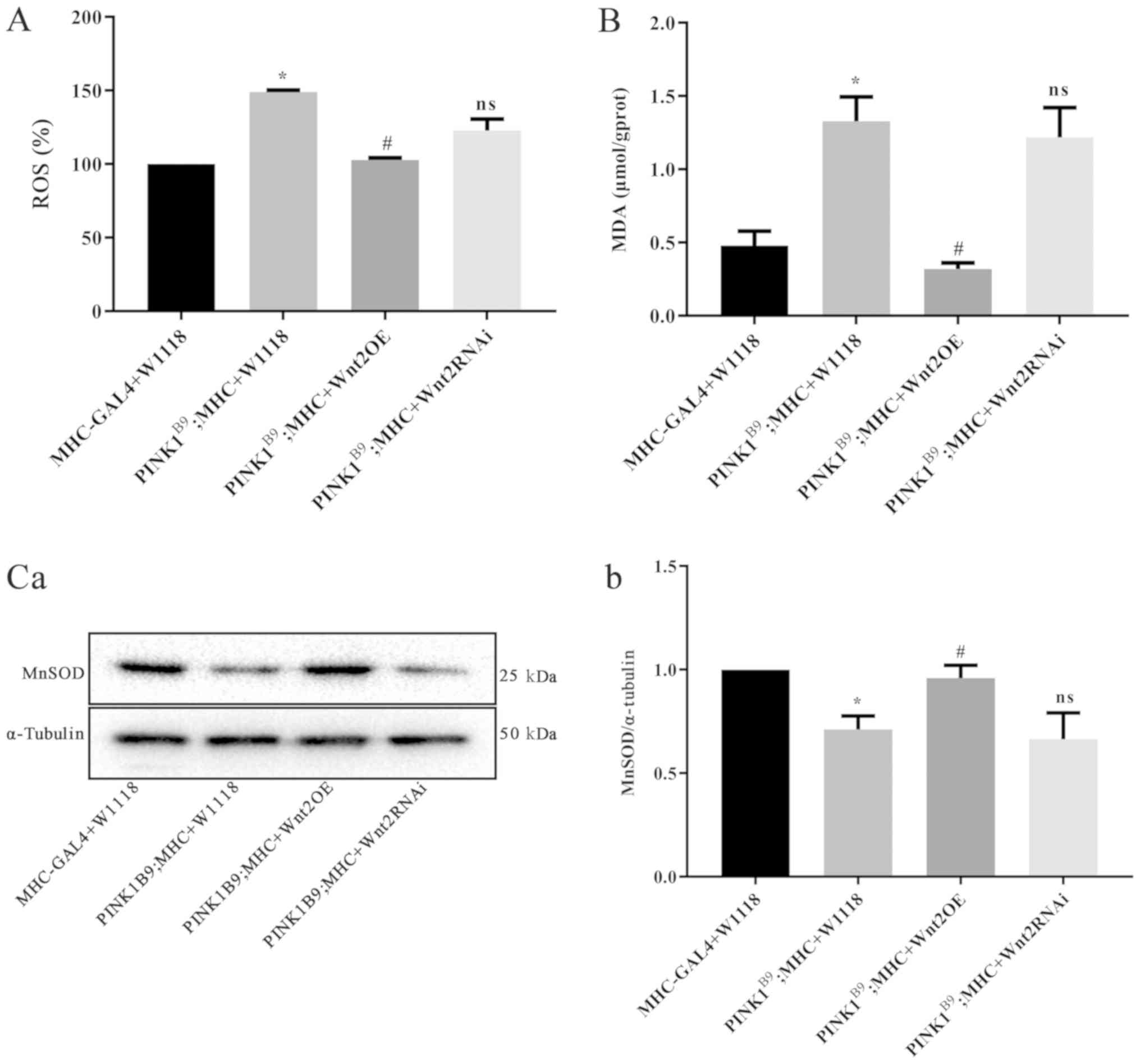
Wnt2OE reduces oxidative stress damage
in PINK1B9 transgenic Drosophila
ROS production in the PINK1B9
disease model group was significantly higher compared with the
normal control group (P<0.05; Fig.
3A). Furthermore, following Wnt2OE intervention in
PINKB9 transgenic Drosophila, ROS
production was significantly reduced (P<0.05) and almost
returned to normal levels. MDA, a commonly used indicator of lipid
peroxidation injury (27), was
significantly increased in the PINK1B9 disease
model group compared with the normal control (P<0.05; Fig. 3B). Moreover, after Wnt2OE
intervention, MDA production was reduced (P<0.05). It was
demonstrated that the content of ROS and MDA were not significantly
different between the Wnt2RNAi intervention groups and the
disease model group (Fig. 3A and
B).
Western blot analysis results revealed that the
protein expression of MnSOD in the PINK1B9 disease model
group was significantly lower compared with the normal control
group (P<0.05; Fig. 3C).
However, the expression of MnSOD was significantly increased
(P<0.05) following Wnt2OE intervention in the
PINK1B9 disease model. Collectively, the present
results indicated that Wnt2 overexpression reduced oxidative
damage in PINK1B9 transgenic
Drosophila.
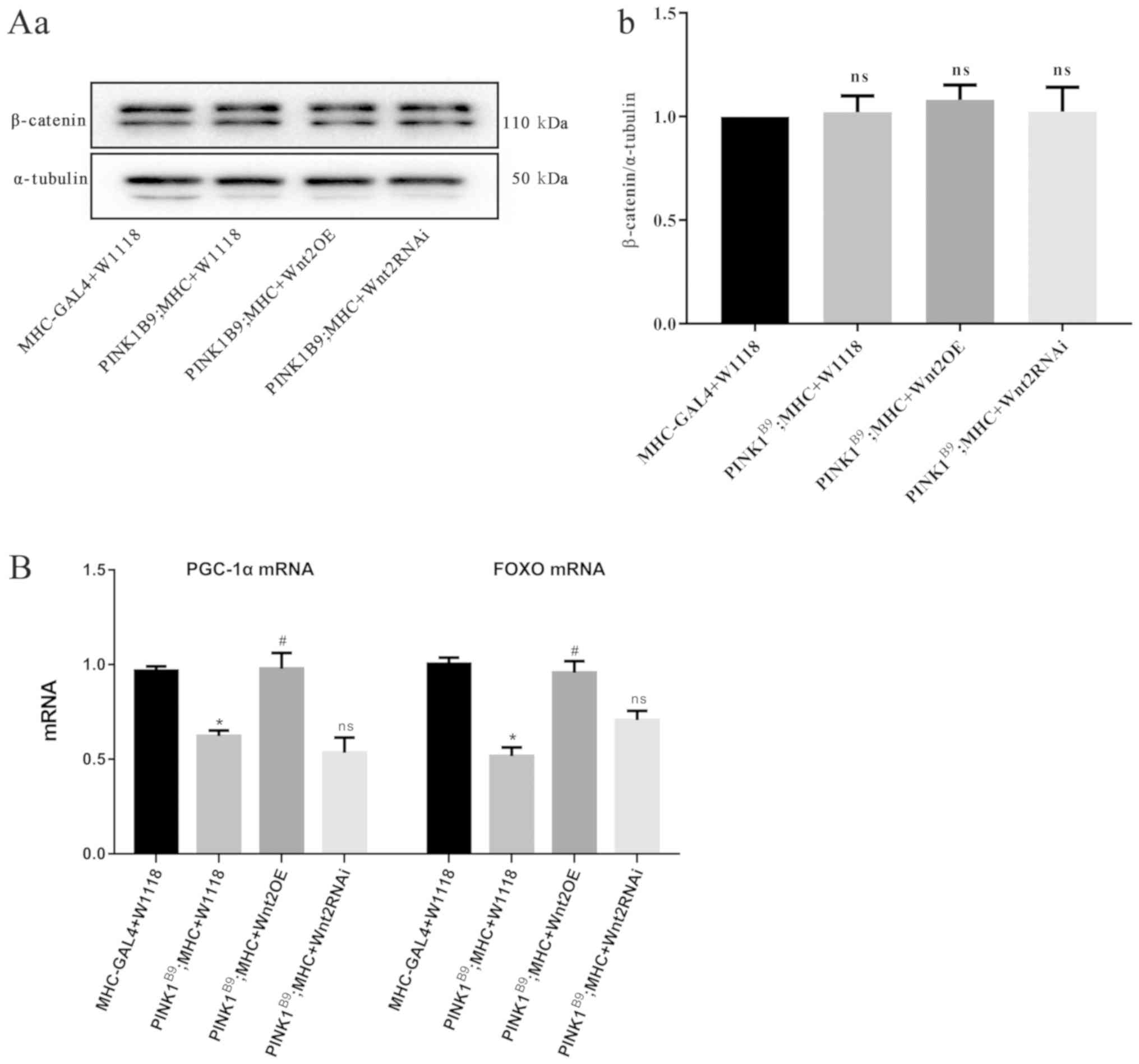
Possible mechanism of Wnt2
overexpression-mediated protection
Western blot analysis results revealed that there
was no significant difference in the protein expression of
β-catenin, a key molecule of the classic Wnt/β-catenin signaling
pathway (28), between each group
(Fig. 4A). Moreover, RT-qPCR
showed that the mRNA expression levels of FOXO and
PGC-1α were decreased in the PINK1B9
disease model group, and were increased following Wnt2OE
intervention in the PINK1B9 disease model
(Fig. 4B). Therefore, it can be
speculated that Wnt2 may activate the FOXO/PGC-1α signaling
pathway to regulate mitochondrial function and inhibit
anti-oxidative stress-induced damage.
Discussion
In Drosophila melanogaster, mutations in
PINK1 causes phenotypic abnormalities and decrease exercise
capacity, resulting in dysfunction of mitochondria and decreased
ATP levels, which leads to selective degeneration of DA neurons and
sensitivity to stress (16,17).
In the present study, the disease model demonstrated a pathological
phenotype that was consistent with previous experimental results
(24,29), including a high abnormal wing rate,
low flight rate, lower mRNA expression of mitochondrial complex
subunits and lower ATP level, which suggested that PINK1
mutations could cause serious damage in mitochondria. Furthermore,
overexpression of Wnt2 gene rescued the phenotype of
PINK1B9 transgenic Drosophila, increased
ATP production level and enhanced the expression of the
mitochondrial biosynthesis-related gene PGC-1α. PGC-1α is a
transcriptional cofactor for numerous mitochondrial proteins, which
can regulate mitochondrial function to meet cellular needs and
protect from oxidative damage (30). Therefore, it is speculated that
Wnt2OE inhibited mitochondrial dysfunction caused by the
PINK1 mutation and eventually protects
PINK1B9 transgenic fruit flies.
When mitochondria are damaged, particularly damage
to the mitochondrial respiratory chain complex, electron chain
leakage may occur and cause the production of superoxide and
hydrogen peroxide (31). These
products, not only participate in the damage of DNA, proteins and
lipids, but also pose a serious threat to cells and tissues
(31). Mitochondrial dysfunction
and oxidative stress play key roles in the development of PD, as
both lead to excessive ROS production (32). This in turn leads to damage and
death of DA neurons (33). A small
level of ROS is essential for normal physiological functions;
however, the accumulation of large amounts of ROS further destroys
mitochondria and exacerbates oxidative stress (34). It has been shown that maintenance
of ATP and inhibition of ROS production protects DA neurons
(35). Reactive oxygen damages
polyunsaturated lipids and forms MDA, which is a marker of lipid
damage in oxidative stress (36).
Furthermore, the present results indicated that the PINK1
mutation increased ROS and MDA levels, but reduced MnSOD protein
expression.
MnSOD, which is mainly distributed in the
mitochondrial matrix, is an important scavenger for the superoxide
anion that is produced during mitochondrial oxidative
phosphorylation (37). Previous
findings have shown that overexpression of MnSOD serves an
important role in the protection of PINK1-mutant PD
Drosophila (38). In the
PINK1 mutants, the present study identified increased ROS
and MDA production, and a reduced MnSOD, thus suggesting that the
PINK1 mutation leads to oxidative stress damage. Moreover,
overexpression of Wnt2 improved mitochondrial function,
which also partially reduced ROS production. However, Wnt2
overexpression also appeared to directly participate in the
regulation of oxidative stress levels in PINK1B9
transgenic Drosophila, as it not only reduced ROS and MDA
production levels, but also increased the protein expression levels
of MnSOD.
The Drosophila gene Wnt2, homologous
to the human gene Wnt7a, is involved in the Wnt/β-catenin
signaling pathway (39). However,
it remains unknown whether the protective effect of Wnt2
overexpression on PINK1B9 transgenic
Drosophila is related to the Wnt/β-catenin signaling
pathway. The present results indicated that Wnt2
overexpression did not increase the protein expression of
β-catenin, a key protein of the Wnt/β-catenin signaling pathway
(28), which suggested that it did
not protect PINK1B9 transgenic flies via the
Wnt/β-catenin signaling pathway. Furthermore, a previous study
found that Wnt2 does not act via the Wnt/β-catenin pathway,
but activates the non-canonical pathway (40). Thus, this raises the question of
how Wnt2 improves mitochondrial function, reduces oxidative
damage and enhances antioxidant capacity. The present study
evaluated the expression of FOXO, a gene associated with
mitochondrial oxidative stress, which regulates the expression
levels of PGC-1α and MnSOD (38,41).
Under conditions of mitochondrial dysfunction and oxidative stress
caused by PINK1 mutation, the present study hypothesized
that the overexpression of Wnt2 directly regulates the
expression of PGC-1α/FOXO/MnSOD, improves mitochondrial function
and improves antioxidant capacity to rescue
PINK1B9 transgenic fruit flies.
The FOXO family members are key regulators
of neuronal processes, such as dendritic structural function and
memory consolidation (42,43). FOXO is also an important
regulator of cellular stress response, which enhances cellular
antioxidant defenses (44). A
previous study revealed that the expression of genes, such as
MnSOD, could be controlled by the forkhead transcription
factor FOXO3a (44).
Moreover, PGC-1α has been shown to regulate FOXO
activity in different systems. For example, it has been reported
that PGC-1α is a positive regulator of fasting-induced
hepatic gluconeogenesis, which is mediated by its interaction with
FOXO1a (45). Similarly,
overexpression of PGC-1α enhances the stimulatory effect of
FOXO1a on selenoprotein P promoter activity and insulin
attenuation (46). PGC-1α
has also been shown to interact with FOXO3a, which regulates
antioxidant gene expression in endothelial cells and skeletal
muscle (47). In addition,
upregulation of PGC-1α and FOXO3a protects against
oxidative stress injury induced by a high-fat diet and inhibits
adipocyte apoptosis (47,48). The Wnt signaling pathway is also
involved in the regulation of mitochondrial energy metabolism and
oxidative stress (49). When ROS
levels exceed the body's ability to scavenge, Wnt and
β-catenin interact with FOXO under stimuli of
oxidative stress (13,50). Furthermore, FOXO1 interacts
with PGC-1α in different systems (45,46).
PGC-1α is a mitochondrial energy-metabolizing enzyme. When ROS are
in excess, the human body can enhance the detoxification ability of
mitochondrial ROS by increasing the expression levels of
FOXO1 and PGC-1α to promote the expression of
downstream antioxidant systems, such as MnSOD (49).
To the best of our knowledge, the condition of
mitochondrial dysfunction caused by PINK1 mutation and
oxidative stress remains to be determined. Although the Wnt
signaling pathway is linked to the FOXO signaling pathway via
β-catenin (50), the present
results suggested that Wnt2 did not change the β-catenin
protein expression level, but did affect the mRNA expression levels
of PGC-1α and FOXO, and the expression levels of
their target protein MnSOD. Based on these results, it can be
speculated that Wnt2 may be directly involved in the
PGC-1α/FOXO/MnSOD signaling pathway; however, the specific
mechanism remains to be investigated. Moreover, the regulation of
signaling pathways is complex and there will be cross-effects
between the pathways (47–48), with both upregulation and
inhibition of the expression of related factors leading to cascade
reactions. Therefore, it will be beneficial to examine how the Wnt2
pathway interacts with or influences the PGC-1α/FOXO/MnSOD pathway
in future experiments.
The present study demonstrated that Wnt2
overexpression protected PINK1B9 transgenic flies
by improving flight muscle and mitochondrial morphology, and
enhancing mitochondrial complex I and II function. Furthermore, it
was found that Wnt2 overexpression exhibited a protective
effect on early mitochondria and oxidative stress-related PD by
improving mitochondrial function and reducing oxidative stress
damage. However, the present study does have some limitations.
First, the model is monotonous and limited to fruit flies, and thus
requires further examination in higher animal models such as mice.
Secondly, further research into the underlying pathways and
mechanisms is required, such as whether it is the experimental
effects that are related to the non-classical Wnt signaling
pathway. Furthermore, how Wnt2 cross-links with the
PGC-1α/FOXO/MnSOD signaling pathway is not fully understood. In the
PINK1 mutant PD Drosophila model of mutation
constructed by Park et al (24), mitochondrial dysfunction and
oxidative stress injury are primarily exhibited in the early stage,
while DA neuronal loss predominantly occurs in the middle and late
stages of the Drosophila, which is after 25 days (24,50).
This is consistent with the progressive DA neuronal loss observed
in clinical patients with PD. Thus, this may indicate that damage
to DA neurons in PD is the result of further neurological damage
caused by these pathogenic factors.
To the best of our knowledge, there are currently
no treatments that mitigate disease progression or prevent
neurodegeneration in all neurodegenerative disease. Therefore, it
is necessary to develop interventions that are effective prior to
severe damage of the neuronal in the early stages of PD. The
present results indicated that Wnt2 overexpression had a
protective effect on early mitochondrial damage and oxidative
stress in PD, improving mitochondrial function and reducing
oxidative stress damage. Due to the limitations of the present
study, the specific mechanism of action of Wnt2 is not fully
understood. However, the improvement of mitochondrial function and
the reduction of oxidative stress damage can support the
experimental results of the Drosophila model, providing a
strong basis for future experiments.
In conclusion, the present results suggested that
the Wnt2 gene may have a protective effect on
PINK1B9 transgenic Drosophila. Therefore,
it can be hypothesized that the reduction of oxidative stress and
the restoration of mitochondrial function via Wnt2 gene
overexpression in the PINK1 mutant transgenic
Drosophila may be related to the PGC-1α/FOXO/MnSOD signaling
pathway.
Acknowledgements
The authors would like to thank Dr Yu-Feng Yang,
from Institute of life Sciences of Fuzhou University, for donating
the PINK1B9 Drosophila model; Dr Ru-Jia Liao, Dr
Jing-Xin Mo and Dr Qing-Tuan Meng, from Guangxi Clinical Research
Center for Neurological Disease of Affliated Hospital of Guilin
Medical University, for sharing their expertise; Miss Liang-Xian
Li, intermediate research assistant, from Guangxi Key Laboratory of
Brain and Cognitive Neuroscience of Guilin Medical University; Miss
Ying Cui and Miss Xiao-Jun Diao, PhD candidate, Department of
Neurology from Xiangya School of Medicine of Central South
University; Miss Wen-Jing Wang, postgraduate student, Department of
Neurology from Guilin Medical University; Miss Fang Shi
(intermediate research assistant), Miss Ning Tian (inter- mediate
research assistant) and Miss Mei-Rong Chen (intermediate research
assistant), from Guangxi Clinical Research Center for Neurological
Disease of Affliated Hospital of Guilin Medical University, for
their help.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81460180 and
31460256), 2017 Youth and Middle School Teachers' Basic Ability
Improvement Project of the Education Department of Guangxi Zhuang
Autonomous Region (grant no. 30606017018) and the Innovation
Project of Guangxi Graduate Education (grant no. YCSW2018206).
Availability of data and materials
The datasets used and/or analyzed during the
present study are available from the corresponding author on
reasonable request.
Authors' contributions
SRX was responsible for construction of the
Drosophila models, western blotting, MDA and ROS level
determination, and data analysis. XYW was responsible for
construction of the Drosophila models, morphological
observation of Drosophila, O2K detection and data
statistics. SRX and XYW wrote the first draft of this article. XLF
was responsible for electron microscopy analysis. XRC performed the
ATP determination experiment. ZWW was responsible for mRNA
expression level detection. QHL and LS undertook project funding,
project design, manuscript revision and quality control. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhou C, Huang Y, Shao Y, May J, Prou D,
Perier C, Dauer W, Schon EA and Przedborski S: The kinase domain of
mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci USA.
105:12022–12027. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hatano Y, Li Y, Sato K, Asakawa S,
Yamamura Y, Tomiyama H, Yoshino H, Asahina M, Kobayashi S,
Hassin-Baer S, et al: Novel PINK1 mutations in early-onset
parkinsonism. Ann Neurol. 56:424–427. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu S, Sawada T, Lee S, Yu W, Silverio G,
Alapatt P, Millan I, Shen A, Saxton W, Kanao T, et al: Parkinson's
disease-associated kinase PINK1 regulates Miro protein level and
axonal transport of mitochondria. PLoS Genet. 8:e10025372012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang W, Wang X, Fujioka H, Hoppel C, Whone
AL, Caldwell MA, Cullen PJ, Liu J and Zhu X: Parkinson's
disease-associated mutant VPS35 causes mitochondrial dysfunction by
recycling DLP1 complexes. Nat Med. 22:54–63. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu Z, Wang Y, Lim J, Liu B, Li Y, Vartak
R, Stankiewicz T, Montgomery S and Lu B: Ubiquitination of ABCE1 by
NOT4 in response to mitochondrial damage links co-translational
quality control to PINK1-directed mitophagy. Cell Metab.
28:130–144.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Oh SE, Park HJ, He L, Skibiel C, Junn E
and Mouradian MM: The Parkinson's disease gene product DJ-1
modulates miR-221 to promote neuronal survival against oxidative
stress. Redox Biol. 19:62–73. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Burbulla LF, Song P, Mazzulli JR, Zampese
E, Wong YC, Jeon S, Santos DP, Blanz J, Obermaier CD, Strojny C, et
al: Dopamine oxidation mediates mitochondrial and lysosomal
dysfunction in Parkinson's disease. Science. 357:1255–1261. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Andersson ER, Saltó C, Villaescusa JC,
Cajanek L, Yang S, Bryjova L, Nagy II, Vainio SJ, Ramirez C, Bryja
V, et al: Wnt5a cooperates with canonical Wnts to generate midbrain
dopaminergic neurons in vivo and in stem cells. Proc Natl Acad Sci
USA. 110:E602–E610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sousa KM, Villaescusa JC, Cajanek L, Ondr
JK, Castelo-Branco G, Hofstra W, Bryja V, Palmberg C, Bergman T,
Wainwright B, et al: Wnt2 regulates progenitor proliferation in the
developing ventral midbrain. J Biol Chem. 285:7246–7253. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Marui T, Funatogawa I, Koishi S, Yamamoto
K, Matsumoto H, Hashimoto O, Jinde S, Nishida H, Sugiyama T, Kasai
K, et al: Association between autism and variants in the
wingless-type MMTV integration site family member 2 (WNT2) gene.
Int J Neuropsychopharmacol. 13:443–449. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang X, Abreu JG, Yokota C, MacDonald BT,
Singh S, Coburn KL, Cheong SM, Zhang MM, Ye QZ, Hang HC, et al:
Tiki1 is required for head formation via Wnt cleavage-oxidation and
inactivation. Cell. 149:1565–1577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kahn M: Can we safely target the WNT
pathway? Nat Rev Drug Discov. 13:513–532. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Parker JA, Vazquez-Manrique RP, Tourette
C, Farina F, Offner N, Mukhopadhyay A, Orfila AM, Darbois A, Menet
S, Tissenbaum HA, et al: Integration of β-catenin, sirtuin, and
FOXO signaling protects from mutant huntingtin toxicity. J
Neurosci. 32:12630–12640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Arrázola MS, Ramos-Fernández E, Cisternas
P, Ordenes D and Inestrosa NC: Wnt signaling prevents the Aβ
oligomer-induced mitochondrial permeability transition pore opening
preserving mitochondrial Structure in hippocampal neurons. PLoS
One. 12:e01688402017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wei L, Ding L, Mo MS, Lei M, Zhang L, Chen
K and Xu P: Wnt3a protects SH-SY5Y cells against 6-hydroxydopamine
toxicity by restoration of mitochondria function. Transl
Neurodegener. 4:112015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang Y, Gehrke S, Imai Y, Huang Z, Ouyang
Y, Wang JW, Yang L, Beal MF, Vogel H and Lu B: Mitochondrial
pathology and muscle and dopaminergic neuron degeneration caused by
inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl
Acad Sci USA. 103:10793–10798. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu W, Acín-Peréz R, Geghman KD, Manfredi
G, Lu B and Li C: Pink1 regulates the oxidative phosphorylation
machinery via mitochondrial fission. Proc Natl Acad Sci USA.
108:12920–12924. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dung VM and Thao DTP: Parkinson's Disease
Model. Adv Exp Med Biol. 1076:41–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bilder D and Irvine KD: Taking stock of
the Drosophila research ecosystem. Genetics. 206:1227–1236. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Barwell T, DeVeale B, Poirier L, Zheng J,
Seroude F and Seroude L: Regulating the UAS/GAL4 system in adult
Drosophila with Tet-off GAL80 transgenes. PeerJ. 5:e41672017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Southall TD, Elliott DA and Brand AH: The
GAL4 system: A versatile toolkit for gene expression in Drosophila.
CSH Protoc. doi:10.1101/pdb.top49.
|
|
22
|
Tu H and Casadaban MJ: The upstream
activating sequence for L-leucine gene regulation in Saccharomyces
cerevisiae. Nucleic Acids Res. 18:3923–3931. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Robertson LK, Dey BK, Campos AR and
Mahaffey JW: Expression of the drosophila gene disconnected using
the UAS/GAL4 system. Genesis. 34:103–106. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Park J, Lee SB, Lee S, Kim Y, Song S, Kim
S, Bae E, Kim J, Shong M and Kim JM: Mitochondrial dysfunction in
Drosophila PINK1 mutants is complemented by parkin. Nature.
441:1157–1161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rogério M, Cardoso C and Pestana C:
Measurement of malondialdehyde in fish: A comparison study between
HPLC methods and the traditional spectrophotometric test. Food
Chem. 112:1038–1045. 2009. View Article : Google Scholar
|
|
27
|
Dragun Z, Filipović Marijić V, Krasnići N,
Ramani S, Valić D, Rebok K, Kostov V, Jordanova M and Erk M:
Malondialdehyde concentrations in the intestine and gills of Vardar
chub (Squalius vardarensis Karaman) as indicator of lipid
peroxidation. Environ Sci Pollut Res Int. 24:16917–16926. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boonchai W, Walsh M, Cummings M and
Chenevix-Trench G: Expression of beta-catenin, a key mediator of
the WNT signaling pathway, in basal cell carcinoma. Arch Dermatol.
136:937–938. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gehrke S, Wu Z, Klinkenberg M, Sun Y,
Auburger G, Guo S and Lu B: PINK1 and Parkin control localized
translation of respiratory chain component mRNAs on mitochondria
outer membrane. Cell Metabolism. 21:95–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsunemi T and La Spada AR: PGC-1α at the
intersection of bioenergetics regulation and neuron function: From
Huntington's disease to Parkinson's disease and beyond. Prog
Neurobiol. 97:142–151. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zorov DB, Juhaszova M and Sollott SJ:
Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS
release. Physiol Rev. 94:909–950. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ray A, Martinez BA, Berkowitz LA, Caldwell
GA and Caldwell KA: Mitochondrial dysfunction, oxidative stress,
and neurodegeneration elicited by a bacterial metabolite in a C.
elegans Parkinson's model. Cell Death Dis. 5:e9842014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Subramaniam SR and Chesselet MF:
Mitochondrial dysfunction and oxidative stress in Parkinson's
disease. Prog Neurobiol 106–107. 17–32. 2013. View Article : Google Scholar
|
|
34
|
Kudryavtseva AV, Krasnov GS, Dmitriev AA,
Alekseev BY, Kardymon OL, Sadritdinova AF, Fedorova MS, Pokrovsky
AV, Melnikova NV, Kaprin AD, et al: Mitochondrial dysfunction and
oxidative stress in aging and cancer. Oncotarget. 7:44879–44905.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zuo L and Motherwell MS: The impact of
reactive oxygen species and genetic mitochondrial mutations in
Parkinson's disease. Gene. 532:18–23. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Weismann D, Hartvigsen K, Lauer N, Bennett
KL, Scholl HP, Charbel Issa P, Cano M, Brandstätter H, Tsimikas S
and Skerka C: Complement factor H binds malondialdehyde epitopes
and protects from oxidative stress. Nature. 478:76–81. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sheshadri P and Kumar A: Managing odds in
stem cells: Insights into the role of mitochondrial antioxidant
enzyme MnSOD. Free Radic Res. 50:570–584. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Koh H, Kim H, Kim MJ, Park J, Lee HJ and
Chung J: Silent information regulator 2 (Sir2) and Forkhead box O
(FOXO) complement mitochondrial dysfunction and dopaminergic neuron
loss in Drosophila PTEN-induced kinase 1 (PINK1) null mutant. J
Biol Chem. 287:12750–12758. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Swarup S and Verheyen EM: Wnt/Wingless
signaling in Drosophila. Cold Spring Harb Perspect Biol.
4:a0079302012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yu M, Ting DT, Stott SL, Wittner BS,
Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D and Gilman AJ:
RNA sequencing of pancreatic circulating tumour cells implicates
WNT signalling in metastasis. Nature. 487:510–513. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mukherjee S and Duttaroy A: Spargel/dPGC-1
is a new downstream effector in the insulin-TOR signaling pathway
in Drosophila. Genetics. 195:433–441. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Salih DA, Rashid AJ, Colas D, de la
Torre-Ubieta L, Zhu RP, Morgan AA, Santo EE, Ucar D, Devarajan K,
Cole CJ, et al: FoxO6 regulates memory consolidation and synaptic
function. Genes Dev. 26:2780–2801. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sears JC and Broihier HT: FoxO regulates
microtubule dynamics and polarity to promote dendrite branching in
Drosophila sensory neurons. Dev Biol. 418:40–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kops GJ, Dansen TB, Polderman PE, Saarloos
I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH and Burgering
BM: Forkhead transcription factor FOXO3a protects quiescent cells
from oxidative stress. Nature. 419:316–321. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Puigserver P, Rhee J, Donovan J, Walkey
CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D,
et al: Insulin-regulated hepatic gluconeogenesis through
FOXO1-PGC-1alpha interaction. Nature. 423:550–555. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Speckmann B, Walter PL, Alili L, Reinehr
R, Sies H, Klotz LO and Steinbrenner H: Selenoprotein P expression
is controlled through interaction of the coactivator PGC-1alpha
with FoxO1a and hepatocyte nuclear factor 4alpha transcription
factors. Hepatology. 48:1998–2006. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chung HW, Lim JH, Kim MY, Shin SJ, Chung
S, Choi BS, Kim HW, Kim YS, Park CW and Chang YS: High-fat
diet-induced renal cell apoptosis and oxidative stress in
spontaneously hypertensive rat are ameliorated by fenofibrate
through the PPARα-FoxO3a-PGC-1α pathway. Nephrol Dial Transplant.
27:2213–2225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Geng T, Li P, Yin X and Yan Z: PGC-1α
promotes nitric oxide antioxidant defenses and inhibits FOXO
signaling against cardiac cachexia in mice. Am J Pathol.
178:1738–1748. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Essers MA, de Vries-Smits LM, Barker N,
Polderman PE, Burgering BM and Korswagen HC: Functional interaction
between beta-catenin and FOXO in oxidative stress signaling.
Science. 308:1181–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yang Y, Cehrke S, Imai Y, Huang Z, Ouyang
Y, Wang JW, Yang L, Beal MF, Vogel H and Lu B: Mitochondrial
pathology and muscle and dopaminergic neuron degeneration caused by
inactivitation of Drosophila Pink1 is rescued by Parkin. Proc Natl
Acad Sci USA. 103:10793–10798. 2006. View Article : Google Scholar : PubMed/NCBI
|