Introduction
Necrotizing enterocolitis (NEC) is an acute
life-threatening gastrointestinal disease that predominantly
affects premature neonates (1,2). The
morbidity of NEC is rising rapidly with the continuing increased
rate of survival of preterm infants (3). Patients that survive severe NEC may
develop serious sequelae, including gastrointestinal complications
and abnormal neurodevelopment (4).
Multiple factors contribute to NEC, including premature delivery,
intestinal bacterial colonization and infant formula feeding
(5). Furthermore, NEC is
characterized by severe intestinal inflammation, impaired mucosal
healing and intestinal necrosis (4). However, the etiology and pathogenesis
of NEC are not fully understood.
Long non-coding RNAs (lncRNAs) are autonomously
transcribed non-coding RNAs, which are >200 nucleotides in
length (6). lncRNAs can act at the
transcriptional, post-transcriptional and epigenetic levels via
cis- or trans-acting pathways (7,8).
Previous studies have reported the potential importance of lncRNAs
in maintaining the intestinal barrier and mediating inflammatory
lesions (9–11). For example, lncRNA Uc.173 was
revealed to enhance intestinal epithelial barrier function by
increasing the expression of claudin 1 (9). However, lncRNA Uc.173 silencing leads
to dysfunction of the intestinal epithelial barrier in vitro
(9). Another study showed that
lncRNA SPRY4-intronic transcript 1 (SPRY4-IT1) regulates the
intestinal epithelial barrier by affecting the expression levels of
claudin-1, junctional adhesion molecule 1 and occludin (10). Moreover, lncRNA SPRY4-IT1
overexpression in intestinal mucosa in mice protects the intestinal
barrier against infection (10).
In addition, lncRNA H19 promotes the proliferation of intestinal
epithelial cells by directly binding to tumor protein p53 protein,
microRNA34a and let-7 (11).
Therefore, these previous results indicated that lncRNAs directly
affect the expression of mRNAs via co-expression networks. However,
the role of lncRNA-mRNA networks in the pathogenesis of NEC is not
fully understood. Thus, the construction of such networks may
increase the understanding of the mechanisms underlying NEC.
In the present study, the expression profiles of
lncRNAs and mRNAs in NEC lesion and adjacent intestinal tissues
obtained from patients with NEC were analyzed by Next Generation
Sequencing (NGS). Additionally, a number of these genes were
validated by reverse transcription-quantitative PCR (RT-qPCR) in
vitro. Moreover, Gene Ontology (GO) and Kyoto Encyclopedia of
Genes and Genomes (KEGG) analyses were performed to assess the
potential functions of the differentially expressed lncRNAs and
mRNAs. In addition, a lncRNA-mRNA network was constructed in order
to identify the potential underlying mechanism of NEC. The present
results suggested that lncRNAs in intestinal tissues may serve an
important role in the development of NEC.
Materials and methods
Sample collection and ethics
statement
Preterm infants (25–36 weeks of age; one female and
two males) with stage III acute NEC undergoing bowel resection were
enrolled in the present study between October 2017 and October 2018
at The Affiliated Wuxi Children's Hospital of Nanjing Medical
University. The diagnosis of NEC was based on Bell's stages
(12). Infants with other serious
intestinal diseases, such as congenital megacolon, were excluded.
Tissues of the small bowel segment with NEC lesions that exhibited
perforation or necrosis, and adjacent healthy tissue [referred to
as the control (CTL)] were resected and stored at −80°C in liquid
nitrogen until analysis. The study protocol was approved by the
Institutional Review Board of The Affiliated Wuxi Children's
Hospital of Nanjing Medical University (approval no.
2017-EYLL-115). Informed consent was obtained from the parents or
guardians of all patients.
RNA isolation, quality control and
RNA-Seq
Total RNA was extracted from the tissues using a
mirVana microRNA isolation kit (cat. no. AM1561; Ambion; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
RNA integrity was subsequently assessed using an Agilent
Bioanalyzer 2100 system (Agilent Technologies, Inc.). RNA was
further purified using an RNAClean XP kit (Beckman Coulter, Inc.)
and an RNase-Free DNase set (Qiagen GmbH). The samples were then
subjected to NGS. The mRNAs and lncRNAs were identified using an
Illumina platform (HiSeq 2500; Illumina, Inc.). The gene abundance
was calculated using StringTie (version no. 1.3.0) (13). NGS, data acquisition and processing
were performed by Shanghai Biotechnology Corporation.
NEC cell model construction
The human normal colorectal FHC cell line (American
Type Culture Collection) was cultured in RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) containing 5% FBS (Gibco; Thermo
Fisher Scientific, Inc.) and 1% penicillin/streptomycin solution at
37°C and 5% CO2. In order to establish the NEC cell
model, FHC cells were treated with 200 µg/ml lipopolysaccharide
(LPS; Sigma-Aldrich; Merck KGaA) at 37°C for 6 h, referring to the
dose and time of LPS treatment described in previous study
(14). LPS used in the present
study was obtained from Escherichia coli O111:B4.
RNA preparation and RT-qPCR
A total of 5 mRNAs and 5 lncRNAs were randomly
selected and further assessed by RT-qPCR. Total RNA from FHC cells
was extracted using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. RT
was performed using a PrimeScript RT reagent kit (Takara Bio, Inc.)
according to the manufacturer's instructions (37°C for 15 min; 85°C
for 5 sec; 4°C for 10 min). qPCR was subsequently performed using a
SYBR Green PCR kit (Takara Bio, Inc.). The following thermocycling
conditions were used: 95°C for 10 min; 95°C for 10 sec, 60°C for 30
sec, 40 cycles. The primer pairs used for qPCR are presented in
Table SI. Gene expression was
calculated using the 2−∆∆Cq method (15), and normalized to the expression of
the internal reference gene GAPDH. Data are presented as the
average of 3 independent experiments.
Protein extraction and western
blotting
Total protein from FHC cells was extracted using
RIPA lysis buffer (Beyotime Institute of Biotechnology) containing
1% phenylmethylsulfonyl fluoride. The protein concentration was
measured by a BCA assay kit (Thermo Fisher Scientific, Inc.). A
total of 20 µg protein/lane was separated via SDS-PAGE on a 10% gel
and transferred onto a PVDF membrane. The membrane was subsequently
blocked with 5% skimmed milk for 1 h at room temperature and
incubated with primary antibodies against Toll-like receptor-4
(TLR4; 1:1,000; cat. no. bs-20594R; BIOSS) and β-actin (1:1,000;
cat. no. CST-4970; Cell Signaling Technology, Inc.) for 12 h at
4°C. Following primary antibody incubation, the membrane was
incubated with a horseradish peroxidase conjugated-secondary
antibody (1:5,000; cat. no. CST-7074; Cell Signaling Technology,
Inc.) for 30 min at room temperature. Proteins were visualized
using an enhanced chemiluminescence detection system (Pierce ECL
Plus Western Blotting Substrate; Thermo Fisher Scientific, Inc.).
Protein bands were analyzed by ImageJ software (version 1.47;
National Institutes of Health).
Hematoxylin and eosin staining
Intestines were fixed in 4% paraformaldehyde
overnight at room temperature, dehydrated with various
concentrations of ethanol (50, 75, 85, 95 and 100% ethanol) and
xylene, and embedded in paraffin. Paraffin-embedded small
intestinal specimens were cut into 5-µm sections. Sections were
then deparaffinized and rehydrated. Following which, the sections
were stained with Mayer's hematoxylin for 30 sec and 1% eosin Y
solution for 10–30 sec at room temperature. After dehydration and
mounting, the sections were examined under a light microscope, at
magnification, ×200, and images were taken.
GO and KEGG analysis
GO (version 14; www.geneontology.org) was used to analyze the
biological processes, cellular components and molecular functions
of the differentially expressed mRNAs and host genes of the
lncRNAs. In addition, KEGG (version 4.0; www.genome.jp/kegg) pathway analysis was performed to
investigate the associated pathways. GO terms and KEGG pathways
were ranked by the enrichment factor.
lncRNA-mRNA co-expression network
A lncRNA-mRNA network was constructed using
Cytoscape software (version 3.4.0; http://cytoscape.org/). A total of 38 differentially
expressed mRNAs and 44 lncRNAs, whose target genes are same to
those mRNAs, were selected. In total, 80 co-expression
relationships were identified.
Statistical analysis
Statistical differences were evaluated using the two
independent samples t-test. Data are presented as the mean ±
standard deviation. The edgeR software package (version 3.28.1)
(16) was used to identify
differentially expressed lncRNAs and mRNAs, between and NEC and CTL
tissues. P<0.05 was considered to indicate a statically
significant difference.
Results
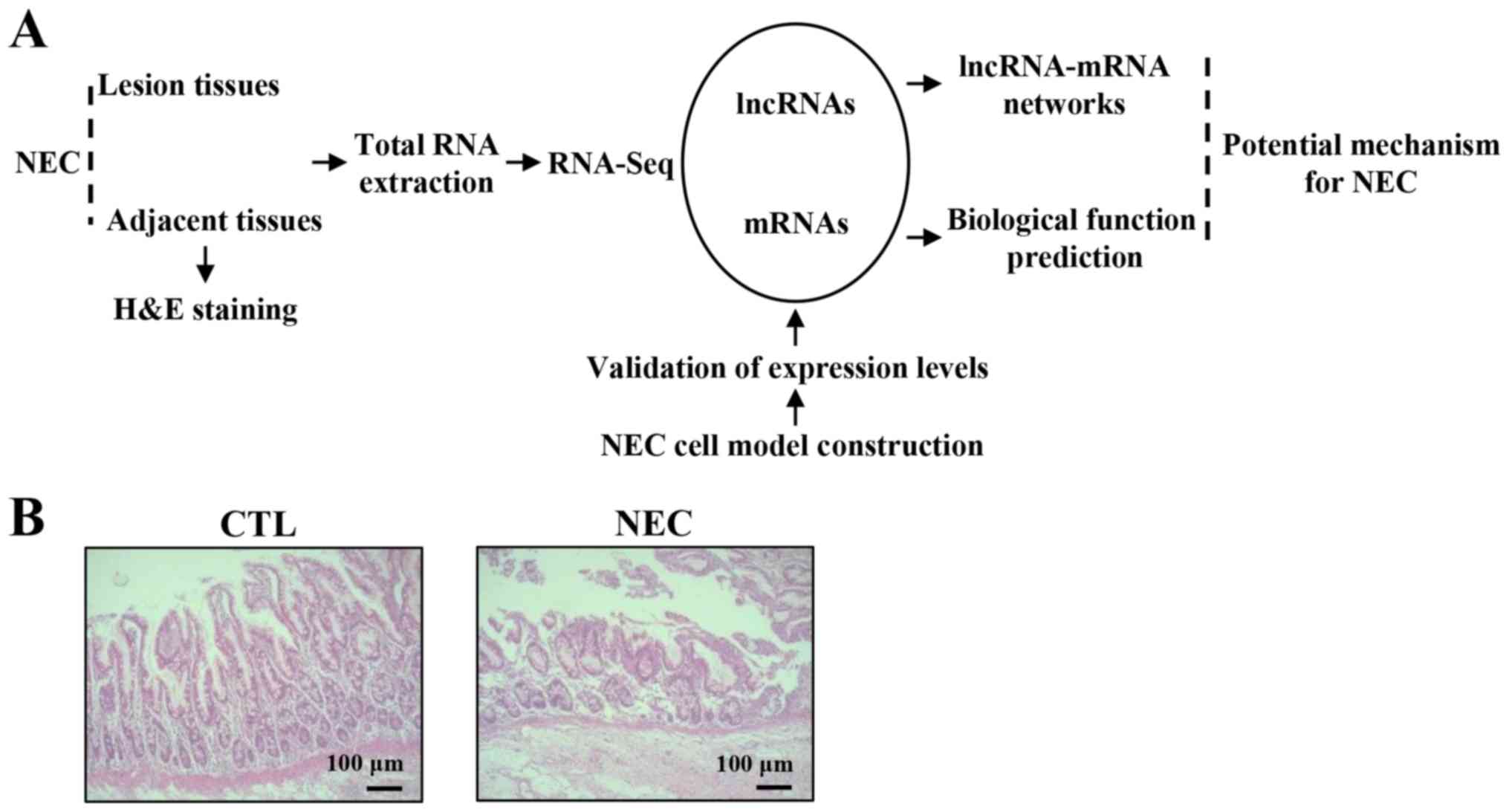
Detection of lncRNA and mRNA profiles
in NEC tissues
NGS was used to detect the expression of mRNAs and
lncRNAs in NEC and CTL tissues. Moreover, differentially expressed
mRNAs and lncRNAs were analyzed using a lncRNA-mRNA co-expression
network (Fig. 1A). Furthermore,
hematoxylin and eosin staining identified a disrupted mucosal
architecture in NEC tissues (Fig.
1B).
Differentially expressed gene
analysis
A total of 4,202 lncRNAs were significantly
differentially expressed between NEC and CTL tissues (fold-change
>2; P<0.05), which included 1,657 upregulated lncRNAs and
2,545 downregulated lncRNAs (Fig.
2A; Table SII). In addition,
it was identified that 7,860 mRNAs were significantly
differentially expressed between NEC and CTL tissues (fold-change
>2; P<0.05), which included 4,241 upregulated mRNAs and 3,619
downregulated mRNAs (Fig. 2B;
Table SIII). The top 50
significantly downregulated and upregulated lncRNAs and mRNAs are
presented in Tables I and II, respectively.
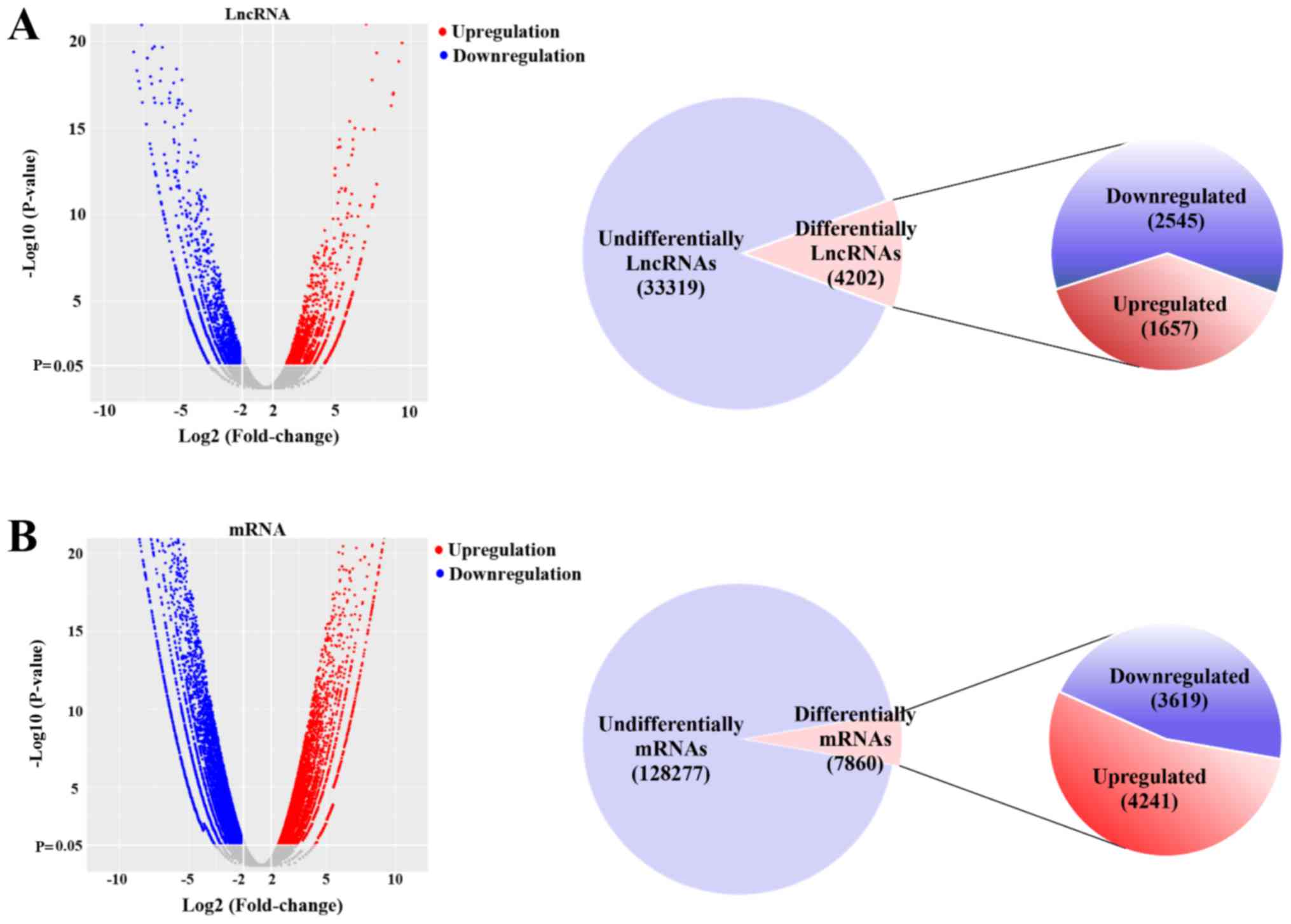 | Figure 2.Differentially expressed mRNAs and
lncRNAs in necrotizing enterocolitis and adjacent tissues. (A) A
volcano plot was used to assess the differentially expressed
lncRNAs. Of the 37,521 non-redundant lncRNAs, 4,202 lncRNAs were
significantly differentially expressed (fold-change >2;
P<0.05). A total of 1,657 and 2,545 lncRNAs were upregulated and
downregulated, respectively. (B) A volcano plot was used to assess
differentially expressed mRNAs. Of the 136,137 non-redundant mRNAs,
7,860 mRNAs were significantly differentially expressed
(fold-change >2; P<0.05). A total of 4,241 and 3,619 mRNAs
were upregulated and downregulated, respectively. lncRNA, long
non-coding RNA. |
 | Table I.Top 50 significantly upregulated and
downregulated long non-coding RNAs. |
Table I.
Top 50 significantly upregulated and
downregulated long non-coding RNAs.
| A, Upregulated |
|---|
|
|---|
| lncRNA_ID | log2FC | P-value | Q-value |
|---|
|
NONHSAT155609.1 | 11.61383132 |
1.43×10−23 |
1.22×10−20 |
|
ENST00000602813 | 11.55252438 |
1.30×10−09 |
7.94×10−08 |
|
NONHSAT059749.2 | 10.91351071 |
3.06×10−22 |
1.92×10−19 |
|
NONHSAT118562.2 | 10.66845241 |
2.81×10−19 |
1.06×10−16 |
|
ENST00000620266 | 10.60925657 |
2.84×10−30 |
5.92×10−27 |
|
NONHSAT209475.1 | 10.52975989 |
1.54×10−17 |
4.47×10−15 |
|
NONHSAT159678.1 | 10.44685758 |
2.78×10−17 |
7.58×10−15 |
|
NONHSAT169788.1 | 10.30207377 |
8.74×10−11 |
6.80×10−09 |
|
NONHSAT123206.2 | 10.20925898 |
2.57×10−10 |
1.81×10−08 |
|
NONHSAT094312.2 | 10.08888094 |
2.95×10−27 |
4.47×10−24 |
|
NONHSAT009758.2 | 9.718282592 |
1.25×10−14 |
2.10×10−12 |
|
ENST00000499624 | 9.681004925 |
6.54×10−25 |
7.20×10−22 |
|
NONHSAT177823.1 | 9.583016227 |
1.06×10−16 |
2.59×10−14 |
|
NONHSAT159417.1 | 9.438955909 |
1.51×10−23 |
1.26×10−20 |
|
NONHSAT173659.1 | 9.328552802 |
1.54×10−17 |
4.47×10−15 |
|
NONHSAT137151.2 | 9.243165734 |
2.22×10−08 |
1.04×10−06 |
|
NONHSAT206983.1 | 9.228339375 |
2.26×10−22 |
1.47×10−19 |
|
NONHSAT178474.1 | 9.086352515 |
1.08×10−10 |
8.23×10−09 |
|
ENST00000628476 | 9.043520963 |
4.70×10−11 |
3.93×10−09 |
|
NONHSAT081637.2 | 9.023472571 |
1.02×10−05 | 0.000234023 |
|
NONHSAT076868.2 | 8.99220343 |
1.31×10−32 |
3.36×10−29 |
|
NONHSAT155904.1 | 8.936911199 |
7.85×10−14 |
1.12×10−11 |
|
NONHSAT180405.1 | 8.90507266 |
3.21×10−07 |
1.13×10−05 |
|
NONHSAT071404.2 | 8.875787946 |
2.04×10−20 |
9.59×10−18 |
|
NONHSAT183239.1 | 8.869833121 |
3.85×10−15 |
6.90×10−13 |
|
NONHSAT083185.2 | 8.855681215 |
2.60×10−20 |
1.20×10−17 |
|
NONHSAT055522.2 | 8.835207264 |
2.66×10−13 |
3.40×10−11 |
|
ENST00000573951 | 8.78394133 |
3.14×10−16 |
6.98×10−14 |
|
NONHSAT142596.2 | 8.728768517 |
1.32×10−19 |
5.25×10−17 |
|
NONHSAT055581.2 | 8.726034495 |
2.06×10−10 |
1.48×10−08 |
|
NONHSAT050230.2 | 8.724625325 |
8.74×10−11 |
6.80×10−09 |
|
NONHSAT056662.2 | 8.670344103 |
2.25×10−16 |
5.17×10−14 |
|
ENST00000505532 | 8.58704266 |
1.64×10−14 |
2.64×10−12 |
|
NONHSAT009555.2 | 8.577747998 |
3.39×10−06 |
9.05×10−05 |
|
ENST00000635687 | 8.488271168 |
1.13×10−18 |
3.91×10−16 |
|
ENST00000523099 | 8.4516672 |
2.38×10−06 |
6.72×10−05 |
|
NONHSAT050198.2 | 8.410023146 |
1.50×10−05 | 0.000327563 |
|
NONHSAT064287.2 | 8.405723447 |
7.03×10−06 | 0.000168779 |
|
ENST00000558776 | 8.383710584 | 0.004874773 | 0.039194634 |
|
NONHSAT010340.2 | 8.380591242 |
3.29×10−05 | 0.00064125 |
|
ENST00000441790 | 8.379233397 |
1.26×10−07 | 4.90
×10−06 |
|
ENST00000426699 | 8.371776115 |
7.03×10−06 | 0.000168779 |
|
ENST00000614899 | 8.359796918 |
1.26×10−07 |
4.90×10−06 |
|
NONHSAT182506.1 | 8.346803842 |
8.05×10−10 |
5.21×10−08 |
|
ENST00000615722 | 8.342773399 |
1.72×10−07 |
6.44×10−06 |
|
NONHSAT215498.1 | 8.331225239 |
7.54×10−09 |
3.93×10−07 |
|
NONHSAT095885.2 | 8.32856903 |
8.52×10−07 |
2.65×10−05 |
|
NONHSAT175395.1 | 8.31998397 |
1.37×10−12 |
1.50×10−10 |
|
NONHSAT108374.2 | 8.314575441 |
2.86×10−14 | 4.35×10-12 |
|
NONHSAT207205.1 | 8.305343553 |
9.36×10−08 |
3.77×10−06 |
|
| B,
Downregulated |
|
|
lncRNA_ID | log2FC | P-value | Q-value |
|
|
NONHSAT183597.1 | −16.48090468 |
8.79×10−58 |
2.93×10−53 |
|
NONHSAT168464.1 | −14.21176751 |
2.63×10−42 |
2.92×10−38 |
|
NONHSAT213758.1 | −12.54912544 |
1.45×10−36 |
8.05×10−33 |
|
NONHSAT185013.1 | −12.48955925 |
8.00×10−31 |
1.90×10−27 |
|
NONHSAT152066.1 | −12.17786195 |
5.62×10−34 |
1.87×10−30 |
|
NONHSAT024359.2 | −11.87327964 |
3.83×10−45 |
6.38×10−41 |
| MSTRG.44916.1 | −11.60824268 |
7.79×10−35 |
3.24×10−31 |
|
NONHSAT181735.1 | −11.43001449 |
6.93×10−27 |
1.00×10−23 |
|
NONHSAT160167.1 | −11.41835274 |
3.88×10−24 |
3.49×10−21 |
|
NONHSAT165927.1 | −11.25071761 |
1.29×10−24 |
1.23×10−21 |
|
NONHSAT159286.1 | −11.10183428 |
9.99×10−26 |
1.33×10−22 |
|
NONHSAT091106.2 | −11.07224496 |
4.78×10−28 |
7.97×10−25 |
|
NONHSAT175707.1 | −11.04329687 |
1.25×10−25 |
1.61×10−22 |
|
ENST00000504773 | −10.84721244 |
4.46×10−23 |
3.30×10−20 |
|
NONHSAT091102.2 | −10.66881414 |
1.16×10−17 |
3.45×10−15 |
|
NONHSAT159894.1 | −10.62188109 |
1.57×10−21 |
9.02×10−19 |
|
NONHSAT031256.2 | −10.60250823 |
1.47×10−37 |
1.22×10−33 |
|
NONHSAT123764.2 | −10.50424036 |
8.82×10−33 |
2.45×10−29 |
|
ENST00000507311 | −10.47456391 |
1.96×10−30 |
4.36×10−27 |
|
NONHSAT171172.1 | −10.41117159 |
9.23×10−23 |
6.28×10−20 |
|
NONHSAT162645.1 | −10.33478733 |
3.39×10−17 |
9.04×10−15 |
|
NONHSAT091107.2 | −10.32292548 |
2.57×10−23 |
2.04×10−20 |
|
NONHSAT139922.2 | −10.31278929 |
5.33×10−19 |
1.97×10−16 |
|
NONHSAT183589.1 | −10.28237601 |
5.84×10−25 |
6.71×10−22 |
|
NONHSAT196167.1 | −10.27748084 |
6.07×10−23 |
4.31×10−20 |
| MSTRG.37793.3 | −10.20152795 |
2.73×10−22 |
1.75×10−19 |
|
NONHSAT188793.1 | −10.11670026 |
1.02×10−09 |
6.43×10−08 |
|
NONHSAT152222.1 | −10.03914993 |
1.25×10−14 |
2.10×10−12 |
|
ENST00000606993 | −10.00747462 |
1.58×10−18 |
5.28×10−16 |
|
NONHSAT115736.2 | −9.985583318 |
2.27×10−13 |
2.94×10−11 |
|
NONHSAT157008.1 | −9.951490231 |
7.85×10−14 |
1.12×10−11 |
|
NONHSAT160671.1 | −9.908059683 |
3.39×10−15 |
6.15×10−13 |
|
ENST00000427901 | −9.906883277 |
4.70×10−11 |
3.93×10−09 |
|
NONHSAT192263.1 | −9.864619513 |
3.19×10−18 |
1.01×10−15 |
|
NONHSAT182621.1 | −9.857221425 |
2.57×10−11 | 2.28
×10−09 |
|
NONHSAT094167.2 | −9.856028479 |
1.67×10−13 |
2.24×10−11 |
|
ENST00000419296 | −9.82059815 |
1.29×10−24 |
1.23×10−21 |
|
NONHSAT041891.2 | −9.794594044 |
2.11×10−09 |
1.24×10−07 |
|
NONHSAT176544.1 | −9.772352515 |
1.65×10−09 |
9.95×10−08 |
|
ENST00000455071 | −9.756925085 |
1.60×10−24 |
1.48×10−21 |
|
NONHSAT208752.1 | −9.667998866 |
2.92×10−18 |
9.36×10−16 |
|
NONHSAT022134.2 | −9.65276041 |
7.91×10−37 |
5.27×10−33 |
|
NONHSAT017148.2 | −9.551998808 |
3.83×10−11 |
3.29×10−09 |
|
NONHSAT017600.2 | −9.549948279 |
1.26×10−07 |
4.90×10−06 |
|
NONHSAT143306.2 | −9.53507668 |
3.95×10−36 |
1.88×10−32 |
|
NONHSAT152860.1 | −9.531346389 |
1.03×10−18 |
3.63×10−16 |
|
NONHSAT091105.2 | −9.513378928 |
1.40×10−19 |
5.47×10−17 |
|
ENST00000584749 | −9.484529301 |
8.05×10−10 |
5.21×10−08 |
|
NONHSAT060224.2 | −9.352889754 |
4.70×10−11 |
3.93×10−09 |
|
NONHSAT032678.2 | −9.348901378 |
1.43×10−13 |
1.95×10−11 |
 | Table II.Top 50 significantly upregulated and
downregulated mRNAs. |
Table II.
Top 50 significantly upregulated and
downregulated mRNAs.
| A, Upregulated |
|---|
|
|---|
| Gene ID | Gene name | log2FC | P-value | Q-value |
|---|
|
ENSG00000228536 | RP11-392O17.1 | 9.13 |
2.33×10−17 |
3.87×10−16 |
|
ENSG00000129988 | LBP | 8.87 |
9.55×10−30 |
3.12×10−28 |
|
ENSG00000121053 | EPX | 8.64 |
1.62×10−33 |
6.40×10−32 |
|
ENSG00000267653 | RP1-193H18.3 | 8.63 |
1.30×10−09 |
1.16×10−08 |
|
ENSG00000244437 | IGKV3-15 | 8.47 |
4.11×10−07 |
2.73×10−06 |
|
ENSG00000145850 | TIMD4 | 8.34 |
7.39×10−23 |
1.69×10−21 |
|
ENSG00000259183 | MED28P6 | 8.33 | 0.001268 | 0.004697 |
|
ENSG00000229308 | AC010084.1 | 8.32 |
7.04×10−08 |
5.18×10−07 |
|
ENSG00000279686 | ECSCR | 8.25 |
1.97×10−12 |
2.29×10−11 |
|
ENSG00000231123 | SPATA20P1 | 8.17 |
8.35×10−06 |
4.60×10−05 |
|
ENSG00000226012 | AP001434.2 | 8.16 | 0.000667 | 0.002621 |
|
ENSG00000230024 | RP11-95P13.1 | 8.13 |
8.35×10−06 |
4.60×10−05 |
|
ENSG00000273153 | RP11-406H21.2 | 8.07 | 0.002421 | 0.008418 |
|
ENSG00000164821 | DEFA4 | 8.05 |
2.27×10−07 |
1.56×10−06 |
|
ENSG00000267436 | AC005786.7 | 8.04 |
7.45×10−07 |
4.79×10−06 |
|
ENSG00000271656 | RP11-114N19.5 | 7.99 | 0.000667 | 0.002621 |
|
ENSG00000211970 | IGHV4-61 | 7.93 |
2.86×10−05 | 0.000146 |
|
ENSG00000211942 | IGHV3-13 | 7.92 |
5.33×10−05 | 0.000258 |
|
ENSG00000259626 | MTND3P12 | 7.83 | 0.001268 | 0.004697 |
|
ENSG00000279903 | RP11-349F21.5 | 7.82 |
4.54×10−06 |
2.60×10−05 |
|
ENSG00000231613 | RP5-943J3.1 | 7.78 |
2.86×10−05 | 0.000146 |
|
ENSG00000235821 | IFITM4P | 7.73 | 0.000187 | 0.00082 |
|
ENSG00000214402 | LCNL1 | 7.71 |
6.67×10−48 |
4.38×10−46 |
|
ENSG00000225938 | RP4-575N6.4 | 7.67 | 0.000667 | 0.002621 |
|
ENSG00000211946 | IGHV3-20 | 7.62 | 0.000667 | 0.002621 |
|
ENSG00000256039 | RP11-291B21.2 | 7.62 |
1.26×10−07 |
8.98×10−07 |
|
ENSG00000158104 | HPD | 7.55 |
2.07×10−21 |
4.43×10−20 |
|
ENSG00000235151 | AC114730.2 | 7.51 |
5.33×10−05 | 0.000258 |
|
ENSG00000239839 | DEFA3 | 7.51 |
8.35×10−06 |
4.60×10−05 |
|
ENSG00000275558 | RN7SKP175 | 7.49 | 0.008941 | 0.02691 |
|
ENSG00000239353 | RP11-492E3.51 | 7.45 | 0.002421 | 0.008418 |
|
ENSG00000149516 | MS4A3 | 7.45 |
2.99×10−27 |
8.62×10−26 |
|
ENSG00000243066 | RN7SL842P | 7.44 | 0.008941 | 0.02691 |
|
ENSG00000196415 | PRTN3 | 7.39 |
8.74×10−55 |
7.16×10−53 |
|
ENSG00000211950 | IGHV1-24 | 7.35 | 0.002421 | 0.008418 |
|
ENSG00000275791 | TRBV10-3 | 7.34 | 0.004643 | 0.014979 |
|
ENSG00000211664 | IGLV2-18 | 7.31 | 0.004643 | 0.014979 |
|
ENSG00000226660 | TRBV2 | 7.31 | 0.002421 | 0.008418 |
|
ENSG00000250579 | CTD-2297D10.2 | 7.31 |
1.36×10−06 |
8.41×10−06 |
|
ENSG00000166819 | PLIN1 | 7.31 |
4.63×10−102 |
1.30×10−99 |
|
ENSG00000273006 | RP11-314C9.2 | 7.31 | 0.017298 | 0.047391 |
|
ENSG00000258082 | RP11-443B7.3 | 7.31 |
4.54×10−06 |
2.60×10−05 |
|
ENSG00000240864 | IGKV1-16 | 7.30 | 0.004643 | 0.014979 |
|
ENSG00000279180 | RP11-417O18.1 | 7.30 |
1.36×10−06 |
8.41×10−06 |
|
ENSG00000184811 | TUSC5 | 7.26 |
2.61×10−40 |
1.33×10−38 |
|
ENSG00000234436 | AC008984.7 | 7.20 | 0.004643 | 0.014979 |
|
ENSG00000232286 | RP11-80K6.2 | 7.19 | 0.002421 | 0.008418 |
|
ENSG00000211747 | TRBV20-1 | 7.17 | 0.004643 | 0.014979 |
|
ENSG00000254673 | RP11-598P20.5 | 7.16 | 0.000187 | 0.00082 |
|
ENSG00000231863 | RP3-428L16.1 | 7.16 | 0.000667 | 0.002621 |
|
| B,
Downregulated |
|
| Gene ID | Gene
name | log2FC | P-value | Q-value |
|
|
ENSG00000147676 | MAL2 | −9.98 |
2.32×10−71 |
3.10×10−69 |
|
ENSG00000140832 | MARVELD3 | −9.97 |
4.72×10−93 |
1.09×10−90 |
|
ENSG00000158125 | XDH | −9.97 |
1.84×10−153 |
1.60×10−150 |
|
ENSG00000272141 | RP11-465B22.8 | −9.97 |
7.32×10−33 |
2.79×10−31 |
|
ENSG00000235122 | RP3-417L20.4 | −9.96 |
2.50×10−16 |
3.88×10−15 |
|
ENSG00000234155 | RP11-30P6.6 | −9.93 |
5.74×10−57 |
4.96×10−55 |
|
ENSG00000160868 | CYP3A4 | −9.92 |
9.78×10−159 |
9.35×10−156 |
|
ENSG00000103534 | TMC5 | −9.89 |
1.11×10−142 |
7.63×10−140 |
|
ENSG00000235523 | RP11-63P12.7 | −9.86 |
9.90×10−15 |
1.37×10−13 |
|
ENSG00000165841 | CYP2C19 | −9.84 |
2.86×10−62 |
2.90×10−60 |
|
ENSG00000253313 | C1orf210 | −9.83 |
1.62×10−47 |
1.05×10−45 |
|
ENSG00000084674 | APOB | −9.82 |
1.30×10−230 |
1.93×10−226 |
|
ENSG00000118094 | TREH | −9.81 |
8.09×10−90 |
1.75×10−87 |
|
ENSG00000172016 | REG3A | −9.80 |
4.24×10−130 |
2.03×10−127 |
|
ENSG00000044012 | GUCA2B | −9.78 |
4.31×10−20 |
8.54×10−19 |
|
ENSG00000081051 | AFP | −9.77 |
4.20×10−70 |
5.49×10−68 |
|
ENSG00000169876 | MUC17 | −9.77 |
1.80×10−195 |
4.11×10−192 |
|
ENSG00000173467 | AGR3 | −9.69 |
3.30×10−36 |
1.48×10−34 |
|
ENSG00000131910 | NR0B2 | −9.68 |
8.08×10−32 |
2.92×10−30 |
|
ENSG00000122711 | SPINK4 | −9.64 |
3.00×10−23 |
7.07×10−22 |
|
ENSG00000165556 | CDX2 | −9.63 |
8.60×10−110 |
2.83×10−107 |
|
ENSG00000224916 | APOC4-APOC2 | −9.63 |
8.05×10−52 |
6.09×10−50 |
|
ENSG00000167183 | PRR15L | −9.53 |
3.86×10−107 |
1.19×10−104 |
|
ENSG00000232400 | RAD17P1 | −9.53 |
1.93×10−42 | 1.06
×10−40 |
|
ENSG00000278505 | C17orf78 | −9.53 |
1.27×10−54 | 1.04
×10−52 |
|
ENSG00000225329 | LHFPL3-AS2 | −9.53 |
7.93×10−49 | 5.37
×10−47 |
|
ENSG00000143278 | F13B | −9.52 |
4.39×10−49 | 3.00
×10−47 |
|
ENSG00000187664 | HAPLN4 | −9.51 |
5.32×10−60 | 5.03
×10−58 |
|
ENSG00000127831 | VIL1 | −9.50 |
1.33×10−163 |
1.65×10−160 |
|
ENSG00000108242 | CYP2C18 | −9.49 |
3.21×10−52 |
2.46×10−50 |
|
ENSG00000136872 | ALDOB | −9.44 |
4.47×10−204 |
1.66×10−200 |
|
ENSG00000160182 | TFF1 | −9.38 |
6.20×10−16 |
9.32×10−15 |
|
ENSG00000105398 | SULT2A1 | −9.38 |
5.95×10−128 |
2.76×10−125 |
|
ENSG00000241388 | HNF1A-AS1 | −9.35 |
2.35×10−62 |
2.40×10−60 |
|
ENSG00000224163 | RP11-309L24.6 | −9.35 |
2.45×10−15 |
3.57×10−14 |
|
ENSG00000257084 | U47924.27 | −9.33 |
3.00×10−10 |
2.85×10−09 |
|
ENSG00000166391 | MOGAT2 | −9.28 |
4.57×10−140 |
2.95×10−137 |
|
ENSG00000140297 | GCNT3 | −9.28 |
4.25×10−76 |
6.45×10−74 |
|
ENSG00000275410 | HNF1B | −9.27 |
6.98×10−76 |
1.06×10−73 |
|
ENSG00000132437 | DDC | −9.27 |
6.68×10−100 |
1.80×10−97 |
|
ENSG00000171431 | KRT20 | −9.26 |
1.15×10−99 |
3.07×10−97 |
|
ENSG00000162460 | TMEM82 | −9.26 |
7.32×10−33 |
2.79×10−31 |
|
ENSG00000197408 | CYP2B6 | −9.25 |
1.08×10−69 |
1.39×10−67 |
|
ENSG00000249948 | GBA3 | −9.25 |
2.62×10−99 |
6.81×10−97 |
|
ENSG00000100604 | CHGA | −9.19 |
1.03×10−130 |
5.09×10−128 |
|
ENSG00000198944 | SOWAHA | −9.18 |
1.05×10−73 |
1.49×10−71 |
|
ENSG00000203858 | HSD3BP2 | −9.17 |
9.98×10−24 |
2.40×10−22 |
|
ENSG00000166869 | CHP2 | −9.17 |
4.15×10−112 |
1.45×10−109 |
|
ENSG00000060566 | CREB3L3 | −9.13 |
9.98×10−136 |
5.58×10−133 |
|
ENSG00000260934 | CTA-363E6.7 | −9.10 |
1.19×10−12 |
1.40×10−11 |
Construction of the NEC cell model and
validation of genes by RT-qPCR in vitro
The pathology underlying NEC development requires
the activation of the innate immune TLR4 on enterocytes (17). Moreover, TLR4 activation by
bacterial LPS inhibits enterocyte proliferation and leads to
disease progression (18,19). It was demonstrated that LPS
exposure increased the protein expression of TLR4 in FHCs, thus
suggesting that it is a reliable cell model for studying NEC in
vitro (Fig. 3A). In total, 5
mRNAs and 5 lncRNAs were randomly selected to assess the NGS
results by RT-qPCR. The present results indicated that the mRNA
expression levels of interleukin (IL)-6 and IL-1β were upregulated,
whereas the expression levels of transmembrane 4 L six family
member 20, solute carrier family 5 member 12 and leucine rich
repeat containing 19 were downregulated (Fig. 3B-F). Furthermore, NONHSAT022134.2
was downregulated, while NONHSAT076868.2, NONHSAT159417.1,
NONHSAT094312.2 and NONHSAT142596.2 were upregulated (Fig. 3G-K). Therefore, the RT-qPCR results
were consistent with the NGS results.
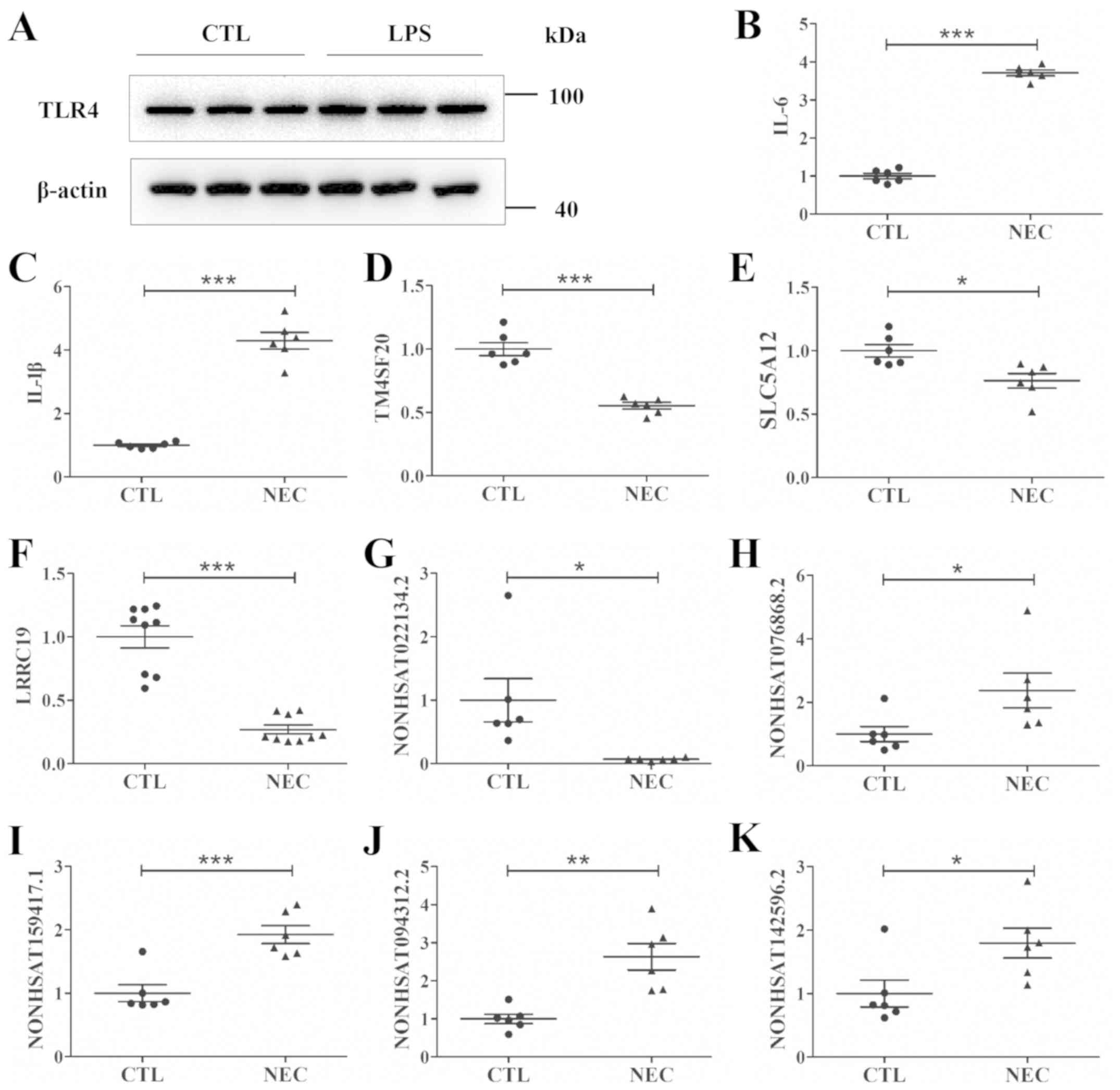 | Figure 3.NEC cell model construction, and
assessment of the dysregulated lncRNAs and mRNAs by reverse
transcription-quantitative PCR in vitro. (A) TLR4 protein
expression in CTL FHC cells, and FHC cells treated with LPS.
Relative mRNA expression levels of (B) IL-6, (C) IL-1β, (D)
TM4SF20, (E) SLC5A12 and (F) LRRC19 in the CTL and NEC groups.
Relative lncRNA expression levels of (G) NONHSAT022134.2, (H)
NONHSAT076868.2, (I) NONHSAT159417.1, (J) NONHSAT094312.2 and (K)
NONHSAT142596.2 in the CTL and NEC groups. Data are presented as
mean ± standard deviation. *P<0.05, **P<0.01, ***P<0.001.
CTL, control; lncRNA, long non-coding RNA; IL, interleukin;
TM4SF20, Transmembrane 4 L six family member 20; SLC5A12, solute
carrier family 5 member 12; LRRC19, leucine rich repeat containing
19; NEC, necrotizing enterocolitis. |
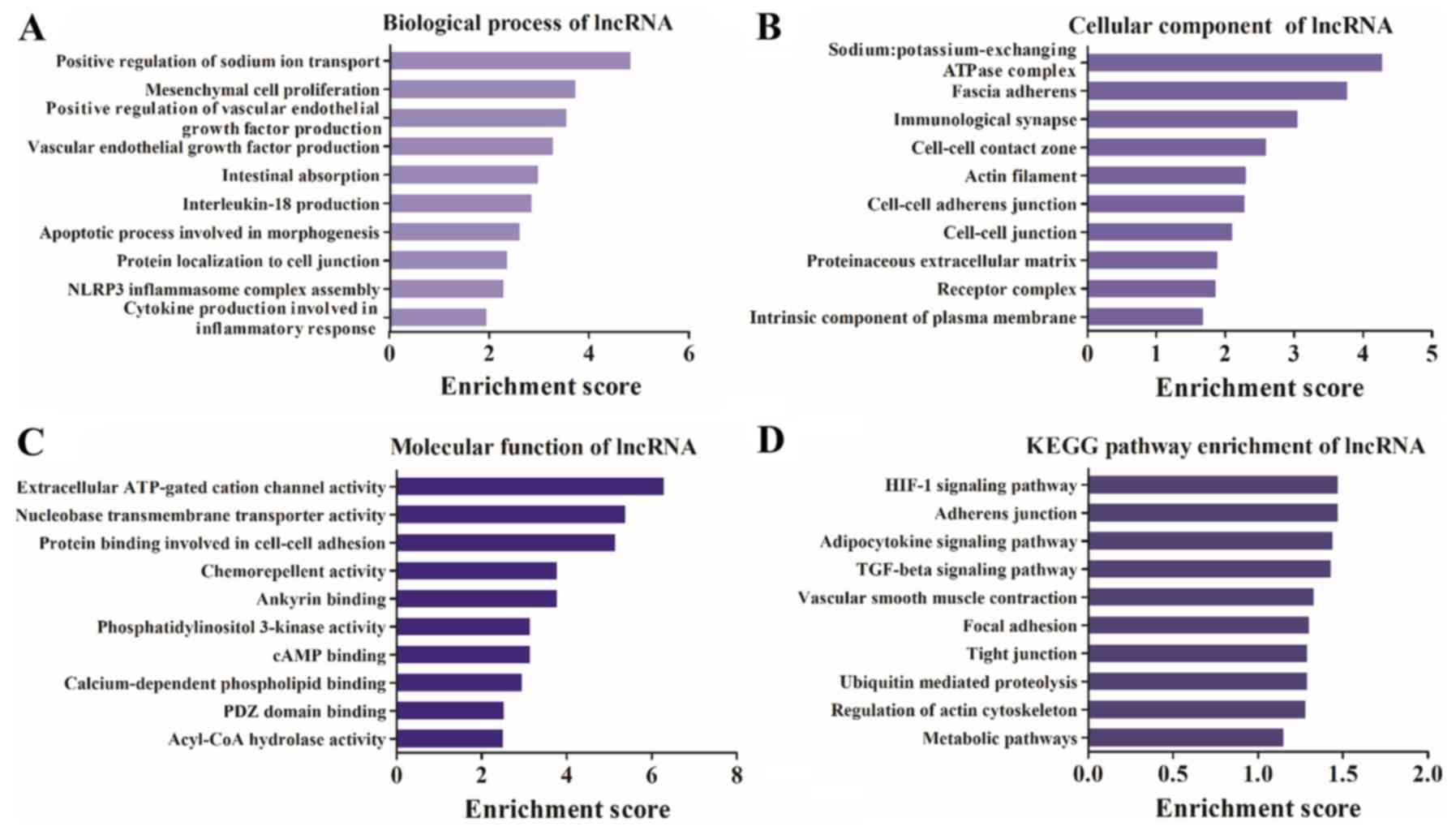
Differentially expressed lncRNAs are
associated with the inflammatory response and the transforming
growth factor-β (TGF-β) signaling pathway
GO and KEGG analyses of the differentially expressed
lncRNAs were performed. It was identified that the terms
‘intestinal absorption’, ‘IL-18 production’, ‘apoptotic process
involved in morphogenesis’ and ‘inflammatory response’ were highly
enriched in the biological process category (Fig. 4A). In addition, ‘immunological
synapse’, ‘actin filament’, ‘cell-cell contact zone’ and ‘receptor’
were highly enriched in the cellular component category (Fig. 4B). Furthermore, it was demonstrated
that ‘extracellular ATP-gated cation channel activity’,
‘chemorepellent activity’, ‘PI3K activity’ and ‘cAMP binding’ were
highly enriched in the molecular function category (Fig. 4C). The KEGG pathway analysis
results identified that the differentially expressed lncRNA host
genes were involved in ‘TGF-β signaling pathway’,
‘hypoxia-inducible factor 1 (HIF-1) signaling pathway’ and ‘tight
junction and regulation of actin cytoskeleton’ (Fig. 4D).
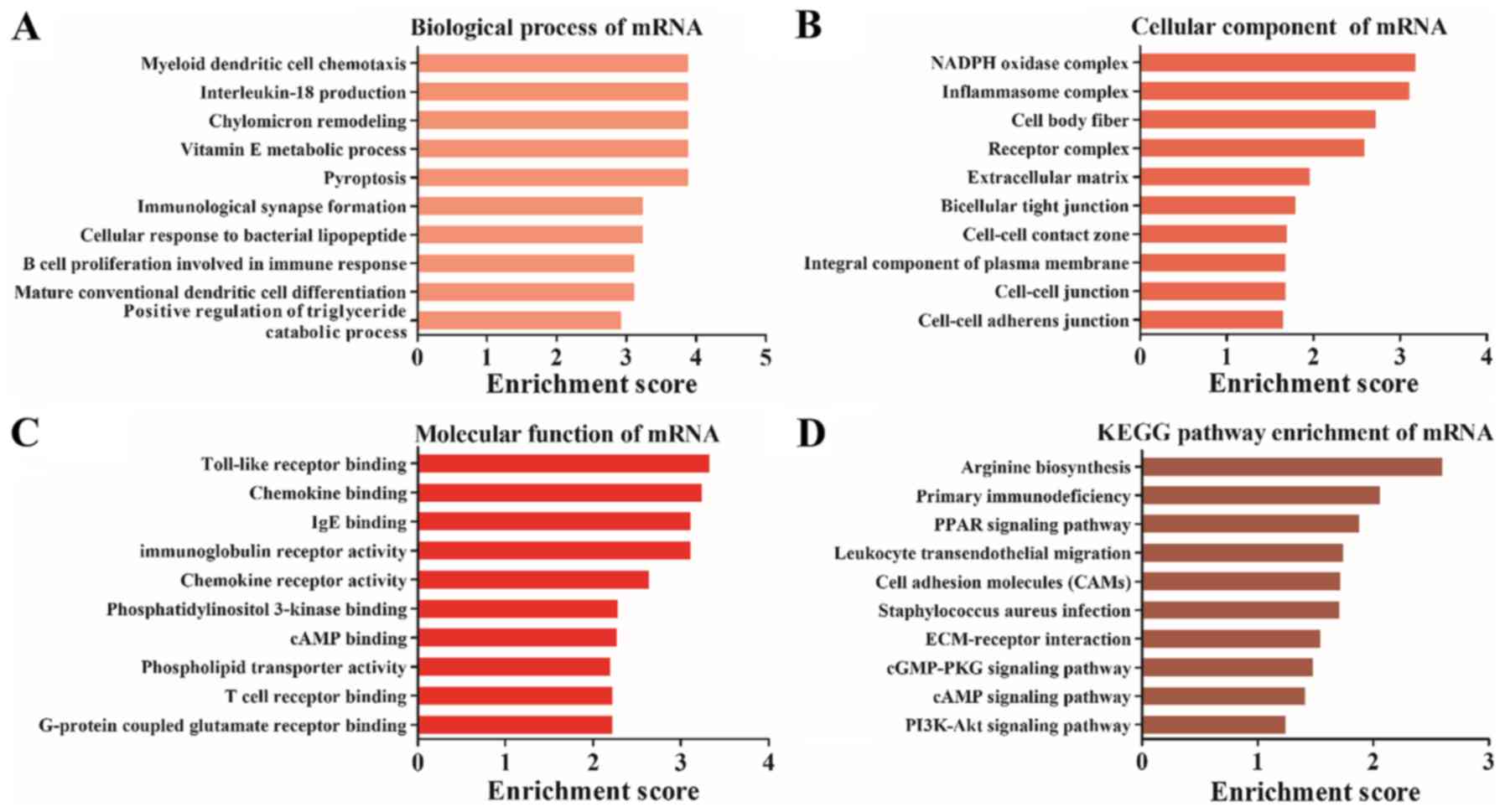
Differentially expressed mRNAs are
involved in TLR binding and the PI3K-Akt signaling pathway
GO and KEGG analysis were also performed for the
differentially expressed mRNAs, which were significantly enriched
in ‘myeloid dendritic cell chemotaxis’ (biological process;
Fig. 5A), ‘inflammasome complex’
(cellular component; Fig. 5B) and
‘TLR binding’ (molecular function; Fig. 5C). Furthermore, KEGG pathway
analysis results demonstrated that the differentially expressed
mRNAs were enriched in ‘peroxisome proliferator-activated receptors
(PPAR) signaling pathway’, ‘leukocyte transendothelial migration’,
‘extracellular matrix-receptor interaction’ and ‘PI3K-Akt signaling
pathway’ (Fig. 5D).
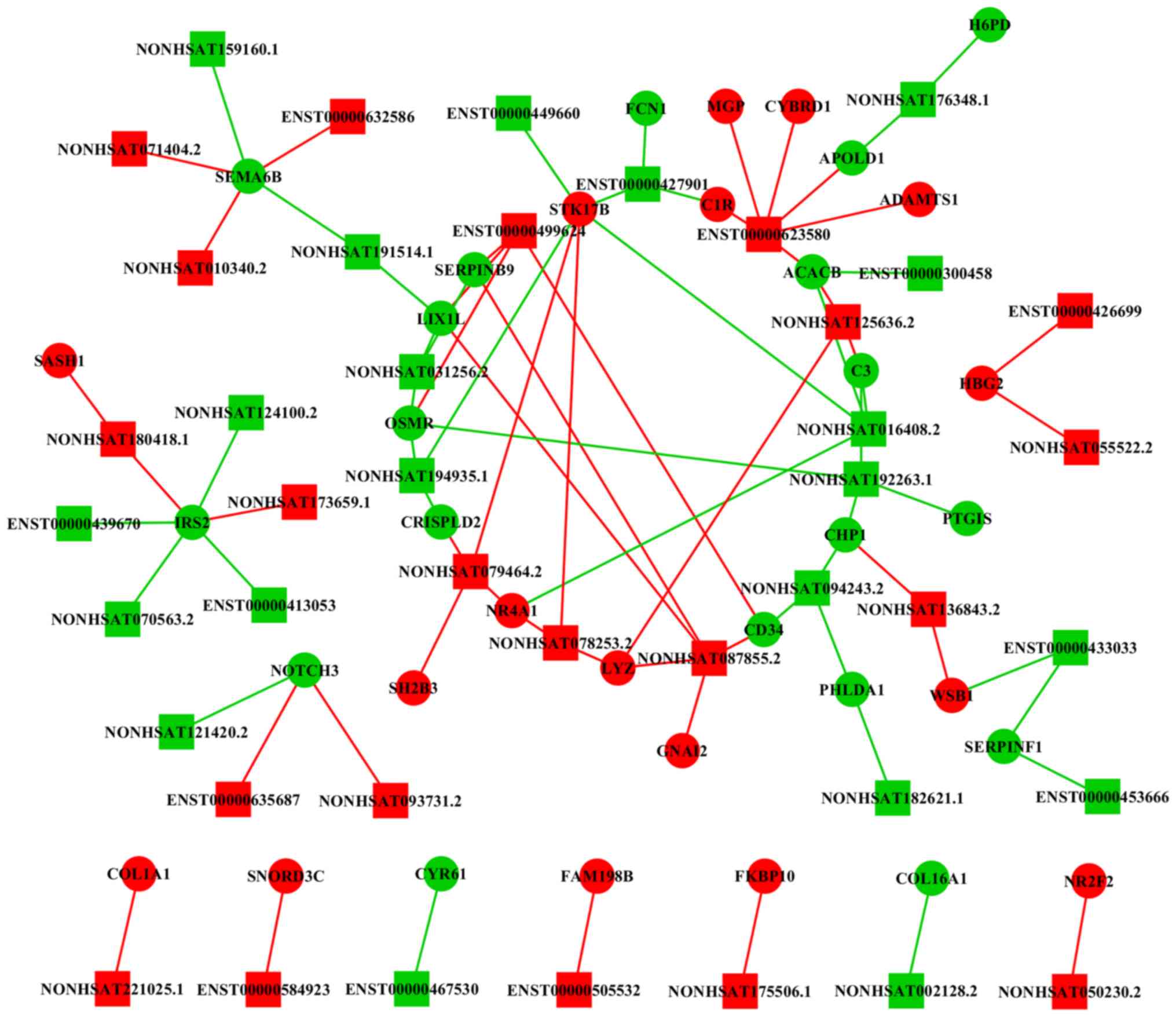
lncRNA-mRNA network reveals a
potential mechanism for NEC
The lncRNA-mRNA co-expression network results
suggested that lncRNAs had a complex interaction with mRNAs
(Fig. 6). A total of 80
associations were identified among the 44 lncRNAs and 38 mRNAs.
Moreover, it was identified that lncRNAs ENST00000623580,
NONHSAT180418.1, NONHSAT125636.2 and NONHSAT087855.2 interacted
with mRNAs that were closely associated with NEC. Therefore, the
present results suggested that these lncRNAs may serve important
roles in the pathogenesis of NEC.
Discussion
To the best of our knowledge, no previous studies
have examined lncRNAs associated with NEC, and there is no
functional evidence from studies investigating lncRNAs with NEC and
intestinal function. The present study analyzed the differentially
expressed lncRNAs and mRNAs in ileal samples obtained from
premature neonates with NEC. Furthermore, an in vitro cell
model of NEC was investigated. The aims of the present study were
to predict the potential functions of the differentially expressed
lncRNAs and mRNAs, and to identify a potential mechanism for NEC
via lncRNA-mRNA network analyses. While the pathophysiological
mechanism of NEC remains unknown, previous studies have reported
that the expression of TLR4 is upregulated in NEC (20,21).
The activation of TLR4 on intestinal epithelial cells induces a
significant proinflammatory response (22). Furthermore, TLR4 activation also
results in increased intestinal necrosis and apoptosis, which is
the main feature of NEC (19). In
addition, it has been shown that LPS-induced reactive oxygen
species accumulation is involved in NF-κB activation and subsequent
release of proinflammation cytokines, including IL-6 and IL-1β, in
the pathogenesis of NEC (23,24).
A previous study revealed that excessive inflammation of the
immature intestine predisposes premature infants to NEC (25). Therefore, elevated levels of IL-6
and IL-1β may be used as an index of successful NEC cell model
construction (26–28). The present results suggested that
the expression levels of IL-1β, IL-6 and TLR4 in FHCs were
significantly increased following LPS exposure, thus indicating
that this was a reliable cell model for studying NEC in
vitro.
GO and KEGG analyses were performed to identify
potential mechanisms underlying NEC. The GO analysis results
demonstrated that both the differentially expressed lncRNA host
genes and mRNAs were associated with inflammation, which is a
typical feature of NEC (29).
Moreover, it was identified that TLR binding was a highly enriched
molecular function, thus indicating the importance of the TLR in
NEC. The results of KEGG pathway analysis identified pathways that
were mainly associated with the progression of NEC. The PPAR
pathway has been shown to affect mucosal immunity, commensal
homeostasis and intestinal inflammation (30). Moreover, PPARα downregulation in
mice contributes to an imbalance of intestinal symbiosis, resulting
in enhanced susceptibility to intestinal inflammation and increased
production of inflammatory cytokines (30). Previous studies have shown that the
PI3K-Akt signaling pathway is altered in an experimental NEC animal
model (31,32). Furthermore, berberine inhibits
apoptosis and inflammation via the PI3K/AKT signaling pathway to
attenuate the progression of NEC (32). Macrophages exhibited increased
Smad7 expression in areas with high bacterial load and severe
tissue damage (33). In addition,
elevated Smad7 expression inhibits TGF-β signaling in intestinal
macrophages, and promotes NF-κB activation and cytokine production
during NEC (33). The PI3K-Akt
signaling pathway has also been reported to regulate HIF-1 to
protect against intestinal injury during NEC (34).
The lncRNA-mRNA network constructed in the present
study revealed the lncRNAs and mRNAs that may be potentially
involved in NEC. lncRNA ENST00000623580 was identified to interact
with 6 mRNAs, including ADAM metallopeptidase with thrombospondin
type 1 motif 1 (ADAMTS1), complement C1R, apolipoprotein L domain
containing 1 (APOLD1), cytochrome B reductase 1 (CYBRD1),
acetyl-CoA carboxylase β (ACACB) and matrix Gla protein (MGP).
Among them, ADAMTS1 is involved in various inflammatory processes
and is required for maintaining organ function, including the
intestine (35,36). Moreover, C1R encodes several
members of the peptidase S1 protein family, which are proteins that
serve as a proteolytic subunit in the complement system C1 complex
(37). The complement system acts
as a mediator in the innate immune response by triggering
phagocytosis, inflammation and rupture of bacterial cell walls,
which may affect the progression of NEC (38). APOLD1 is an endothelial cell early
response protein that affects vascular function and regulates
endothelial cell signaling, which are closely associated with the
NEC process (39). As a member of
the cytochrome family, CYBRD1 encodes an iron-regulated protein and
exhibits ferric reductase activity (40). Furthermore, CYBRD1 has been
demonstrated to serve a key role in dietary iron absorption
(41). It has been shown that NEC
may affect the expression of CYBRD1 in the duodenal brush border
membrane and contributes to the severe anemia associated with NEC
(42). ACACB is currently
considered to be one of the potential targets for regulating human
disease in diabetes, obesity and cancer (43). It was demonstrated that continuous
oxidation of fatty acid in ACACB knockout mice can lead to high
insulin sensitivity (44).
Moreover, a change in tissue ACACB levels via transcriptional
regulation may be important in the affected intestine in NEC
(45). MGP encodes a member of the
osteocalcin/matrix Gla family of proteins (46). The encoded vitamin K-dependent
protein is secreted by chondrocytes and vascular smooth muscle
cells, and functions as a physiological inhibitor of ectopic tissue
calcification (46). Furthermore,
the present results suggested that lncRNA ENST00000623580 may be
involved in the pathogenesis of NEC by regulating the expression of
the aforementioned mRNAs.
SAM and SH3 Domain Containing 1 (SASH1) was
identified to be associated with lncRNA NONHSAT180418.1. SASH1 is
expressed on all microvascular beds and serves as a scaffold
molecule to bind TBGF-β-activated kinase 1 (TAK1), TNF receptor
associated factor 6 (TRAF6), IκB kinase α and IκB kinase β
(47). This interaction promotes
the ubiquitination of TAK1 and TRAF6, and promotes LPS-induced
activation of NF-κB, p38 and JNK, which leads to increased
expression of proinflammatory cytokines and increased endothelial
migration (47). Moreover, SASH1
has been shown to be involved in downstream signaling of TLR4 and
to activate early endothelial responses to receptor activation
(48). Therefore, lncRNA
NONHSAT180418.1 was hypothesized to regulate the expression of
SASH1 to affect TLR4 and the progression of NEC. In addition,
lncRNAs NONHSAT125636.2 and NONHSAT087855.2 were both associated
with LYZ. LYZ encodes human lysozyme, whose natural substrate is
the bacterial cell wall component peptidoglycan (49). It was reported that increasing the
expression of LYZ may control the microbial load in the intestine
(50). Therefore, lncRNA
NONHSAT125636.2 and NONHSAT087855.2 may exert protective effects in
NEC. It has been revealed that lncRNAs can function as competing
endogenous RNAs to indirectly regulate the expression levels of
target genes via sponging microRNAs, and they are implicated in
numerous biological process (51,52).
Thus, further studies on the lncRNA-associated competing endogenous
RNA regulatory network may be useful for understanding the
pathogenesis of NEC.
In conclusion, the present study identified
differentially expressed lncRNAs and mRNAs between NEC and adjacent
intestinal tissues by NGS. A number of these genes were randomly
selected and assessed by RT-qPCR in vitro. GO and KEGG
analyses were performed, and indicated that lncRNAs
ENST00000623580, NONHSAT180418.1, NONHSAT125636.2 and
NONHSAT087855.2 may have bioactive effects in NEC. Therefore, the
present study may provide novel insights into the pathological
mechanisms underlying NEC. However, further functional
investigation is required to confirm these results.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
Wuxi Key Medical Disciplines (grant no. ZDXK12), the Medical
Innovation Team of Jiangsu Province (grant no. CXTDB2017016), the
Wuxi Medical Development Discipline (grant no. FZXK001), the Wuxi
Young Medical Talents (grant nos. QNRC094 and QNRC089), the Wuxi
Hospital Management Centre Key Project (grant no. YGZXZ1513) and
the Wuxi Hospital Management Centre Joint Research Project (grant
no. YGZXL1319). The study was also support by the Young Project of
Wuxi Health and Family Planning Commission (grant no. Q201815), the
National Natural Science Foundation of China (grant nos. 81741089
and 81971427), the Jiangsu Maternal and Child Health Research
Project (grant no. F2018525), the National Natural Science
Foundation for Youth Scholars of China (grant nos. 81501296,
81901512 and 81901517) and the Nanjing Medical Science and
Technological Development Program (grant no. YKK17176).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WC, XY, TT, RY, XW, ZY and YL collected and analyzed
the data. LZ and SH designed the study and wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was approved by the Institutional
Review Board of The Affiliated Wuxi Children's Hospital of Nanjing
Medical University (approval no. 2017-EYLL-115). Informed consent
was obtained from the parents or guardians of all patients.
Patient consent for publication
Informed consent was obtained from the parents or
guardians of all patients.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Fitzgibbons SC, Ching Y, Yu D, Carpenter
J, Kenny M, Weldon C, Lillehei C, Valim C, Horbar JD and Jaksic T:
Mortality of necrotizing enterocolitis expressed by birth weight
categories. J Pediatr Surg. 44:1072–1076. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kastenberg ZJ, Lee HC, Profit J, Gould JB
and Sylvester KG: Effect of deregionalized care on mortality in
very low-birth-weight infants with necrotizing enterocolitis. JAMA
Pediatr. 169:26–32. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dukleska K, Devin CL, Martin AE, Miller
JM, Sullivan KM, Levy C, Prestowitz S, Flathers K, Vinocur CD and
Berman L: Necrotizing enterocolitis totalis: High mortality in the
absence of an aggressive surgical approach. Surgery. 165:1176–1181.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JS and Polin RA: Treatment and
prevention of necrotizing enterocolitis. Semin Neonatol. 8:449–459.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shulhan J, Dicken B, Hartling L and Larsen
BM: Current Knowledge of Necrotizing Enterocolitis in Preterm
Infants and the Impact of Different Types of Enteral Nutrition
Products. Adv Nutr. 8:80–91. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim ED and Sung S: Long noncoding RNA:
Unveiling hidden layer of gene regulatory networks. Trends Plant
Sci. 17:16–21. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Goff LA and Rinn JL: Linking RNA biology
to lncRNAs. Genome Res. 25:1456–1465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han Y, Hong Y, Li L, Li T, Zhang Z, Wang
J, Xia H, Tang Y, Shi Z, Han X, et al: A Transcriptome-Level Study
Identifies Changing Expression Profiles for Ossification of the
Ligamentum Flavum of the Spine. Mol Ther Nucleic Acids. 12:872–883.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim YJ, Hwang KC, Kim SW and Lee YC:
Potential miRNA-target interactions for the screening of gastric
carcinoma development in gastric adenoma/dysplasia. Int J Med Sci.
15:610–616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao L, Rao JN, Cao S, Liu L, Chung HK,
Zhang Y, Zhang J, Liu Y, Gorospe M and Wang JY: Long noncoding RNA
SPRY4-IT1 regulates intestinal epithelial barrier function by
modulating the expression levels of tight junction proteins. Mol
Biol Cell. 27:617–626. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Geng H, Bu HF, Liu F, Wu L, Pfeifer K,
Chou PM, Wang X, Sun J, Lu L, Pandey A, et al: In Inflamed
Intestinal Tissues and Epithelial Cells, Interleukin 22 Signaling
Increases Expression of H19 Long Noncoding RNA, Which Promotes
Mucosal Regeneration. Gastroenterology. 155:144–155. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bell MJ, Ternberg JL, Feigin RD, Keating
JP, Marshall R, Barton L and Brotherton T: Neonatal necrotizing
enterocolitis. Therapeutic decisions based upon clinical staging.
Ann Surg. 187:1–7. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mortazavi A, Williams BA, McCue K,
Schaeffer L and Wold B: Mapping and quantifying mammalian
transcriptomes by RNA-Seq. Nat Methods. 5:621–628. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu M, Song S, Li H, Jiang X, Yin P, Wan
C, Liu X, Liu F and Xu J: The protective effect of caffeic acid
against inflammation injury of primary bovine mammary epithelial
cells induced by lipopolysaccharide. J Dairy Sci. 97:2856–2865.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yazji I, Sodhi CP, Lee EK, Good M, Egan
CE, Afrazi A, Neal MD, Jia H, Lin J, Ma C, et al: Endothelial TLR4
activation impairs intestinal microcirculatory perfusion in
necrotizing enterocolitis via eNOS-NO-nitrite signaling. Proc Natl
Acad Sci USA. 110:9451–9456. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pang Y, Du X, Xu X, Wang M and Li Z:
Monocyte activation and inflammation can exacerbate Treg/Th17
imbalance in infants with neonatal necrotizing enterocolitis. Int
Immunopharmacol. 59:354–360. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Good M, Siggers RH, Sodhi CP, Afrazi A,
Alkhudari F, Egan CE, Neal MD, Yazji I, Jia H, Lin J, et al:
Amniotic fluid inhibits Toll-like receptor 4 signaling in the fetal
and neonatal intestinal epithelium. Proc Natl Acad Sci USA.
109:11330–11335. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jilling T, Simon D, Lu J, Meng FJ, Li D,
Schy R, Thomson RB, Soliman A, Arditi M and Caplan MS: The roles of
bacteria and TLR4 in rat and murine models of necrotizing
enterocolitis. J Immunol. 177:3273–3282. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sharma R, Tepas JJ III, Hudak ML, Mollitt
DL, Wludyka PS, Teng RJ and Premachandra BR: Neonatal gut barrier
and multiple organ failure: Role of endotoxin and proinflammatory
cytokines in sepsis and necrotizing enterocolitis. J Pediatr Surg.
42:454–461. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bonora M, Wieckowsk MR, Chinopoulos C,
Kepp O, Kroemer G, Galluzzi L and Pinton P: Molecular mechanisms of
cell death: Central implication of ATP synthase in mitochondrial
permeability transition. Oncogene. 34:16082015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang L, Fan J, He J, Chen W, Jin W, Zhu
Y, Sun H, Li Y, Shi Y, Jing Y, et al: Regulation of ROS-NF-κB axis
by tuna backbone derived peptide ameliorates inflammation in
necrotizing enterocolitis. J Cell Physiol. 234:14330–14338. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Good M, Sodhi CP, Egan CE, Afrazi A, Jia
H, Yamaguchi Y, Lu P, Branca MF, Ma C, Prindle T Jr, et al: Breast
milk protects against the development of necrotizing enterocolitis
through inhibition of Toll-like receptor 4 in the intestinal
epithelium via activation of the epidermal growth factor receptor.
Mucosal Immunol. 8:1166–1179. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wijendran V, Brenna JT, Wang DH, Zhu W,
Meng D, Ganguli K, Kothapalli KS, Requena P, Innis S and Walker WA:
Long-chain polyunsaturated fatty acids attenuate the IL-1β-induced
proinflammatory response in human fetal intestinal epithelial
cells. Pediatr Res. 78:626–633. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng Z, Zhou H, Ma S, Guan X, Chen L,
Huang J, Gui H, Miao X, Yu S, Wang JH and Wang J: FTY720 attenuates
intestinal injury and suppresses inflammation in experimental
necrotizing enterocolitis via modulating CXCL5/CXCR2 axis. Biochem
Biophys Res Commun. 505:1032–1037. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chan KY, Leung FW, Lam HS, Tam YH, To KF,
Cheung HM, Leung KT, Poon TC, Lee KH, Li K, et al: Immunoregulatory
protein profiles of necrotizing enterocolitis versus spontaneous
intestinal perforation in preterm infants. PLoS One. 7:e369772012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sheng Q, Lv Z, Cai W, Song H, Qian L, Mu
H, Shi J and Wang X: Human β-defensin-3 promotes intestinal
epithelial cell migration and reduces the development of
necrotizing enterocolitis in a neonatal rat model. Pediatr Res.
76:269–279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kandasamy J, Huda S, Ambalavanan N and
Jilling T: Inflammatory signals that regulate intestinal epithelial
renewal, differentiation, migration and cell death: Implications
for necrotizing enterocolitis. Pathophysiology. 21:67–80. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Manoharan I, Suryawanshi A, Hong Y,
Ranganathan P, Shanmugam A, Ahmad S, Swafford D, Manicassamy B,
Ramesh G, Koni PA, et al: Homeostatic PPARalpha Signaling Limits
Inflammatory Responses to Commensal Microbiota in the Intestine. J
Immunol. 196:4739–4749. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Shiou SR, Yu Y, Chen S, Ciancio MJ, Petrof
EO, Sun J and Claud EC: Erythropoietin protects intestinal
epithelial barrier function and lowers the incidence of
experimental neonatal necrotizing enterocolitis. J Biol Chem.
286:12123–12132. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fang C, Xie L, Liu C, Fu C, Ye W, Liu H
and Zhang B: Berberine ameliorates neonatal necrotizing
enterocolitis by activating the phosphoinositide 3-kinase/protein
kinase B signaling pathway. Exp Ther Med. 15:3530–3536.
2018.PubMed/NCBI
|
|
33
|
MohanKumar K, Namachivayam K,
Chapalamadugu KC, Garzon SA, Premkumar MH, Tipparaju SM and
Maheshwari A: Smad7 interrupts TGF-β signaling in intestinal
macrophages and promotes inflammatory activation of these cells
during necrotizing enterocolitis. Pediatr Res. 79:951–961. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baregamian N, Rychahou PG, Hawkins HK,
Evers BM and Chung DH: Phosphatidylinositol 3-kinase pathway
regulates hypoxia-inducible factor-1 to protect from intestinal
injury during necrotizing enterocolitis. Surgery. 142:295–302.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Günther W, Skaftnesmo KO, Arnold H,
Bjerkvig R and Terzis AJ: Distribution patterns of the
anti-angiogenic protein ADAMTS-1 during rat development. Acta
Histochem. 107:121–131. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hummitzsch L, Zitta K, Berndt R, Wong YL,
Rusch R, Hess K, Wedel T, Gruenewald M, Cremer J, Steinfath M, et
al: Remote ischemic preconditioning attenuates intestinal mucosal
damage: Insight from a rat model of ischemia-reperfusion injury. J
Transl Med. 17:1362019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kardos J, Harmat V, Palló A, Barabás O,
Szilágyi K, Gráf L, Náray-Szabó G, Goto Y, Závodszky P and Gál P:
Revisiting the mechanism of the autoactivation of the complement
protease C1r in the C1 complex: Structure of the active catalytic
region of C1r. Mol Immunol. 45:1752–1760. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kobayashi H, Albarracin L, Sato N, Kanmani
P, Kober AK, Ikeda-Ohtsubo W, Suda Y, Nochi T, Aso H, Makino S, et
al: Modulation of porcine intestinal epitheliocytes
immunetranscriptome response by Lactobacillus jensenii
TL2937. Benef Microbes. 7:769–782. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Regard JB, Scheek S, Borbiev T, Lanahan
AA, Schneider A, Demetriades AM, Hiemisch H, Barnes CA, Verin AD
and Worley PF: Verge: a novel vascular early response gene. J
Neurosci. 24:4092–4103. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Oakhill JS, Marritt SJ, Gareta EG, Cammack
R and McKie AT: Functional characterization of human duodenal
cytochrome B (Cybrd1): Redox properties in relation to iron and
ascorbate metabolism. Biochim Biophys Acta. 1777:260–268. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schlottmann F, Vera-Aviles M and
Latunde-Dada GO: Duodenal cytochrome B (Cybrd1) ferric reductase
functional studies in cells. Metallomics. 9:1389–1393. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Patel RM, Knezevic A, Shenvi N, Hinkes M,
Keene S, Roback JD, Easley KA and Josephson CD: Association of Red
Blood Cell Transfusion, Anemia, and Necrotizing Enterocolitis in
Very Low-Birth-Weight Infants. JAMA. 315:889–897. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Glorieux C, Zamocky M, Sandoval JM, Verrax
J and Calderon PB: Regulation of catalase expression in healthy and
cancerous cells. Free Radic Biol Med. 87:84–97. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ma L, Mondal AK, Murea M, Sharma NK,
Tönjes A, Langberg KA, Das SK, Franks PW, Kovacs P, Antinozzi PA,
et al: The effect of ACACB cis-variants on gene expression
and metabolic traits. PLoS One. 6:e238602011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen G, Li Y, Su Y, Zhou L, Zhang H, Shen
Q, Du C, Li H, Wen Z, Xia Y, et al: Identification of candidate
genes for necrotizing enterocolitis based on microarray data. Gene.
661:152–159. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Caiado H, Conceição N, Tiago D, Marreiros
A, Vicente S, Enriquez JL, Vaz AM, Antunes A, Guerreiro H, Caldeira
P, et al: Evaluation of MGP gene expression in colorectal cancer.
Gene. 723:1441202020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dauphinee SM, Clayton A, Hussainkhel A,
Yang C, Park YJ, Fuller ME, Blonder J, Veenstra TD and Karsan A:
SASH1 is a scaffold molecule in endothelial TLR4 signaling. J
Immunol. 191:892–901. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Coulombe P, Paliouras GN, Clayton A,
Hussainkhel A, Fuller M, Jovanovic V, Dauphinee S, Umlandt P, Xiang
P, Kyle AH, et al: Endothelial Sash1 Is Required for Lung
Maturation through Nitric Oxide Signaling. Cell Rep.
27:1769–1780.e1764. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Um SH, Kim JS, Kim K, Kim N, Cho HS and Ha
NC: Structural basis for the inhibition of human lysozyme by PliC
from Brucella abortus. Biochemistry. 52:9385–9393. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Singh SB, Wilson M, Ritz N and Lin HC:
Autophagy Genes of Host Responds to Disruption of Gut Microbial
Community by Antibiotics. Dig Dis Sci. 62:1486–1497. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fan Z, Gao S, Chen Y, Xu B, Yu C, Yue M
and Tan X: Integrative analysis of competing endogenous RNA
networks reveals the functional lncRNAs in heart failure. J Cell
Mol Med. 22:4818–4829. 2018. View Article : Google Scholar : PubMed/NCBI
|