Introduction
Glaucoma represents a group of chronic and
progressive optic neuropathies with distinctive pattern of
progressive visual field loss and optic disc damage, and is the
leading cause of irreversible blindness worldwide. This enigmatic
and heterogeneous disease has affected 3.5% of the world's
population with approximately 5.7 million people visually impaired
and 3.1 million blind, and an estimate to affect 111.8 million
people by 2040 (1). Primary
open-angle glaucoma (POAG), the most common representative, is
characterized by a normal, open anterior chamber angle and elevated
intraocular pressure (IOP) or even normal IOP, named normal tension
glaucoma (NTG). Two hallmark theories attempt to explain the
pathway of the disease; the mechanical and vascular theory.
Although both present postulated mechanisms of progressive retinal
ganglion cell (RGC) damage and eventual optic neuropathy, the exact
etiology still remains unknown (2). The well recognised risk factors for
POAG include higher age, high IOP, decreased central corneal
thickness (CCT), African descent, high myopia and a positive family
history (3,4). Amongst them, high IOP remains the
only modifiable risk factor for the development and prognostic
factor for the progression of POAG (3).
POAG has long shown a strong genetic component with
60% of patients with a positive family history and family based
studies identifying a 10-fold increased risk of POAG for first
degree relatives (5). Heritability
of POAG can be divided into two major categories: direct
association (increased POAG risk) and indirect (increased risk for
a component of the disease). The first one deals with several genes
linked to POAG through family-based genetic linkage analysis, with
major examples being myocilin (MYOC), optineurin (OPTN), WD repeat
domain 36 (WDR36), cytochrome P450, family 1, subfamily B,
polypeptide 1 (CYP1B1) and neurotrophin 4 (NTF4) (6–10).
With heritability ranging from 0 (no genetic component) to 1
(phenotype determined by genes), these genes attribute to POAG
heritability at ~0.81 (11). The
second factor refers to the endophenotype traits related to POAG
pathogenesis, highly heritable and polymorphic. Notable examples
are IOP (0.55), vertical cup-to-disc ratio (VCDR=0.48–0.66), CCT
(0.72), cup area (CA=0.75) and disc area (DA=0.72) (12,13).
Genome-wide association studies (GWAS) have been
used in the past decade in an attempt to uncover genetic variants
of complex diseases. For POAG, over 70 single-nucleotide
polymorphisms (SNPs) have been identified and linked to POAG or its
endophenotypes, changing our understanding of the molecular
pathways of the disease (14).
Identification of the genes associated with the disease may also
provide useful information for both patients and their physicians,
by enabling the design of genetic screening tests that may help
physicians assess the risk of patients for disease as well as to
differentiate between clinically similar disorders. Valuable
research available has provided details for many SNPs related to
POAG. In this review the authors summarize the most common
variants, present their possible clinical correlations and
highlight their interactions.
Search strategy
A MEDLINE/PubMed search was performed for articles
in English from January 2010 to January 2020. Keywords included
glaucoma, primary open angle glaucoma, genetics, genetic
association studies, GWAS and SNP. Further search was performed in
the database upon relative findings in the articles.
Genomic loci
Genetic linkage analysis was the first genetic
decoding of glaucoma. Classically used for Mendelian traits or
mutations in a single gene, this method reveals chromosomal regions
associated with a specific phenotype. Followed by GWAS in various
populations, a number of variants and interactions unfolded, thus
forming the complicated image of POAG. These genomic loci are
marked GLC1A through GLC1P and show high heterogeneity (14,15)
(Table I).
 | Table I.Glaucoma genomic loci (GLC1A-P),
candidate genes and locations and their associated glaucoma
type. |
Table I.
Glaucoma genomic loci (GLC1A-P),
candidate genes and locations and their associated glaucoma
type.
| Locus | Gene | Location | Glaucoma type |
|---|
| GLC1A | MYOC | 1q24.3-q25.2 | POAG/JOAG |
| GLC1B | – | 2cen-q13 | POAG, early
onset |
| GLC1C | – | 3q21-q24 | POAG |
| GLC1D | – | 8q23 | POAG, early
onset |
| GLC1E | OPTN | 10p13 | POAG/NTG |
| GLC1F | ASB10 | 7q36 | POAG |
| GLC1G | WDR36 | 5q22 | POAG |
| GLC1H | – | 2p16-p15 | POAG |
| GLC1I | – | 15q11-q13 | POAG |
| GLC1J | – | 9q22 | JOAG |
| GLC1K | – | 20p12 | JOAG |
| GLC1L | – | 3p22-p21 | POAG |
| GLC1M | – | 5q22 | JOAG |
| GLC1N | – | 15q22-q24 | JOAG |
| GLC1O | NTF4 | 19q13.3 | POAG |
| GLC1P | Possibly
TBK1 | 12q24 | POAG |
Common variants
Although a number of SNPs has been linked to POAG,
those summarized in Table II,
appear to be the most commonly studied in the literature.
 | Table II.Candidate genes and their possible
association to glaucoma pathogenesis. |
Table II.
Candidate genes and their possible
association to glaucoma pathogenesis.
| Gene | Location | Possible glaucoma
mechanism/effect |
|---|
|
MYOC/TIGR | 1q24 | TM,
outflowa |
| CYP1B1 | 2p22.2 | Anterior segment
dysgenesis |
| OPTN | 10p13 | Autophagy,
apoptosis |
| TBK1 | 12q14 | Autophagy,
inflammatory response |
| WDR36 | 5q22 | TM via
apoptosis |
| NTF4 | 19q13.33 | Impaired neuronal
survival |
| ASB10 | 7q36 | Outflow |
| ATOH7 | 10q21 | DA, VCDR, CA, POAG
risk |
|
CAV1/CAV2 | 7q31 | IOP changes,
outflow |
|
CDKN2B-AS1 | 9p21 | VCDR |
| SIX6 | 14q23 | VCDR, myopia,
reduced RNFL thickness |
| TMCO1 | 1q24.1 | IOP changes,
increased POAG risk |
| ABCA1 | 9q31 | Inflammatory
response, neurodegeneration |
| AFAP1 | 4p16.1 | Unknown |
|
ARHGEF12 | 11q23.3 | IOP changes,
outflow |
| TXNRD2 | 22q11 | Oxidative
stress |
| FOXC1 | 6p25.3 | Anterior segment
dysgenesis |
| ATXN2 | 12q24.12 |
Neurodegeneration |
| GAS7 | 17p13 | CA, IOP
changes |
| GALC | 14q31.3 | Increased POAG
risk |
MYOC/trabecular meshwork-induced glucocorticoid
response protein (TIGR). Located on chromosome 1q24, MYOC
encodes protein MYOC, which is believed to have a role in
cytoskeletal function and represents the first gene to be
associated with POAG (14).
Mutations in MYOC (alternatively TIGR) are linked to
the GLC1A locus (14,16). Over 100 POAG-associated mutations
have been identified in the MYOC gene making it accountable
for 3–5% of POAG cases (15).
Primarily associated with juvenile-onset POAG, expresses an
autosomal dominant and highly penetrant severe phenotype, with
highly elevated IOP which often requires surgical management
(10,17). A less severe form is observed in
the adult onset form, the most prevalent one (10). Other associations include primary
congenital glaucoma (18). The
effect of MYOC mutations appears to be on the trabecular
meshwork (TM) and aqueous humor outflow (19).
CYP1B1
Located on chromosome 2p22.2, CYP1B1 belongs to the
cytochrome P450 superfamily of enzymes. Mutations in CYP1B1,
although classically linked to anterior segment dysgenesis and
congenital glaucoma, may also contribute to the GLC1A phenotype
through a digenic inheritance, acting as a modifier for MYOC
(20).
OPTN
Located on chromosome 10p13, OPTN encodes the
coiled-coil containing protein OPTN that plays an important role in
the maintenance of the Golgi complex, in membrane trafficking and
exocytosis. Mutations in OPTN are linked to the GLC1E locus.
Even though variable mutations have been recognised, E50K has been
strongly linked to glaucoma (21).
Primarily associated with NTG, it has also been reported in
patients with amyotrophic lateral sclerosis (ALS) (22). A variety of interactions and
cellular effects have been reported for OPTN, notably its
role in autophagy and apoptosis (23,24).
It has been reported that although MYOC overexpression has
no effect on OPTN expression, OPTN overexpression
upregulates endogenous MYOC in TM cells (25). Noteworthy, the most notable is the
interaction of OPTN with TANK-binding kinase 1 (TBK1)
as they may share a common pathway (23,26).
TBK1
Located on chromosome 12q14, mutations in TBK1 are
linked to the GLC1P locus (10).
Responsible for approximately 1% of NTG through a copy number
variant (CNV) consisting of duplications in the TBK1,
mutations have also been reported in patients with ALS and other
central nervous system disorders (22,27,28).
TBK1 is a kinase that regulates the expression of genes in the
NF-κB signaling pathway, playing an essential role in regulating
inflammatory responses to foreign agents and is also implicated in
autophagy (29).
WDR36
Located on chromosome 5q22, WDR36 encodes a
member of the WD repeat protein family that is involved in T-cell
activation. Mutations in WDR36 are linked to the GLC1G locus
(10). Linked to an increased risk
for POAG, mutations in WDR36 were initially reported to be
responsible for 1.6 to 17% of POAG (8,30,31).
WDR36 appears to negatively affect TM cells via apoptotic
cell death (32).
NTF4
Located on chromosome 19q13.33, NTF4 encodes
a neurotrophic factor that signals predominantly through the
tyrosine kinase receptor B (TrkB) receptor tyrosine kinase.
Mutations in NTF4 are linked to the GLC1O locus. Negatively
affecting the activation of TrkB, NTF4 mutations have been
found in 1.7% of POAG patients in a study of European patients
(9).
Ankyrin repeat and SOCS box containing
10 (ASB10)
Located on chromosome 7q36, ASB10 encodes a
protein that is a member of the ASB family of proteins. Mutations
in ASB10 are linked to the GLC1F locus (33). ASB10 transcription appears
to reduce aqueous humor outflow facility (34).
Atonal bHLH transcription factor 7
(ATOH7)
Located on chromosome 10q21, ATOH7 encodes a
transcription factor that appears to play a role in the
differentiation of the RGCs and determination of DA, possibly
during embryogenesis. It is associated with DA, VCDR, CA and POAG
risk (35,36).
Caveolin1/caveolin2 (CAV1/CAV2)
Located on chromosome 7q31, CAVs are involved in a
number of cellular processes such as transcellular transport, cell
proliferation and signal transduction (37). CAV1 and CAV2 are
found to be expressed in ciliary muscle, TM, Schlemm's canal and
retinal cells, and variants in the CAV1/CAV2 region
are associated with IOP changes, possibly through alterations of
aqueous outflow resistance (37–39).
A possible direct, cell type specific interaction of CAV1
with ATP-binding cassette, subfamily A, member 1 (ABCA1),
also supports a possible role in POAG (40).
Cyclin-dependent kinase inhibitor 2B
antisense RNA 1 (CDKN2B-AS1)
Located on chromosome 9p21, in the
CDKN2B-CDKN2A gene cluster, CDKN2B protein is
expressed in the inner nuclear layers of the retina and the
ganglion cell layer. CDKN2B-AS1 is detected in the retina,
ciliary body and optic nerve and variants have been associated with
VCDR changes and NTG especially in women, as well as various
non-ocular disorders, namely myocardial infarction and intracranial
aneurysms; an association of POAG risk and these conditions has not
been made however (41–47).
Six homeobox 6 (SIX6)
Located on chromosome 14q23, SIX6 gene
encodes homeobox proteins, critical for ocular development. The
protein encoded by this gene is a homeobox protein that is similar
to the Drosophila ‘sine oculis’ gene product and is
thought to be involved in eye development. Variants in this locus
have been linked to POAG through VCDR, myopia and reduced retinal
nerve fiber layer (RNFL) thickness. An interesting interaction of
the POAG-risk His141 SIX6 variant with CDKN2B-AS1 has been
proposed as the former has been shown to elevate expression of the
latter. A possibly protective Asn141 SIX6 variant remains to
be further observed (44,47,48–52).
Transmembrane and coiled-coil domains
protein 1 (TMCO1)
Located on chromosome 1q24.1, TMCO1 encodes a
transmembrane protein playing a key role in calcium homeostasis. It
is expressed, among other ocular tissues, in the TM and retina.
Sequence variants have been associated with IOP and increased POAG
risk to a degree of 1.7-fold per risk allele, making it a
significant quantitative trait (46,50,53–55).
ABCA1
Located on chromosome 9q31, ABCA1 encodes a
protein complex associated with extracellular and intracellular
membrane transport. Although the locus is mainly associated with
familial high-density lipoprotein deficiency and Tangier's disease,
ABCA1 is also expressed in numerous ocular tissues, namely
TM, ciliary body and RGCs. Sequence variants have been linked to
POAG and IOP, possibly through an inflammatory and
neurodegenerative mechanism (56,57).
Actin filament-associated protein 1
(AFAP1)
Located on chromosome 4p16.1, AFAP1 encodes
an adaptor protein that modulates changes in actin filament
integrity in response to cellular signals. It is expressed, among
others, in the retina and TM cells. Sequence variants have been
associated with POAG through a still unknown mechanism (41,57).
RHO guanine nucleotide exchange factor
12 (ARHGEF12)
Located on chromosome 11q23.3, ARHGEF12
encodes a guanine nucleotide exchange factor (GEF) that may form a
complex with G proteins and stimulate Rho-dependent signals. An
intronic variant has been associated with POAG and IOP, possibly
through activation of RhoA and subsequent ROCK activation, leading
to decreased outflow and elevated IOP. An interesting interaction
is that with ABCA1 protein, with ARHGEF12 preventing
ABCA1 degradation (58–60).
Thioredoxin reductase 2 (TXNRD2)
Located on chromosome 22q11, TXNRD2 gene
encodes a mitochondrial protein important for scavenging reactive
oxygen species in mitochondria. It belongs to the family of
flavoenzymes that catalyze redox reactions, thus controlling the
levels of reactive oxygen species. TXNRD2 is found, among
others, in the retina and optic nerve and sequence variants have
been associated with POAG, possibly through a protective effect of
TXNRD2 against oxidative stress of the RGCs (41,61,62).
Forkhead box C1 (FOXC1)
Located on chromosome 6p25.3, FOXC1 is found
next to GDP-mannose 4,6-dehydratase (GMDS) gene. It encodes
a protein that has been shown to play a role in the regulation of
embryonic and ocular development. Although mainly linked to
anterior segment dysgenesis and Axenfeld-Rieger syndrome, sequence
variants within the GMDS gene as well as variants located
upstream of FOXC1 have been associated with POAG (41,57).
Ataxin 2 (ATXN2)
Located on chromosome 12q24.12, ATXN2 is
involved in regulating mRNA translation through its interactions
with the poly (A)-binding protein. It is also involved in the
formation of stress granules and P-bodies, which also play roles in
RNA regulation (63). ATXN2
is expressed, among others, in the ciliary body, retina, and optic
nerve. Although mainly linked to spinocerebellar ataxia and ALS,
variants in ATXN2 have been associated with POAG possibly
through neurodegeneration (41,64).
Growth arrest-specific 7 (GAS7)
Located on chromosome 17p13, GAS7 encodes a
protein that plays a putative role in neuronal development as well
as maturation and is primarily expressed in vivo, in
terminally differentiated brain cells. GAS7 has been
associated with increased cup volume and IOP (47,54,55,65).
Galactosylceramidase (GALC)
Located on chromosome 14q31.3, GALC encodes a
lysosomal enzyme called galactosylceramidase, which is mainly
linked to Krabbe disease, a rare disorder of the myelin sheath with
progressive optic neuropathy. GALC has been associated with
a 5-fold increased risk of POAG when heterozygous for a CNV
deletion (66,67).
Additional genes associated with POAG
Single SNPs in the MYOC, COL8A2, COL1A1 and
ZNF469 gene regions have shown marginal association with
POAG in a study conducted in South Africa (68). Although the number of the subjects
enrolled in the study was small, it was demonstrated that the main
POAG-associated susceptibility alleles found in other populations,
play a reduced role in populations of African ancestry. Moreover, a
GWAS performed in India identified a novel candidate gene for POAG,
called membrane palmitoylated protein 7 (MPP7), which is
dysregulated under cyclic mechanical stress in the TM, thus causing
dysfunction of cell to cell interaction (69), while another study in India led to
an novel association of POAG with Opticin (OPTC) gene
(70).
Endophenotypes and clinical association
Endophenotypes are stable, heritable quantitative
traits, disease independent, that are usually helpful in analysing
heterogeneous or variably phenotypic diseases. For POAG, these
traits include IOP, CCT, VCDR, DA and RNFL thickness and as
mentioned, contribute separately in the heritability of the disease
(14). Studying the Endophenotypes
helped dissect the disease and focus on separate components that
are linked to clinical practice; nonetheless the variety of POAG
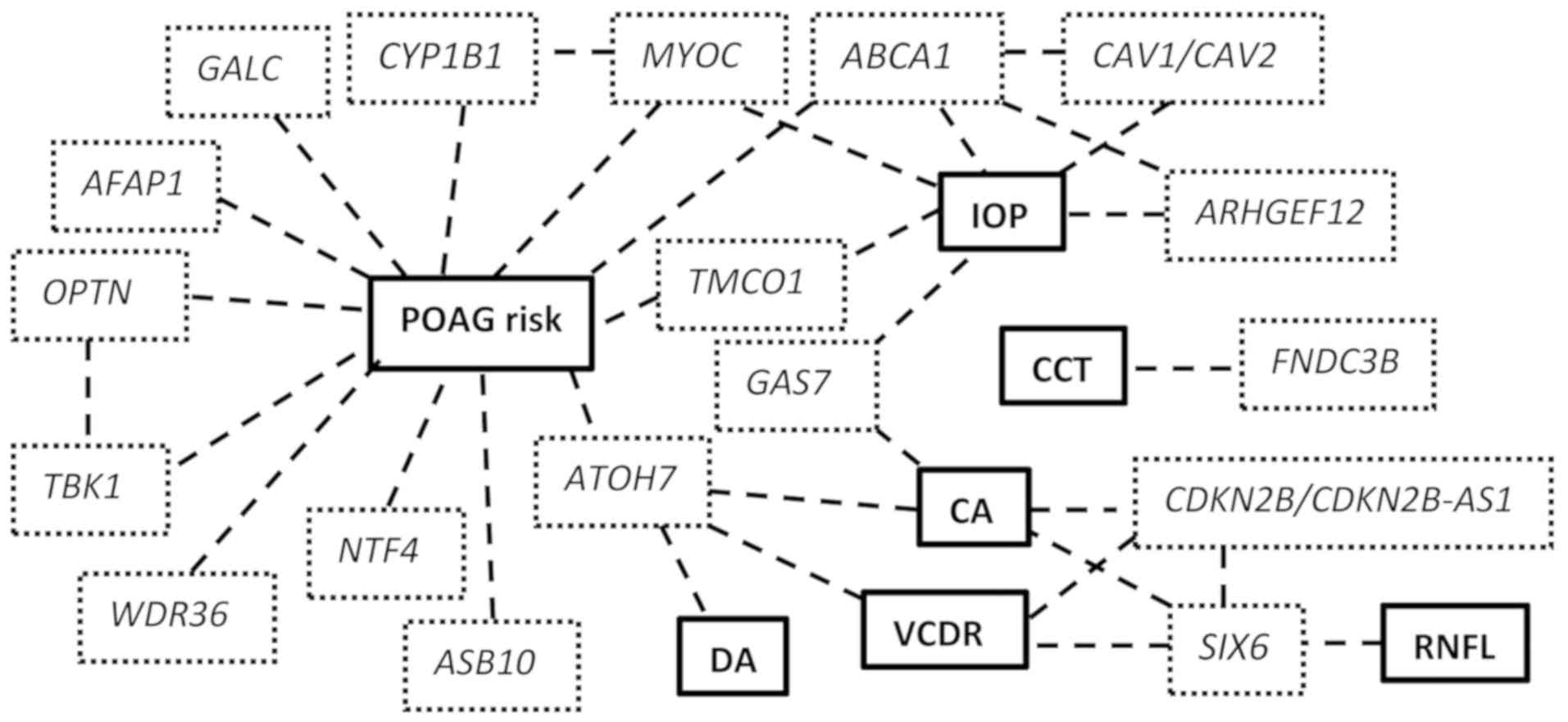
endophenotypes highlights the complexity of the disease (Fig. 1).
IOP
Independent of the genetic component, IOP remains
the only modifiable risk factor and IOP reduction the most commonly
used treatment of POAG. Of the aforementioned genetic variants,
TMCO1, CAV1/CAV2, ARHGEF12, ABCA1 and GAS7
have been linked to IOP through GWAS, among other susceptibility
loci (43,54,55).
Of great interest is that the combined heritability of these loci
regarding IOP is less than 2%, meaning that the majority is still
to be explained (55). This
observation could mean that further investigating the IOP
endophenotype could clarify its role in glaucoma both as a
contributing factor and a therapeutic target.
CCT
Well recognized as an independent risk factor for
POAG, over 26 loci have been identified for CCT (3,71).
With a highly heritable trait, CCT shows association with
ethnicity, with lower values in populations of African descent
(11,72). Of the many sequence variants
identified for CCT, of interest is fibronectin type III
domain-containing protein 3B (FNDC3B) which appears to be
involved also in IOP changes (71).
VCDR
Increased VCDR is one of the cardinal clinical signs
of glaucomatous optic neuropathy. Of the above mentioned genetic
variants, ATOH7, CDKN2B-AS1 and SIX6 have been linked
to VCDR, among other susceptibility loci (73,74).
DA and CA
For DA 14 loci have been identified, 5 of which are
also associated with VCDR. Of these, perhaps the most interesting
is ATOH7, associated with DA, CA and VCDR, that appears to
affect the differentiation of the RGCs (35,73,74).
Approximately 27 variants have been associated with CA, notably
ATOH7, CDKN2B-AS1 and SIX6 which, as mentioned
previously, have also been linked to VCDR (49,75).
RNFL
Two susceptibility loci have been identified for
RNFL thickness through linkage analysis. As previously discussed, a
SIX6 coding variant is one of them (51,76).
Conclusion
Since the turn of the century there has been an
explosion of research using genetic and next generation,
high-throughput DNA sequencing technologies and it is
unquestionably expected that they will be extensively employed also
for further investigation and diagnosis of glaucoma. We aimed to
present the basic role of the most studied sequence variants
regarding POAG and their complex interaction. Although many more
have been identified and many remain to be further clarified,
linkage analysis initially and, recently, GWAS, have opened the
road to the admittedly enigmatic and heterogenic disease that is
POAG. As regards the issue of genetic counselling, genetic testing
for glaucoma may obviously be helpful in some specific situations,
such as screening of family members with a known genetic mutation
in autosomal dominant POAG of early onset, although at a population
level this is not presently justified (77).
With various components and differential clinical
expression, the identification of genetic factors either directly
linked to the disease or endophenotype-related, brings us closer to
understanding the molecular and cellular mechanisms and,
eventually, to the development of efficient therapeutic approaches
targeted at the root cause of the condition.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
AT, ETD, GNG, DAS and MIZ conceived and designed the
study. AT, GNG and ETD researched the literature, performed
critical analysis and review of the literature and drafted the
manuscript. DAS and MIZ critically revised the article for
important intellectual content. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests. DAS is the Editor-in-Chief for the journal, but had no
personal involvement in the reviewing process, or any influence in
terms of adjudicating on the final decision, for this article.
References
|
1
|
Tham YC, Li X, Wong TY, Quigley HA, Aung T
and Cheng CY: Global prevalence of glaucoma and projections of
glaucoma burden through 2040: A systematic review and
meta-analysis. Ophthalmology. 121:2081–2090. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kwon YH, Fingert JH, Kuehn MH and Alward
WL: Primary open-angle glaucoma. N Engl J Med. 360:1113–1124. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gordon MO, Beiser JA, Brandt JD, Heuer DK,
Higginbotham EJ, Johnson CA, Keltner JL, Miller JP, Parrish RK II,
Wilson MR, et al: The Ocular Hypertension Treatment Study: Baseline
factors that predict the onset of primary open-angle glaucoma. Arch
Ophthalmol. 120:714–720, discussion 829–830. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fan BJ, Leung YF, Wang N, Lam SC, Liu Y,
Tam OS and Pang CP: Genetic and environmental risk factors for
primary open-angle glaucoma. Chin Med J (Engl). 117:706–710.
2004.PubMed/NCBI
|
|
5
|
Wolfs RC, Klaver CC, Ramrattan RS, van
Duijn CM, Hofman A and de Jong PT: Genetic risk of primary
open-angle glaucoma. Population-based familial aggregation study.
Arch Ophthalmol. 116:1640–1645. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Stone EM, Fingert JH, Alward WL, Nguyen
TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A,
Nichols BE, et al: Identification of a gene that causes primary
open angle glaucoma. Science. 275:668–670. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rezaie T, Child A, Hitchings R, Brice G,
Miller L, Coca-Prados M, Héon E, Krupin T, Ritch R, Kreutzer D, et
al: Adult-onset primary open-angle glaucoma caused by mutations in
optineurin. Science. 295:1077–1079. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Monemi S, Spaeth G, DaSilva A, Popinchalk
S, Ilitchev E, Liebmann J, Ritch R, Héon E, Crick RP, Child A, et
al: Identification of a novel adult-onset primary open-angle
glaucoma (POAG) gene on 5q22.1. Hum Mol Genet. 14:725–733. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pasutto F, Matsumoto T, Mardin CY, Sticht
H, Brandstätter JH, Michels-Rautenstrauss K, Weisschuh N, Gramer E,
Ramdas WD, van Koolwijk LM, et al: Heterozygous NTF4 mutations
impairing neurotrophin-4 signaling in patients with primary
open-angle glaucoma. Am J Hum Genet. 85:447–456. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fingert JH: Primary open-angle glaucoma
genes. Eye (Lond). 25:587–595. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Charlesworth J, Kramer PL, Dyer T, Diego
V, Samples JR, Craig JE, Mackey DA, Hewitt AW, Blangero J and Wirtz
MK: The path to open-angle glaucoma gene discovery: Endophenotypic
status of intraocular pressure, cup-to-disc ratio, and central
corneal thickness. Invest Ophthalmol Vis Sci. 51:3509–3514. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Klein BE, Klein R and Lee KE: Heritability
of risk factors for primary open-angle glaucoma: The Beaver Dam Eye
Study. Invest Ophthalmol Vis Sci. 45:59–62. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sanfilippo PG, Hewitt AW, Hammond CJ and
Mackey DA: The heritability of ocular traits. Surv Ophthalmol.
55:561–583. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Y and Allingham RR: Major review:
Molecular genetics of primary open-angle glaucoma. Exp Eye Res.
160:62–84. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y and Allingham RR: Molecular genetics
in glaucoma. Exp Eye Res. 93:331–339. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheffield VC, Stone EM, Alward WL, Drack
AV, Johnson AT, Streb LM and Nichols BE: Genetic linkage of
familial open angle glaucoma to chromosome 1q21-q31. Nat Genet.
4:47–50. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fingert JH, Héon E, Liebmann JM, Yamamoto
T, Craig JE, Rait J, Kawase K, Hoh ST, Buys YM, Dickinson J, et al:
Analysis of myocilin mutations in 1703 glaucoma patients from five
different populations. Hum Mol Genet. 8:899–905. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kaur K, Reddy AB, Mukhopadhyay A, Mandal
AK, Hasnain SE, Ray K, Thomas R, Balasubramanian D and Chakrabarti
S: Myocilin gene implicated in primary congenital glaucoma. Clin
Genet. 67:335–340. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dismuke WM, Challa P, Navarro I, Stamer WD
and Liu Y: Human aqueous humor exosomes. Exp Eye Res. 132:73–77.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kaur K, Mandal AK and Chakrabarti S:
Primary congenital glaucoma and the involvement of CYP1B1. Middle
East Afr J Ophthalmol. 18:7–16. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aung T, Rezaie T, Okada K, Viswanathan AC,
Child AH, Brice G, Bhattacharya SS, Lehmann OJ, Sarfarazi M and
Hitchings RA: Clinical features and course of patients with
glaucoma with the E50K mutation in the optineurin gene. Invest
Ophthalmol Vis Sci. 46:2816–2822. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cirulli ET, Lasseigne BN, Petrovski S,
Sapp PC, Dion PA, Leblond CS, Couthouis J, Lu YF, Wang Q, Krueger
BJ, et al FALS Sequencing Consortium, : Exome sequencing in
amyotrophic lateral sclerosis identifies risk genes and pathways.
Science. 347:1436–1441. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heo JM, Ordureau A, Paulo JA, Rinehart J
and Harper JW: The PINK1-PARKIN mitochondrial ubiquitylation
pathway drives a program of OPTN/NDP52 recruitment and TBK1
activation to promote mitophagy. Mol Cell. 60:7–20. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Slowicka K, Vereecke L and van Loo G:
Cellular functions of optineurin in health and disease. Trends
Immunol. 37:621–633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Park BC, Tibudan M, Samaraweera M, Shen X
and Yue BYJT: Interaction between two glaucoma genes, optineurin
and myocilin. Genes Cells. 12:969–979. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Morton S, Hesson L, Peggie M and Cohen P:
Enhanced binding of TBK1 by an optineurin mutant that causes a
familial form of primary open angle glaucoma. FEBS Lett.
582:997–1002. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Garrett ME, Yaspan BL, Bailey JC,
Loomis SJ, Brilliant M, Budenz DL, Christen WG, Fingert JH,
Gaasterland D, et al: DNA copy number variants of known glaucoma
genes in relation to primary open-angle glaucoma. Invest Ophthalmol
Vis Sci. 55:8251–8258. 2014b. View Article : Google Scholar
|
|
28
|
Gijselinck I, Van Mossevelde S, van der
Zee J, Sieben A, Philtjens S, Heeman B, Engelborghs S, Vandenbulcke
M, De Baets G, Bäumer V, et al BELNEU Consortium, : Loss of TBK1 is
a frequent cause of frontotemporal dementia in a Belgian cohort.
Neurology. 85:2116–2125. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsumoto G, Shimogori T, Hattori N and
Nukina N: TBK1 controls autophagosomal engulfment of
polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation.
Hum Mol Genet. 24:4429–4442. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Janssen SF, Gorgels TG, Ramdas WD, Klaver
CC, van Duijn CM, Jansonius NM and Bergen AA: The vast complexity
of primary open angle glaucoma: Disease genes, risks, molecular
mechanisms and pathobiology. Prog Retin Eye Res. 37:31–67. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kramer PL, Samples JR, Monemi S, Sykes R,
Sarfarazi M and Wirtz MK: The role of the WDR36 gene on chromosome
5q22.1 in a large family with primary open-angle glaucoma mapped to
this region. Arch Ophthalmol. 124:1328–1331. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gallenberger M, Meinel DM, Kroeber M,
Wegner M, Milkereit P, Bösl MR and Tamm ER: Lack of WDR36 leads to
preimplantation embryonic lethality in mice and delays the
formation of small subunit ribosomal RNA in human cells in vitro.
Hum Mol Genet. 20:422–435. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Murakami K, Meguro A, Ota M, Shiota T,
Nomura N, Kashiwagi K, Mabuchi F, Iijima H, Kawase K, Yamamoto T,
et al: Analysis of microsatellite polymorphisms within the GLC1F
locus in Japanese patients with normal tension glaucoma. Mol Vis.
16:462–466. 2010.PubMed/NCBI
|
|
34
|
Pasutto F, Keller KE, Weisschuh N, Sticht
H, Samples JR, Yang YF, Zenkel M, Schlötzer-Schrehardt U, Mardin
CY, Frezzotti P, et al: Variants in ASB10 are associated with
open-angle glaucoma. Hum Mol Genet. 21:1336–1349. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Macgregor S, Hewitt AW, Hysi PG, Ruddle
JB, Medland SE, Henders AK, Gordon SD, Andrew T, McEvoy B,
Sanfilippo PG, et al: Genome-wide association identifies ATOH7 as a
major gene determining human optic disc size. Hum Mol Genet.
19:2716–2724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Venturini C, Nag A, Hysi PG, Wang JJ, Wong
TY, Healey PR, Mitchell P, Hammond CJ and Viswanathan AC; Wellcome
Trust Case Control Consortium 2 and BMES GWAS Group, : Clarifying
the role of ATOH7 in glaucoma endophenotypes. Br J Ophthalmol.
98:562–566. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gu X, Reagan AM, McClellan ME and Elliott
MH: Caveolins and caveolae in ocular physiology and
pathophysiology. Prog Retin Eye Res. 56:84–106. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen F, Klein AP, Klein BE, Lee KE, Truitt
B, Klein R, Iyengar SK and Duggal P: Exome array analysis
identifies CAV1/CAV2 as a susceptibility locus for intraocular
pressure. Invest Ophthalmol Vis Sci. 56:544–551. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Aga M, Bradley JM, Wanchu R, Yang YF,
Acott TS and Keller KE: Differential effects of caveolin-1 and −2
knockdown on aqueous outflow and altered extracellular matrix
turnover in caveolin-silenced trabecular meshwork cells. Invest
Ophthalmol Vis Sci. 55:5497–5509. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kuo CY, Lin YC, Yang JJ and Yang VC:
Interaction abolishment between mutant caveolin-1(Δ62-100) and
ABCA1 reduces HDL-mediated cellular cholesterol efflux. Biochem
Biophys Res Commun. 414:337–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bailey JN, Loomis SJ, Kang JH, Allingham
RR, Gharahkhani P, Khor CC, Burdon KP, Aschard H, Chasman DI, Igo
RP Jr, et al ANZRAG Consortium, : Genome-wide association analysis
identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for
primary open-angle glaucoma. Nat Genet. 48:189–194. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Burdon KP, Crawford A, Casson RJ, Hewitt
AW, Landers J, Danoy P, Mackey DA, Mitchell P, Healey PR and Craig
JE: Glaucoma risk alleles at CDKN2B-AS1 are associated with lower
intraocular pressure, normal-tension glaucoma, and advanced
glaucoma. Ophthalmology. 119:1539–1545. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen Y, Hughes G, Chen X, Qian S, Cao W,
Wang L, Wang M and Sun X: Genetic variants associated with
different risks for high tension glaucoma and normal tension
glaucoma in a Chinese population. Invest Ophthalmol Vis Sci.
56:2595–2600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wiggs JL, Yaspan BL, Hauser MA, Kang JH,
Allingham RR, Olson LM, Abdrabou W, Fan BJ, Wang DY, Brodeur W, et
al: Common variants at 9p21 and 8q22 are associated with increased
susceptibility to optic nerve degeneration in glaucoma. PLoS Genet.
8:e10026542012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kathiresan S, Voight BF, Purcell S,
Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani
NJ, Schunkert H, et al Wellcome Trust Case Control Consortium, :
Genome-wide association of early-onset myocardial infarction with
single nucleotide polymorphisms and copy number variants. Nat
Genet. 41:334–341. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
46
|
Burdon KP, Macgregor S, Hewitt AW, Sharma
S, Chidlow G, Mills RA, Danoy P, Casson R, Viswanathan AC, Liu JZ,
et al: Genome-wide association study identifies susceptibility loci
for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet.
43:574–578. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shiga Y, Nishiguchi KM, Kawai Y, Kojima K,
Sato K, Fujita K, Takahashi M, Omodaka K, Araie M, Kashiwagi K, et
al: Genetic analysis of Japanese primary open-angle glaucoma
patients and clinical characterization of risk alleles near
CDKN2B-AS1, SIX6 and GAS7. PLoS One. 12:e01866782017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Carnes MU, Liu YP, Allingham RR, Whigham
BT, Havens S, Garrett ME, Qiao C, Katsanis N, Wiggs JL, Pasquale
LR, et al NEIGHBORHOOD Consortium Investigators, : Discovery and
functional annotation of SIX6 variants in primary open-angle
glaucoma. PLoS Genet. 10:e10043722014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Iglesias AI, Springelkamp H, van der Linde
H, Severijnen LA, Amin N, Oostra B, Kockx CE, van den Hout MC, van
Ijcken WF, Hofman A, et al: Exome sequencing and functional
analyses suggest that SIX6 is a gene involved in an altered
proliferation-differentiation balance early in life and optic nerve
degeneration at old age. Hum Mol Genet. 23:1320–1332. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Burdon KP, Mitchell P, Lee A, Healey PR,
White AJ, Rochtchina E, Thomas PB, Wang JJ and Craig JE:
Association of open-angle glaucoma loci with incident glaucoma in
the Blue Mountains Eye study. Am J Ophthalmol. 159:31–36.e1. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kuo JZ, Zangwill LM, Medeiros FA, Liebmann
JM, Girkin CA, Hammel N, Rotter JI and Weinreb RN: Quantitative
trait locus analysis of SIX1-SIX6 with retinal nerve fiber layer
thickness in individuals of European descent. Am J Ophthalmol.
160:123–130.e1. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Skowronska-Krawczyk D, Zhao L, Zhu J,
Weinreb RN, Cao G, Luo J, Flagg K, Patel S, Wen C, Krupa M, et al:
P16INK4a Upregulation mediated by SIX6 defines retinal ganglion
cell pathogenesis in glaucoma. Mol Cell. 59:931–940. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Scheetz TE, Faga B, Ortega L, Roos BR,
Gordon MO, Kass MA, Wang K and Fingert JH: Glaucoma risk alleles in
the ocular hypertension treatment study. Ophthalmology.
123:2527–2536. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
van Koolwijk LM, Ramdas WD, Ikram MK,
Jansonius NM, Pasutto F, Hysi PG, Macgregor S, Janssen SF, Hewitt
AW, Viswanathan AC, et al DCCT/EDIC Research Group; Wellcome Trust
Case Control Consortium 2, : Common genetic determinants of
intraocular pressure and primary open-angle glaucoma. PLoS Genet.
8:e10026112012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hysi PG, Cheng CY, Springelkamp H,
Macgregor S, Bailey JNC, Wojciechowski R, Vitart V, Nag A, Hewitt
AW, Höhn R, et al BMES GWAS Group; NEIGHBORHOOD Consortium;
Wellcome Trust Case Control Consortium 2, : Genome-wide analysis of
multi-ancestry cohorts identifies new loci influencing intraocular
pressure and susceptibility to glaucoma. Nat Genet. 46:1126–1130.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen Y, Lin Y, Vithana EN, Jia L, Zuo X,
Wong TY, Chen LJ, Zhu X, Tam PO, Gong B, et al: Common variants
near ABCA1 and in PMM2 are associated with primary open-angle
glaucoma. Nat Genet. 46:1115–1119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gharahkhani P, Burdon KP, Fogarty R,
Sharma S, Hewitt AW, Martin S, Law MH, Cremin K, Bailey JNC, Loomis
SJ, et al Wellcome Trust Case Control Consortium 2, NEIGHBORHOOD
consortium, : Common variants near ABCA1, AFAP1 and GMDS confer
risk of primary open-angle glaucoma. Nat Genet. 46:1120–1125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Springelkamp H, Iglesias AI,
Cuellar-Partida G, Amin N, Burdon KP, van Leeuwen EM, Gharahkhani
P, Mishra A, van der Lee SJ, Hewitt AW, et al: ARHGEF12 influences
the risk of glaucoma by increasing intraocular pressure. Hum Mol
Genet. 24:2689–2699. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lessey-Morillon EC, Osborne LD,
Monaghan-Benson E, Guilluy C, O'Brien ET, Superfine R and Burridge
K: The RhoA guanine nucleotide exchange factor, LARG, mediates
ICAM-1-dependent mechanotransduction in endothelial cells to
stimulate transendothelial migration. J Immunol. 192:3390–3398.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Okuhira K, Fitzgerald ML, Tamehiro N,
Ohoka N, Suzuki K, Sawada J, Naito M and Nishimaki-Mogami T:
Binding of PDZ-RhoGEF to ATP-binding cassette transporter A1
(ABCA1) induces cholesterol efflux through RhoA activation and
prevention of transporter degradation. J Biol Chem.
285:16369–16377. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chen Y, Cai J and Jones DP: Mitochondrial
thioredoxin in regulation of oxidant-induced cell death. FEBS Lett.
580:6596–6602. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Caprioli J, Munemasa Y, Kwong JM and Piri
N: Overexpression of thioredoxins 1 and 2 increases retinal
ganglion cell survival after pharmacologically induced oxidative
stress, optic nerve transection, and in experimental glaucoma.
Trans Am Ophthalmol Soc. 107:161–165. 2009.PubMed/NCBI
|
|
63
|
Orr HT: Cell biology of spinocerebellar
ataxia. J Cell Biol. 197:167–177. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lattante S, Millecamps S, Stevanin G,
Rivaud-Péchoux S, Moigneu C, Camuzat A, Da Barroca S, Mundwiller E,
Couarch P, Salachas F, et al French Research Network on FTD and
FTD-ALS, : Contribution of ATXN2 intermediary polyQ expansions in a
spectrum of neurodegenerative disorders. Neurology. 83:990–995.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Ju YT, Chang ACY, She BR, Tsaur ML, Hwang
HM, Chao CCK, Cohen SN and Lin-Chao S: gas7: A gene expressed
preferentially in growth-arrested fibroblasts and terminally
differentiated Purkinje neurons affects neurite formation. Proc
Natl Acad Sci USA. 95:11423–11428. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wenger DA, Rafi MA, Luzi P, Datto J and
Costantino-Ceccarini E: Krabbe disease: Genetic aspects and
progress toward therapy. Mol Genet Metab. 70:1–9. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Liu Y, Gibson J, Wheeler J, Kwee LC,
Santiago-Turla CM, Akafo SK, Lichter PR, Gaasterland DE, Moroi SE,
Challa P, et al: GALC deletions increase the risk of primary
open-angle glaucoma: The role of Mendelian variants in complex
disease. PLoS One. 6:e271342011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Williams SE, Carmichael TR, Allingham RR,
Hauser M and Ramsay M: The genetics of POAG in black South
Africans: A candidate gene association study. Sci Rep. 5:83782015.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Vishal M, Sharma A, Kaurani L, Alfano G,
Mookherjee S, Narta K, Agrawal J, Bhattacharya I, Roychoudhury S,
Ray J, et al: Genetic association and stress mediated
down-regulation in trabecular meshwork implicates MPP7 as a novel
candidate gene in primary open angle glaucoma. BMC Med Genomics.
9:152016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Acharya M, Mookherjee S, Bhattacharjee A,
Thakur SK, Bandyopadhyay AK, Sen A, Chakrabarti S and Ray K:
Evaluation of the OPTC gene in primary open angle glaucoma:
functional significance of a silent change. BMC Mol Biol. 8:212007.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lu Y, Vitart V, Burdon KP, Khor CC,
Bykhovskaya Y, Mirshahi A, Hewitt AW, Koehn D, Hysi PG, Ramdas WD,
et al NEIGHBOR Consortium, : Genome-wide association analyses
identify multiple loci associated with central corneal thickness
and keratoconus. Nat Genet. 45:155–163. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Chua J, Tham YC, Liao J, Zheng Y, Aung T,
Wong TY and Cheng CY: Ethnic differences of intraocular pressure
and central corneal thickness: The Singapore Epidemiology of Eye
Diseases study. Ophthalmology. 121:2013–2022. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ramdas WD, van Koolwijk LME, Ikram MK,
Jansonius NM, de Jong PTVM, Bergen AAB, Isaacs A, Amin N, Aulchenko
YS, Wolfs RC, et al: A genome-wide association study of optic disc
parameters. PLoS Genet. 6:e10009782010. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Springelkamp H, Höhn R, Mishra A, Hysi PG,
Khor CC, Loomis SJ, Bailey JN, Gibson J, Thorleifsson G, Janssen
SF, et al Blue Mountains Eye Study-GWAS group; NEIGHBORHOOD
Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2), :
Meta-analysis of genome-wide association studies identifies novel
loci that influence cupping and the glaucomatous process. Nat
Commun. 5:48832014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Springelkamp H, Iglesias AI, Mishra A,
Höhn R, Wojciechowski R, Khawaja AP, Nag A, Wang YX, Wang JJ,
Cuellar-Partida G, et al NEIGHBORHOOD Consortium, : New insights
into the genetics of primary open-angle glaucoma based on
meta-analyses of intraocular pressure and optic disc
characteristics. Hum Mol Genet. 26:438–453. 2017.PubMed/NCBI
|
|
76
|
Axenovich T, Zorkoltseva I, Belonogova N,
van Koolwijk LM, Borodin P, Kirichenko A, Babenko V, Ramdas WD,
Amin N, Despriet DD, et al: Linkage and association analyses of
glaucoma related traits in a large pedigree from a Dutch
genetically isolated population. J Med Genet. 48:802–809. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Khawaja AP and Viswanathan AC: Are we
ready for genetic testing for primary open-angle glaucoma? Eye
(Lond). 32:877–883. 2018. View Article : Google Scholar : PubMed/NCBI
|