Introduction
Hepatocellular carcinoma (HCC) is one of the most
common cancers and the third leading cause of cancer mortality
worldwide (1). HCC has a high
degree of malignancy and develops rapidly, with patients usually
having already reached the middle or late stage of the disease at
diagnosis (2,3). If patients with HCC are not actively
treated, their natural disease course is short and their 5-year
survival rate is low (4).
Therefore, HCC poses a serious threat to the health and lives of
people (5). Actively searching for
early-warning markers for HCC prognosis and further studying the
molecular mechanism of HCC may provide important theoretical
guidance for early intervention for patients with HCC.
C-type lectin domain family 4 member M (CLEC4M),
also known as DC-SIGNR, is a Ca2+-dependent C-type
lectin that has been reported to be associated with tumor
progression (6). For example, a
previous study demonstrated that the level of serum CLEC4M in
patients with colon cancer was higher compared with healthy
controls (7). In addition, studies
have also revealed that CLEC4M may promote the occurrence of liver
metastases in colon cancer (8) and
gastric cancer (9). However, the
opposite results were observed in lung cancer and serum CLEC4M
levels were lower in patients compared with healthy controls
(10). These results indicated a
controversial role for CLEC4M in tumor progression. Additionally,
while it has been verified that Janus kinase 1/signal transducer
and activator of transcription 3 (JAK1/STAT3) pathway serves
crucial roles in HCC (11,12), whether there is a link between
CLEC4M and this process is still unknown.
Based on the abovementioned problems, the present
study investigated the novel role and mechanism of CLEC4M in HCC
progression; thus, the present study may provide a theoretical
basis for improving the survival of patients with HCC.
Materials and methods
Bioinformatic analysis
As described previously (13), published data in the Oncomine™
database (oncomine.org/resource/main.html) was investigated to
analyze the CLEC4M mRNA levels in unpaired non-tumor liver tissue
samples and HCC tissue samples to determine the clinical importance
of CLEC4M in HCC. The threshold settings were set to P<0.0001,
fold change ≥2 and Gene rank=top 10%. The following four datasets
were selected for analysis: GSE14520_GPL571 (non-tumor=21,
tumor=22) (14), GSE14520_GPL3921
(non-tumor=220, tumor=225) (14),
GSE14323_GPL571 (non-tumor=19, tumor=38) (15) and GSE6764 (non-tumor-10, tumor=35)
(16).
Additionally, the KMplot™ database
(kmplot.com/analysis) was used to analyze the 5-year prognostic
value of CLEC4M. The ‘Auto select best cutoff’ setting was used to
divide patients with HCC into two groups: The high and low CLEC4M
expression groups. Follow-up time was set to 60 months and patients
who were still alive at 60 months were censored. Finally, the
association between CLEC4M expression and the 5-year overall
survival (OS; low expression=90, high expression=274), relapse-free
survival (RFS; low expression=88, high expression=228),
progression-free survival (PFS; low expression=103, high
expression=267) and disease-specific survival (DSS; low
expression=89, high expression=273) was assessed.
Cell lines and cell culture
Huh7 and PLC/PRF/5 cells were acquired from the Cell
Bank of Type Culture Collection of the Chinese Academy of Sciences.
Huh7 cells were cultured in DMEM (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher Scientific,
Inc.) and 1% penicillin/streptomycin (P/S; Gibco; Thermo Fisher
Scientific, Inc.). PLC/PRF/5 cells were cultured in MEM (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS and 1%
P/S. All cells were incubated at 37°C and 5% CO2.
Lentivirus-mediated CLEC4M
overexpression
The lentiviruses were purchased from Shanghai
GeneChem, Inc. and the lentivirus-mediated CLEC4M overexpression in
Huh7 and PLC/PRF/5 cells was performed according to the
manufacturer's protocol. Briefly, a total of 4 µl of lentivirus
titer (1×108 TU/ml) with CLEC4M was added to
1×106 HCC cells at 37°C for 12 h (CLEC4M overexpression
group). Lentivirus without CLEC4M served as the negative control
(vector group). The supernatants were then replaced with normal
culture medium (DMEM/MEM supplemented with 10% FBS and 1% P/S) and
these HCC cells were cultured at 37°C for 72 h for subsequent
experiments.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
RT-qPCR analysis was performed as previously
described (17). Briefly, total
RNA was extracted from Huh7 and PLC/PRF/5 cells using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Reverse transcription was
performed using a PrimeScript™ RT kit (Takara Bio, Inc.), according
to the manufacturer's protocol. SYBR Premix EX Taq™ (Takara Bio,
Inc.) was used for qPCR on an ABI 7900 Prism HT (Applied
Biosystems; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. The PCR primers were as follows: CLEC4M
forward, 5′-TGTCCAAGGTCCCCAGCTCCC-3′ and reverse,
5′-GAACTCACCAAATGCAGTCTTCAAATC-3′; and GAPDH forward,
5′-AACAGCCTCAAGATCATCAGCA-3′ and reverse,
5′-CATGAGTCCTTCCACGATACCA-3′. Gene expression was calculated using
the 2−ΔΔCq method (18).
Western blotting
Western blotting was performed as previously
described (19). Briefly, protein
lysates (25 µg) were separated by using SDS-PAGE and target
proteins were detected by western blotting with appropriate primary
antibodies. A rabbit anti-CLEC4M antibody was purchased from Abcam
(1:1,000; cat. no. ab232709). Anti-GAPDH (1:1,000; cat. no. 2118),
anti-phospho-JAK1(p-JAK1; 1:1,000; cat. no. 3331), anti-JAK1
(1:1,000; cat. no. 3332), anti-p-STAT3(p-STAT3; 1:2,000; cat. no.
9145) anti-STAT3 (1:2,000; cat. no. 4904) and the horseradish
peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody
(1:10,000; cat. no. 7074) were purchased from Cell Signaling
Technology, Inc.
Cell counting kit-8 (CCK-8)
proliferation assay
Huh7 and PLC/PRF/5 cells stably infected with
lentivirus (2×103/well) were seeded in 96-well plates
and incubated at 37°C overnight. These HCC cells were then
incubated at 37°C for 24, 48, 72 or 96 h. CCK-8 (Dojindo Molecular
Technologies, Inc.) was used to determine the viability of
proliferating cells, according to the manufacturer's protocol.
Briefly, HCC cells were incubated at 37°C for 2 h after adding 10
µl/well CCK8 reagent, and the absorbance was measured at 450 nm
using an Infinite M200 Pro Multifunctional microplate reader (Tecan
Group Ltd).
5-Ethynyl-2′-deoxyuridine (EdU)
assay
EdU (Guangzhou RiboBio Co., Ltd.) was used to
determine the proliferative ability of Huh7 and PLC/PRF/5 cells,
according to the manufacturer's protocol. Briefly, HCC cells stably
infected with lentivirus were seeded in 96-well plates and treated
with EdU at 37°C for 2 h. Subsequently, Hoechst 33342
(Sigma-Aldrich; Merck KGaA) was added to each well and incubated at
room temperature for 30 min in the dark. Fluorescence microscopy
(Olympus IX50; Olympus Corporation) was used to determine the
proportion of EdU-positive cells.
Flow cytometric assay
An Annexin V-FITC/propidium iodide (PI) Cell
Apoptosis kit (Nanjing KeyGen Biotech, Co., Ltd.) was used to
identify apoptosis of Huh7 and PLC/PRF/5 cells of the CLEC4M or
Vector group, according to the manufacturer's protocol. Briefly,
100 µl suspension containing 5×105 HCC cells were
incubated with 5 µl Annexin V and 1 µl PI in the dark at room
temperature for 15 min. The apoptotic rate (early + late apoptotic)
was determined using a BD FACSCalibur flow cytometre (BD
Biosciences) and the data was analyzed using FlowJo software
(version 7.6.1; FlowJo LLC).
Statistical analysis
All data analyses were conducted using GraphPad
Prism software (version 7.0; GraphPad Software, Inc.). Unpaired
Student's t-test was used for the comparison of parameters between
two groups. Survival analyses were performed using Kaplan-Meier
curves and survival was compared using the log-rank test. Data are
presented as the means ± standard deviations. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were performed in triplicate.
Results
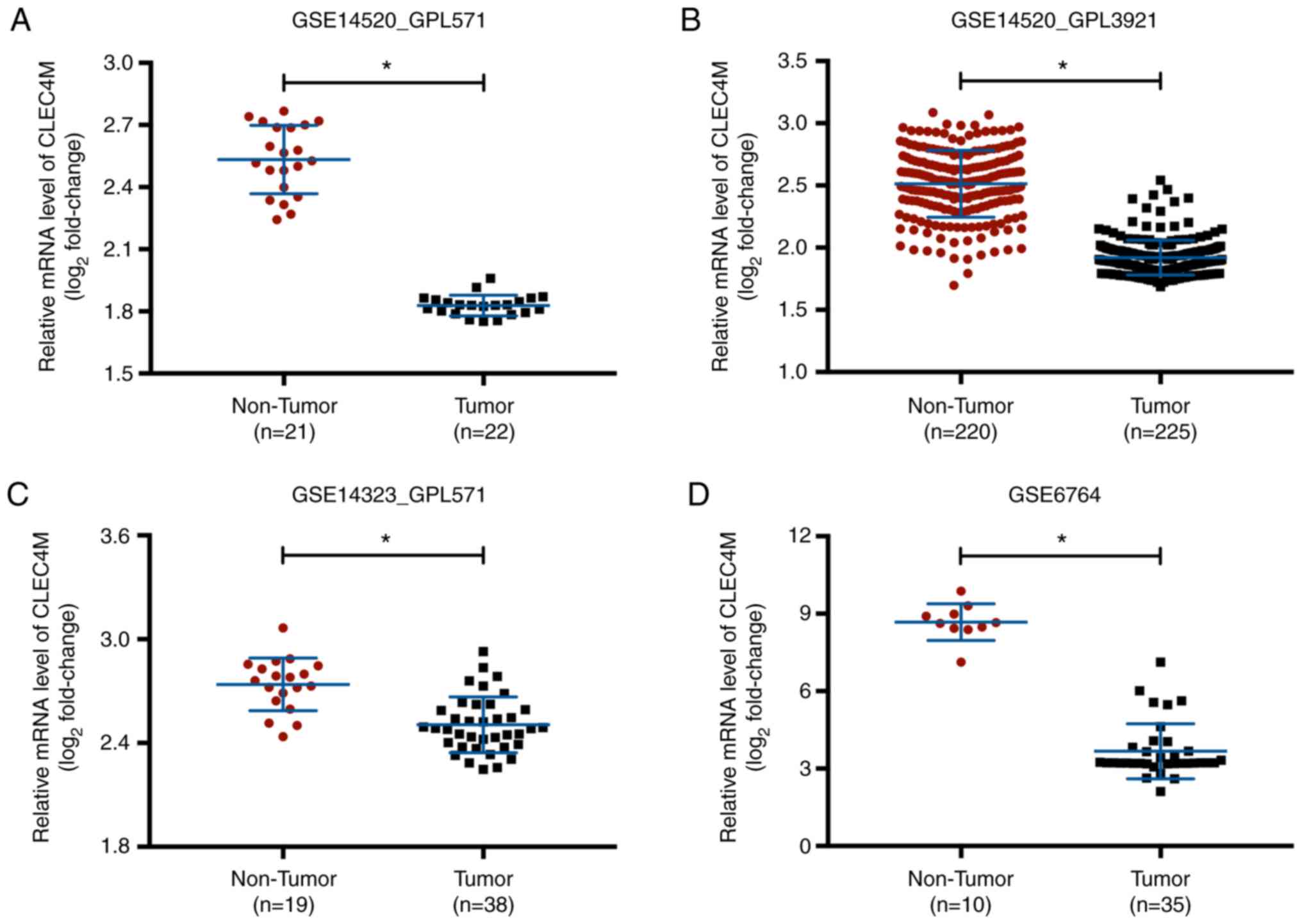
CLEC4M overexpression in unpaired
non-tumor liver tissue samples is compared to HCC tissue
samples
CLEC4M mRNA expression levels were collected from
four published HCC datasets in the Oncomine™ database. CLEC4M mRNA
levels were significantly higher in the unpaired non-tumor liver
tissue samples compared with HCC tissue samples in all datasets
(P<0.05; Fig. 1).
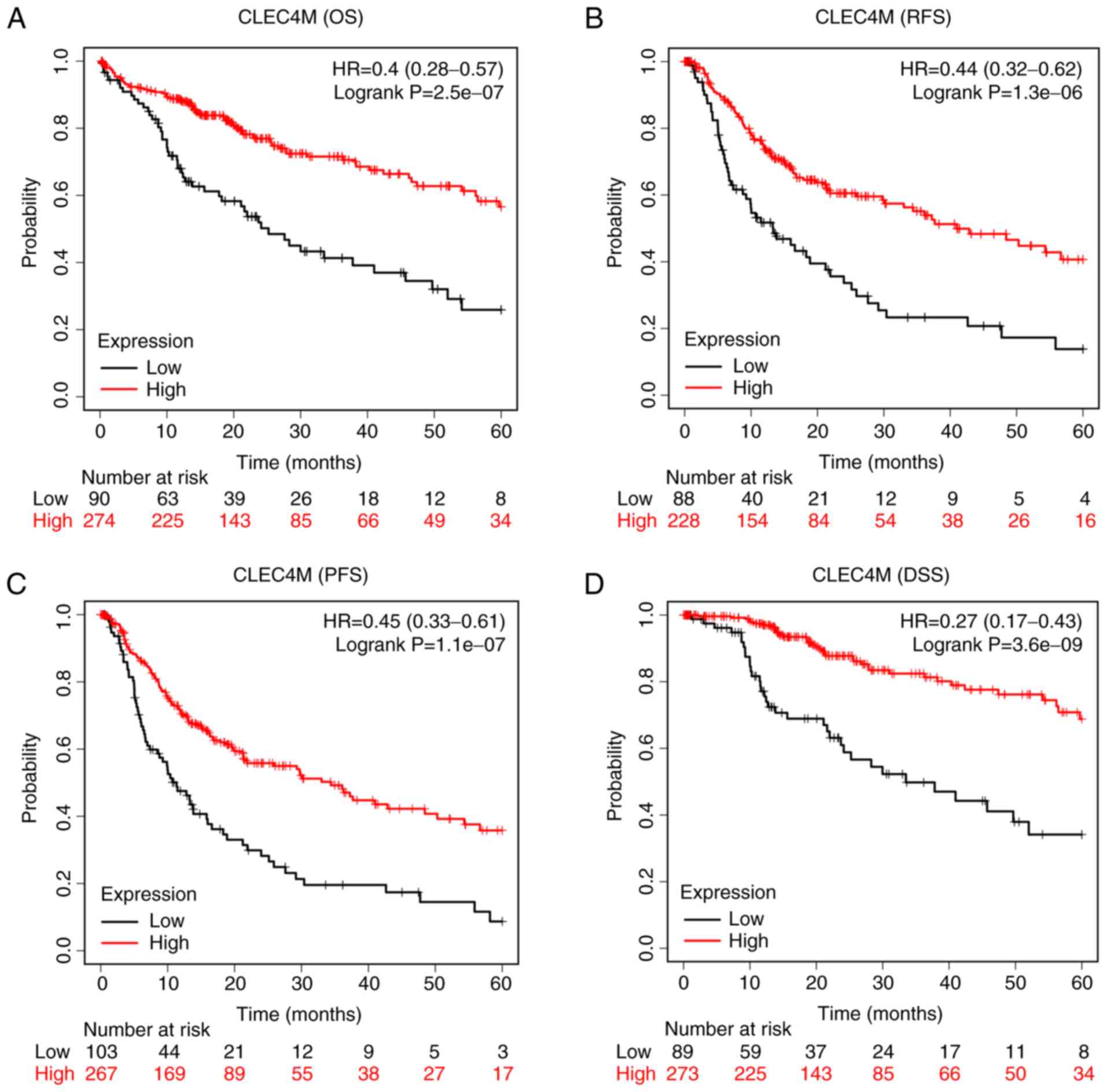
CLEC4M overexpression is associated
with a favorable prognosis
Data from the KMplot™ database demonstrated the
CLEC4M overexpression group had prolonged OS [P<0.05; hazard
ratio (HR), 0.4; 95% confidence interval (CI), 0.28–0.57; Fig. 2A] and RFS (P<0.05; HR, 0.44; 95%
CI, 0.32–0.62; Fig. 2B) times
compared with the low-expression group. Similar results were
reported for PFS and DSS. The CLEC4M overexpression group had
longer PFS (P<0.05; HR, 0.45; 95% CI, 0.33–0.61; Fig. 2C) and DSS (P<0.05; HR, 0.27; 95%
CI, 0.17–0.43; Fig. 2D) times
compared with the low-expression group.
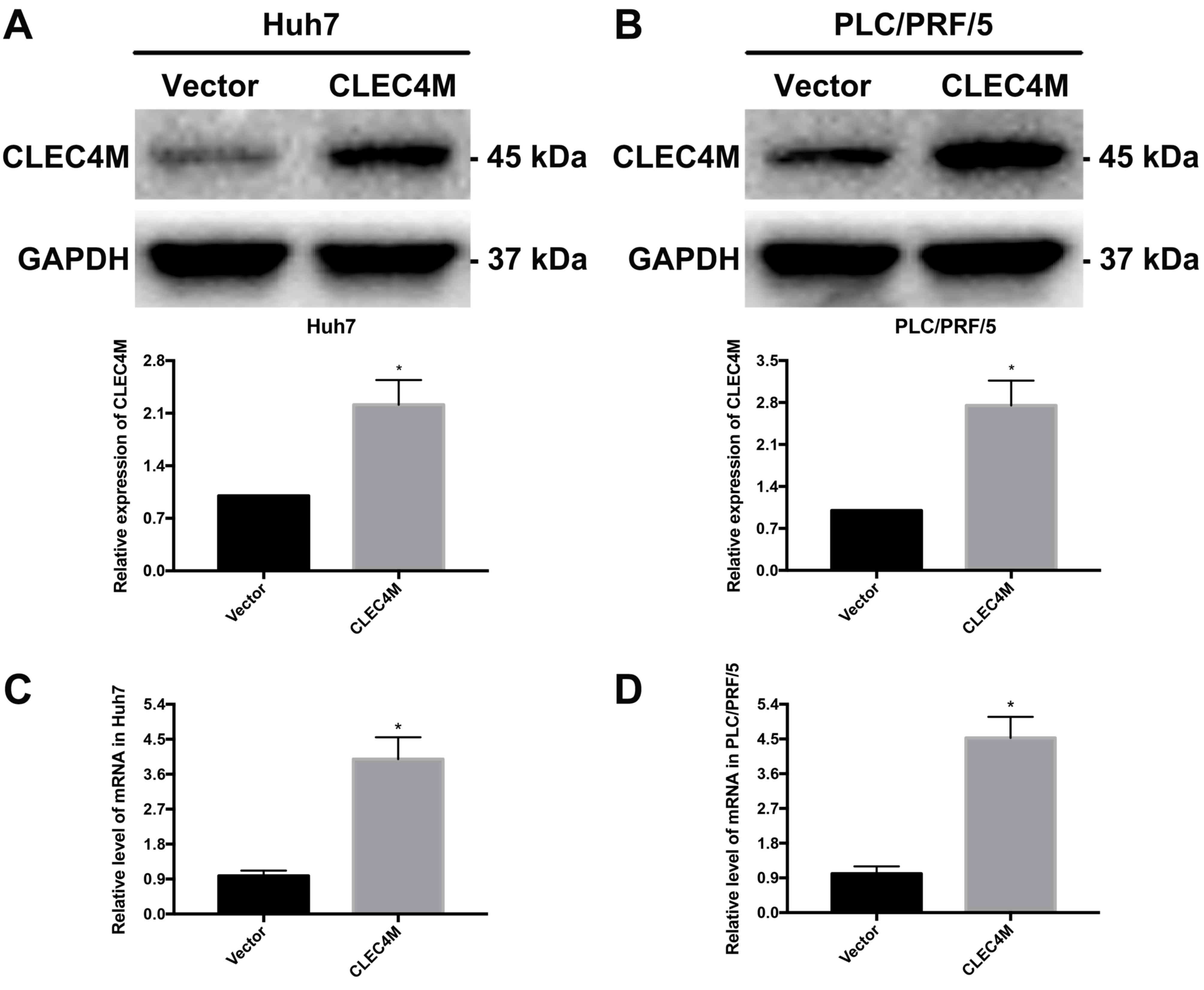
Construction of HCC cell lines with
stable overexpression of CLEC4M
The liver cancer cell lines exhibited significant
increases in CLEC4M expression at the protein (P<0.05; Fig. 3A and B) and mRNA (P<0.05;
Fig. 3C and D) levels in the
CLEC4M overexpression group compared with the vector group,
indicating that CLEC4M was successfully overexpressed. Stable
CLEC4M-overexpressing HCC cell lines were used for subsequent
experiments.
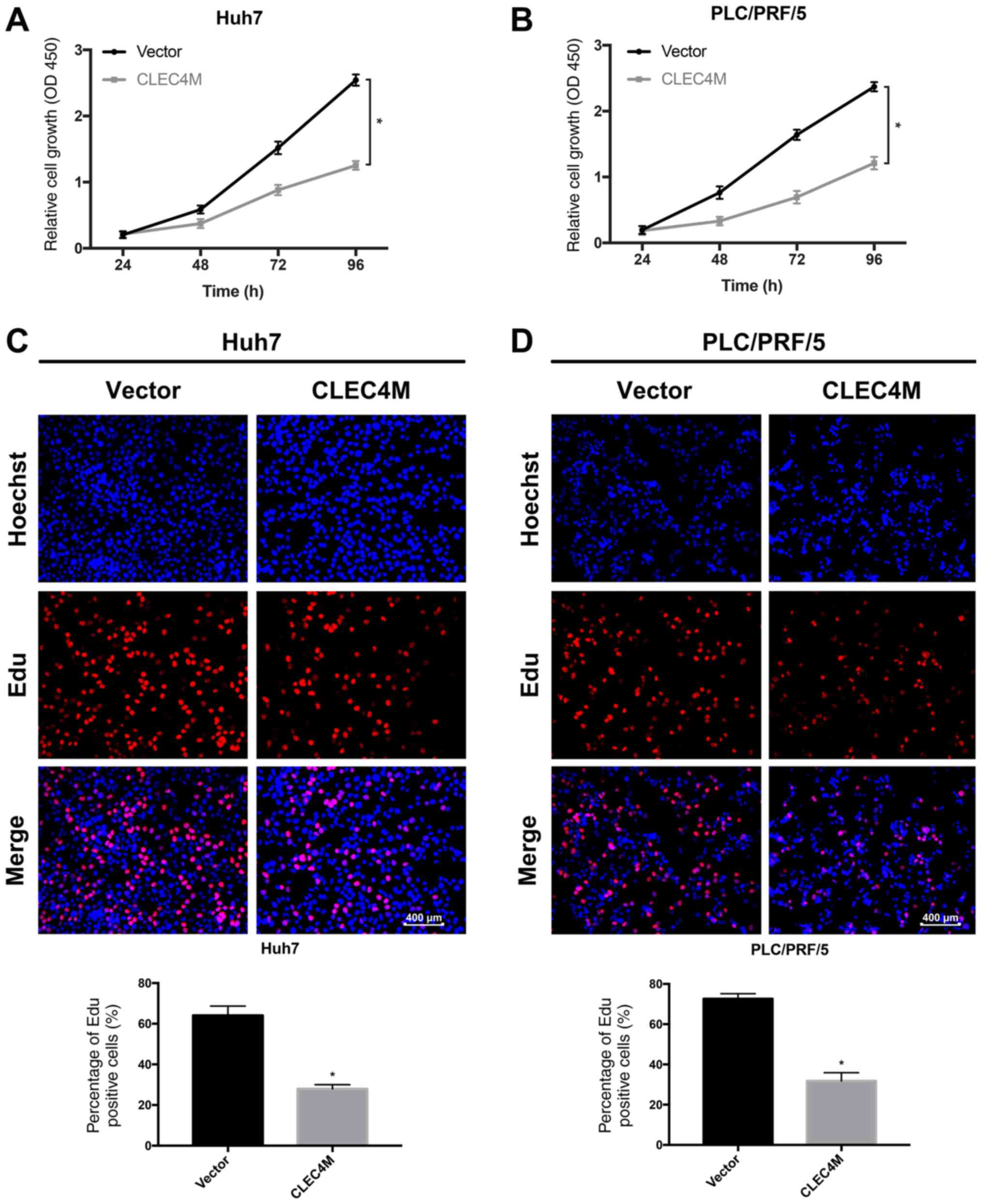
CLEC4M overexpression inhibits the
proliferation of HCC cells
Whether CLEC4M influences the viability of
proliferating HCC cell lines was examined. Huh7 (P<0.05;
Fig. 4A) and PLC/PRF/5 (P<0.05;
Fig. 4B) cell viability was
significantly reduced by CLEC4M overexpression, as evidenced by
CCK-8 assays. Additionally, HCC cell proliferation was evaluated
using EdU assays. Huh7 cell proliferation was significantly
decreased by CLEC4M overexpression (P<0.05; Fig. 4C), as was PLC/PRF/5 cell
proliferation (P<0.05; Fig.
4D).
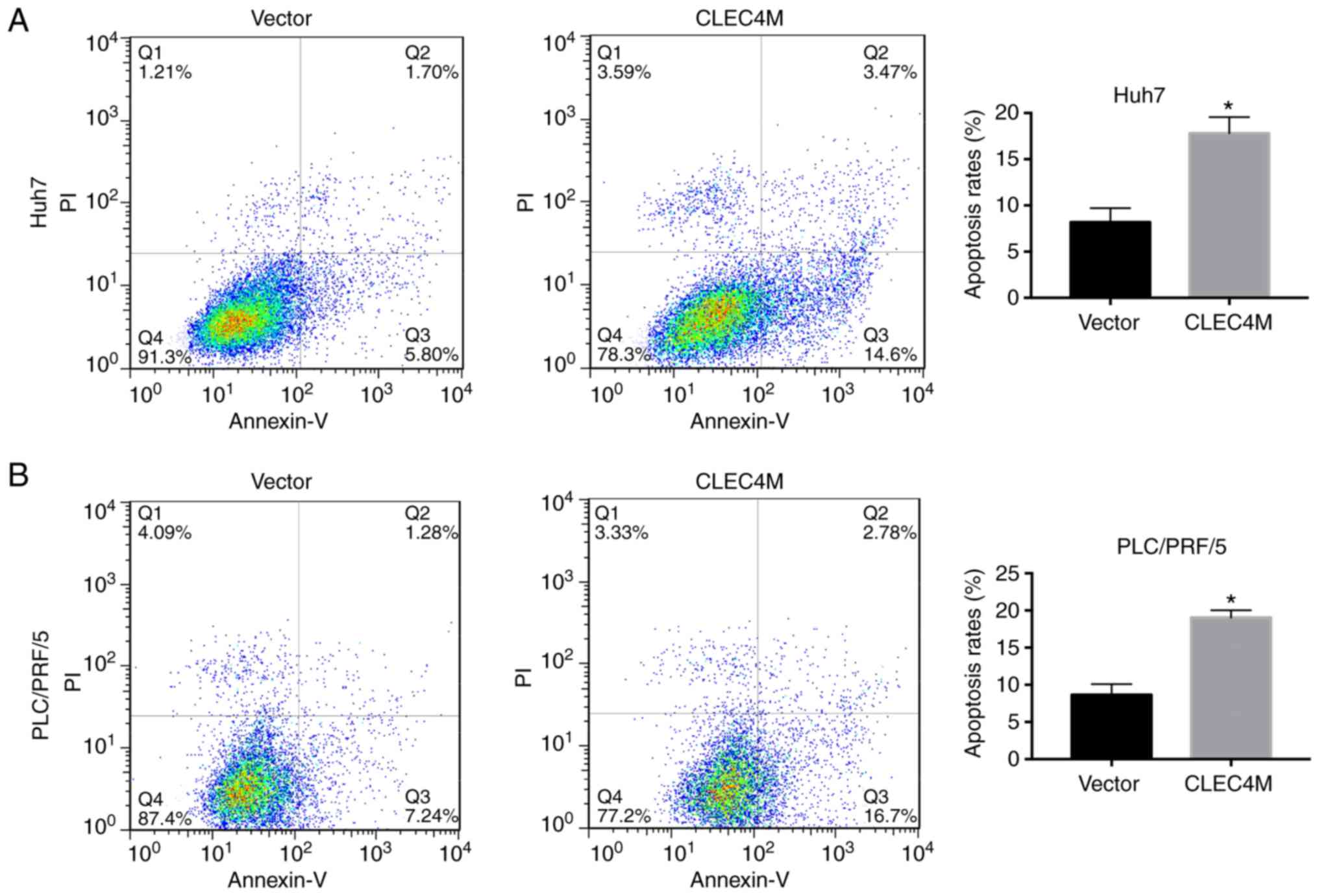
CLEC4M overexpression induces
apoptosis in HCC cells
Additionally, whether CLEC4M overexpression induced
apoptosis in HCC cell lines was examined. The results revealed that
CLEC4M overexpression significantly triggered apoptosis in Huh7
(P<0.05; Fig. 5A) and PLC/PRF/5
(P<0.05; Fig. 5B) cells.
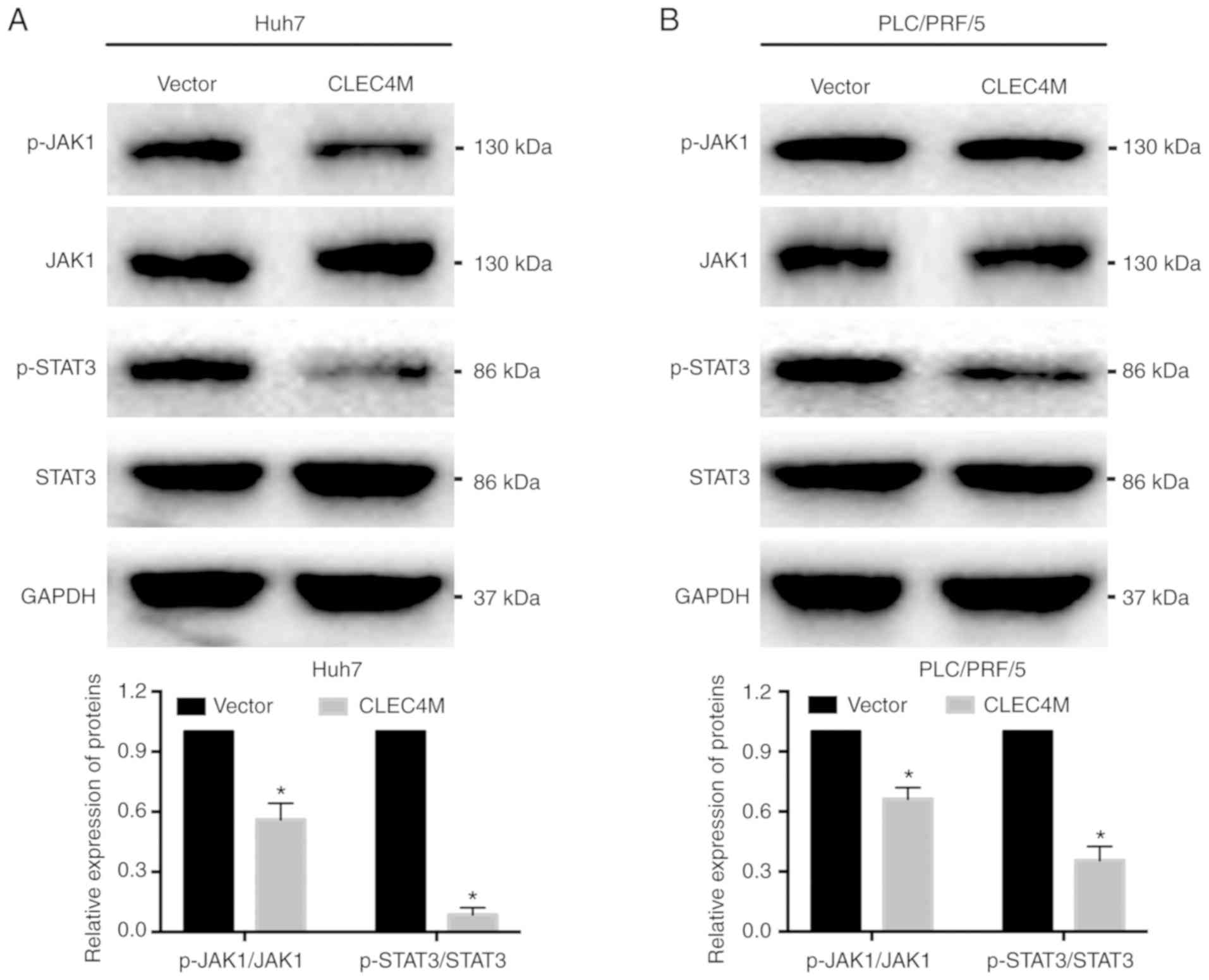
CLEC4M overexpression inhibits the
JAK1/STAT3 pathway
Since the JAK1/STAT3 pathway has been revealed to
serve a critical role in the growth of malignant cells (20,21),
the impact of CLEC4M on the JAK1/STAT3 pathway was assessed. The
results demonstrated that the protein levels of p-JAK1 and p-STAT3
in Huh7 (P<0.05; Fig. 6A) and
PLC/PRF/5 (P<0.05; Fig. 6B)
cells were significantly reduced in the CLEC4M overexpression group
compared with the vector group. These results indicated that CLEC4M
overexpression inhibited the JAK1/STAT3 pathway, which may inhibit
HCC progression.
Discussion
It is critical to investigate the genes responsible
for HCC progression and elucidate the molecular pathogenesis of
HCC. The present study primarily investigated the biological
functions of CLEC4M on HCC and the results revealed significantly
decreased expression of CLEC4M in HCC tissue samples compared with
unpaired non-tumor liver tissue samples. Furthermore, CLEC4M
overexpression was associated with favorable patient OS, RFS, PFS
and DSS. Moreover, the results for the Huh7 and PLC/PRF/5 cell
lines confirmed that CLEC4M inhibited proliferation and enhanced
apoptosis in HCC cells, at least in part via the JAK1/STAT3
pathway. These results indicated that the favorable clinical
outcomes of patients with HCC and increased CLEC4M expression may
result from proliferation inhibition and apoptosis enhancement.
C-type lectin CLEC4M is mainly localized in the
endothelial cells of the liver, lungs and lymph nodes (6). Although the clinical significance of
CLEC4M in HCC has been investigated previously, this previous study
was limited to OS (22) and the
other biological functions of CLEC4M in the context of HCC remain
unclear. Similar to a previous result (22), prognostic analysis of patients with
HCC observed that those with higher expression of CLEC4M exhibited
prolonged OS, RPS, PFS and DSS compared with patients with lower
expression in the current study. These results indicated that
CLEC4M may lead to improved clinical outcomes in patients with HCC.
Due to these results, the role of CLEC4M in HCC was further
investigated. The results revealed that CLEC4M overexpression
significantly inhibited proliferation in the HCC cell lines and
enhanced cell apoptosis, indicating a new tumor-suppressive effect
of CLEC4M in HCC. These results were similar to those reported by
Liu et al (10), who
reported low CLEC4M expression in serum samples from patients with
lung cancer (10). Moreover,
lymphoid tissue samples from patients with non-Hodgkin's lymphoma
have been demonstrated to be negative for CLEC4M by
immunohistochemistry (23).
However, the results of these studies are in contrast to previous
research results for colon cancer (8) and gastric cancer (9). This may be due to tumor complexity
and the functions of CLEC4M varying among different tumor types and
samples (24).
Reportedly, among the JAK/STAT family members,
JAK1/STAT3 serves crucial roles in numerous biological processes,
including cell growth, apoptosis, migration and invasion (25–27).
For instance, miR-34e was revealed to suppress HCC cell
proliferation and invasion by regulating the JAK1/STAT3 pathway
(12) and inhibition of the
JAK1/STAT3 pathway suppressed HCC cell growth in vivo
(11). Thus, the JAK1/STAT3
pathway may be a promising molecular target for the treatment of
HCC. The results of the present study demonstrated that CLEC4M
overexpression inhibited the JAK1/STAT3 pathway in HCC.
Specifically, CLEC4M overexpression inhibited JAK1 and STAT3
phosphorylation in HCC cells. Therefore, CLEC4M may hinder HCC
progression by inhibiting the JAK1/STAT3 pathway. However, the
present study also has some limitations; for example, whether
CLEC4M overexpression has the same tumor suppressor function in
vivo remains unknown and further animal experiments may better
reflect the role of CLEC4M in HCC.
In conclusion, the results of the present study
reported that CLEC4M overexpression was associated with a favorable
HCC prognosis and described a novel role for CLEC4M in HCC.
Moreover, although no detailed mechanism was demonstrated in this
study, the possible process by which JAK1/STAT3 signaling was
regulated by CLEC4M was also elucidated. These results revealed
that CLEC4M overexpression contributed to the inhibition of HCC
progression and that CLEC4M may be a novel HCC biomarker that
modulates JAK1 and STAT3.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Hunan Provincial
Natural and Science Foundation (grant no. 2018JJ6126).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KG designed the present study. QY and KG performed
the experiments and analyzed the data. QY wrote the manuscript.
Both authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Llovet JM, Burroughs A and Bruix J:
Hepatocellular carcinoma. Lancet. 362:1907–1917. 2004. View Article : Google Scholar
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J and Llovet JM: Major achievements
in hepatocellular carcinoma. Lancet. 373:614–616. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen C and Wang G: Mechanisms of
hepatocellular carcinoma and challenges and opportunities for
molecular targeted therapy. World J Hepatol. 7:1964–1970. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang F, Ren S and Zuo Y: DC-SIGN,
DC-SIGNR and LSECtin: C-type lectins for infection. Int Rev
Immunol. 33:54–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang Y, Zhang C, Chen K, Chen Z, Sun Z,
Zhang Z, Ding D, Ren S and Zuo Y: The clinical significance of
DC-SIGN and DC-SIGNR, which are novel markers expressed in human
colon cancer. PLoS One. 9:e1147482014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Na H, Liu X, Li X, Zhang X, Wang Y, Wang
Z, Yuan M, Zhang Y, Ren S and Zuo Y: Novel roles of DC-SIGNR in
colon cancer cell adhesion, migration, invasion, and liver
metastasis. J Hematol Oncol. 10:282017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z,
Zhang J, Zhou X, Zhang Y, Lin F, Na H, et al: DC-SIGNR by
influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4
in gastric cancer liver metastasis. Mol Cancer. 16:782017.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu X, Zhang H, Su L, Yang P, Xin Z, Zou
J, Ren S and Zuo Y: Low expression of dendritic cell-specific
intercellular adhesion molecule-grabbing nonintegrin-related
protein in lung cancer and significant correlations with brain
metastasis and natural killer cells. Mol Cell Biochem. 407:151–160.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liao J, Xu T, Zheng JX, Lin JM, Cai QY, Yu
DB and Peng J: Nitidine chloride inhibits hepatocellular carcinoma
cell growth in vivo through the suppression of the
JAK1/STAT3 signaling pathway. Int J Mol Med. 32:79–84. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mao J, Hu X, Pang P, Zhou B, Li D and Shan
H: miR-30e acts as a tumor suppressor in hepatocellular carcinoma
partly via JAK1/STAT3 pathway. Oncol Rep. 38:393–401. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cao L, Cheng H, Jiang Q, Li H and Wu Z:
APEX1 is a novel diagnostic and prognostic biomarker for
hepatocellular carcinoma. Aging. 12:4573–4591. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Roessler S, Jia HL, Budhu A, Forgues M, Ye
QH, Lee JS, Thorgeirsson SS, Sun Z, Tang ZY, Qin LX and Wang XW: A
unique metastasis gene signature enables prediction of tumor
relapse in early-stage hepatocellular carcinoma patients. Cancer
Res. 70:10202–10212. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mas VR, Maluf DG, Archer KJ, Yanek K, Kong
X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P and Fisher
R: Genes involved in viral carcinogenesis and tumor initiation in
hepatitis C virus-induced hepatocellular carcinoma. Mol Med.
15:85–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wurmbach E, Chen YB, Khitrov G, Zhang W,
Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, et
al: Genome-wide molecular profiles of HCV-induced dysplasia and
hepatocellular carcinoma. Hepatology. 45:938–947. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z,
Zhang J, Zhou X, Zhang Y, Lin F, Na H, et al: DC-SIGNR by
influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4
in gastric cancer liver metastasis. Mol Cancer. 16:782017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan LM, Li X, Qiu CF, Zhu T, Hu CP, Yin
JY, Zhang W, Zhou HH and Liu ZQ: CLEC4M is associated with poor
prognosis and promotes cisplatin resistance in NSCLC patients. J
Cancer. 10:6374–6383. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wen W, Liang W, Wu J, Kowolik CM, Buettner
R, Scuto A, Hsieh MY, Hong H, Brown CE, Forman SJ, et al: Targeting
JAK1/STAT3 signaling suppresses tumor progression and metastasis in
a peritoneal model of human ovarian cancer. Mol Cancer Ther.
13:3037–3048. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yan CM, Zhao YL, Cai HY, Miao GY and Ma W:
Blockage of PTPRJ promotes cell growth and resistance to 5-FU
through activation of JAK1/STAT3 in the cervical carcinoma cell
line C33A. Oncol Rep. 33:1737–1744. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xia HB, Wang HJ, Song SS, Zhang JG, He XL,
Hu ZM, Zhang CW, Huang DS and Mou XZ: Decreased DC-SIGNR expression
in hepatocellular carcinoma predicts poor patient prognosis. Oncol
Lett. 19:69–76. 2020.PubMed/NCBI
|
|
23
|
Zhang Z, Chen K, Yan L, Yang Z, Zhu Z,
Chen C, Zeng J, Wei W, Qi X, Ren S and Zuo Y: Low expression of
dendritic cell-specific intercellular adhesion molecule-grabbing
nonintegrin-related protein in non-Hodgkin lymphoma and significant
correlations with lactic acid dehydrogenase and β2-microglobulin.
Biochem Cell Biol. 91:214–220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dulak AM, Schumacher SE, van Lieshout J,
Imamura Y, Fox C, Shim B, Ramos AH, Saksena G, Baca SC, Baselga J,
et al: Gastrointestinal adenocarcinomas of the esophagus, stomach,
and colon exhibit distinct patterns of genome instability and
oncogenesis. Cancer Res. 72:4383–4393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao W, Liu Y, Zhang R, Zhang B, Wang T,
Zhu X, Mei L, Chen H, Zhang H, Ming P and Huang L:
Homoharringtonine induces apoptosis and inhibits STAT3 via
IL-6/JAK1/STAT3 signal pathway in Gefitinib-resistant lung cancer
cells. Sci Rep. 5:84772015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tactacan CM, Phua YW, Liu L, Zhang L,
Humphrey ES, Cowley M, Pinese M, Biankin AV and Daly RJ: The
pseudokinase SgK223 promotes invasion of pancreatic ductal
epithelial cells through JAK1/Stat3 signaling. Mol Cancer.
14:1392015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van der Zee M, Sacchetti A, Cansoy M,
Joosten R, Teeuwssen M, Heijmans-Antonissen C, Ewing-Graham PC,
Burger CW, Blok LJ and Fodde R: IL6/JAK1/STAT3 signaling blockade
in endometrial cancer affects the ALDHhi/CD126+ Stem-like component
and reduces tumor burden. Cancer Res. 75:3608–3622. 2015.
View Article : Google Scholar : PubMed/NCBI
|