Introduction
Peripheral arterial disease (PAD) is a common
peripheral circulatory problem, in which narrowed arteries occur,
especially in the lower extremities (1,2). PAD
has been reported as a highly age-related vascular disease,
typically occurring in patients aged 60–70 years, which leads to
health problems (3). Surgical and
catheter-based revascularization that target primary occluded macro
vessels are the primary conventional treatment strategies for PAD.
Unfortunately, these high-cost methods result in limited
improvement in the microcirculation, due to complications that
occur in patients with PAD (4).
Angiogenesis is a process of the endothelium, by which a new vessel
is formed from the old one (5).
Therapeutic angiogenesis has been studied for two decades, focusing
on the overexpression of growth factors, which have been identified
as factors that evoke angiogenesis in vitro in PAD,
including vascular endothelial growth factor (VEGF) and hepatocyte
growth factor (6,7). However, almost none of the trials met
the expected goal, including significant clinical remission, which
has led researchers to investigate other molecules that are
responsible for the limited effects.
MicroRNAs (miRNAs/miRs), a class of ~22
nucleotide-long, small non-coding RNAs, regulate gene expression by
repressing post-transcription to modulate cell fate decisions
(8,9). miRNAs were first defined as
endogenous regulators for a variety of cellular physiologic and
pathologic processes (10). To
date, 2,654 mature human miRNAs have been identified (11). Recently, certain miRNAs have been
identified to be associated with ischemia-reperfusion injury
(12,13). A number of identified miRNAs have
been reported to serve as endogenous negative regulators of
angiogenesis. For example, the antiangiogenesis function of miR-92a
has been verified in vitro and in vivo (14,15).
However, whether other miRNAs serve the same role as miR-92a is not
completely understood. miR-124-3p has been reported to regulate
glioma angiogenesis (16,17) and participate in ischemic diseases
(18,19). Nevertheless, there are limited
studies on the role of miR-124-3p in PAD. Therefore, the present
study investigated the effect of miR-124-3p on endothelial cells
(ECs) and PAD.
In the present study, the role of miR-124-3p in PAD
was investigated in the blood of patients with PAD, as well as in
the ECs and in the hindlimb ischemia model. In addition, the target
of miR-124-3p was identified in the present study.
Materials and methods
Cell culture, stimulation and
transfection
Human umbilical vein ECs (HUVECs; cat. no. 8000;
ScienCell Research Laboratories, Inc.) were cultured and stimulated
at 37°C in 5% CO2 in Endothelial Cell Medium (cat. no.
1001; ScienCell Research Laboratories, Inc.) supplemented with 5%
FBS (cat. no. 0025; ScienCell Research Laboratories, Inc.), 1%
endothelial cell growth supplement (cat. no. 1052; ScienCell
Research Laboratories, Inc.) and 1% penicillin/streptomycin
solution (cat. no. 0503; ScienCell Research Laboratories, Inc.).
Cobalt (II) chloride hexahydrate (500 µM, cat. no. C8661;
Sigma-Aldrich; Merck KGaA) was used to stimulate HUVECs at 37°C in
5% CO2 for 24 or 48 h when cells reach 90% confluence.
At ~80% confluence, HUVECs were transfected with mimic (50 nM;
Guangzhou RiboBio Co., Ltd.), inhibitor (100 nM; Guangzhou RiboBio
Co., Ltd.), small interfering (si)RNA (25 nM; Shanghai GenePharma
Co., Ltd.) or corresponding negative controls (NCs, 25 nM; Shanghai
GenePharma Co., Ltd.) for 36 h using Lipofectamine® 2000
(cat. no. 11668019; Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. The sequences of the
mimics, inhibitors and siRNAs were as follows: NC mimic forward,
5′-UUUGUACUACACAAAAGUACUG-3′ and reverse,
5′-CAGUACUUUUGUGUAGUACAAA-3′; miR-124-3p mimic forward,
5′-UAAGGCACGCGGUGAAUGCC-3′ and reverse, 5′-GGCAUUCACCGCGUGCCUUA-3′;
NC inhibitor, 5′-CAGUACUUUUGUGUAGUACAAA-3′; miR-124-3p inhibitor,
5′-GGCAUUCACCGCGUGCCUUA-3′; si-NC forward,
5′-UUCUCCGAACGUGUCACGU-3′ and reverse, 5′-ACGUGACACGUUCGGAGAA-3′;
si-STAT3 forward, 5′-CCCGGAAAUUUAACAUUCU-3′ and reverse,
5′-AGAAUGUUAAAUUUCCGGG-3′.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from human blood, HUVECs,
gastrocnemius and the blood of HLI model mice using
TRIzol® (cat. no. 15596026; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Total RNA (1 µg) was reverse transcribed into cDNA using the
PrimeScript RT Reagent kit (cat. no. RR037A; Takara Bio, Inc.) as
follows: 37°C for 15 min, 85°C for 5 sec and 4°C thereafter.
Subsequently, qPCR was performed using KAPA SYBR FAST qPCR Master
Mix (cat. no. KM4101; Roche Diagnostics) as follows: 95°C for 3
min, then 40 cycles of 95°C for 10 sec and 60°C for 30 sec. miRNA
and mRNA expression levels were quantified using the
2−∆∆Cq method (20) and
normalized to the internal reference genes U6 and GAPDH,
respectively. Stem-loop primers of miR-124-3p and U6 were purchased
from Guangzhou RiboBio Co., Ltd. The primer sequences of
miR-124-3p, U6, STAT3 and GAPDH were as follows: miR-124-3p
forward, 5′-TCTTTAAGGCACGCGGTG-3′ and reverse,
TATGGTTTTGACGACTGTGTGAT; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′ and
reverse, 5′-AACGCTTCACGAATTTGCGT-3′; STAT3 forward,
5′-CAGCAGCTTGACACACGGTA-3′ and reverse,
5′-AAACACCAAAGTGGCATGTGA-3′; and GAPDH forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′.
Proliferation assay
Cell proliferation was determined using the Edu Cell
Proliferation kit (cat. no. C10339; Invitrogen; Thermo Fisher
Scientific, Inc.). Following 12 h serum starvation, HUVECs were
incubated with Edu-labeling mixture (10 µM) in combination with
recombinant human VEGFA-165 (50 ng/ml; cat. no. 100-20; PeproTech,
Inc.) for 12 h at 37°C in 5% CO2. Images (four pictures
of each group) were captured by an Olympus IX83 fluorescence
microscope (Olympus Corporation) at 10× magnification. The rate of
cell proliferation was calculated using the following formula:
Number of Edu+ cells/total number of cells in each
field.
Wound healing assay
At ~90% confluence, the limit of HUVEC
proliferation, a scratch was made in the center of each well using
the tip of a 200 µl pipette. Subsequently, serum-starved HUVECs
were cultured with VEGFA-165 (50 ng/ml). Images of the wounds were
captured at 0 and 24 h by an Olympus CKX53 inverted microscope at
4× magnification (Olympus Corporation). Cell migration was analyzed
using ImageJ software (version 1.52a; National Institutes of
Health).
Tube formation assay
For the assay, 96-well plates were pre-coated with
Matrigel (cat. no. 354230; Corning, Inc.) at 37°C for 30 min, and
then HUVECs were seeded (2×104 cells/well) into the
Matrigel. Following culture with VEGFA-165 (50 ng/ml) for 8 h at
37°C in 5% CO2, images were captured to detect tube
formation by an Olympus CKX53 inverted microscope at 4×
magnification (Olympus Corporation). The total tube length was
assessed using ImageJ software (version 1.52a; National Institutes
of Health).
Dual-luciferase reporter assay
The target of miR-124-3p was predicted using
TargetScan (version 7.1, http://www.targetscan.org). The 3′-untranslated region
(3′-UTR) luciferase reporter construct of STAT3 was cloned
downstream of the Renilla luciferase gene in the pSI-check2
vector (Hanbio Biotechnology Co., Ltd.). 293T cells (cat. no.
CRL-11268; American Type Culture Collection) were seeded into a
96-well plate with density of 80%. Subsequently, 293T cells were
co-transfected with 5 pmol miR-124-3p mimic or NC mimic and 0.16 µg
STAT3-wild-type (WT) or STAT3-mutant (Mut) using
Lipofectamine® 2000 (cat. no. 11668019; Invitrogen;
Thermo Fisher Scientific, Inc.). At 48 h post-transfection,
luciferase activities were detected using the Dual-Luciferase
Reporter Assay System (cat. no. E1910; Promega Corporation).
Firefly luciferase activities were normalized to Renilla
luciferase activities.
Protein extraction and western
blotting
Total protein was extracted from HUVECs using Cell
Lysis Buffer (cat. no. 9803s; Cell Signaling Technology, Inc.) with
protease inhibitors (cat. no. 04693159001; Roche Diagnostics). The
bicinchoninic acid method was used for protein determination.
Protein (10 µg) was separated via 8% SDS-PAGE and transferred to
PVDF membranes, which were blocked with 5% bovine serum albumin at
room temperature for 1 h. Subsequently, the membranes were
incubated overnight at 4°C with the following primary antibodies:
STAT3 (1:1,000; cat. no. 9139; Cell Signaling Technology, Inc.) and
GAPDH (1:5,000; cat. no. 60004-1-Ig; ProteinTech Group, Inc.).
Following primary incubation, the membranes were incubated with
horseradish peroxidase-conjugated secondary antibodies (anti-mouse
IgG, 1:2,000, cat. no. ab205719, Abcam) at room temperature for 1
h. Protein bands were visualized using chemiluminescence (cat. no.
180-5001; Tanon Science and Technology Co., Ltd.) and detected
using the Amersham Imager 600 system (GE Healthcare Life Sciences).
Protein expression was semi-quantified using ImageJ software
(version 1.52a; National Institutes of Health) with GAPDH as the
loading control.
HLI model and detection of perfusion
recovery
A total of 50 eight-week-old male mice (18 to 22 g)
were used for HLI model. The HLI model was established as
previously described (21).
Briefly, mice were anesthetized with an intraperitoneal injection
of ketamine (80 mg/kg) and xylazine (5 mg/kg). Excision and
ligation were performed on the left femoral artery. For the sham
operation, excision was performed on the contralateral hindlimb.
miR-124-3p was overexpressed using agomiR-124-3p and inhibited
using antagomiR-124-3p. Accordingly, mice were injected with 5 nmol
agomiR-124-3p or agomiR-NC (Guangzhou RiboBio Co., Ltd.) into two
sites of the gastrocnemius and one site of the tibialis anterior
muscle on day 0, 7 and 14 post-HLI. In addition, 8 mg/kg
antagomiR-124-3p or antagomiR-NC (Guangzhou RiboBio Co., Ltd.) were
injected via a tail vein injection on day 0, 7 and 14 post-HLI. The
method was adapted from Hazarika et al (22). Perfusion recovery was detected via
laser Doppler imaging (Moor Instruments Ltd.) on day 0, 3, 7, 14
and 21 post-HLI and quantified using moorLDI Image Processing
software (version 5.3; Moor Instruments Ltd.). Perfusion in the
ischemic limb was normalized to the sham limb for each mouse. The
mice were anesthetized by isoflurane (5%) inhalation and euthanized
by CO2 (100%) inhalation at a rate of 30% volume/minute.
Animal studies were conducted in compliance with the Guide for the
Care and Use of Laboratory Animals published by the NIH (23) and approved by the Animal Care and
Use Committees of the Shanghai Tenth People's Hospital (approval
no. SHDSYY-2019-2149).
The agomir and antagomir sequences were as follows:
agomiR-NC forward, 5′-UUCUCCGAACGUGUCACGU3′ and reverse,
5′-ACGUGACACGUUCGGAGAA-3′; agomiR-124-3p forward,
5′-UAAGGCACGCGGUGAAUGCC-3′ and reverse, 5′-GGCAUUCACCGCGUGCCUUA-3′;
antagomiR-NC, 5′-CAGUACUUUUGUGUAGUACAA-3′; and antagomiR-124-3p,
5′-GGCAUUCACCGCGUGCCUUA-3′.
Immunofluorescence
Gastrocnemius tissues of mice were harvested and
processed in optimal cutting temperature compound (cat. no. 4583,
Sakura Finetek USA, Inc.). The treated tissues were sliced into 5
µm-thick sections. Subsequently, sections were fixed in 4%
formaldehyde for 15 min, incubated in 10% normal goat serum (cat.
no. AR0009, Wuhan Boster Biological Technology, Ltd.) and 0.5%
Triton X-100 for 30 min, all at room temperature. The sections were
incubated with a rat anti-mouse CD31 antibody (1:100; cat. no.
557355; BD Biosciences) overnight at 4°C. Following washing with
PBS, the sections were incubated with a secondary antibody (Alexa
Fluor 594-conjugated donkey anti-rat, 1:200, cat. no. 34412ES60;
Shanghai Yeasen Biotechnology Co., Ltd.) at 37°C for 1 h in the
dark. Nuclear staining was performed using
4,6-amidine-2-phenylindoles (cat. no. 28718-90-3; Merck KGaA) for
30 min at room temperature. Fluorescence images were obtained using
an Olympus IX83 fluorescence microscope at 10× magnification
(Olympus Corporation) and quantified using ImageJ software (version
1.52a; National Institutes of Health).
Study population
A total of 49 patients with PAD and 47 sex- and
age-matched participants without PAD were recruited at the
Department of Endocrinology, Xinhua Hospital Affiliated to Shanghai
Jiaotong University School of Medicine and Department of
Cardiology, Shanghai Tenth People's Hospital between August 2019
and July 2020. According to the 2016 AHA/ACC guidelines on PAD,
ankle-brachial index (ABI) was used to diagnose and evaluate the
severity of PAD. PAD was diagnosed in patients with an ABI <0.9
(24), which was measured and
calculated according to the method described by Aboyans et
al (25). Patients with
cancer, acute myocardial infarction, severe kidney failure, acute
infection or connective tissue disease were excluded from the
present study. It is recognized that patients with hypertension or
diabetes are at higher risk of PAD (3,4).
Hypertension was defined as systolic or diastolic blood pressure
≥140/90 mmHg, or normal blood pressure following the admission of
antihypertensive medications prior to recruitment. Diabetes was
defined as a fasting blood glucose ≥7 mmol/l, non-fasting plasma
glucose level of ≥11.1 mmol/l or normal blood glucose levels
following known treatment for diabetes. Blood samples (4 ml) were
collected from participants at the time of admission. Written
informed consent was obtained from all patients. The present study
was approved by the Ethics Committee of the Shanghai Tenth People's
Hospital (approval no. 2019-K-153).
Statistical analysis
Data are presented as the mean ± standard deviation.
Comparisons between two groups were analyzed using the unpaired
Student's t-test. Comparisons among multiple groups were analyzed
using one-way or two-way ANOVA followed by Bonferroni's post hoc
test. The parameters from baseline characteristics of patients with
PAD and non-PAD individuals were analyzed using the χ2
test (age, BMI and blood lipid were excluded). Pearson's
correlation analysis was performed to investigated the correction
between ABI and the expression level of miR-124-3p. Statistical
analyses were performed using GraphPad Prism software (version
6.01; GraphPad Software, Inc.). P<0.05 was considered to
indicate a statistically significant difference. All experiments
were repeated ≥3 times.
Results
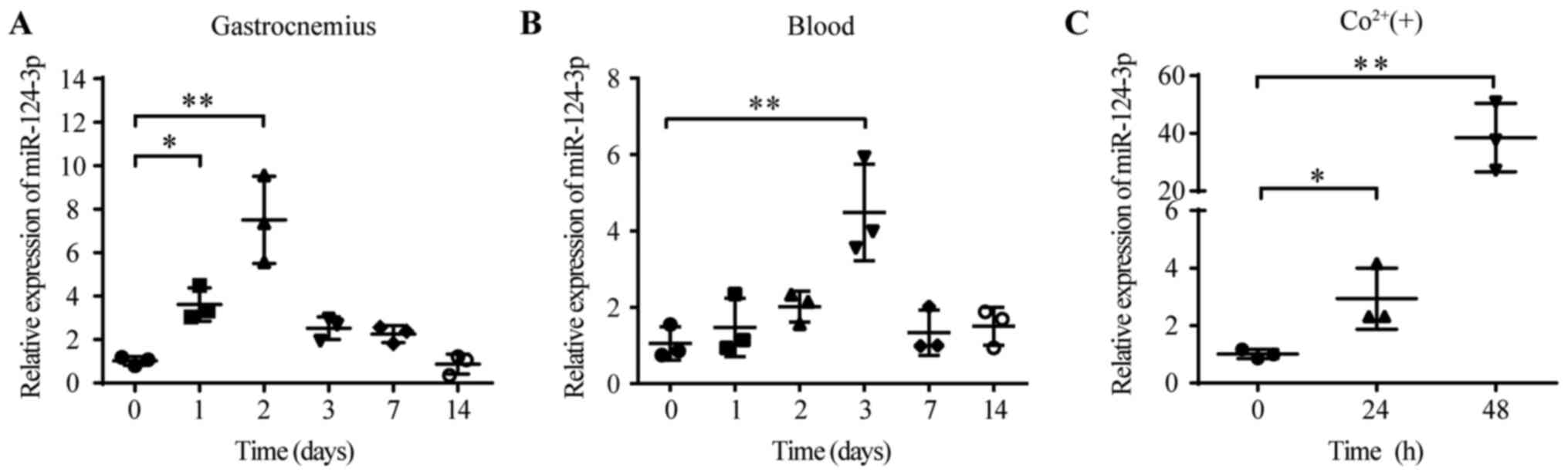
Expression levels of miR-124-3p are
increased in the HLI model and hypoxic HUVECs
To identify the role of miR-124-3p in the HLI model,
the dynamic expression of miR-124-3p was detected in the ligated
ischemic gastrocnemius. Total RNA was extracted from the
gastrocnemius and blood of HLI model mice. The RT-qPCR results
suggested that the levels of miR-124-3p in the ischemic
gastrocnemius were significantly upregulated on day 2 compared with
day 0 (day 2, 7.50±2.00 vs. day 0, 1.01±0.20; Fig. 1A). However, the levels of
miR-124-3p in the blood peaked on day 3 (day 3, 4.48±1.26 vs. day
0, 1.05±0.43; Fig. 1B).
Hypoxia-inducible factor-α (HIF-α) is a powerful inducer of
angiogenesis (26,27). Cobalt has been reported to mimic
hypoxia by preventing the degradation of HIF-α (28). Therefore, cobalt was used to mimic
hypoxia in HUVECs. The levels of miR-124-3p were significantly
increased following cobalt-induced hypoxia (500 µM) at 48 h
compared with 0 h (48 h, 38.54±11.88 vs. 0 h, 1.01±0.16; Fig. 1C). Collectively, the results
indicated that miR-124-3p levels were increased in HUVECs and
tissues under hypoxic conditions, which suggested that miR-124-3p
might be essential for the progression of HLI.
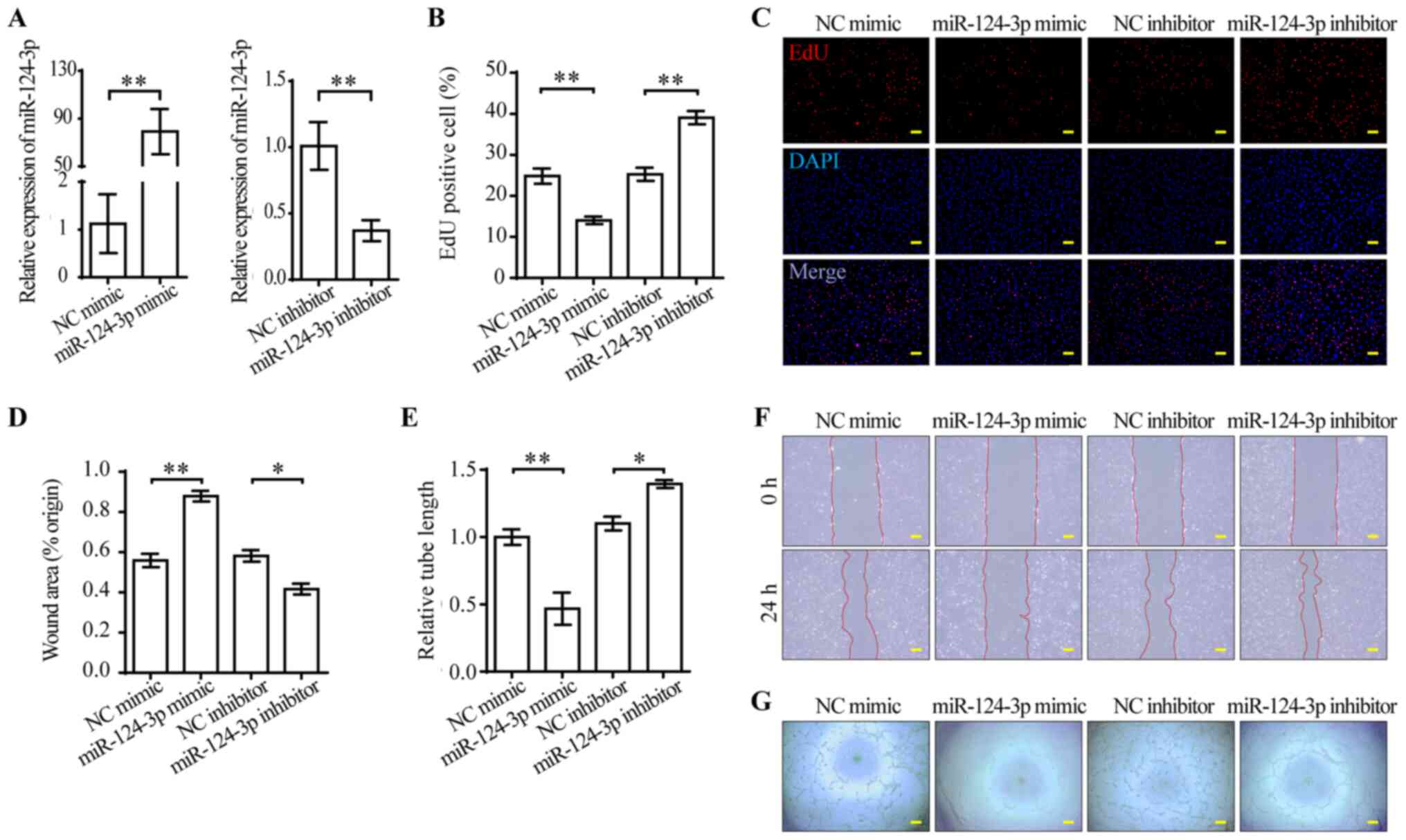
miR-124-3p impairs the functions of
VEGFA-165-treated HUVECs
VEGFA-165-stimulated ECs display enhanced
proliferation, migration and stability for angiogenesis (29). Moreover, the aforementioned results
provided evidence for investigating the functions of miR-124-3p in
ECs (Fig. 1C). miR-124-3p mimic
was used to overexpress miR-124-3p, whereas miR-124-3p inhibitor
was used to knock down miR-124-3p. At 48 h post-transfection, the
expression levels of miR-124-3p were measured via RT-qPCR. Compared
with the corresponding NCs, miR-124-3p was successfully
overexpressed by mimic and knocked down by inhibitor in HUVECs
(Fig. 2A). To investigate the
effect of miR-124-3p mimic and inhibitor, alterations to the cell
phenotype were examined. miR-124-3p mimic significantly inhibited
HUVEC proliferation compared with NC mimic, whereas miR-124-3p
inhibitor significantly enhanced HUVEC proliferation compared with
NC inhibitor (Fig. 2B and C).
Subsequently, the effect of miR-124-3p on HUVEC migration was
analyzed. The wound healing assay results indicated that HUVEC
migration was significantly inhibited by miR-124-3p mimic compared
with NC mimic, but significantly enhanced by miR-124-3p inhibitor
compared with NC inhibitor (Fig. 2D
and F). The tube formation assay was conducted to evaluate the
effect of miR-124-3p on angiogenesis. miR-124-3p mimic
significantly inhibited tube formation compared with NC mimic,
whereas miR-124-3p inhibitor significantly enhanced tube formation
compared with NC inhibitor (Fig. 2E
and G). Therefore, the results suggested that miR-124-3p
impaired HUVEC functions in vitro.
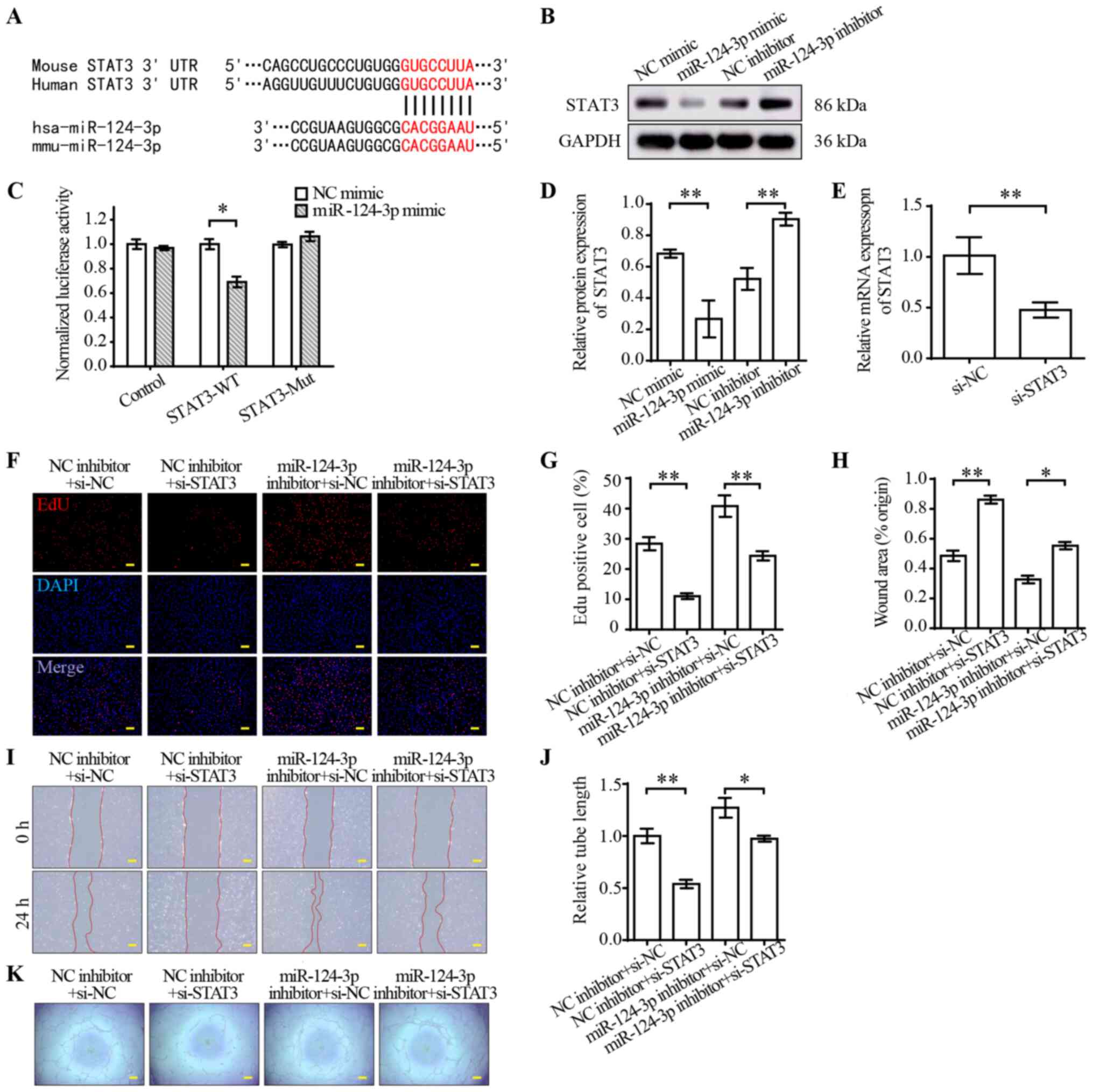
STAT3 is a target of miR-124-3p
TargetScan indicated that STAT3 was a potential
target of miR-124-3p (Fig. 3A).
STAT3 is a member of the STAT protein family, and emerging evidence
has suggested that it is a regulator of angiogenesis (30). To verify STAT3 as a target of
miR-124-3p, luciferase reporter plasmids containing miR-124-3p
binding sites in the 3′-UTRs of STAT3 were constructed. The
dual-luciferase reporter assay results indicated that miR-124-3p
mimic significantly decreased the luciferase activities of STAT3-WT
compared with NC mimic (Fig. 3C).
Furthermore, the protein expression levels of STAT3 were measured
via western blotting. STAT3 protein expression levels were
significantly decreased in the miR-124-3p mimic group compared with
the NC mimic group, but significantly increased in the miR-124-3p
inhibitor group compared with the NC inhibitor group (Fig. 3B and D). To further investigate the
effect of miR-124-3p on STAT3, a rescue experiment was conducted.
In brief, STAT3 knockdown was performed in combination with
miR-124-3p inhibitor transfection in HUVECs. Subsequently,
proliferation, wound healing and tube formation assays were
preformed to assess angiogenesis in HUVECs. The RT-qPCR results
indicated that STAT3 mRNA expression levels were significantly
decreased by si-STAT3 compared with si-NC (Fig. 3E). Moreover, STAT3 knockdown
significantly suppressed EC proliferation (Fig. 3F and G), migration (Fig. 3H and I) and tube formation
(Fig. 3J and K) compared with the
NC group. miR-124-3p inhibitor reversed STAT3 knockdown-mediated
inhibition of angiogenesis in ECs. Collectively, the results
indicated that miR-124-3p regulated the functions of HUVECs by
targeting STAT3.
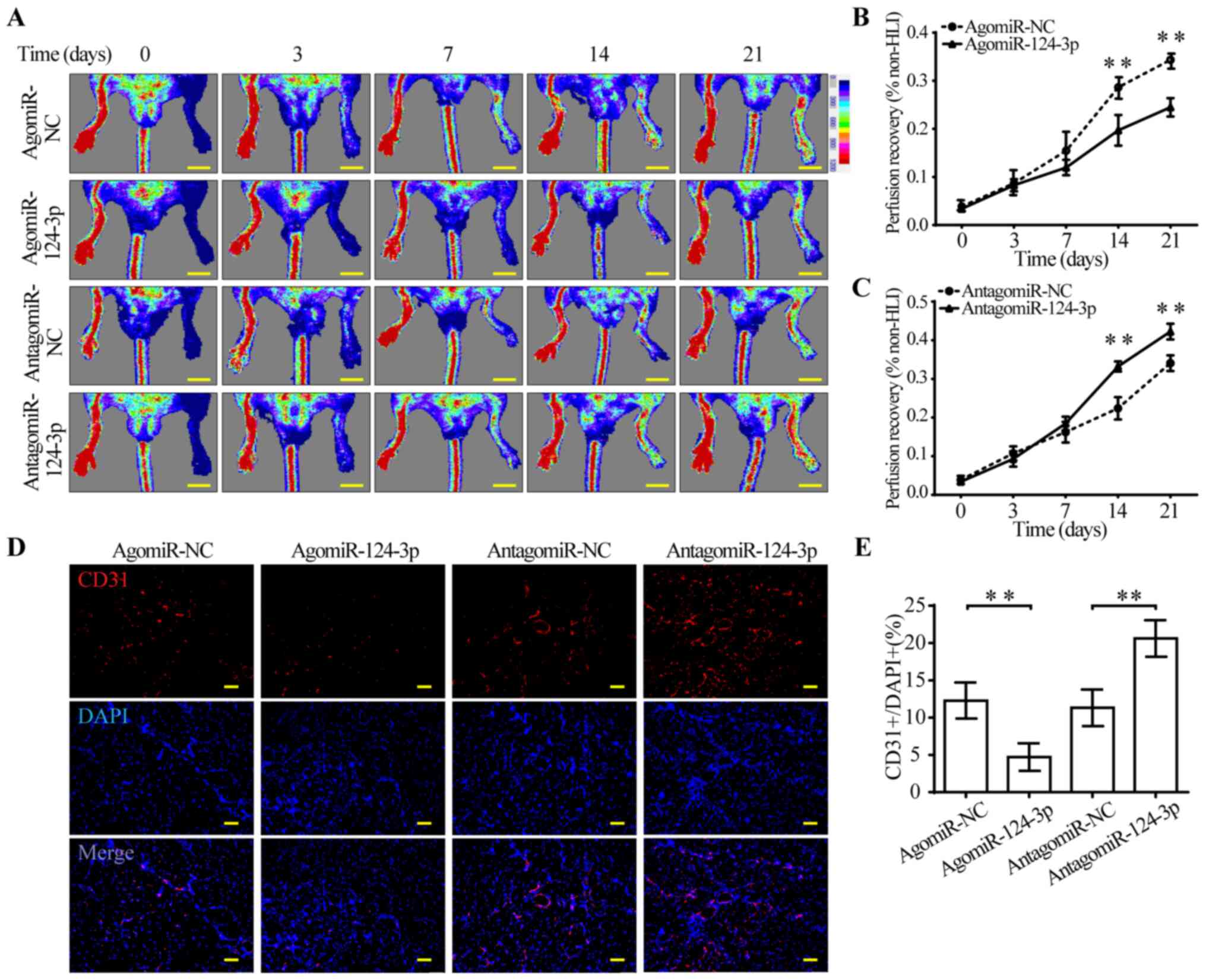
miR-124-3p impairs perfusion recovery
and capillary density in the HLI model
To investigate whether miR-124-3p directly modulated
perfusion recovery following HLI, agomiR-124-3p and
antagomiR-124-3p were injected in femoral artery-ligated mice. The
results suggested that agomiR-124-3p-treated mice displayed
significantly impaired perfusion recovery from day 14 post-HLI
compared with agomiR-NC-treated mice (Fig. 4A and B). By contrast,
antagomiR-124-3p significantly improved perfusion recovery from day
14 post-HLI compared with antagomiR-NC (Fig. 4A and C). Consistent with blood
perfusion recovery, capillary density of ischemic muscles displayed
the same tendency. AgomiR-124-3p-treated mice displayed
significantly fewer CD31+ cells in the gastrocnemius
compared with agomiR-NC-treated mice, which suggested that
agomiR-124-3p might inhibit angiogenesis in ischemic hindlimbs. By
contrast, antagomiR-124-3p significantly increased the number of
CD31+ cells compared with antagomiR-NC, suggesting
activation of angiogenesis in gastrocnemius (Fig. 4D and E). In combination, the
results indicated that miR-124-3p regulated angiogenesis following
ischemic injury in mouse hindlimbs.
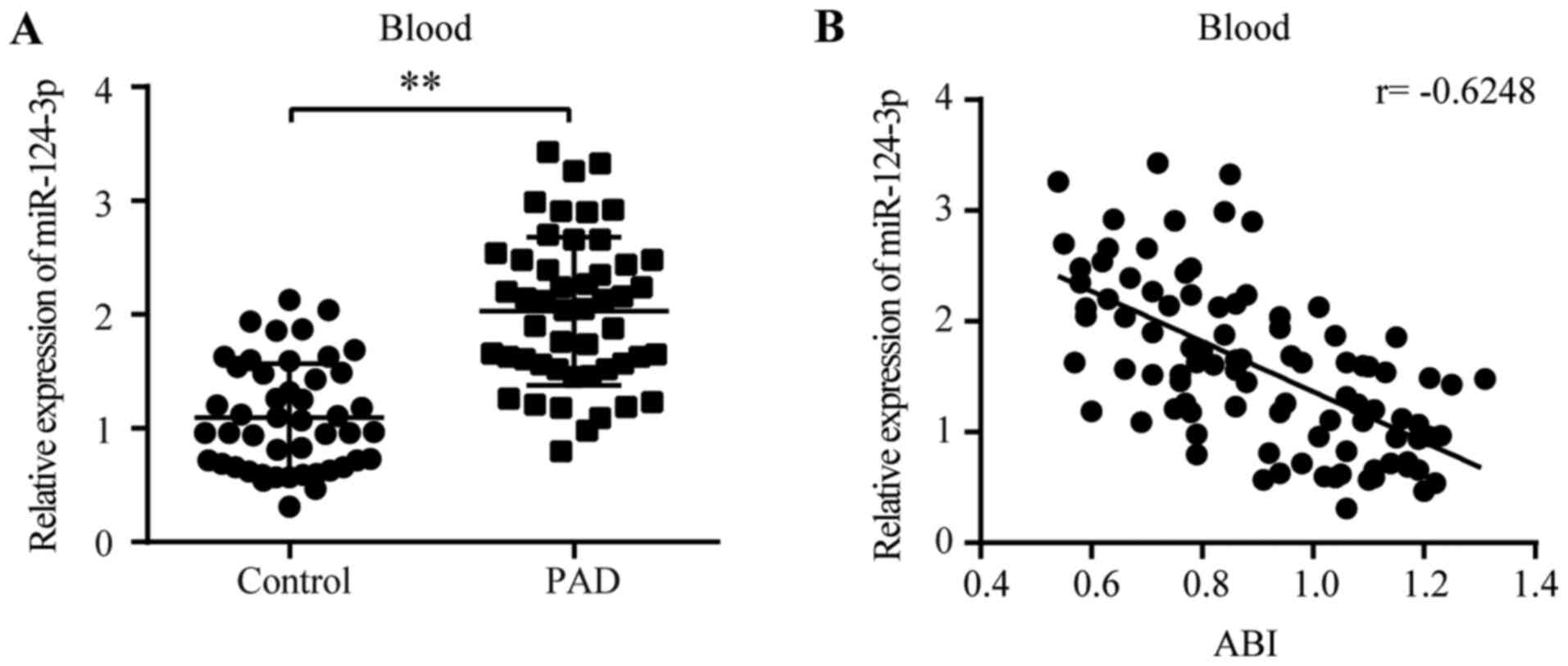
miR-124-3p expression is increased in
patients with PAD
The aforementioned results indicated that the
expression of miR-124-3p in the tissues and blood of the HLI model
displayed a similar peak time, and increased miR-124-3p expression
markedly inhibited angiogenesis in ligated legs, suggesting that
blood-derived miR-124-3p might serve as a marker of serious PAD. To
investigate the hypothesis, 49 patients with PAD and 47 healthy
individuals were enrolled in the present study. The baseline
characteristics of the patients are presented in Table I. RT-qPCR was conducted to detect
the levels of miR-124-3p in human blood. The levels of miR-124-3p
in patients with PAD were significantly higher compared with
non-PAD individuals (1.85-fold; PAD, 2.03±0.65 vs. non-PAD,
1.10±0.47; Fig. 5A). ABI is an
index for the assessment of the severity of PAD (31,32).
Pearson's correlation analysis indicated that the levels of
miR-124-3p in the blood were negatively correlated with ABI scores
(r=−0.6248; Fig. 5B), suggesting
that miR-124-3p expression levels were positively correlated with
the severity of PAD.
 | Table I.Baseline characteristics of patients
with PAD and non-PAD individuals. |
Table I.
Baseline characteristics of patients
with PAD and non-PAD individuals.
| Variable | Non-PAD (n=47) | PAD (n=49) | P-value |
|---|
| Age (years) |
62.60±11.88 | 65.90±8.00 | 0.104 |
| Gender (male) | 22 (46.81%) | 29 (59.18%) | 0.225 |
| BMI
(kg/m2) | 24.16±2.87 | 24.28±1.87 | 0.801 |
| Smoking | 11 (23.40%) | 28 (57.14%) | 0.001 |
| Blood lipid |
|
|
|
| Total
cholesterol (mmol/l) | 4.22±1.02 | 4.70±1.45 | 0.059 |
|
Triglyceride (mmol/l) | 1.99±0.84 | 1.70±0.87 | 0.092 |
|
High-density lipoprotein
(mmol/l) | 1.21±0.27 | 1.43±0.50 | 0.008 |
|
Low-density lipoprotein
(mmol/l) | 2.74±0.86 | 2.89±0.98 | 0.413 |
| Medical
history |
|
|
|
|
Coronary heart disease | 9
(19.14%) | 24 (48.98%) | 0.003 |
|
Diabetes | 31 (65.96%) | 46 (93.88%) | 0.001 |
|
Hypertension | 24 (51.06%) | 33 (67.35%) | 0.146 |
|
Statins | 18 (38.30%) | 28 (57.14%) | 0.071 |
Discussion
The concept of therapeutic angiogenesis for PAD has
been around for decades, but the advances made thus far fall far
below expectations. A potential reason for the lack of effective
results from therapeutic angiogenesis could be the complicated
self-regulation of cells in the microenvironment (33,34).
miRNAs are a type of small molecule that can serve as endogenous
regulators of cells (8,9). Although miRNAs have been reported to
be widely involved in the regulation of diseases, such as cancers
(35), autoimmune diseases
(36), central nervous system
injuries (37) and heart diseases
(38), there is limited
information on the involvement of miRNAs in PAD. miR-124-3p was
initially reported to be highly expressed in brain tissues
(39), serving a critical role in
neuronal differentiation (40).
Further studies investigated other functions of miR-124-3p.
According to Ando et al (15), miR-124-3p suppressed tumor
development by inhibiting angiogenesis. Shi et al (16) also demonstrated that miR-124-3p
might predict acute myocardial infarction, suggesting that
miR-124-3p might serve as a regulator of angiogenesis. Therefore,
the aforementioned studies highlighted the importance of
investigating the correlation between miR-124-3p and angiogenesis
in PAD.
In the present study, the results indicated that
miR-124-3p was upregulated under hypoxic conditions both in
vivo and in vitro compared with the corresponding
control groups. Moreover, compared with the NC groups, miR-124-3p
overexpression significantly suppressed HUVEC functions and
impaired perfusion recovery in the HLI model. STAT3 has been
recognized as a regulator of angiogenesis beyond inflammation
(41). The functions of STAT3 are
precisely regulated by multiple chaperonins under specific
conditions. For example, canonical STAT3 signaling is associated
with JAK-STAT signaling, whereby STAT3 is phosphorylated on
tyrosine 750 (Y750), facilitating STAT3 homodimerization, nuclear
translation, DNA binding and initiation of transcription (42). The noncanonical nuclear activities,
including acetylation (43),
alkylation (44), methylation
(45), ubiquitination (46) and glutathionylation (44), have been implicated in STAT3
transcriptional activity in various cells. Certain studies have
reported the axis of miR-124-3p/STAT3 (47–49),
but to the best of our knowledge, no previous study has focused on
the functions of STAT3 in EC proliferation. Inhibition of the STAT3
signaling pathway impairs angiogenesis and perfusion recovery in
the muscles of patients with PAD (50). In addition, an increasing number of
studies have verified that STAT3 was involved in the regulation of
tumor angiogenesis by modulating the expression of VEGF (51,52).
The results of the present study indicated that miR-124-3p
overexpression significantly decreased STAT3 protein expression
levels and inhibited HUVEC proliferation compared with NC mimic.
Furthermore, it has been reported that STAT3 could bind with
Yes-associated protein to regulate the mRNA expression levels of
angiopoietin-2 in ECs (53,54).
Therefore, the aforementioned results suggested that miR-124-3p
regulated angiogenesis following ischemic injury in mouse hindlimbs
by targeting STAT3.
Another interesting finding of the present study was
that the levels of circulating miR-124-3p were negatively
correlated with ABI, the index for PAD severity. ABI is a
non-invasive physical index that provides the standard for the
evaluation of PAD severity (31).
ABI is less sensitive in conditions associated with vessel
stiffness (55); therefore, the
expression of miR-124-3p in the blood might serve as an improved
marker for screening patients than ABI. However, the role of
miR-124-3p in the progression of PAD requires further investigation
with additional samples. In addition, the long-term outcome of
patients with increased miR-124-3p expression requires further
investigation.
Although previous studies have reported possible
roles of miR-124-3p in ischemic diseases (27–28),
there were several novel aspects of the present study. First, the
potential role of miR-124-3p was identified in the HLI model.
Secondly, the results indicated that the levels of miR-124-3p in
human blood were positively correlated with the severity of PAD,
which suggested that miR-124-3p might serve as a strong potential
target for the evaluation and treatment of PAD. Therefore, the
aforementioned findings may aid with the clinical translation of
the present study.
In conclusion, the present study provided evidence
for the link between miR-124-3p and PAD. miR-124 regulated
angiogenesis by decreasing STAT3 expression. Although miRNA-based
therapeutics are still being developed, the results of the present
study are encouraging and suggested the potential of miR-124 as a
diagnostic, prognostic and therapeutic target for PAD in the
future.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81670746, 81670230
and 91939101).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WP and WJ designed the study. YS and XX performed
the experiments, analyzed the data and wrote the manuscript. PL,
WK, ML, QY, JZ and YX were responsible for collecting blood
samples, baseline characteristics of patients and analyzing the
data of population study. All authors read and approved the final
manuscript, and agreed to be accountable for the work in ensuring
that questions related to the integrity of any part of the work
were appropriately investigated and resolved.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients. The present study was approved by the Ethics Committee of
Shanghai Tenth People's Hospital, Shanghai, China (approval no.
2019-K-153). Animal experiments were approved by the Laboratory
Animal Ethics Committee of Shanghai Tenth People's Hospital,
Shanghai, China approval no. SHDSYY-2019-2149).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Abdulhannan P, Russell DA and
Homer-Vanniasinkam S: Peripheral arterial disease: A literature
review. Br Med Bull. 104:21–39. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fowkes FG, Rudan D, Rudan I, Aboyans V,
Denenberg JO, McDermott MM, Norman PE, Sampson UK, Williams LJ,
Mensah GA and Criqui MH: Comparison of global estimates of
prevalence and risk factors for peripheral artery disease in 2000
and 2010: A systematic review and analysis. Lancet. 382:1329–1340.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Criqui MH and Aboyans V: Epidemiology of
peripheral artery disease. Circ Res. 116:1509–1526. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Collinson DJ and Donnelly R: Therapeutic
angiogenesis in peripheral arterial disease: Can biotechnology
produce an effective collateral circulation? Eur J Vasc Endovasc
Surg. 28:9–23. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vandekeere S, Dewerchin M and Carmeliet P:
Angiogenesis revisited: An overlooked role of endothelial cell
metabolism in vessel sprouting. Microcirculation. 22:509–517. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taniyama Y, Azuma J, Rakugi H and
Morishita R: Plasmid DNA-based gene transfer with ultrasound and
microbubbles. Curr Gene Ther. 11:485–490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Forster R, Liew A, Bhattacharya V, Shaw J
and Stansby G: Gene therapy for peripheral arterial disease.
Cochrane Database Syst Rev. 10:CD0120582018.PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
miRBase. Release 22.1. simplehttp://www.mirbase.orgOctober. 2018
|
|
12
|
Zhao J, Li X, Hu J, Chen F, Qiao S, Sun X,
Gao L, Xie J and Xu B: Mesenchymal stromal cell-derived exosomes
attenuate myocardial ischaemia-reperfusion injury through
miR-182-regulated macrophage polarization. Cardiovasc Res.
115:1205–1216. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hou Z, Qin X, Hu Y, Zhang X, Li G, Wu J,
Li J, Sha J, Chen J, Xia J, et al: Longterm exercise-derived
exosomal miR-342-5p: A novel exerkine for cardioprotection. Circ
Res. 124:1386–1400. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu X, Tian L and Zhang Z: Triptolide
inhibits angiogenesis in microvascular endothelial cells through
regulation of miR-92a. J Physiol Biochem. 75:573–583. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ando H, Okamoto A, Yokota M, Shimizu K,
Asai T, Dewa T and Oku N: Development of a miR-92a delivery system
for anti-angiogenesis-based cancer therapy. J Gene Med. 15:20–27.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi Z, Chen Q, Li C, Wang L, Qian X, Jiang
C, Liu X, Wang X, Li H, Kang C, et al: MiR-124 governs glioma
growth and angiogenesis and enhances chemosensitivity by targeting
R-Ras and N-Ras. Neuro Oncol. 16:1341–1353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang G, Chen L, Khan AA, Li B, Gu B, Lin
F, Su X and Yan J: miRNA-124-3p/neuropilin-1(NRP-1) axis plays an
important role in mediating glioblastoma growth and angiogenesis.
Int J Cancer. 143:635–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo ML, Guo LL and Weng YQ: Implication of
peripheral blood miRNA-124 in predicting acute myocardial
infarction. Eur Rev Med Pharmacol Sci. 21:1054–1059.
2017.PubMed/NCBI
|
|
19
|
Xu SY, Jiang XL, Liu Q, Xu J, Huang J, Gan
SW, Lu WT, Zhuo F, Yang M and Sun SQ: Role of rno-miR-124-3p in
regulating MCT1 expression in rat brain after permanent focal
cerebral ischemia. Genes Dis. 6:398–406. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
He Y, Luo Y, Tang S, Rajantie I, Salven P,
Heil M, Zhang R, Luo D, Li X, Chi H, et al: Critical function of
Bmx/Etk in ischemia-mediated arteriogenesis and angiogenesis. J
Clin Invest. 116:2344–2355. 2006.PubMed/NCBI
|
|
22
|
Hazarika S, Farber CR, Dokun AO,
Pitsillides AN, Wang T, Lye RJ and Annex BH: MicroRNA-93 controls
perfusion recovery after hindlimb ischemia by modulating expression
of multiple genes in the cell cycle pathway. Circulation.
127:1818–1828. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
National Reaearch Council (US) Committee
for the Guide for the Care and Use of Laboratory Animals: Guide for
the Care and Use of Laboratory Animals. 8th edition. National
Academies Press (US); Washington, DC: 2011
|
|
24
|
Gerhard-Herman MD, Gornik HL, Barrett C,
Barshes NR, Corriere MA, Drachman DE, Fleisher LA, Fowkes FGR,
Hamburg NM, Kinlay S, et al: 2016 AHA/ACC Guideline on the
Management of Patients With Lower Extremity Peripheral Artery
Disease: A Report of the American College of Cardiology/American
Heart Association Task Force on Clinical Practice Guidelines. J Am
Coll Cardiol. 69:e71–e126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aboyans V, Criqui MH, Abraham P, Allison
MA, Creager MA, Diehm C, Fowkes FG, Hiatt WR, Jönsson B, Lacroix P,
et al: Measurement and interpretation of the ankle-brachial index:
A scientific statement from the American Heart Association.
Circulation. 126:2890–2909. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu D, Potluri N, Lu J, Kim Y and
Rastinejad F: Structural integration in hypoxia-inducible factors.
Nature. 524:303–308. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Semenza GL: Life with oxygen. Science.
318:62–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Muñoz-Sánchez J and Chánez-Cárdenas ME:
The use of cobalt chloride as a chemical hypoxia model. J Appl
Toxicol. 39:556–570. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chung AS and Ferrara N: Developmental and
pathological angiogenesis. Annu Rev Cell Dev Biol. 27:563–584.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miyazaki T, Taketomi Y, Saito Y, Hosono T,
Lei XF, Kim-Kaneyama J, Arata S, Takahashi H, Murakami M and
Miyazaki A: Calpastatin counteracts pathological angiogenesis by
inhibiting suppressor of cytokine signaling 3 degradation in
vascular endothelial cells. Circ Res. 116:1170–1181. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin JS, Olson CM, Johnson ES and Whitlock
EP: The ankle-brachial index for peripheral artery disease
screening and cardiovascular disease prediction among asymptomatic
adults: A systematic evidence review for the U.S. Preventive
Services Task Force. Ann Intern Med. 159:333–341. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kravos A and Bubnic-Sotosek K:
Ankle-brachial index screening for peripheral artery disease in
asymptomatic patients between 50 and 70 years of age. J Int Med
Res. 37:1611–1619. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Veith AP, Henderson K, Spencer A, Sligar
AD and Baker AB: Therapeutic strategies for enhancing angiogenesis
in wound healing. Adv Drug Deliv Rev. 146:97–125. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mitsos S, Katsanos K, Koletsis E, Kagadis
GC, Anastasiou N, Diamantopoulos A, Karnabatidis D and Dougenis D:
Therapeutic angiogenesis for myocardial ischemia revisited: Basic
biological concepts and focus on latest clinical trials.
Angiogenesis. 15:1–22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rupaimoole R, Calin GA, Lopez-Berestein G
and Sood AK: miRNA deregulation in cancer cells and the tumor
microenvironment. Cancer Discov. 6:235–246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mehta A and Baltimore D: MicroRNAs as
regulatory elements in immune system logic. Nat Rev Immunol.
16:279–294. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bhalala OG, Srikanth M and Kessler JA: The
emerging roles of microRNAs in CNS injuries. Nat Rev Neurol.
9:328–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Katz MG, Fargnoli AS, Kendle AP, Hajjar RJ
and Bridges CR: The role of microRNAs in cardiac development and
regenerative capacity. Am J Physiol Heart Circ Physiol.
310:H528–541. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sempere LF, Freemantle S, Pitha-Rowe I,
Moss E, Dmitrovsky E and Ambros V: Expression profiling of
mammalian microRNAs uncovers a subset of brain-expressed microRNAs
with possible roles in murine and human neuronal differentiation.
Genome Biol. 5:R132004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoo AS, Sun AX, Li L, Shcheglovitov A,
Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW and Crabtree
GR: MicroRNA-mediated conversion of human fibroblasts to neurons.
Nature. 476:228–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen Z and Han ZC: STAT3: A critical
transcription activator in angiogenesis. Med Res Rev. 28:185–200.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Koo MY, Park J, Lim JM, Joo SY, Shin SP,
Shim HB, Chung J, Kang D, Woo HA and Rhee SG: Selective inhibition
of the function of tyrosine-phosphorylated STAT3 with a
phosphorylation site-specific intrabody. Proc Natl Acad Sci USA.
111:6269–6274. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Yuan ZL, Guan YJ, Chatterjee D and Chin
YE: Stat3 dimerization regulated by reversible acetylation of a
single lysine residue. Science. 307:269–273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Buettner R, Corzano R, Rashid R, Lin J,
Senthil M, Hedvat M, Schroeder A, Mao A, Herrmann A, Yim J, et al:
Alkylation of cysteine 468 in Stat3 defines a novel site for
therapeutic development. ACS Chem Biol. 6:432–443. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Stark GR, Kerr IM, Williams BR, Silverman
RH and Schreiber RD: How cells respond to interferons. Annu Rev
Biochem. 67:227–264. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stark GR, Wang Y and Lu T: Lysine
methylation of promoter-bound transcription factors and relevance
to cancer. Cell Res. 21:375–380. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhou YL, Zhang L, Zhou Z, Liu W, Lu Y, He
S, Cui Y, Qin Y and Hua M: Antibody modified nanoparticle-mediated
delivery of miR-124 regulates apoptosis via repression the Stat3
signal in mycobacterial-infected microglia. J Biomed Nanotechnol.
14:2185–2197. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang Y, Li X, Zhang J and Liang H:
Natural killer T cell cytotoxic activity in cervical cancer is
facilitated by the LINC00240/microRNA-124-3p/STAT3/MICA axis.
Cancer Lett. 474:63–73. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vuokila N, Aronica E, Korotkov A, van
Vliet EA, Nuzhat S, Puhakka N and Pitkanen A: Chronic regulation of
miR-124-3p in the perilesional cortex after experimental and human
TBI. Int J Mol Sci. 21:24182020. View Article : Google Scholar
|
|
50
|
Ganta VC, Choi M, Kutateladze A and Annex
BH: VEGF165b modulates endothelial VEGFR1-STAT3 signaling pathway
and angiogenesis in human and experimental peripheral arterial
disease. Circ Res. 120:282–295. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Banerjee K and Resat H: Constitutive
activation of STAT3 in breast cancer cells: A review. Int J Cancer.
138:2570–2578. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tartour E, Pere H, Maillere B, Terme M,
Merillon N, Taieb J, Sandoval F, Quintin-Colonna F, Lacerda K,
Karadimou A, et al: Angiogenesis and immunity: A bidirectional link
potentially relevant for the monitoring of antiangiogenic therapy
and the development of novel therapeutic combination with
immunotherapy. Cancer Metastasis Rev. 30:83–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He J, Bao Q, Zhang Y, Liu M, Lv H, Liu Y,
Yao L, Li B, Zhang C, He S, et al: Yes-associated protein promotes
angiogenesis via signal transducer and activator of transcription 3
in endothelial cells. Circ Res. 122:591–605. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wan L, Zhang Q, Wang S, Gao Y, Chen X,
Zhao Y and Qian X: Gambogic acid impairs tumor angiogenesis by
targeting YAP/STAT3 signaling axis. Phytother Res. 33:1579–1591.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Suominen V, Uurto I, Saarinen J, Venermo M
and Salenius J: PAD as a risk factor for mortality among patients
with elevated ABI-A clinical study. Eur J Vasc Endovasc Surg.
39:316–322. 2010. View Article : Google Scholar : PubMed/NCBI
|