Introduction
Intracranial aneurysm (IA) rupture-induced
subarachnoid hemorrhage (SAH) is an acute, non-traumatic and
devastating condition that accounts for ~5% of cerebrovascular
strokes (1,2), and can result in 45% of mortality,
30% of disability and 50% of cognitive impairment morbidity cases
in patients suffering from SAH (3–6).
Thus, understanding the molecular mechanism underlying IA
rupture-induced systemic damage is critical for public health.
Changes in gene expression profiles in response to
disease onset reflect crucial processes involved in disease
pathogenesis, development and treatment. The identification of
circulating genetic and metabolic biomarkers is of great value for
early diagnosis and timely intervention (7,8). For
example, a nonsense truncated mutation (R450X, 13:52952757, c.1348
C>T, NM_018676.3) in thrombospondin type 1 domain containing
protein 1 (THSD1) is associated with SAH morbidity (9). Furthermore, in zebrafish and mice,
THSD1 loss-of-function impaired the focal adhesion of endothelial
cells to the basement membrane, resulting in cerebral bleeding and
mortality (9). The NF-κB
transcription factor (TF), a key factor in inflammation, has been
identified to be associated with IA rupture-induced SAH (10,11).
In addition, higher expression levels of Toll-like receptor (TLR)4
in the peripheral blood mononuclear cells (PBMCs) of patients with
SAH was associated with a larger SAH volume, delayed cerebral
infarction and worse functional recovery (12). A therapeutic role for inactivation
of TLR4 in neuroinflammation post-SAH has been reported (13). However, these limited reports on
recognized genes in response to IA rupture-induced SAH cannot
explain the underlying complex neuroinflammation and immune
mechanisms post-SAH. IA rupture-induced systemic damage requires
improved understanding. The more genes that are changed in response
to SAH that can be identified, the clearer the molecular mechanisms
underlying the development of cerebral disability and cognitive
impairment post-SAH will be.
The present study was conducted using a microarray
dataset previously reported by Pera et al (14) with the goal of identifying more
circulating biomarkers, including long non-coding RNAs (lncRNAs)
and genes in the blood, as well as the pathways associated with
them, via systemic and rigorous bioinformatics analyses. Taken
together, the study aimed to provide more information on the
molecular mechanism underlying SAH-induced cerebral disability and
cognitive impairment.
Materials and methods
Data selection
A microarray dataset reporting lncRNA and gene
expression profiles in patients with SAH from ruptured IA
(GSE36791) (14) was downloaded
from the National Center for Biotechnology Information Gene
Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The GSE36791
dataset was generated using the GPL10558 Illumina Human HT-12 V4.0
expression beadchip platform and comprises 4,055 lncRNAs and 19,198
protein-coding genes. The dataset includes 61 samples of venous
whole blood, including 43 samples from patients with SAH from
ruptured IA (48.0±13.9 years old and 48.8% male) and 18 samples
from age-matched healthy controls (46.1±17.5 years old and 72.2%
male) suffering from headaches. The study by Pera et al
(14) was approved by the ethics
committee at their institution and conducted with informed consent
from all subjects.
Data processing and gene expression
profiling
All txt files in the GSE36791 dataset were
downloaded and processed using the Limma package (version 3.34.0;
http://bioconductor.org/packages/release/bioc/html/limma.html)
(15) for data conversion
(log2), background correction and data normalization.
The transcript IDs and RefSeq IDs of the lncRNAs and genes in the
dataset were annotated in the HUGO Gene Nomenclature Committee
database (http://www.genenames.org/) (16). The differentially expressed lncRNAs
(DElncRNAs) and genes (DEGs) in the blood samples were screened
with the criteria of false discovery rate <0.05 and
|log2fold-change (FC)|≥0.263. Hierarchical clustering of
the screened DElncRNAs and DEGs in the GSE36791 dataset was
performed using pheatmap (version 1.0.8; http://cran.r-project.org/package=pheatmap)
(17).
Weighted gene co-expression network
analysis (WGCNA) and module identification
To screen lncRNAs and genes with similar expression
profiles in patients with SAH from ruptured IA compared with
controls, the gene co-expression network and co-expressed gene
modules were identified using the WGCNA integrated algorithm
(version 1.61; http://CRAN.R-project.org/package=WGCNA) (18,19).
All algorithms were conducted following the approximate scale-free
features. The co-expression correlation matrix and adjacency
function (soft-thresholding parameter and intramodular
connectivity) were defined, and the dissimilarity coefficients
between genes were calculated. Accordingly, the modules associated
with disease traits (phenotypes) were identified and correlation
parameters (Pearson's r and P-value) were calculated. Modules of
eigengenes (≥50 genes) that are highly associated with disease
traits (P<0.05) were identified with the following criteria:
cutHeight=0.995 and significant stability correlation (P<0.05).
The eigengenes in modules that significantly associated with
disease traits were subjected to enrichment analysis.
Enrichment analysis
Gene enrichment was performed to identify the Gene
Ontology (20,21) (GO) biological processes (BPs) that
were significantly associated with the DEGs in significant modules.
The GO BP terms in the Database for Annotation, Visualization and
Integrated Discovery (DAVID; v6.8; http://david.ncifcrf.gov/) (22) were picked with the criterion of
P<0.05.
lncRNA-mRNA regulatory network
construction
The Pearson's correlation coefficients (r) between
the DElncRNAs and DEGs in the significant WGCNA modules were
calculated using the Cor function (23). The lncRNA-mRNA pairs with r≥0.6
were retained and used as candidates for the construction of a
lncRNA-mRNA network. Cytoscape software (version 3.6.1; http://www.cytoscape.org/) (24) was used for the construction of the
lncRNA-mRNA network. DEGs in the lncRNA-mRNA network were further
exploited for Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathway (25) enrichment analysis
with the criterion of P<0.05 for inclusion.
Screening of SAH-associated biomarkers
and pathways
Finally, the genes and KEGG pathways associated with
IA rupture-induced SAH were screened in the Comparative
Toxicogenomics Database (CTD, 2019 update; http://ctd.mdibl.org/) (26) with the key phrase ‘Intracranial
Aneurysms’. The overlapping SAH-associated pathways and related
genes between the DAVID and CTD databases were selected and
regarded as candidate biomarkers or pathways. The DElncRNAs that
interacted with the candidate DEGs were selected, and the network
involving SAH-associated pathways, DEGs and DElncRNAs was
constructed using Cytoscape.
Validation of SAH-associated
biomarkers
The expression profiles of the SAH-associated
candidates in the GSE36791 dataset were then presented. The
expression levels of these DEGs in IA samples and controls were
calculated and compared. In addition, the expression profiles of
these DEGs in another dataset, GSE13353 (27) [GPL570(HG-U133_Plus_2) Affymetrix
Human Genome U133 Plus 2.0 Array], which consisted of 19 wall
samples (including 11 ruptured and 8 unruptured IA) were
verified.
Statistical analysis
The expression levels of the SAH-associated genes in
the GSE36791 and GSE13353 datasets were expressed as the mean ±
standard deviation. The differences were identified using a t-test
in the Limma package. GraphPad Prism 6.4 software (GraphPad
Software, Inc.) was used to generate histograms. P<0.05 was
considered to indicate a statistically significant difference.
Results
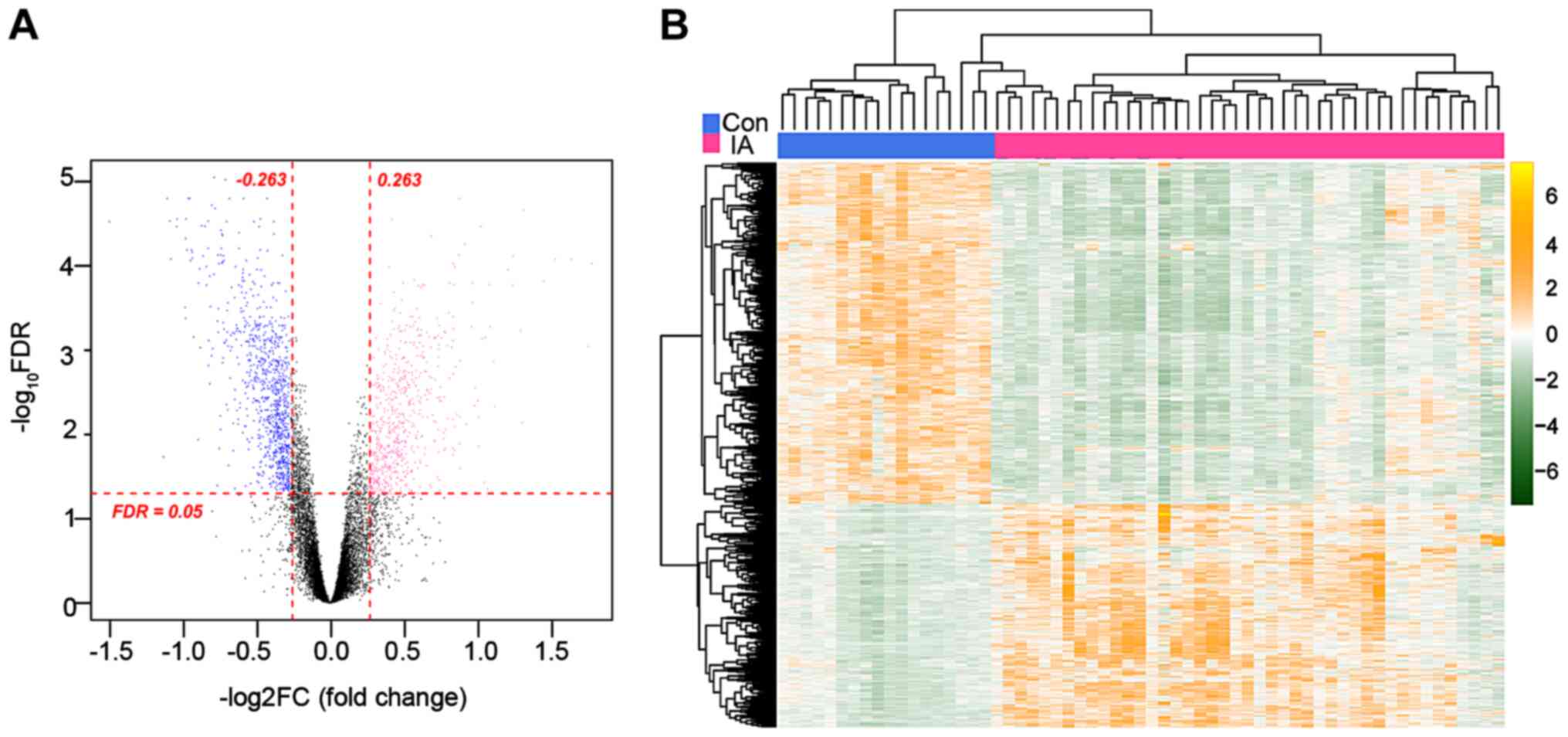
DElncRNA and DEG statistics in
patients with IA
A total of 25 DElncRNAs (13 downregulated and 12
upregulated) and 1,979 DEGs (1,198 downregulated and 781
upregulated) were screened from patients with IA rupture-induced
SAH compared with the controls in the GSE36791 dataset (Fig. 1A). The clustering heatmap showed
the distinct expression profiles of up- and downregulated DElncRNAs
and DEGs in the dataset (Fig.
1B).
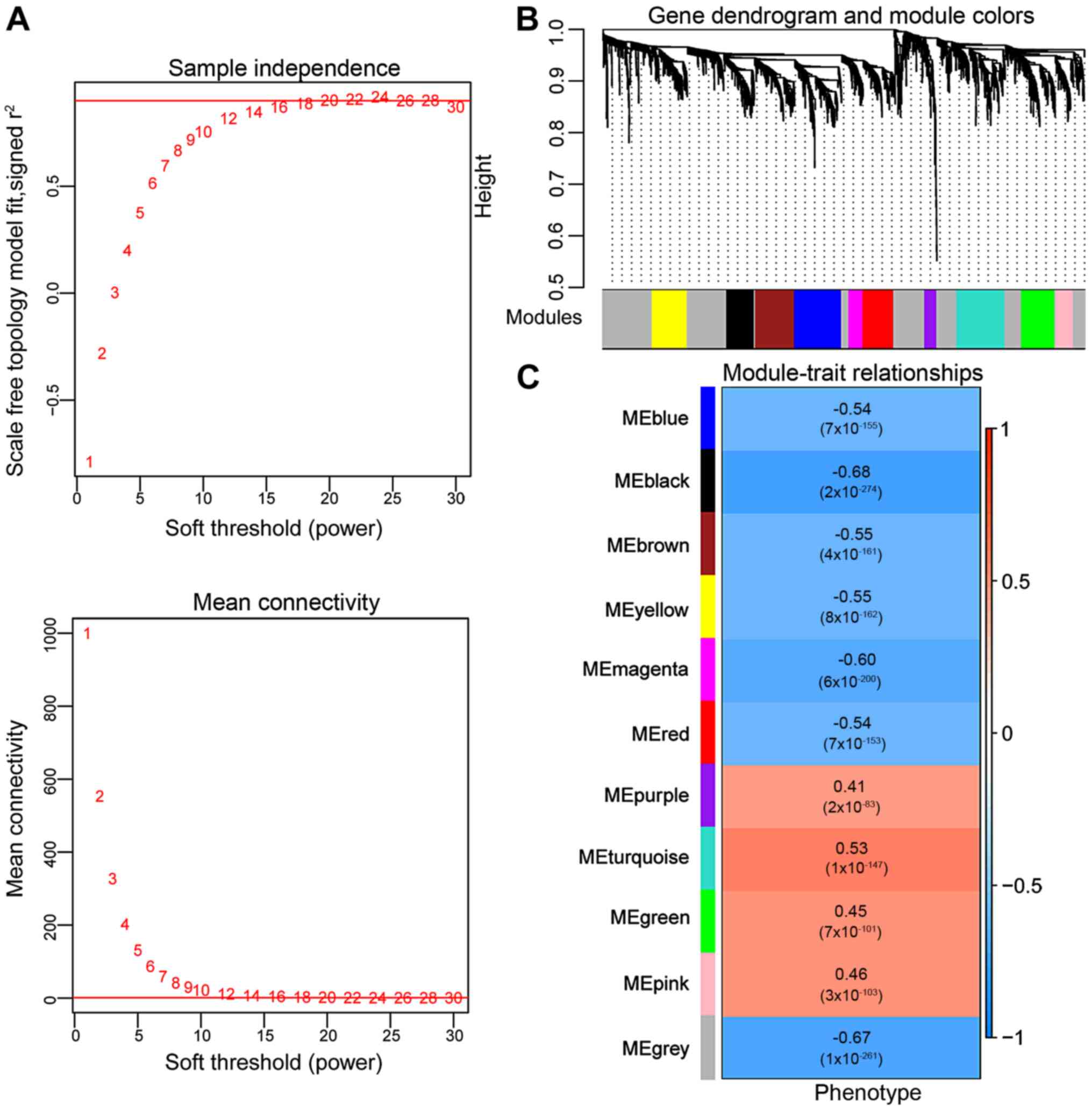
Identification of SAH-associated WGCNA
modules
WGCNA was conducted according to the approximate
scale-free features. Accordingly, an eigengene correlation
coefficient square (r2) of 0.9, a soft threshold power
of 20, and an intramodular connectivity of 1 were selected as the
adjacency function parameters (Fig.
2A). A total of 11 WGCNA modules were identified following the
adjacency function parameters in combination with the criteria of
eigengenes ≥50 and cutHeight=0.995 (Fig. 2B). Four (purple, turquoise, green
and pink) of the 11 WGCNA modules displayed significant positive
correlations with disease traits (IA phenotypes; P<0.05 and
r>0; Fig. 2C). These modules
included 50, 201, 140 and 76 DElncRNAs and/or DEGs, respectively
(467 in total; Table SI). One
module (black; r=−0.68, n=115) showed a relatively higher negative
correlation with disease traits than the gray module (r=−0.67;
Fig. 2C). Since most of the DEGs
related to IA rupture-induced SAH were upregulated, the present
study mainly focused on the selection of upregulated biomarkers;
consequently, the downregulated genes in the black module were
excluded from analysis. The expression profiles of the upregulated
DElncRNAs and DEGs in the four WGCNA modules are shown in Fig. S1.
Enrichment analysis for DEGs in
SAH-associated modules
In order to identify the biological functions
associated with the DEGs in the four SAH-associated modules
(purple, turquoise, green and pink), the DEG clusters were
individually processed using DAVID, and the DEG-associated GO BP
terms were selected. The results indicated that the DEGs in the
green, pink and purple modules were associated with GO BP terms
related to programmed cell death (including GO:0043067, regulation
of programmed cell death; GO:0012501, programmed cell death;
GO:0008219, cell death; GO:0012502, induction of programmed cell
death; GO:0043068, positive regulation of programmed cell death)
and apoptosis (including GO:0043065, positive regulation of
apoptosis; GO:0006917, induction of apoptosis; GO:0042981,
regulation of apoptosis and GO:0006915, apoptosis). DEGs in the
green and purple modules were associated with ‘GO:0009611, response
to wounding’, ‘GO:0006952, defense response’ and ‘GO:0010033,
response to organic substance’. DEGs in the pink and turquoise
modules were associated with ‘GO:0006796, phosphate metabolic
process’ and ‘GO:0006468, protein amino acid phosphorylation’. All
modules were associated with ‘GO:0006955, immune response’
(Table SII).
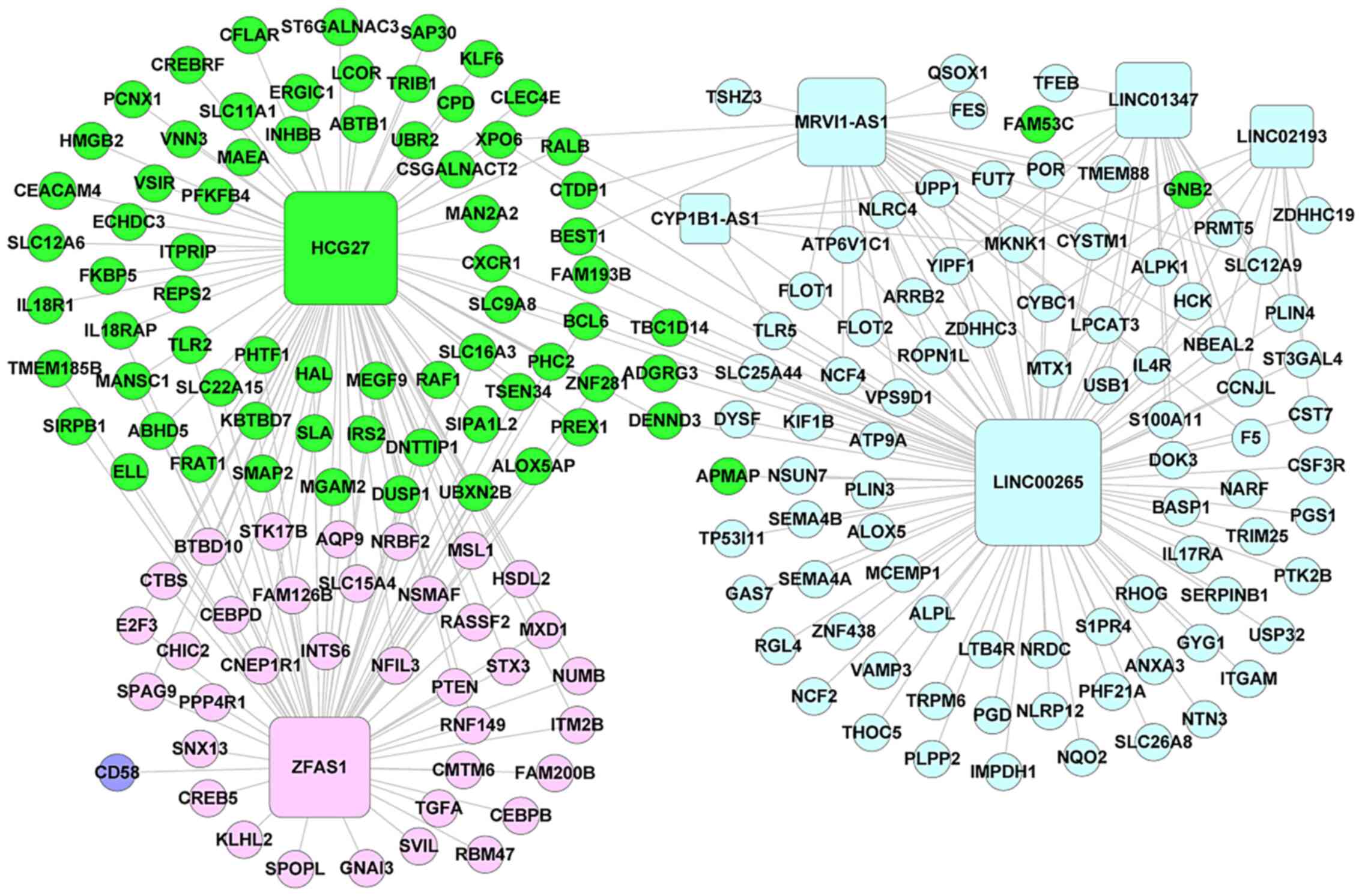
lncRNA-mRNA regulatory network
construction
All r values between the 467 DElncRNAs and DEGs in
SAH-associated modules were calculated, and a total of 1,020
lncRNA-mRNA pairs with r≥0.6 were retained for the construction of
the lncRNA-mRNA regulatory network. The network consisted of 1,020
lines (interactions) and 382 nodes (DElncRNAs and DEGs), including
135, 71, 2 and 174 nodes in the green, pink, purple and turquoise
module, respectively (Fig. S2). A
total of seven DElncRNAs were included, including one green (HCG27,
degree=198), one pink [ZNFX1 antisense RNA 1 (ZFAS1), degree=212],
no purple and five turquoise [long intergenic non-protein coding
RNA (LINC)00265, degree=214; murine retrovirus integration site 1
homolog (MRVI1)-antisense RNA 1 (AS1), degree=128; cytochrome P450
1B1 (CYP1B1)-AS1, degree=115; LINC01347, degree=94; LINC02193,
degree=62]. The results demonstrated that mitogen-activated protein
kinase (MAPK)-interacting serine/threonine-protein kinase 1 (MKNK1)
was regulated by all seven DElncRNAs, whereas Rho GTPase-activating
protein 24, BCL6, CXC chemokine receptor 1 (CXCR1), C-terminal
domain phosphatase 1, DENN/MADD domain containing 3, NOP2/Sun
domain family, member 7 and exportin 6 were regulated by all
lncRNAs except LINC02193. To further select the potential hub genes
and lncRNAs associated with IA rupture-induced SAH, the lncRNA-mRNA
network involving lncRNA-mRNA pairs with r≥0.7 was constructed,
which consisted of 197 nodes (190 genes and seven lncRNAs) and 297
lncRNA-mRNA pairs (Fig. 3). Among
these pairs, MKNK1 was regulated by four lncRNAs, including
MRVI1-AS1, LINC01347, LINC00265 and CYP1B1-AS1. A total of 88
mRNAs, including CXCR1, neutrophil cytosolic factor (NCF)2 and
NCF4, were targeted by LINC00265 (Fig.
3).
KEGG pathway enrichment analysis for
DEGs in the lncRNA-mRNA regulatory network
Next, the KEGG pathways associated with all the 375
DEGs in the lncRNA-mRNA regulatory network were screened. DEGs were
observed to be enriched in six pathways, including ‘hsa04062:
Chemokine signaling pathway’ (CXCR1; CXCR2; VAV3; MAPK1 and
α-inhibiting activity polypeptide 3), ‘hsa04060: Cytokine-cytokine
receptor interaction’ (CXCR1; CXCR2 and interleukin-4 receptor
gene), ‘hsa04010: MAPK signaling pathway’ (MKNK1, MAPK1 and
MAPK14), ‘hsa04670: leukocyte transendothelial migration’ (MAPK14,
VAV3, NCF2 and NCF4), ‘hsa04620: TLR signaling pathway’ (MAPK1,
MAPK14, TLR2, TLR4 and TLR5) and ‘hsa04666: Fc gamma R-mediated
phagocytosis’ (MAPK1 and VAV3; Table
I).
 | Table I.KEGG pathways that are associated
with differentially expressed genes in the long non-coding RNA-mRNA
regulatory network. |
Table I.
KEGG pathways that are associated
with differentially expressed genes in the long non-coding RNA-mRNA
regulatory network.
| Term | Count | P-value | Gene names |
|---|
| hsa04062: Chemokine
signaling pathway | 14 |
1.35×10−3 | VAV3, GNAI3, LYN,
PREX1, HCK, RAF1, CXCR1, CXCR2, STAT3, MAPK1, ARRB2, GNB2, PTK2B,
GNG5 |
| hsa04060:
Cytokine-cytokine receptor interaction | 16 |
4.00×10−3 | IL18R1, IL1R2,
IL1R1, IL18RAP, CXCR1, TNFSF14, CXCR2, IL17RA, IFNAR1, INHBB,
ACVR1B, TNFRSF1A, TNFRSF10C, IL4R, CSF3R, CSF2RA |
| hsa04010: MAPK
signaling pathway | 15 |
1.14×10−2 | IL1R2, IL1R1,
MKNK1, RAF1, MAPKAPK2, ACVR1B, TNFRSF1A, MAP4K4, MAPK1, ARRB2,
MAP3K3, DUSP1, RPS6KA1, RASGRP4, MAPK14 |
| hsa04670: Leukocyte
transendothelial migration | 9 |
1.32×10−2 | GNAI3, VAV3, NCF2,
PTK2B, MAPK14, MMP9, NCF4, VASP, ITGAM |
| hsa04620: Toll-like
receptor signaling pathway | 8 |
1.79×10−2 | MAPK1, MYD88,
MAPK14, TLR2, FADD, TLR4, TLR5, IFNAR1 |
| hsa04666: Fc
γ-R-mediated phagocytosis | 7 |
4.09×10−2 | MAPK1, VAV3, LYN,
HCK, RAF1, FCGR2A, VASP |
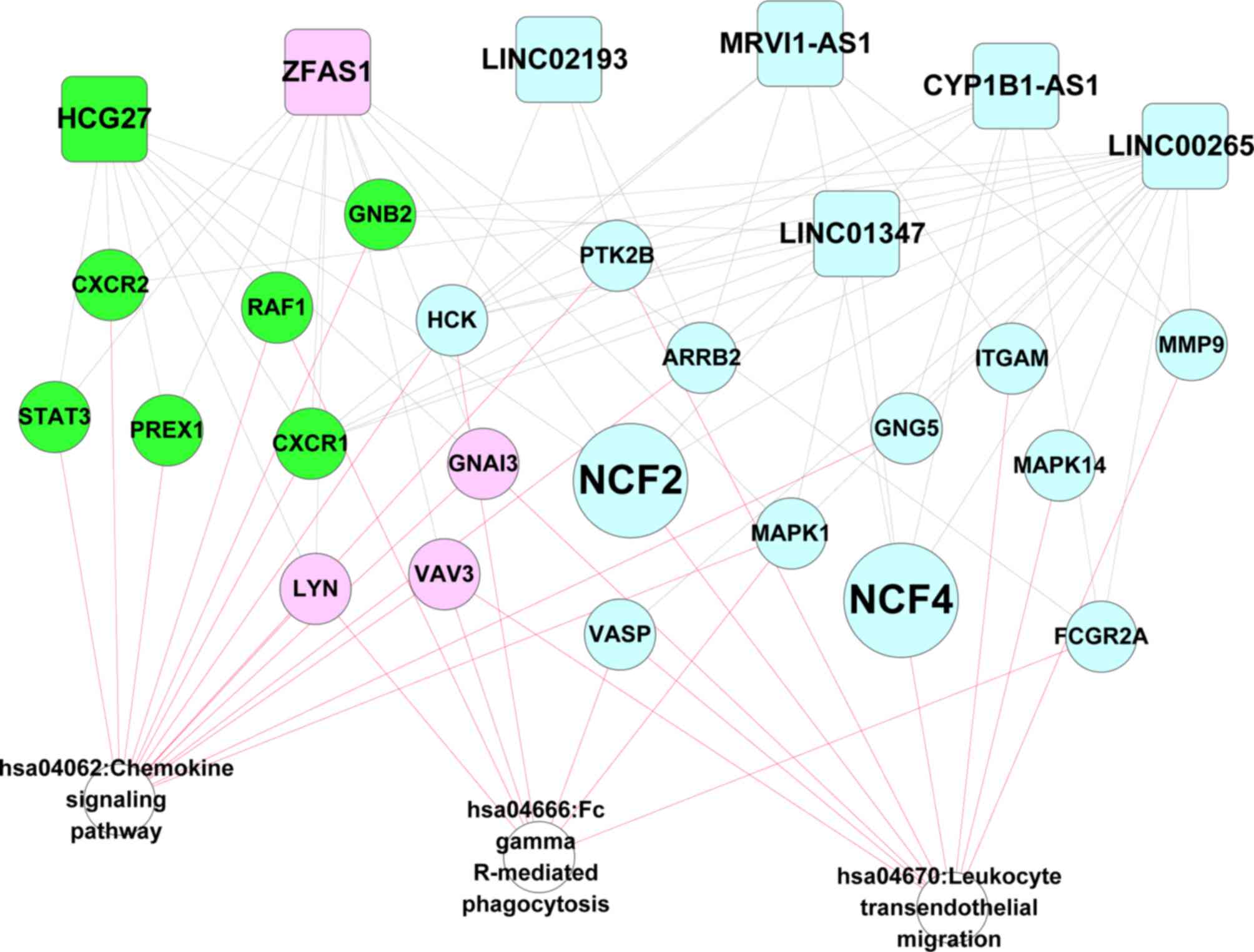
Identification of SAH-associated
biomarkers and KEGG pathways
The recognized genes and KEGG pathways associated
with IA rupture-induced SAH were screened out from CTD using the
search phrase ‘Intracranial Aneurysms’. One gene (NCF1) and seven
KEGG pathways (Table II) were
identified, including three common KEGG pathways between CTD and
Table I (hsa04062, hsa04670 and
hsa04666; DAVID enrichment analysis). Using the related DElncRNAs,
DEGs, and the three common KEGG pathways, an lncRNA-mRNA-pathway
network was constructed, which included all the seven DElncRNAs and
21 DEGs [including CXCR2, CXCR1, MAPK1, MAPK14, matrix
metalloproteinase 9 (MMP9), NCF2 and NCF4; Fig. 4].
 | Table II.Intracranial aneurysm
(MESH:D002532)-associated genes and KEGG pathways in the
Comparative Toxicogenomics Database. |
Table II.
Intracranial aneurysm
(MESH:D002532)-associated genes and KEGG pathways in the
Comparative Toxicogenomics Database.
| KEGG pathway | ID | Gene | Overlap |
|---|
| Chemokine signaling
pathway | hsa04062 | NCF1 | Yes |
| Phagosome | hsa04145 | NCF1 | No |
| Osteoclast
differentiation | hsa04380 | NCF1 | No |
| Fc gamma R-mediated
phagocytosis | hsa04666 | NCF1 | Yes |
| Leukocyte
transendothelial migration | hsa04670 | NCF1 | Yes |
| Leishmaniasis | hsa05140 | NCF1 | No |
| Fluid shear stress
and atherosclerosis | hsa05418 | NCF1 | No |
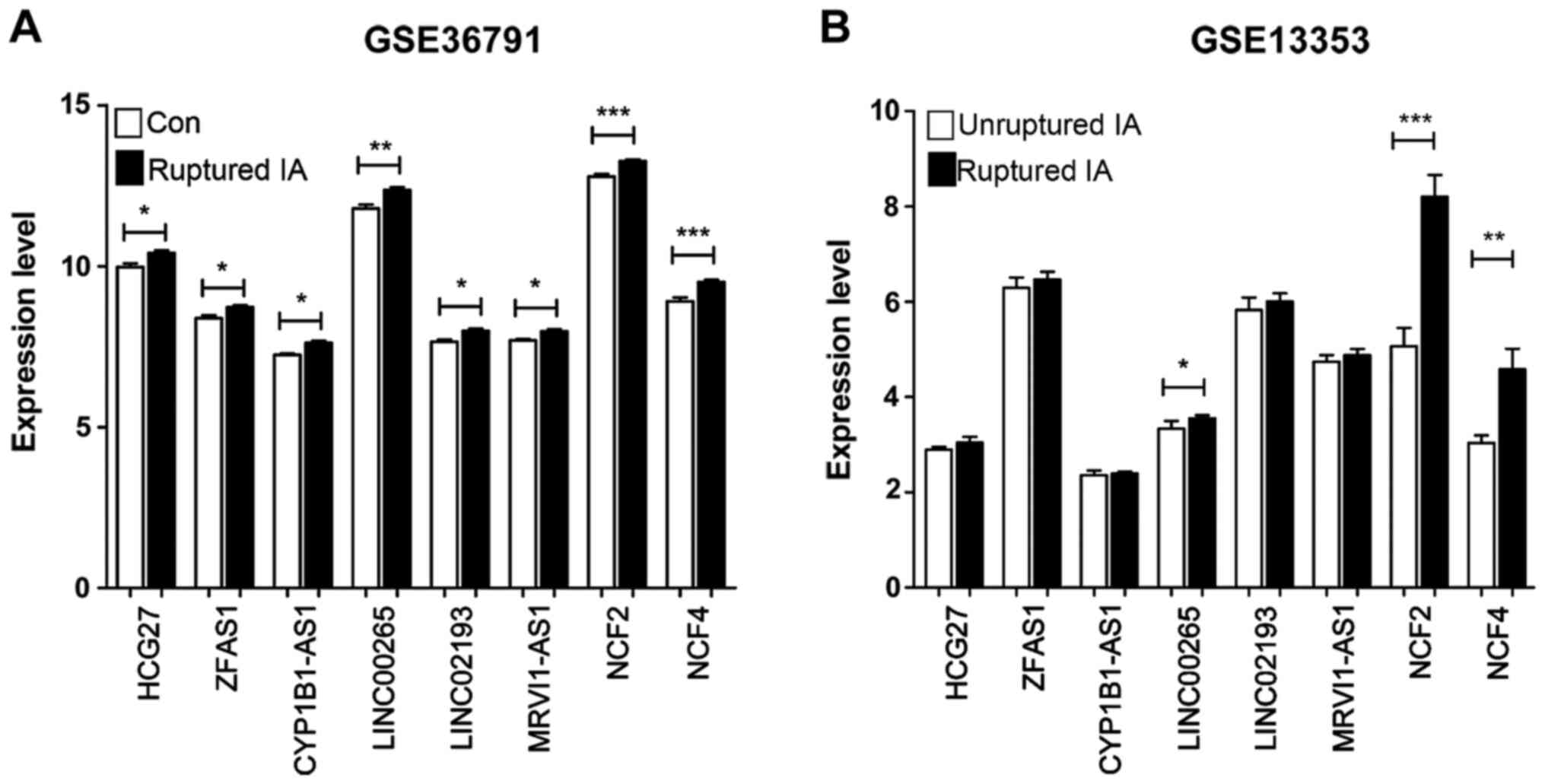
Expression profiles of SAH-associated
DElncRNAs and DEGs
Finally, the expression profiles of SAH-associated
DElncRNAs and DEGs in the GSE36791 dataset were extracted. The
GSE13353 dataset, including the gene profiles in ruptured (n=11)
and unruptured (n=8) saccular IA wall samples, was also downloaded.
The six DElncRNAs (all but LINC01347) and DEGs (including NCF2 and
NCF4) in the GSE36791 dataset were all identified in the GSE13353
dataset. The significant upregulation of these eight factors in the
ruptured IA blood samples compared with controls in the GSE36791
dataset is shown in Fig. 5A.
Significant upregulations of LINC00265, NCF2 and NCF4 were verified
in the ruptured IA wall samples compared with unruptured ones in
the GSE13353 dataset (Fig. 5B).
These findings suggested that LINC00265, NCF2 and NCF4 may serve
important roles in post-rupture processes.
Discussion
It has been reported that damage or injury to the
cerebral and central nervous system is enlarged and prolonged by
SAH-induced inflammation (10,28–30).
The inhibition of SAH-induced inflammation attenuates brain injury
(31). The present study
demonstrated that IA rupture-induced SAH was accompanied by changes
in numerous lncRNAs and genes, including HCG2, ZFAS1, LINC00265,
MRVI1-AS1, CYP1B1-AS1, LINC01347, LINC02193, CXCR1, CXCR2, TLR2,
TLR4, NCF2 and NCF4, as well as pathways, including ‘chemokine
signaling pathway’, ‘leukocyte transendothelial migration’ and ‘Fc
gamma R-mediated phagocytosis’, which suggested that there are
potential roles for these factors in the immune response to
SAH.
NCF2 and NCF4 were two upregulated SAH-associated
genes. Both NCF2 and NCF4 encode nicotinamide adenine dinucleotide
phosphate (NADPH) oxidases (32).
Mutations in NCF2, NCF4 and other genes encoding NAPDH oxidases are
associated with reduced levels of reactive oxygen species (ROS),
higher proportions of neutrophils and incremental perianal disease
rate in patients with Crohn's disease (32). Moreover, the NCF2 H389Q mutation
predisposes individuals to developing lupus (33), and the mutation in the binding site
of NCF4 to phosphatidylinositol 3-phosphate (R58A) promotes ROS
reduction and autoimmune arthritis in mice (34). However, NCF2 is induced by tumor
necrosis factor-α (TNF-α) (35)
and type II interferon-γ (IFN-γ), and is mapped to the IFN
signaling pathway (36). The
expression of NCF2 is regulated by the TF activator protein 1
(AP-1) by binding to the promoter of NCF2, which could be
upregulated by TNF-α (35,37–39).
These findings suggest that TNF-α may induce NCF2, which promotes
ROS production, activates MMPs (including MMP9 and MMP2) and the
p38 MAPK pathway, and enhances inflammation (40–42).
The present study identified the upregulation of NCF2 and NCF4, as
well as MMP9 and MAPK1, in patients with IA rupture-induced SAH
compared with controls and patients with unruptured IA. Thus, the
upregulation of NCF2 and NCF4 may indicate enhanced ROS production
and inflammatory status in patients with IA rupture-induced
SAH.
AP-1 is a switch for various signaling factors
(39), which includes NF-κB, TLR,
MAPK and JNK (43,44). Activation of ERK, JNK, NF-κB and
AP-1, as well as ROS production, in vascular smooth muscular cells
could be induced by the pro-inflammatory cytokine TNF-α (44), which has been shown to be elevated
in the cerebrospinal fluid of patients with SAH (45). The production of interleukin-6 and
TNF-α activates the NF-κB and MAPK pathways, which are associated
with an increased percentage of neuronal apoptosis (46–48).
It has been reported that suppression of TNF-α, as well as the
NF-κB and MAPK pathways, prevents inflammatory arthritis (49), asthma (50) and nervous system injury (46–48).
These observations suggest that the upregulation of NCF2, NCF4 and
MAPK in patients with IA rupture-induced SAH may indicate their
respective roles in SAH-induced inflammation and nervous system
injury.
The present study found that NCF2 and NCF4 were
regulated by ZFAS1, MRVI1-AS1, LINC00265, HCG27 and CYP1B1-AS1.
Among these lncRNAs, LINC00265 has been reported to promote
inflammation in abdominal aortic aneurysm (51). LINC00265 knockdown in abdominal
artery reduces the formation of aneurysm and inflammation (51). The upregulation of CYP1B1 during
inflammation has previously been observed in cancer cells (52), and this link is mediated by the
MAPK pathway (53). A study of
lncRNA microarray expression profiles in human umbilical vein
endothelial cells exposed to high atheroprotective shear stress
showed prominent upregulation of CYP1B1-AS1, which was found to
have associations with mitotic nuclear division and cell migration
(54). lncRNA HCG27 regulates
MAPK1 expression by sponging microRNA in gestational diabetes
(55). Moreover, ZFAS1 was
increased in patients with rheumatoid arthritis, and promoted
fibroblast-like synoviocyte migration and invasion (56). However, none of their roles have
been reported in inflammation. Their interactions with CXCR1 and
CXCR2, two inflammatory chemokine receptors (57–59),
indicate roles for these lncRNAs in inflammation processes.
Moreover, it has been reported that induction of CXCR1 and CXCR2
results in neuronal death and apoptosis (60,61),
whereas the reduction of CXCR1 and CXCR2 attenuates inflammation
and neuropathic pain (62,63). These reports suggest that these
lncRNAs may serve as therapeutic targets for inflammation post-IA
rupture-induced SAH.
In summary, the present study revealed potential
roles for lncRNAs and genes in SAH-induced inflammation. The
upregulation of lncRNAs (including HCG2, ZFAS1, LINC00265,
MRVI1-AS1, CYP1B1-AS1, LINC01347 and LINC02193) and genes
(including CXCR1, CXCR2, NCF2 and NCF4) in peripheral blood from
patients with IA rupture-induced SAH compared with the controls
suggested that they may have potential as biomarkers of SAH or
therapeutic targets for inhibiting SAH-induced nervous system
injury. However, this hypothesis should be validated using both
in vitro and in vivo experiments.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81873186).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The datasets (GSE36791 and
GSE13353) analyzed during the current study are available in the
the National Center for Biotechnology Information Gene Expression
Omnibus (https://www.ncbi.nlm.nih.gov/geo/).
Authors' contributions
The study was conceived and designed by XZ and LH.
Acquisition, analysis and interpretation of data were conducted by
YL, XL and ZC. LH drafted the manuscript. XL revised the manuscript
for important intellectual content. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Garg K, Sinha S, Kale SS, Chandra PS, Suri
A, Singh MM, Kumar R, Sharma MS, Pandey RM, Sharma BS and Mahapatra
AK: Role of simvastatin in prevention of vasospasm and improving
functional outcome after aneurysmal sub-arachnoid hemorrhage: A
prospective, randomized, double-blind, placebo-controlled pilot
trial. Br J Neurosurg. 27:181–186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wesali S, Persson HC, Cederin B and
Sunnerhagen KS: Improved survival after non-traumatic subarachnoid
haemorrhage with structured care pathways and modern intensive
care. Clin Neurol Neurosurg. 138:52–58. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Banki NM, Alexander K, Dae MW, Miss J,
Tung P, Lawton MT, Drew BJ, Foster E, Smith W, Parmley WW and
Zaroff JG: Acute neurocardiogenic injury after subarachnoid
hemorrhage. Circulation. 112:3314–3319. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnston SC, Selvin S and Gress DR: The
burden, trends, and demographics of mortality from subarachnoid
hemorrhage. Neurology. 50:1413–1418. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Suarez JI, Tarr RW and Selman WR:
Aneurysmal subarachnoid hemorrhage. N Engl J Med. 354:387–396.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zumofen DW, Roethlisberger M, Achermann R,
Bawarjan S, Stienen MN, Fung C, D'Alonzo D, Maldaner N, Ferrari A,
Corniola MV, et al: Factors associated with clinical and
radiological status on admission in patients with aneurysmal
subarachnoid hemorrhage. Neurosurg Rev. 41:1059–1069. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hendrix P, Foreman PM, Harrigan MR, Fisher
WS Rd, Vyas NA, Lipsky RH, Lin M, Walters BC, Tubbs RS, Shoja MM,
et al: The role of endothelial nitric oxide synthase-786 T/C
polymorphism in cardiac instability following aneurysmal
subarachnoid hemorrhage. Nitric Oxide. 71:52–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liang Z, Chi YJ, Lin GQ, Xiao LF, Su GL
and Yang LM: LncRNA MEG3 participates in neuronal cell injury
induced by subarachnoid hemorrhage via inhibiting the Pi3k/Akt
pathway. Eur Rev Med Pharmacol Sci. 22:2824–2831. 2018.PubMed/NCBI
|
|
9
|
Santiago-Sim T, Fang X, Hennessy ML,
Nalbach SV, DePalma SR, Lee MS, Greenway SC, McDonough B,
Hergenroeder GW, Patek KJ, et al: THSD1 (thrombospondin type 1
domain containing protein 1) mutation in the pathogenesis of
intracranial aneurysm and subarachnoid hemorrhage. Stroke.
47:3005–3013. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pawlowska E, Szczepanska J, Wisniewski K,
Tokarz P, Jaskólski DJ and Blasiak J: NF-κB-mediated inflammation
in the pathogenesis of intracranial aneurysm and subarachnoid
hemorrhage. Does autophagy play a role? Int J Mol Sci.
19:12452018.
|
|
11
|
Zhao L, Liu H, Yue L, Zhang J, Li X, Wang
B, Lin Y and Qu Y: Melatonin attenuates early brain injury via the
melatonin receptor/Sirt1/NF-κB signaling pathway following
subarachnoid hemorrhage in mice. Mol Neurobiol. 54:1612–1621. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma C, Zhou W, Yan Z, Qu M and Bu X:
Toll-like receptor 4 (TLR4) is associated with cerebral vasospasm
and delayed cerebral ischemia in aneurysmal subarachnoid
hemorrhage. Neurol Med Chir (Tokyo). 55:878–884. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Okada T and Suzuki H: Toll-like receptor 4
as a possible therapeutic target for delayed brain injuries after
aneurysmal subarachnoid hemorrhage. Neural Regen Res. 12:193–196.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pera J, Korostynski M, Golda S, Piechota
M, Dzbek J, Krzyszkowski T, Dziedzic T, Moskala M, Przewlocki R,
Szczudlik A and Slowik A: Gene expression profiling of blood in
ruptured intracranial aneurysms: In search of biomarkers. J Cereb
Blood Flow Metab. 33:1025–1031. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yates B, Braschi B, Gray KA, Seal RL,
Tweedie S and Bruford EA: Genenames.org: The HGNC and VGNC
resources in 2017. Nucleic Acids Res. 45(D1): D619–D625. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Szekely GJ and Rizzo ML: Hierarchical
clustering via joint between-within distances: Extending ward's
minimum variance method. J Classif. 22:151–183. 2005. View Article : Google Scholar
|
|
18
|
Wright RM, Aglyamova GV, Meyer E and Matz
MV: Variance stabilized gene expression data for WGCNA. Comput
Graph. 29:805–817. 2015.
|
|
19
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
The Gene Ontology Consortium: The gene
ontology resource: 20 Years and still GOing strong. Nucleic Acids
Res. 47(D1): D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: 2015
|
|
24
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davis AP, Grondin CJ, Johnson RJ, Sciaky
D, McMorran R, Wiegers J, Wiegers TC and Mattingly CJ: The
comparative toxicogenomics database: Update 2019. Nucleic Acids
Res. 47(D1): D948–D954. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kurki MI, Häkkinen SK, Frösen J, Tulamo R,
von und zu Fraunberg M, Wong G, Tromp G, Niemelä M, Hernesniemi J,
Jääskeläinen JE and Ylä-Herttuala S: Upregulated signaling pathways
in ruptured human saccular intracranial aneurysm wall: An emerging
regulative role of Toll-like receptor signaling and nuclear
factor-κB, hypoxia-inducible factor-1A, and ETS transcription
factors. Neurosurgery. 68:1667–1676. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guo J, Qi J, He K, Wu J, Bai S, Zhang T,
Zhao J and Wang Z: The Asian corn borer Ostrinia furnacalis feeding
increases the direct and indirect defense of mid-whorl stage
commercial maize in the field. Plant Biotechnol J. 17:88–102. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Diringer MN and Zazulia AR: Aneurysmal
subarachnoid hemorrhage: Strategies for preventing vasospasm in the
intensive care unit. Semin Respir Crit Care Med. 38:760–767. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Leclerc JL, Blackburn S, Neal D, Mendez
NV, Wharton JA, Waters MF and Doré S: Haptoglobin phenotype
predicts the development of focal and global cerebral vasospasm and
may influence outcomes after aneurysmal subarachnoid hemorrhage.
Proc Natl Acad Sci USA. 112:1155–1160. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li YC, Zhao L, Wu JP, Qu CX, Song QK and
Wang RB: Cytokine-induced killer cell infusion combined with
conventional treatments produced better prognosis for
hepatocellular carcinoma patients with barcelona clinic liver
cancer B or earlier stage: A systematic review and meta-analysis.
Cytotherapy. 18:1525–1531. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Denson LA, Jurickova I, Karns R, Shaw KA,
Cutler DJ, Okou DT, Dodd A, Quinn K, Mondal K, Aronow BJ, et al:
Clinical and genomic correlates of neutrophil reactive oxygen
species production in pediatric patients with Crohn's disease.
Gastroenterology. 154:2097–2110. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Armstrong DL, Miriam E, Raphael Z and
Jacob CO: Systemic lupus erythematosus-associated neutrophil
cytosolic factor 2 mutation affects the structure of NADPH oxidase
complex. J Biol Chem. 290:12595–12602. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Winter S, Hultqvist Hopkins M, Laulund F
and Holmdahl R: A reduction in intracellular reactive oxygen
species due to a mutation in NCF4 promotes autoimmune arthritis in
mice. Antioxid Redox Signal. 25:983–996. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gauss KA, Bunger PL, Larson TC, Young CJ,
Nelson-Overton LK, Siemsen DW and Quinn MT: Identification of a
novel tumor necrosis factor alpha-responsive region in the NCF2
promoter. J Leukoc Biol. 77:267–278. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cunninghame Graham DS, Morris DL, Bhangale
TR, Criswell LA, Syvänen AC, Rönnblom L, Behrens TW, Graham RR and
Vyse TJ: Association of NCF2, IKZF1, IRF8, IFIH1, and TYK2 with
systemic lupus erythematosus. PLoS Genet. 7:e10023412011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li SL, Valente AJ, Wang L, Gamez MJ and
Clark RA: Transcriptional regulation of the p67phox gene: Role of
AP-1 in concert with myeloid-specific transcription factors. J Biol
Chem. 276:39368–39378. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gupta JW, Kubin M, Hartman L, Cassatella M
and Trinchieri G: Induction of expression of genes encoding
components of the respiratory burst oxidase during differentiation
of human myeloid cell lines induced by tumor necrosis factor and
gamma-interferon. Cancer Res. 52:2530–2537. 1992.PubMed/NCBI
|
|
39
|
Wisdom R: AP-1: One switch for many
signals. Exp Cell Res. 253:180–185. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Cohen M, Meisser A, Haenggeli L and
Bischof P: Involvement of MAPK pathway in TNF-alpha-induced MMP-9
expression in human trophoblastic cells. Mol Hum Reprod.
12:225–232. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Robinson SC, Scott KA and Balkwill FR:
Chemokine stimulation of monocyte matrix metalloproteinase-9
requires endogenous TNF-alpha. Eur J Immunol. 32:404–412. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Soumyarani VS and Jayakumari N:
Oxidatively modified high density lipoprotein promotes inflammatory
response in human monocytes-macrophages by enhanced production of
ROS, TNF-α, MMP-9, and MMP-2. Mol Cell Biochem. 366:277–285. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen J, Yang X, Zhang W, Peng D, Xia Y, Lu
Y, Han X, Song G, Zhu J and Liu R: Therapeutic effects of
resveratrol in a mouse model of LPS and cigarette smoke-induced
COPD. Inflammation. 39:1949–1959. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Higgins KA, Park D, Lee GY, Curran WJ and
Deng X: Exercise-induced lung cancer regression: Mechanistic
findings from a mouse model. Cancer. 120:3302–3310. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li BS, Zuo QF, Zhao YL, Xiao B, Zhuang Y,
Mao XH, Wu C, Yang SM, Zeng H, Zou QM and Guo G: MicroRNA-25
promotes gastric cancer migration, invasion and proliferation by
directly targeting transducer of ERBB2, 1 and correlates with poor
survival. Oncogene. 34:2556–2565. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qian Y, Cao L, Guan T, Chen L, Xin H, Li
Y, Zheng R and Yu D: Protection by genistein on cortical neurons
against oxidative stress injury via inhibition of NF-kappaB, JNK
and ERK signaling pathway. Pharm Biol. 53:1124–1132. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wu JS, Tsai HD, Cheung WM, Hsu CY and Lin
TN: PPAR-γ ameliorates neuronal apoptosis and ischemic brain injury
via suppressing NF-κB-driven p22phox transcription. Mol Neurobiol.
53:3626–3645. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pan W, Lin L, Zhang N, Yuan F, Hua X, Wang
Y and Mo L: Neuroprotective effects of dexmedetomidine against
hypoxia-induced nervous system injury are related to inhibition of
NF-κB/COX-2 pathways. Cell Mol Neurobiol. 36:1179–1188. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Joosten LA, Helsen MM, Saxne T, van De Loo
FA, Heinegard D and van Den Berg WB: IL-1 alpha beta blockade
prevents cartilage and bone destruction in murine type II
collagen-induced arthritis, whereas TNF-alpha blockade only
ameliorates joint inflammation. J Immunol. 163:5049–5055.
1999.PubMed/NCBI
|
|
50
|
Charrad R, Berraïes A, Hamdi B, Ammar J,
Hamzaoui K and Hamzaoui A: Anti-inflammatory activity of IL-37 in
asthmatic children: Correlation with inflammatory cytokines TNF-α,
IL-β, IL-6 and IL-17A. Immunobiology. 221:182–187. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Su Y, Hu J, Lan H, Chen Y and Wei X: Long
non-coding RNA LINC00265 promotes inflammation via sponging
miR-let-7a in abdominal aortic aneurysm. Aging. 11:4463–4477. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Malaplate-Armand C, Ferrari L, Masson C,
Siest G and Batt AM: Astroglial CYP1B1 up-regulation in
inflammatory/oxidative toxic conditions: IL-1beta effect and
protection by N-acetylcysteine. Toxicol Lett. 138:243–251. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Smerdová L, Šmerdová J, Kabátková M,
Kohoutek J, Blažek D, Machala M and Vondráček J: Upregulation of
CYP1B1 expression by inflammatory cytokines is mediated by the p38
MAP kinase signal transduction pathway. Carcinogenesis.
35:2534–2543. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fan W, Fnag R, Feng M, Dai G and Wu G:
Microarray expression profile of long non-coding RNAs in human
endothelial cells exposed to atheroprotective shear stress. J Am
Coll Cardiol. 66:C67–C68. 2015. View Article : Google Scholar
|
|
55
|
Leng L, Zhang C, Ren L and Li Q:
Construction of a long non-coding RNA-mediated competitive
endogenous RNA network reveals global patterns and regulatory
markers in gestational diabetes. Int J Mol Med. 43:927–935.
2019.PubMed/NCBI
|
|
56
|
Ye Y, Gao X and Yang N: LncRNA ZFAS1
promotes cell migration and invasion of fibroblast-like
synoviocytes by suppression of miR-27a in rheumatoid arthritis.
Human Cell. 31:14–21. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Marcos V, Zhou Z, Yildirim AO, Bohla A,
Hector A, Vitkov L, Wiedenbauer EM, Krautgartner WD, Stoiber W,
Belohradsky BH, et al: CXCR2 mediates NADPH oxidase-independent
neutrophil extracellular trap formation in cystic fibrosis airway
inflammation. Nat Med. 16:1018–1023. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bertini R, Allegretti M, Bizzarri C,
Moriconi A, Locati M, Zampella G, Cervellera MN, Di Cioccio V,
Cesta MC, Galliera E, et al: Noncompetitive allosteric inhibitors
of the inflammatory chemokine receptors CXCR1 and CXCR2: Prevention
of reperfusion injury. Proc Natl Acad Sci USA. 101:11791–11796.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wuyts A, Van Osselaer N, Haelens A, Samson
I, Herdewijn P, Ben-Baruch A, Oppenheim JJ, Proost P and Van Damme
J: Characterization of synthetic human granulocyte chemotactic
protein 2: Usage of chemokine receptors CXCR1 and CXCR2 and in vivo
inflammatory properties. Biochemistry. 36:2716–2723. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
De Paola M, Buanne P, Biordi L, Bertini R,
Ghezzi P and Mennini T: Chemokine MIP-2/CXCL2, acting on CXCR2,
induces motor neuron death in primary cultures.
Neuroimmunomodulation. 14:310–316. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Du SH, Zhang W, Yue X, Luo XQ, Tan XH, Liu
C, Qiao DF and Wang H: Role of CXCR1 and interleukin-8 in
methamphetamine-induced neuronal apoptosis. Front Cell Neurosci.
12:2302018. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhou Y, Li RJ, Li M, Liu X, Zhu HY, Ju Z,
Miao X and Xu GY: Overexpression of GRK6 attenuates neuropathic
pain via suppression of CXCR2 in rat dorsal root ganglion. Mol
Pain. 12:17448069166463812016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lopes AH, Brandolini L, Aramini A,
Bianchini G, Silva RL, Zaperlon AC, Verri WA Jr, Alves-Filho JC,
Cunha FQ, Teixeira MM, et al: DF2755A, a novel non-competitive
allosteric inhibitor of CXCR1/2, reduces inflammatory and
post-operative pain. Pharmacol Res. 103:69–79. 2016. View Article : Google Scholar : PubMed/NCBI
|