Introduction
Systemic lupus erythematosus (SLE) is an autoimmune
disease characterized by the production of auto-antibodies, forming
immune complexes and potentially causing life-threatening renal,
cardiac or brain damage (1,2). Due to the heterogeneous clinical
manifestations and unpredictable disease course, accurate diagnosis
is important for correct treatment and good prognosis of patients
with SLE; however, the accurate diagnosis of SLE is difficult due
to this heterogeneity of clinical manifestations and the ambiguity
of the pathogenesis (3–6). Currently, there is a lack of sensitive
and specific diagnostic methods for SLE, but there have been an
increased number of studies investigating novel biomarkers for
improved SLE diagnosis (7–9).
Circular RNAs (circRNAs) are a type of closed
circular non-coding RNA (10,11).
Moreover, as circRNAs do not have 5′ or 3′ ends, they are resistant
to exonuclease-mediated degradation and are more stable compared
with most linear RNAs (12).
Recently, circRNAs have received increasing interest due to their
potential in regulating gene expression, mainly by acting as
‘microRNA (miRNA/miR) sponges’ to sequester target miRNAs (13–15).
Furthermore, aberrant expression of circRNAs has been revealed to
occur in numerous diseases, such as atherosclerotic vascular
disease, neurological disorders, prion diseases, cancer and
autoimmune diseases (16–19). While the ‘sponge’ function of
circRNAs has been the focus of research, several other circRNA
roles have also been studied.
Previous studies have reported that high levels of
circRNAs are widely distributed in the cytoplasm, nucleus and a
variety of body fluids, including saliva and blood (20,21),
and often demonstrate tissue and developmental stage-specific
expression (13,22,23).
Due to their high abundance, stability, tissue-specific expression
and easily availability, circRNAs possess the potential to serve as
biomarkers for diseases diagnosis. For example, Zhao et al
(24) revealed that
hsa_circ_0054633 in peripheral blood could be used as a new
biomarker for pre-diabetes and type 2 diabetes mellitus. Moreover,
Zhao et al (25) identified
that peripheral blood hsa_circ_0124644 can be used as a diagnostic
biomarker of coronary artery disease.
Our previous study showed that there are several
differentially expressed circRNAs in peripheral blood mononuclear
cells (PBMCs) between patients with SLE and healthy controls (HCs),
and that certain differentially expressed circRNAs may have roles
in the pathogenesis of SLE (26).
Therefore, the aim of the present study was to assess the potential
of circRNAs as biomarkers for SLE diagnosis in peripheral blood
samples, which is a sample that is relatively simple for collection
and preprocessing.
Materials and methods
Patients and ethics statement
Patients who fulfilled the revised American College
of Rheumatology criteria for SLE (27) were recruited from The First
Affiliated Hospital of Nanchang University between November 2016
and September 2017. Disease activity was assessed using the SLE
disease activity index (SLEDAI) (28). Patients with SLE were classified
into an inactive group (SLEDAI, 0–9) and an active group (SLEDAI,
≥10) according to the SLEDAI (28).
Healthy volunteers unrelated to the patients with SLE and who had
no inflammatory or autoimmune diseases were recruited as HCs. As an
autoimmune disease control, patients with rheumatoid arthritis (RA)
who fulfilled the revised American College of Rheumatology criteria
for RA (29) were enrolled from The
First Affiliated Hospital of Nanchang University between November
2016 and September 2017. The samples for this study were stored
(immediately after collection) in the Department of Clinical
Laboratory, The First Affiliated Hospital of Nanchang
University.
This study was approved by the Ethics Committee of
The First Affiliated Hospital of Nanchang University (approval no.
2014003) and was performed in compliance with the Helsinki
Declaration (30).
Samples collection and total RNA
extraction
Blood sample collection was performed as follows:
After overnight fasting, 2 ml blood was collected from the median
cubital vein of each subject and then stored in EDTA anticoagulant
vacutainers. Total RNA was extracted within 4 h using
TRIzol® (Invitrogen, Inc.; Thermo Fisher Scientific,
Inc.) reagent according to the manufacturer's instruction. The
concentration and quality of the RNA were assessed by absorbance
spectrometry measuring absorbance ratios of A260/A280 and
A260/A230, respectively, using a NanoDrop ND-1,000 spectrophotomete
(Agilent Technologies, Inc.).
Microarray analysis
Equal amounts of RNA from three patients with SLE
were collected in a SLE sample for microarray experiment. Equal
amounts of RNA from three HCs were also collected in a HC sample
for microarray experiment. Sample labeling and array hybridization
were performed according to the manufacturer's protocol (Arraystar,
Inc.). Total RNA was digested with RNase R (Epicentre, Inc.) to
remove linear RNAs and enrich circRNAs. The enriched circRNAs were
amplified and transcribed into fluorescent circRNAs using a random
priming method (Arraystar Super RNA Labeling kit; Arraystar, Inc.).
Then, the labeled circRNAs were hybridized to the Arraystar Human
circRNA Microarray (version 2.0; 8 × 15K; Arraystar, Inc). After
washing the slides, the arrays were scanned with an Agilent Scanner
G2505C (Agilent Technologies, Inc.). Then, Agilent Feature
Extraction software (version 11.0.1.1) (Agilent Technologies, Inc.)
(19) was used to analyze the
acquired array images. Quantile normalization and subsequent data
processing were performed using R software package (R version
3.1.2) (Agilent Technologies, Inc.) (19). Ggplot2 (version R-3.3.2; r-project.org/) was used to create a heat map. The
microarray work was performed by KangChen Bio-Tech (Shanghai,
China).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was reverse transcribed into cDNA using a
PrimeScript™ RT reagent kit (Takara Bio, Inc.). The RT reaction was
performed in a 10 µl reaction containing 5X PrimeScriptTM Buffer,
1.0 µl RT specific primer, 0.5 µl PrimeScriptTM RT Enzyme Mix and
5.0 µg total RNA. The RT assay was set at an initial denaturation
step at 37°C for 15 min, followed by 85°C for 5 sec. qPCR was then
performed on an ABI 7,500 RT PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.), using SYBR® Premix Ex Taq™ II
(Takara Bio, Inc.). The following PCR conditions were used: Initial
denaturation at 95°C for 5 min, followed by 40 cycles at 95°C for
15 sec and 60°C for 1 min, then a melt curve was detected to assess
the specificity of amplification and lack of primer dimers. The
primers used for RT-qPCR are presented in Table I. β-actin was used as an internal
control. After the reactions, the Cq values were determined using
the fixed threshold settings. The relative expression of circRNAs
was calculated using the 2−∆∆Cq method (31) normalized to endogenous control, with
∆Cq=Cqtarget-Cqreference.
 | Table I.Specific circRNA primers used for
reverse transcription-quantitative PCR. |
Table I.
Specific circRNA primers used for
reverse transcription-quantitative PCR.
| Name | Sequence |
|---|
| hsa_circ_
0008675 | Forward:
5′-GGAAGCCTTGCAGTTTGCTC-3′ |
|
| Reverse:
5′-AGCATTGGCTGGTGGGTTAT-3′ |
| hsa_circ_
0082689 | Forward:
5′-GTCCCCAAACACTCTTAGCCA-3′ |
|
| Reverse:
5′-CACACTCAGGTTGTGTTCGG-3′ |
| hsa_circ_
0082688 | Forward:
5′-TGCCGTATCGATGGCAATTC-3′ |
|
| Reverse:
5′-ATAGCTCAGGTGGTCAACGC-3′ |
| β-actin | Forward:
CATGTACGTTGCTATCCAGGC |
|
| Reverse:
CTCCTTAATGTCACGCACGAT |
Blood routine, serum inflammatory
indicators and autoantibodies determination
The concentrations of serum C3, C4 and C-reactive
protein (CRP) were determined by nephelometry methods, according to
the manufacturer's instructions (IMMUNE800; Beckman Coulter, Inc.).
Erythrocyte sedimentation rate (ESR) and blood routine were also
determined according to the manufacturer's instructions.
The antinuclear antibody was detected using an
indirect immunofluorescence method according to the manufacturer's
instructions (Euroimmun AG). Anti-dsDNA of IgG in serum was
determined by using both an indirect immunofluorescence method
(Euroimmun AG) and ELISA kits (cat. no. KX-E-DSD01096; Shanghai
Kexin Biotech Co., Ltd.). Anti-extractable nuclear antigens (ENAs)
antibodies including anti-sjögren syndrome A antigen antibody
(anti-SSA), anti-sjögren syndrome B antigen antibody (anti-SSB),
anti-Ro52, anti-Smith antibody (anti-Sm), anti-nuclear
ribonucleoprotein/Smith antibody (anti-nRNP/Sm), anti-ribosomal
protein P (anti-RIB-P), anti-histone and anti-nucleosome antibody
were determined using line immunoassays kits (cat. no. DL
1590-6401-3G; Euroimmun AG) according to the manufacturer's
instructions. The results of anti-ENAs detection were presented as
negative (−) or positive (+, ++, +++) using EuroBlot One (Euroimmun
AG).
Statistical analysis
Statistical analysis and graphic presentation were
conducted with GraphPad Prism version 5.0 (GraphPad Software, Inc.)
and SPSS version 16.0 (IBM Corp.). Student's t-test was used with
normalized data, while the non-parametric Mann-Whitney test was
used to analyze data that did not pass the normality test. A
Kruskal-Wallis test was used in comparisons between three groups
and Dunns post hoc test was used following the Kruskal-Wallis test.
Moreover, the Pearson's method or the non-parametric Spearman's
method were used for correlation analysis. The cut-off values were
determined by receiver operating characteristic (ROC) curves
analysis using GraphPad Prism version 5.0; ROC curves were
performed to evaluate the diagnostic value of circRNAs that were
dysregulated in the peripheral blood of patients with SLE compared
to HCs. A parallel model was used to evaluate the diagnostic
efficiency of the hsa_circ_0082688-hsa_circ_0082689 combination
model (32); if one of the multiple
indicators is positive, the disease can be diagnosed. P<0.05 was
considered to indicate a statistically significant difference.
Results
Characteristics of the study
population
A total of 185 participants were enrolled in the
present study, including 76 patients with SLE, 76 HCs and 33
patients with RA. Among the patients with SLE, 13 were newly
diagnosed patients with no history of corticosteroids or
immunosuppressive drugs use before registration. The demographic
characteristics of the study population are presented in Tables II and III.
 | Table II.Demographic characteristics of the
study population. |
Table II.
Demographic characteristics of the
study population.
| Study set | Categories | SLE | HC | RA |
|---|
| Discovery set |
| 3 | 3 |
|
|
| Females | 3 (100.00) | 3 (100.00) |
|
|
| Age, years | 29.00±15.40 | 34.00±7.21 |
|
|
| SLEDAI score |
18.33±3.79a | – |
|
| Training set |
| 50 | 50 |
|
|
| Females | 46 (92.00) | 43 (86.00) |
|
|
| Age, years | 42.70±14.80 | 44.77±12.90 |
|
|
| SLEDAI score | 7.69±4.60 | – |
|
| Blind testing
set |
| 23 | 23 | 33 |
|
| Females | 21 (91.30) | 18 (78.26) | 24 (72.73) |
|
| Age, years | 35.70±12.52 | 41.30±12.70 |
58.00±10.15b |
|
| SLEDAI score | 5.90±3.94 | – | – |
 | Table III.Clinical characteristics of patients
with SLE. |
Table III.
Clinical characteristics of patients
with SLE.
| Categories | Patients with SLE
(n=76) |
|---|
| Females | 70 (90.91) |
| Age, years | 40.00±14.61 |
| SLEDAI score (67
patients) | 7.61±4.98 |
| ds-DNA, IU/ml (71
patients) | 243.80±407.82 |
| Anti-ENA (68
patients) |
|
|
Anti-Sm | 21 (30.88) |
|
Anti-Ro52 | 45 (66.18) |
|
Anti-nRNP/Sm | 33 (48.53) |
|
Anti-RIB-P | 26 (38.24) |
|
Anti-nucleosome | 22 (32.35) |
|
Anti-SSA | 44 (64.71) |
|
Anti-SSB | 11 (16.18) |
|
Anti-histone | 22 (32.35) |
| C3, g/l (71
patients) | 0.62±0.20 |
| C4, g/l (71
patients) | 0.14±0.06 |
| IgG, g/l (68
patients) | 13.65±5.29 |
| ESR, mm/h (65
patients) | 35.49±34.80 |
| CRP, mg/l (71
patients) | 12.31±23.30 |
| WBC,
109/l | 6.78±3.61 |
| RBC,
1012/l | 3.96±0.82 |
| HGB, g/l | 113.12±25.61 |
| HCT, l/l) | 0.34±0.07 |
| PLT,
109/l | 214.77±85.97 |
| Lymphocytes,
109/l | 1.56±1.10 |
| Monocytes,
109/l | 0.47±0.27 |
| Neutrophils,
109/l | 4.60±2.94 |
| Clinical
features |
|
| Fever
(74 patients) | 7 (9.50) |
|
Cutaneous manifestations (74
patients) | 12 (16.22) |
|
Arthritis (74 patients) | 13 (17.57) |
|
Effusion (74 patients) | 11 (14.86) |
|
Hematuresis (68 patients) | 14 (20.59) |
| Pyuria
(68 patients) | 10 (14.71) |
|
Proteinuria (68 patients) | 25 (36.76) |
In the discovery set, there were three patients with
newly diagnosed SLE and three age-matched and sex-matched HCs. An
additional 50 patients with SLE and 50 HCs were included in the
training set for the validation of differentially expressed
circRNAs and diagnostic model construction. In this patient set,
ten patients had newly diagnosed SLE with no history of
corticosteroids or immunosuppressive drugs use before registration.
Moreover, the other patients with SLE were recurrent patients and
had received therapy with corticosteroids or immunosuppressive
drugs for ≥1 month before registration. An independent cohort
consisting of 23 patients with SLE, 33 patients with RA and 23 HCs
were enrolled in the blind testing set for clinical evaluation of
circRNAs in SLE diagnosis. It was demonstrated that there were no
significant differences in age and sex between the SLE and HCs
groups. Moreover, there were no significant differences in sex
between the RA, SLE and HCs groups. Due to the difference in the
ages of high incidence occurrence between RA and SLE (the incidence
of RA is high in individuals who are 50–60 years, while the
incidence of SLE is high in 20–40 years), patients with RA and SLE
were not age-matched in the present study.
CircRNA expression profiling in
peripheral blood from patients with SLE
The overall distribution of microarray data of these
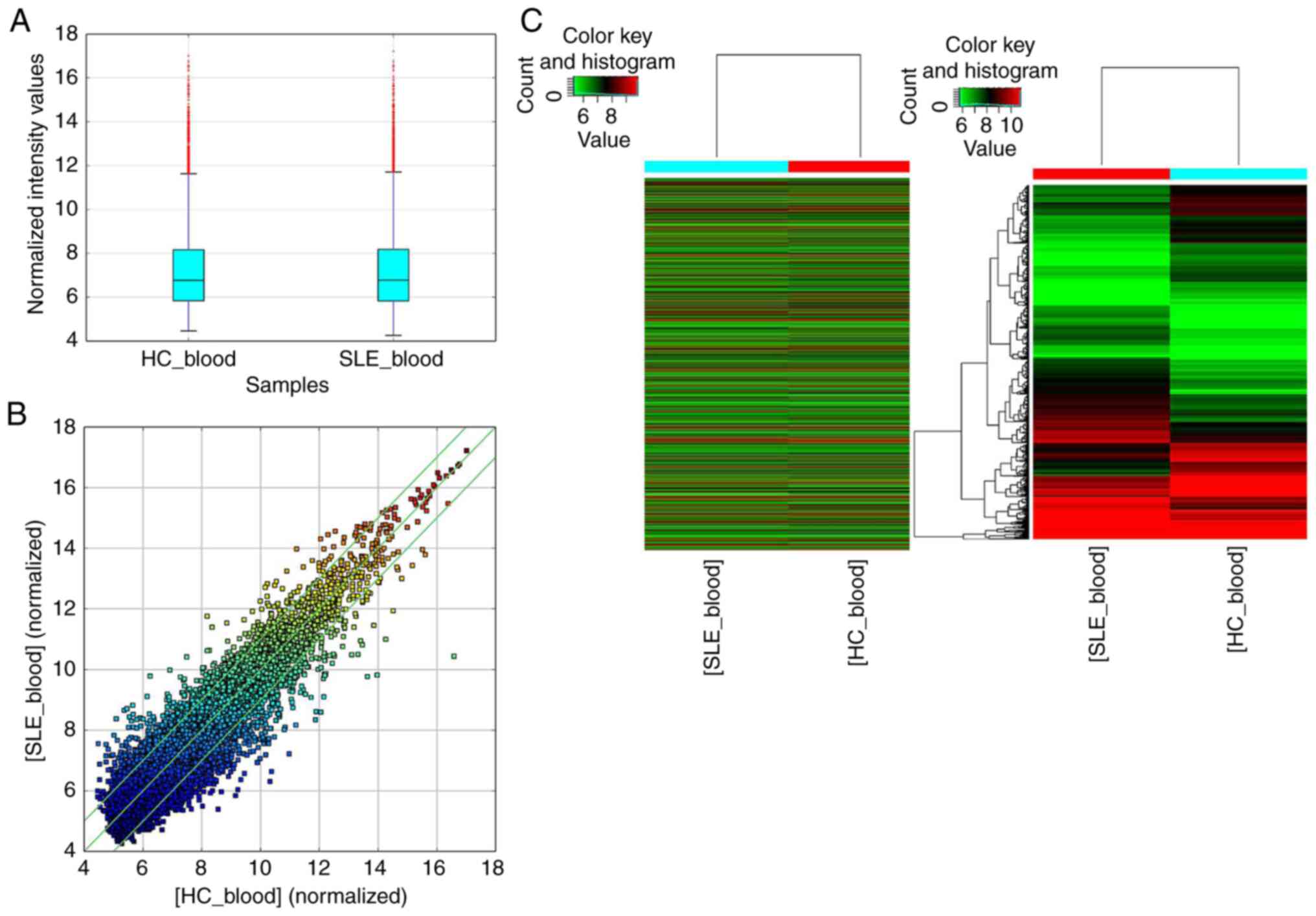
two groups is presented as a box plot (Fig. 1A) and scatter plot (Fig. 1B), and significant differences in
the expression levels of circRNAs between patients with SLE and HCs
were screened with >2.0 fold change and P<0.05. The results
indicated that, compared with the HC group, 753 circRNAs were
significantly upregulated, while 813 circRNAs were significantly
downregulated in the SLE group. Furthermore, a heat map was created
to group the circRNAs based on their expression levels among the
samples (Fig. 1C).
Validation of circRNA expression in
the training set
As the main objective of the study was to identify
diagnostic markers of SLE in peripheral blood, the focus was on the
upregulated circRNAs in patients with SLE. To assess the microarray
data, three circRNAs (hsa_circ_0082688, hsa_circ_0082689 and
hsa_circ_0008675) that were not only listed in the top 50 most
significant upregulated circRNAs in patients with SLE in this
circRNA microarray, but also identified to be upregulated in PBMCs
from patients with SLE in our previous study (26), were selected for validation by
RT-qPCR in the training set, which included 50 patients with SLE
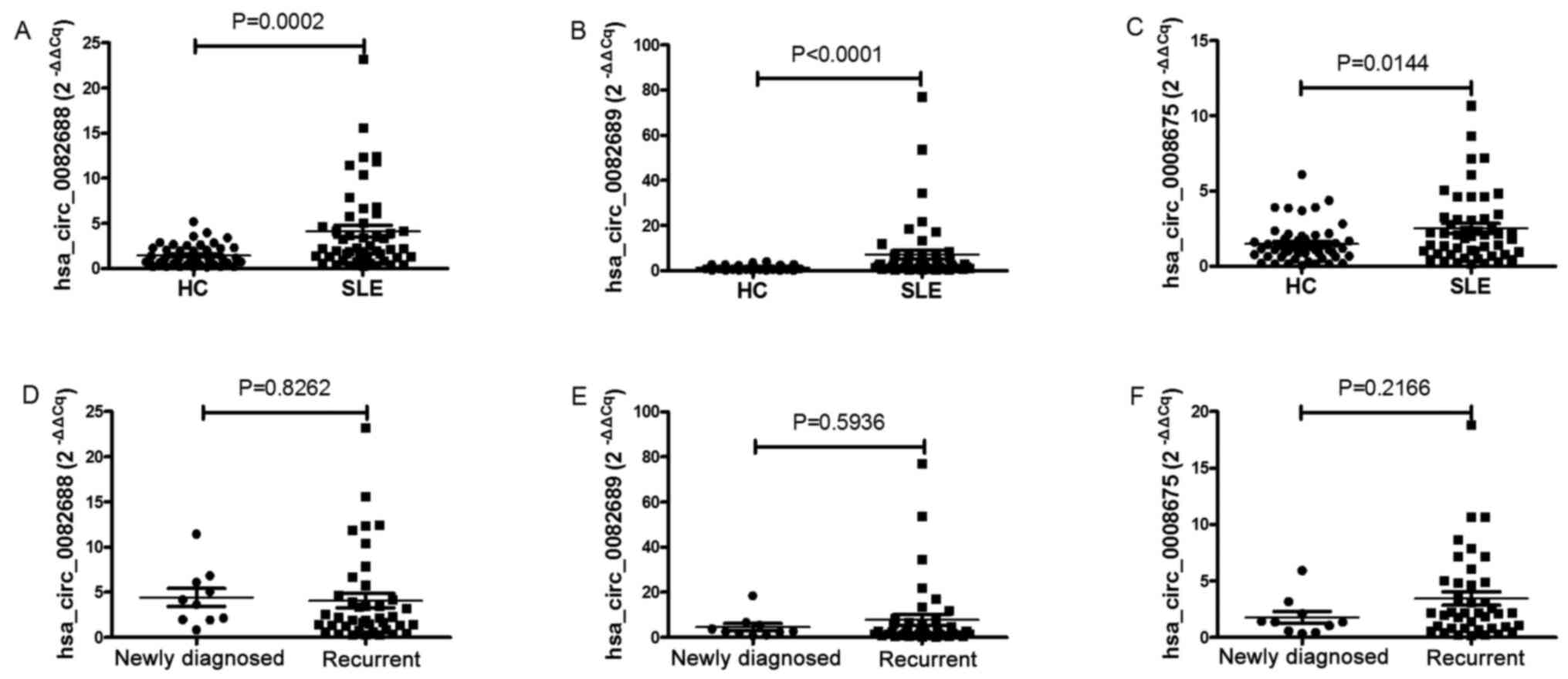
and 50 HC. The results of RT-qPCR were consistent with the circRNA
microarray data, in that all three circRNAs were significantly
upregulated in the SLE group (Fig.
2). Moreover, the expression levels of circRNAs between
patients with newly diagnosed SLE or recurrent SLE patients were
also compared, but no significant difference was demonstrated
(Fig. 2). In addition, there was no
correlation between circRNAs expression levels and age or sex in
the SLE, RA and HC groups (data not shown).
ROC curve analysis of identified
peripheral blood circRNAs among patients with SLE
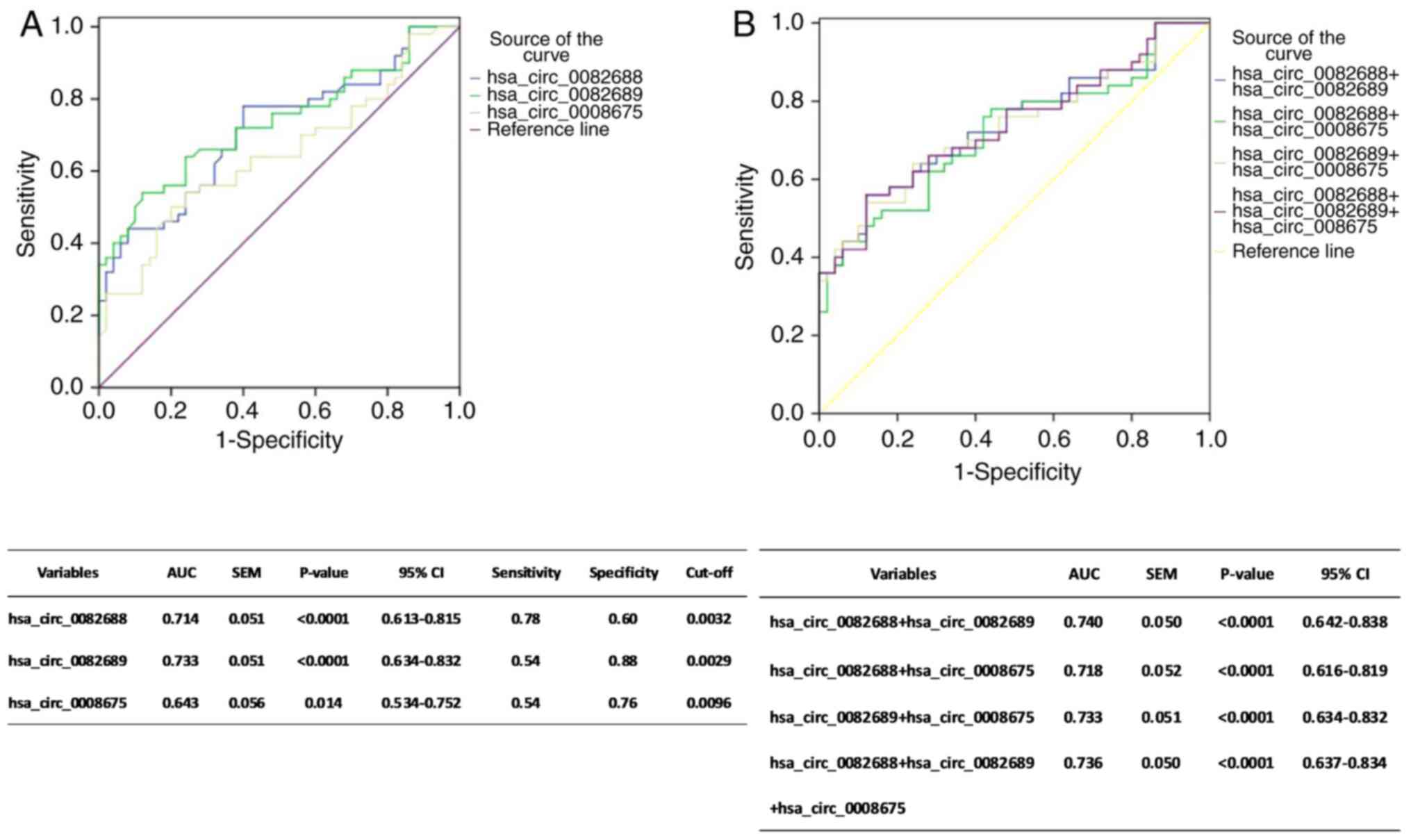
To further evaluate the potential value of the three
circRNAs (hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675)
in SLE diagnosis, ROC curve analysis was performed. The area under
the curve (AUC) values demonstrated that the levels of
hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675 in
peripheral blood could separate the patients with SLE from the HCs.
Moreover, the highest AUC was identified for hsa_circ_0082689 (AUC:
0.733, 95% CI, 0.634–0.832, P<0.0001, Sensitivity: 54.0%,
Specificity: 88.0%, Cut-off: 0.0029), followed by hsa_circ_0082688
(AUC: 0.714, 95% CI, 0.613–0.815, P<0.0001, Sensitivity: 78.0%,
Specificity: 60.0%, Cut-off: 0.0032) and hsa_circ_0008675 (AUC:
0.643, 95% CI, 0.534–0.752, P=0.0140, Sensitivity: 54.0%,
Specificity: 76.0%, Cut-off: 0.0096; Fig. 3A).
To evaluate the cumulative performances of the three
circRNAs in discriminating SLE from HC, a binary logistic
regression was performed. The logistic regression model indicated
that the combination of hsa_circ_0082688 and hsa_circ_0082689 could
provide the best diagnostic accuracy, with an AUC of 0.740 (95% CI,
0.642–0.838, P<0.0001; Fig. 3B).
Furthermore, the combination of all these three circRNAs (Fig. 3B) and any two circRNAs (data no
shown) had no improvement in SLE diagnosis, compared with the
aforementioned combination of the two circRNAs.
The diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 combination model was then
evaluated using the paralleling model, according to their optimal
cutoff value (hsa_circ_0082688: 0.0032, hsa_circ_0082689: 0.0029).
As shown in Table IV, it was
demonstrated that the combination model of
hsa_circ_0082688-hsa_circ_0082689 could effectively discriminate
patients with SLE from HCs, with a sensitivity of 86.00% (43/50), a
specificity of 88.00% (44/50) and an accuracy of 87.0%
(87/100).
 | Table IV.Diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 combination model by the parallel
model in the training set. |
Table IV.
Diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 combination model by the parallel
model in the training set.
| Category | hsa_circ_0082688
> 0.0032 or hsa_circ_0082689 > 0.0029 | hsa_circ_0082688
< 0.0032 or hsa_circ_0082689 < 0.0029 | Sensitivity | Specificity | Accuracy |
|---|
| SLE (50) | 43 | 7 | 86.00% (43/50) | 88.00% (44/50) | 87.0% (87/100) |
| HC (50) | 6 | 44 |
|
|
|
Blind test of the diagnostic value of
differentially expressed circRNAs
To further evaluate the value of
hsa_circ_0082688-hsa_circ_0082689 combination model in SLE
diagnosis, an independent cohort consisting of 23 patients with
SLE, 33 patients with RA and 23 HCs were enrolled and their
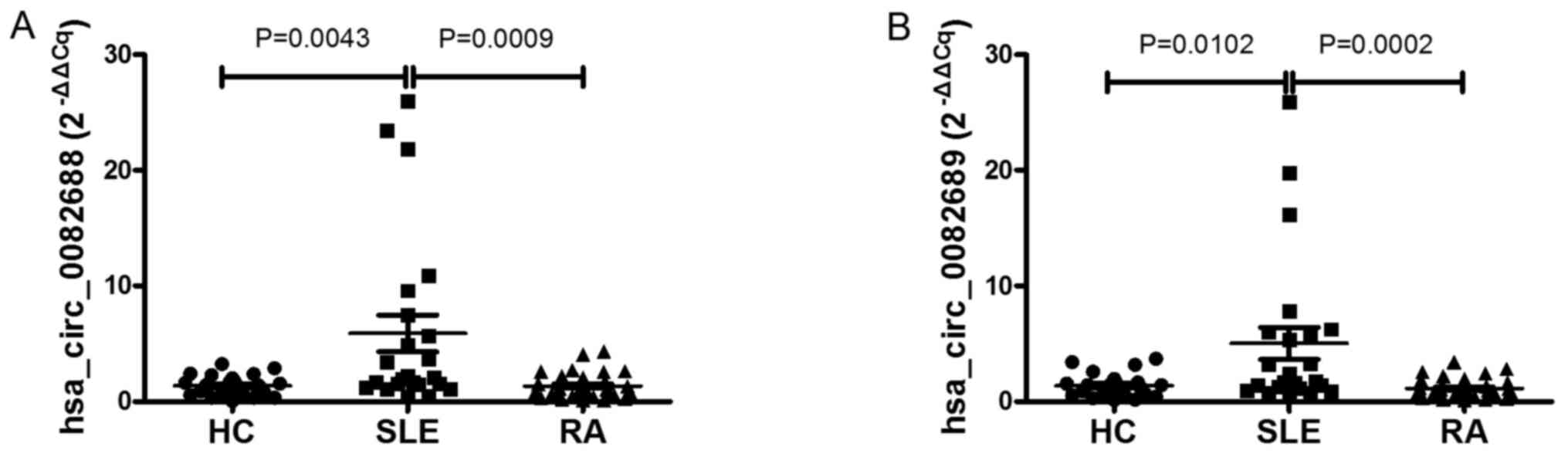
circRNAs expression levels were measured. Similar to the training
set, it was identified that the expression levels of
hsa_circ_0082688 and hsa_circ_0082689 were increased significantly
in patients with SLE compared with RA and HC groups (all P<0.05;
Fig. 4). According to the optimal
cutoff value found in the training stage, As shown in Table V, the combination model of
hsa_circ_0082688-hsa_circ_0082689 could effectively discriminate
between the SLE group and the other two control groups, with a
sensitivity of 91.30% (21/23), a specificity of 78.57% (44/56) and
an accuracy of 82.28% (65/79). Furthermore, this diagnostic model
presented a sensitivity of 91.30% (21/23), a specificity of 78.79%
(26/33) and an accuracy of 83.93% (47/56) in discriminating the SLE
group from the RA group, and a sensitivity of 91.30% (21/23), a
specificity of 78.26% (18/23) and an accuracy of 84.78% (39/46) in
discriminating the SLE group from the HC group.
 | Table V.Diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 combination model by the parallel
model in blind testing set. |
Table V.
Diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 combination model by the parallel
model in blind testing set.
| Category | hsa_circ_0082688
> 0.0032 or hsa_circ_0082689 > 0.0029 | hsa_circ_0082688
< 0.0032 or hsa_circ_0082689 < 0.0029 | Sensitivity | Specificity | Accuracy |
|---|
| SLE vs. RA +
HC |
| SLE (23) | 21 | 2 | 91.30% (21/23) | 78.57% (44/56) | 82.28% (65/79) |
| RA + HC (56) | 12 | 44 |
|
|
|
| SLE vs. RA |
| SLE (23) | 21 | 2 | 91.30% (21/23) | 78.79% (26/33) | 83.93% (47/56) |
| RA (33) | 7 | 26 |
|
|
|
| SLE vs. HC |
| SLE (23) | 21 | 2 | 91.30% (21/23) | 78.26% (18/23) | 84.78% (39/46) |
| HC (23) | 5 | 18 |
|
|
|
Anti-dsDNA is a traditional and most commonly used
diagnostic marker for SLE (33).
The aforementioned results demonstrated that the
hsa_circ_0082688-hsa_circ_0082689 combination model may be used as
a novel biomarker for the diagnosis of SLE. Thus, the present study
evaluated the value of hsa_circ_0082688-hsa_circ_0082689 +
anti-dsDNA combination model in SLE diagnosis. According to the
optimal cutoff value of hsa_circ_0082688-hsa_circ_0082689
identified in the aforementioned results
(hsa_circ_0082688>0.0032, hsa_circ_0082689>0.0029), As shown
in Table VI, the
hsa_circ_0082688-hsa_circ_0082689 + anti-dsDNA combination model
could effectively discriminated the SLE group from the control
groups (RA + HC), with a sensitivity of 95.65% (22/23), a
specificity of 100.00% (56/56) and an accuracy of 98.73% (78/79) in
distinguishing the patients with SLE from both control groups.
Moreover, the sensitivity, specificity and accuracy of
hsa_circ_0082688-hsa_circ_0082689 + anti-dsDNA combination model
were increased compared with hsa_circ_0082688-hsa_circ_0082689
[sensitivity=91.30% (21/23), specificity=78.57% (44/56),
accuracy=82.28% (65/79)] (Table V)
and anti-dsDNA [sensitivity=47.83% (11/23), specificity=100.00%
(56/56), accuracy=79.85% (67/79)] (Table VI).
 | Table VI.Diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 + anti-dsDNA combination model by
the parallel model in blind testing set. |
Table VI.
Diagnostic efficiency of the
hsa_circ_0082688-hsa_circ_0082689 + anti-dsDNA combination model by
the parallel model in blind testing set.
| Category | Positive | Negative | Sensitivity | Specificity | Accuracy |
|---|
| hsa_circ_0082688
> 0.0032 or hsa_circ_0082689 > 0.0029 or
anti-dsDNA>100 |
| SLE (23) | 22 | 1 | 95.65% (22/23) | 100.00%
(56/56) | 98.73% (78/79) |
| RA + HC (56) | 0 | 56 |
|
|
|
|
anti-dsDNA>100 |
| SLE (23) | 11 | 12 | 47.83% (11/23) | 100.00%
(56/56) | 79.85% (67/79) |
| RA + HC (56) | 0 | 56 |
|
|
|
Association of hsa_circ_0082688,
hsa_circ_0082689 and hsa_circ_0008675 expression levels in
peripheral blood with SLE clinical characteristics
To determine whether the aforementioned
differentially expressed circRNAs in the peripheral blood could
serve as relevant biomarkers for the severity of SLE, the clinical
indicators related to inflammation were collected and the SLEDAIs
of all patients with SLE were calculated (Table III), and the correlations between
this dataset and the expression levels of the specific
differentially expressed circRNAs were analyzed. The results
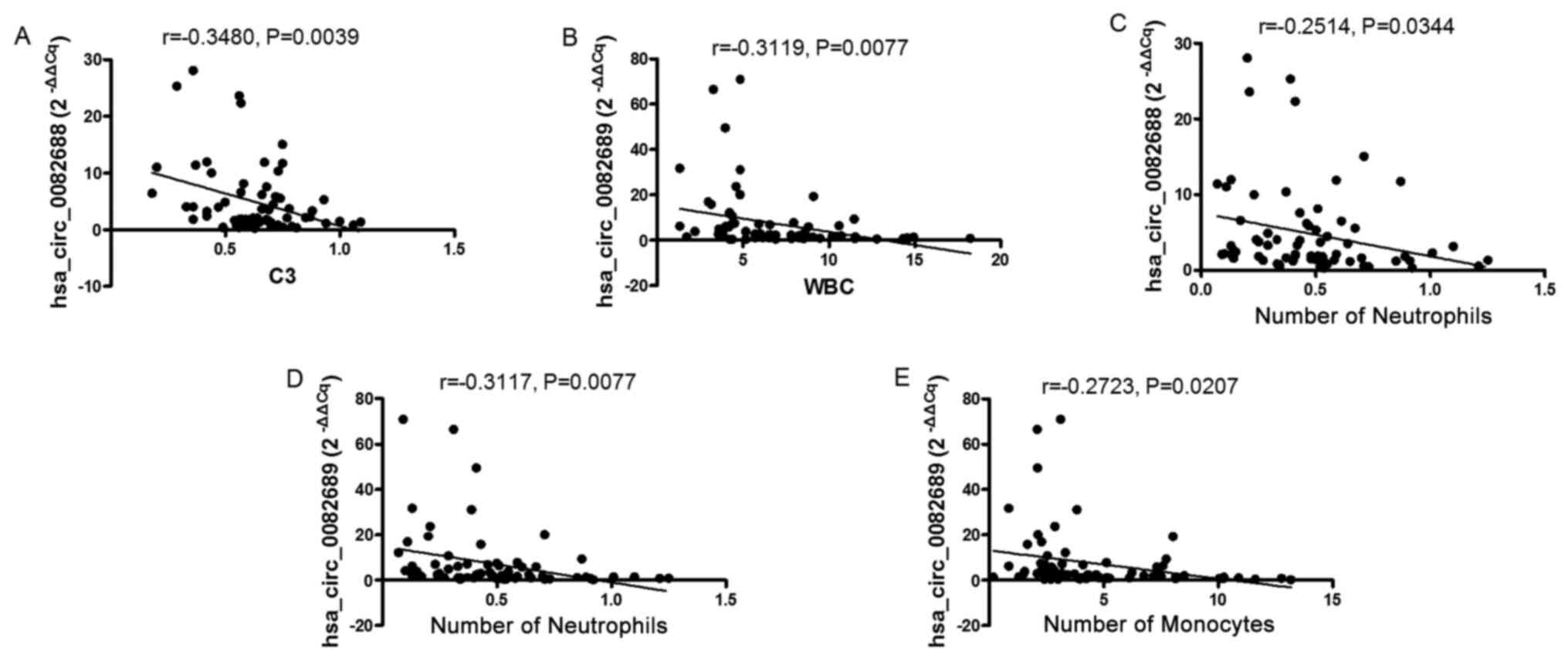
indicated that, while hsa_circ_0082688 expression was negatively
associated with C3 in patients with SLE (r=−0.3480, P=0.0039;
Fig. 5A), there was no correlation
between the expression levels of the other confirmed circRNAs and
C3 (data not shown). Furthermore, the expression levels of all
identified circRNAs in the peripheral blood from patients with SLE
did not correlate with SLEDAI, CRP, ESR or C4, which also reflect
the severity of the disease (data not shown).
SLE is a complex autoimmune disease characterized by
multiple organ system damages, such as the hematological system and
skin. In the present study, the correlations between SLE-related
clinical features and the expression levels of circRNAs were
analyzed. It was demonstrated that the expression of
hsa_circ_0082689 was negatively associated with white blood cell
(WBC) number (r=−0.3119, P=0.0077; Fig.
5B), monocyte number (r=−0.3117, P=0.0077; Fig. 5D) and neutrophil number in patients
with SLE (r=−0.2723, P=0.0207; Fig.
5E). In addition, the expression of hsa_circ_0082688 was
negatively associated with monocytes number in patients with SLE
(r=−0.2514, P=0.0344; Fig. 5C).
However, no significant difference was identified between the
expression levels of circRNAs and other clinical features.
Production of multiple auto-antibodies, such as
anti-dsDNA and anti-ENAs, is an important characteristic of SLE.
Thus, the present study investigated the correlation between the
expression levels of circRNAs and auto-antibodies in patients with
SLE, but no significant difference was found (data not shown).
However, all identified circRNAs did correlate with each other. For
example, the expression of hsa_circ_0082688 correlated with the
expression of hsa_circ_0082689 (r=0.5967; P<0.0001; data not
shown).
Discussion
Previous studies have reported the feasibility of
using circulating miRNAs and long non-coding (lnc) RNA as potential
biomarkers of SLE (9,34,35).
Similar to miRNA and lncRNA, the potential of circulating circRNAs
as powerful and non-invasive biomarkers in a number of diseases,
including cancer (36), RA
(19) and cardiovascular diseases
(25), has been revealed. Compared
with miRNAs and lncRNAs, circRNAs are more stable in mammalian
cells (37) and their expression
levels can be ≥10-fold compared with those of their linear isomers
(12). These properties indicate
that the potential of circRNAs to be ideal biomarkers for human
diseases (21). Moreover, the
diagnostic performance of plasma or PBMC circRNA has been examined
in SLE (38,39). However, to the best of our
knowledge, the expression profiles and diagnostic performance of
circulating circRNAs in peripheral blood from patients with SLE
have been rarely reported, and there was previously only one paper
investigating circRNAs expression in peripheral blood from patients
with SLE (40). In this study, Li
et al (40) assessed the
microarray profile of circRNAs in peripheral blood to identify the
changes in the expression of circRNAs between pediatric patients
with SLE and healthy children, and revealed that the expression
levels of hsa_circ_0057762 and hsa_circ_0003090 could differentiate
the pediatric patients with SLE from the healthy children. To the
best of our knowledge, the present study was the first to performed
a microarray analysis to investigate the changes in expression of
circRNAs in peripheral blood from adult patients with SLE, by
comparing with those in adult HCs. Microarray data identified a
total of 1,566 circRNAs (753 were upregulated) that were
significantly dysregulated in patients with SLE compared with HCs.
Thus, it was speculated that circRNA may be a novel biomarkers for
SLE diagnosis or disease process monitoring in adult patients.
However, possibly due to age differences, the expression levels of
hsa_circ_0057762 and hsa_circ_0003090 in peripheral blood were not
significantly different between adult patients with SLE and
HCs.
To determine whether differentially expressed
circRNAs can be diagnostic biomarkers for SLE, three circRNAs
hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675, which were
listed in the top 50 most significant upregulated circRNAs in
peripheral blood of patients with SLE in this circRNA microarray,
and also identified to be upregulated in PBMCs from patients with
SLE in our previous study (26),
were chosen for validation by RT-qPCR in a training set. The
results suggested that the expression levels of all three circRNAs
increased significantly in patients with SLE. Furthermore, ROC
curve analysis demonstrated that these circRNAs had the potential
to distinguish between SLE and HCs. ROC curve analysis also
indicated that the combination of hsa_circ_0082688 and
hsa_circ_0082689 could provide the best diagnostic accuracy, with
an AUC of 0.740. Moreover, the results from the further blind
testing set suggested its good performance, not only in
distinguishing between SLE and HCs groups, but also in
distinguishing between patients with SLE and those with RA.
Therefore, the present results indicated that this diagnostic model
may be promising for SLE diagnosis.
In addition, the present results demonstrated that
the combination model of hsa_circ_0082688-hsa_circ_0082689 +
anti-dsDNA could more effectively discriminated the SLE group from
the control groups (RA + HC), with a sensitivity of 95.65% (22/23),
a specificity of 100.00% (56/56) and an accuracy of 98.73% (78/79),
which were superior to hsa_circ_0082688-hsa_circ_0082689 and
anti-dsDNA. Collectively, it was speculated that the combination of
circRNAs and traditional biomarkers could further improve the
diagnostic value.
The field of circRNAs is recently discovered, and to
the best of our knowledge, no previous study has definitively
demonstrated the function of hsa_circ_0082688, hsa_circ_0082689 and
hsa_circ_0008675. It has been reported that circRNAs can function
as miRNA ‘sponges’ to sequester and competitively suppress miRNA
activity. Moreover, their interaction with disease-associated
miRNAs suggests that circRNAs are important for regulating
diseases. Therefore, to investigate the possible function of these
candidate circRNAs, the present study searched for potential miRNA
targets of these circRNAs using Arraystar miRNA target prediction
software, and numerous target miRNAs were identified. Among these
target miRNAs, hsa-miR-506-3p, hsa-miR-127-5p and hsa-miR-153-3p
were previously reported to be involved in the pathogenesis of SLE.
For example, hsa-miR-506-3p has the potential to regulate the
expression of Beclin1 (41), a
protein that had been shown to regulate autophagy in SLE (42). In addition, hsa-miR-127-5p is
involved in cell proliferation via the PI3K/Akt pathway (43), and thus may play a role in the
senescence of mesenchymal stem cells and the development of SLE
(44). hsa-miR-153-3p has also been
reported to be involved in the development of lupus nephritis
(45). Therefore, it was
hypothesized that these candidate circRNAs, including
hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675, may
function in SLE pathogenesis.
The present study also investigated the correlations
between the expression levels of these candidate circRNAs and the
severity of SLE. The results demonstrated that, while the
expression of hsa_circ_0082688 was negatively associated with C3 in
patients with SLE, no other circRNAs expression levels correlated
with C3. Moreover, the expression levels of all identified
differentially expressed circRNAs in the peripheral blood from
patients with SLE did not correlate with SLEDAI, CRP, ESR or C4.
Thus, the results demonstrated the expression levels of
hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675 were not
potential biomarkers for the severity of SLE. Furthermore, the
expression levels of these circRNAs between patients with newly
diagnosed and recurrent SLE were not significantly different, and
this also corroborates the aforementioned conclusion. However, it
was found that the expression levels of hsa_circ_0082688,
hsa_circ_0082689 and hsa_circ_0008675 were negatively associated
with the total WBC number or the number of certain subclasses of
WBC in patients with SLE, which indicated that the expression
levels of hsa_circ_0082688, hsa_circ_0082689 and hsa_circ_0008675
in peripheral blood were associated with hematological system
damage of SLE to some extent.
In addition, the present results suggested that the
expressions levels of hsa_circ_0082688, hsa_circ_0082689 and
hsa_circ_0008675 were correlated with each other, and similar
findings have been reported in a previous study (23). Thus, these differentially expressed
circRNAs may interact with each other directly or indirectly,
although further experiments are required to test this
hypothesis.
However, there are several limitations in this study
that should be acknowledged. First is the relatively small sample
size, especially the sample size of patients with newly diagnosed
SLE, which may restrict the generalizability of the present
results. Second, the exact role of these candidate circRNAs in SLE
pathogenesis was not investigated in this study. Therefore, these
are focuses of future studies.
In conclusion, to the best of our knowledge, the
present study was the first to demonstrate the circRNA expression
profiles in peripheral blood from adult patients with SLE, and
identify that circRNAs may serve as novel biomarkers for SLE
diagnosis. Furthermore, it was found that the combination of
hsa_circ_0082688 and hsa_circ_0082689 had a relatively good
capacity in discriminating the SLE groups from both HCs and RA
groups. Therefore, the present results provide a novel potential
diagnostic biomarker for SLE diagnosis, and may facilitate improved
understanding of the hematological system damage of SLE.
Acknowledgements
The authors would like to acknowledge the help from
Dr RuiWu from the Department of Rheumatology, The First Affiliated
Hospital of Nanchang University (Nanchang, China).
Funding
This study was supported by the Science and
Technology Plan Project of the Education Department of Jiangxi
Province (grant no. GJJ170008), the National Natural Science
Foundation of China (grant nos. 81360459, 81560344 and 81660277),
the Jiangxi Provincial Natural Science Foundation of China (grant
nos. 20171BAB205113 and 20171ACB20032), the Science and Technology
Project of Health and Family Planning Commission of Jiangxi
Province of China (grant no. 20165094) and the Foundation for
Distinguished Young Scientists of Jiangxi Province of China (grant
no. 20171BCB23087).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JL and QL conceived the project and wrote the study.
JL, QL, XL and ZH designed the experiments. JL, QL, XL, LZ and BF
collected the study subjects, performed the experiments and
analyzed the data. QL, XL, LZ, CQ, LF, YG and ZH performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was authorized by the Ethics
Committee of The First Affiliated Hospital of Nanchang University.
All participants provided written informed consent prior to the
initiation of the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tsokos GC: Systemic lupus erythematosus. N
Engl J Med. 365:2110–2121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ruiz-Irastorza G, Ramos-Casals M,
Brito-Zeron P and Khamashta MA: Clinical efficacy and side effects
of antimalarials in systemic lupus erythematosus: A systematic
review. Ann Rheum Dis. 69:20–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang NH, Li TT, Kim JJ,
Landolt-Marticorena C, Fortin PR, Gladman DD, Urowitz MB and Wither
JE: Interferon alpha induces altered transitional B cell signaling
and function in systemic lupus erythematosus. J Autoimmun.
58:100–110. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng
W, Zhang Q, Zhang P, Yu X, Xia Y, et al: DNA methylation and mRNA
and microRNA expression of SLE CD4+ T cells correlate with disease
phenotype. J Autoimmun. 54:127–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
O'Gorman WE, Hsieh EW, Savig ES,
Gherardini PF, Hernandez JD, Hansmann L, Balboni IM, Utz PJ,
Bendall SC, Fantl WJ, et al: Single-cell systems-level analysis of
human Toll-like receptor activation defines a chemokine signature
in patients with systemic lupus erythematosus. J Allergy Clin
Immunol. 136:1326–1336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lech M, Kantner C, Kulkarni OP, Ryu M,
Vlasova E, Heesemann J, Anz D, Endres S, Kobayashi KS, Flavell RA,
et al: Interleukin-1 receptor-associated kinase-M suppresses
systemic lupus erythematosus. Ann Rheum Dis. 70:2207–2217. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Z, Shi Y, Cai B, Wang L, Wu Y, Ying
B, Qin L, Hu C and Li Y: MALDI-TOF MS combined with magnetic beads
for detecting serum protein biomarkers and establishment of
boosting decision tree model for diagnosis of systemic lupus
erythematosus. Rheumatology (Oxford). 48:626–631. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ferreira TAR, de Andrade HM, de Pádua PM,
Carvalho MDG, Pires SDF, Oliveira IHR, Lima BSS, Fialho Júnior LC,
Cicarini WB, Chapeourouge DA, et al: Identification of potential
biomarkers for systemic lupus erythematosus diagnosis using
two-dimensional differential gel electrophoresis (2D-DIGE) and mass
spectrometry. Autoimmunity. 50:247–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu GC, Li J, Leng RX, Li XP, Li XM, Wang
DG, Pan HF and Ye DQ: Identification of long non-coding RNAs GAS5,
linc0597 and lnc-DC in plasma as novel biomarkers for systemic
lupus erythematosus. Oncotarget. 8:23650–23663. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Barrett SP, Wang PL and Salzman J:
Circular RNA biogenesis can proceed through an exon-containing
lariat precursor. Elife. 4:e075402015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li F, Zhang L, Li W, Deng J, Zheng J, An
M, Lu J and Zhou Y: Circular RNA ITCH has inhibitory effect on ESCC
by suppressing the Wnt/β-catenin pathway. Oncotarget. 6:6001–6013.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R,
Lou A, Zhu D, Shi GP and Yang M: Microarray expression profile of
circular RNAs in peripheral blood mononuclear cells from rheumatoid
arthritis patients. Cell Physiol Biochem. 42:651–659. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lin X, Lo HC, Wong DT and Xiao X:
Noncoding RNAs in human saliva as potential disease biomarkers.
Front Genet. 6:1752015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs As
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhao Z, Li X, Jian D, Hao P, Rao L and Li
M: Hsa_circ_0054633 in peripheral blood can be used as a diagnostic
biomarker of pre-diabetes and type 2 diabetes mellitus. Acta
Diabetol. 54:237–245. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L
and Li M: Peripheral blood circular RNA hsa_circ_0124644 can be
used as a diagnostic biomarker of coronary artery disease. Sci Rep.
7:399182017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Luo Q, Zhang L, Li X, Fu B, Guo Y, Huang Z
and Li J: Identification of circular RNAs hsa_circ_0044235 and
hsa_circ_0068367 as novel biomarkers for systemic lupus
erythematosus. Int J Mol Med. 44:1462–1472. 2019.PubMed/NCBI
|
|
27
|
Hochberg MC: Updating the American college
of rheumatology revised criteria for the classification of systemic
lupus erythematosus. Arthritis Rheum. 40:17251997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bombardier C, Gladman DD, Urowitz MB,
Caron D and Chang CH: Derivation of the SLEDAI. A disease activity
index for lupus patients. The committee on prognosis studies in
SLE. Arthritis Rheum. 35:630–640. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Arnett FC, Edworthy SM, Bloch DA, McShane
DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS,
et al: The American rheumatism association 1987 revised criteria
for the classification of rheumatoid arthritis. Arthr Rhuem.
31:315–324. 1988. View Article : Google Scholar
|
|
30
|
Luo Q, Li X, Xu C, Zeng L, Ye J, Guo Y,
Huang Z and Li J: Integrative analysis of long non-coding RNAs and
messenger RNA expression profiles in systemic lupus erythematosus.
Mol Med Rep. 17:3489–3496. 2018.PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Luo Q, Zhang L, Fang L, Fu B, Guo Y, Huang
Z and Li J: Circular RNAs hsa_circ_0000479 in peripheral blood
mononuclear cells as novel biomarkers for systemic lupus
erythematosus. Autoimmunity. 53:167–176. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Soliman S and Mohan C: Lupus nephritis
biomarkers. Clin Immunol. 185:10–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stypinska B and Paradowska-Gorycka A:
Cytokines and MicroRNAs as candidate biomarkers for systemic lupus
erythematosus. Int J Mol Sci. 16:24194–24218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu Y, Zhang F, Ma J, Zhang X, Wu L, Qu B,
Xia S, Chen S, Tang Y and Shen N: Association of large intergenic
noncoding RNA expression with disease activity and organ damage in
systemic lupus erythematosus. Arthritis Res Ther. 17:1312015.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Guo JN, Li J, Zhu CL, Feng WT, Shao JX,
Wan L, Huang MD and He JD: Comprehensive profile of differentially
expressed circular RNAs reveals that hsa_circ_0000069 is
upregulated and promotes cell proliferation, migration, and
invasion in colorectal cancer. Onco Targets Ther. 9:7451–7458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP
and Yang M: Using plasma circRNA_002453 as a novel biomarker in the
diagnosis of lupus nephritis. Mol Immunol. 101:531–538. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang MY, Wang JB, Zhu ZW, Li LJ, Liu RS,
Yang XK, Leng RX, Li XM, Pan HF and Ye DQ: Differentially expressed
circular RNAs in systemic lupus erythematosus and their clinical
significance. Biomed Pharmacother. 107:1720–1727. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li S, Zhang J, Tan X, Deng J, Li Y, Piao
Y, Li C, Yang W, Mo W, Sun J, et al: Microarray expression profile
of circular RNAs and mRNAs in children with systemic lupus
erythematosus. Clin Rheumatol. 38:1339–1350. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yi F, Hao Y, Chong X and Zhong W:
Overexpression of microRNA-506-3p aggravates the injury of vascular
endothelial cells in patients with hypertension by downregulating
Beclin1 expression. Exp Ther Med. 15:2844–2850. 2018.PubMed/NCBI
|
|
42
|
Pan Q, Gao C, Chen Y, Feng Y, Liu WJ and
Liu HF: Update on the role of autophagy in systemic lupus
erythematosus: A novel therapeutic target. Biomed Pharmacother.
71:190–193. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liang J, Xu L, Zhou F, Liu AM, Ge HX, Chen
YY and Tu M: MALAT1/miR-127-5p regulates osteopontin (OPN)-mediated
proliferation of human chondrocytes through PI3K/Akt pathway. J
Cell Biochem. 119:431–439. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen H, Shi B, Feng X, Kong W, Chen W,
Geng L, Chen J, Liu R, Li X, Chen W, et al: Leptin and
neutrophil-activating peptide 2 promote mesenchymal stem cell
senescence through activation of the phosphatidylinositol
3-kinase/Akt pathway in patients with systemic lupus erythematosus.
Arthritis Rheumatol. 67:2383–2393. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Navarro-Quiroz E, Pacheco-Lugo L,
Navarro-Quiroz R, Lorenzi H, España-Puccini P, Díaz-Olmos Y,
Almendrales L, Olave V, Gonzalez-Torres H, Diaz-Perez A, et al:
Profiling analysis of circulating microRNA in peripheral blood of
patients with class IV lupus nephritis. PLoS One. 12:e01879732017.
View Article : Google Scholar : PubMed/NCBI
|