Introduction
Macrophages have been reported to have a significant
role in contributing to poor clinical outcomes in sepsis and
infections caused by gram-negative bacteria (1,2).
During infection, lipopolysaccharide (LPS), an important component
of gram-negative bacteria, binds to the Toll-like receptor 4
(TLR4)/myeloid differentiation factor 2 (MD2) complex in
macrophages, which leads to the induction of immune exacerbation
(3). An uncontrolled macrophage
inflammatory response can cause tissue damage, systemic
inflammation and various chronic inflammatory diseases (4). The potentiation of TLR4/MD2 by LPS
drives activation of nuclear factor-κB (NF-κB) and constitutive
inflammatory signalling by binding to the promoter of inflammatory
genes (5). Activation of NF-κB
occurs through the inducible degradation of its inhibitor, IκBα,
which is triggered through the phosphorylation of IKK complexes,
thereby causing the ubiquitination and proteasomal degradation of
IκBα. Degradation of IκBα results in a rapid and transient nuclear
translocation of the NF-κB transcription factor (6). Numerous NF-κB target genes are
involved in the inflammatory response, including inducible nitric
oxide synthase (iNOS), cyclooxygenase (COX)-2 and inflammatory
cytokines (7,8). Inhibition of the LPS-stimulated
formation of the TLR4/MD2 complex and NF-κB activation in
macrophages has been proposed as a target of intervention for acute
and chronic inflammatory diseases (9,10).
Several natural compounds derived from the
Curcuma species have been revealed to be promising compounds
for the development of novel drugs. Among them, turmeric
(Curcuma longa L.) is a species of plant belonging to the
Zingiberaceae family and has been investigated as a rich source of
bioactive compounds (11).
Curcuminoids are multifunctional polyphenolic compounds found in
the rhizome of turmeric (12). The
natural trienone analogue of
curcumin,1,7-bis(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one
(HMPH), has previously been isolated as a minor component from
C. longa (13). Few studies
have investigated the biological activities of trienones. Trienones
have previously been shown to possess cytotoxicity against KB oral
cancer cells (14), PC-3 prostate
cancer cells (15) and oral
squamous cell carcinoma cells (16), as well as inhibitory effects on
steroid 5-α reductase activity (17) and TNF-α production (18). To the best of our knowledge, the
anti-inflammatory activity of trienones in LPS-activated RAW264.7
macrophages has not yet been precisely elucidated by comparing the
results with curcumin. The anti-inflammatory activity of curcumin
has been reported to be associated with the suppression of iNOS
(19) and COX-2 (20) in LPS-activated RAW264.7 cells by
targeting various transcription factors, including NF-κB signalling
proteins. In addition, curcumin has been shown to interfere with
the binding of LPS to the TLR4/MD2 complex, consequently
suppressing activation of the inflammatory pathway (21). However, the drawbacks of curcumin
are its low oral bioavailability, instability at physiological pH,
low solubility in water and rapid metabolism (22,23).
This has prompted researchers to search for novel curcuminoid
analogues with powerful therapeutic potential to use as
anti-inflammatory agents for the treatment of various inflammatory
diseases, or to use this class of compounds as the backbone for
designing new drugs.
The present study aimed to investigate the
anti-inflammatory effect of HMPH, which is structurally related to
curcumin from C. longa in LPS-activated macrophages. In
order to ensure a sufficient supply of HMPH, a novel method of
synthesis of this compound from curcumin was developed.
Subsequently, the anti-inflammatory role of HMPH on iNOS/nitric
oxide (NO), COX-2 and pro-inflammatory cytokine production was
investigated. Furthermore, to clarify the mechanism of action of
HMPH, the critical regulatory protein p65 NF-κB and the interaction
of HMPH with MD2 were additionally evaluated using an in
silico modelling study.
Materials and methods
Synthesis of HMPH
Curcumin was obtained from C. longa as
described previously (24).
Curcumin (1.2 g, 3.24 mmol) was dissolved in tetrahydrofuran (THF;
30 ml) and NaBH4 (50 mg, 1.32 mmol) was added. The
reaction mixture was stirred at room temperature for 4 h. Water
(200 ml) was then added and the mixture was extracted with EtOAc.
The organic phase was washed with water, dried over anhydrous
Na2SO4 and the solvent was evaporated until
dry. The obtained crude product was dissolved in THF (30 ml) and
p-toluenesulfonic acid monohydrate (50 mg) was then added. The
reaction mixture was refluxed for 3 h, water was added and the
mixture was extracted with EtOAc (2×100 ml). The combined organic
phase was washed with water and dried over anhydrous
Na2SO4; the solvent was evaporated and the
residue was chromatographed on a silica column (particle size,
<0.063 mm; silica gel 60; EMD Millipore) using hexane-EtOAc
(3:2) and further purified on a Sephadex LH-20 column eluted with
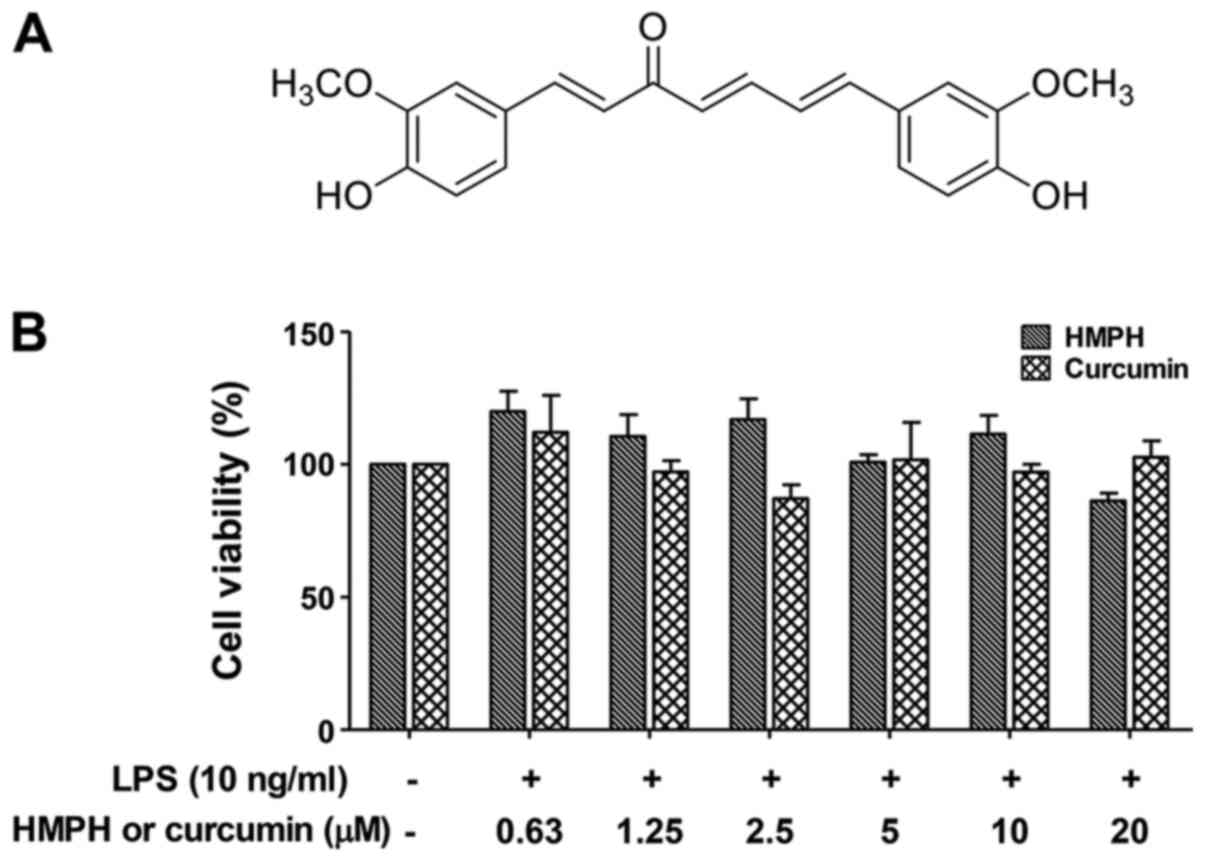
MeOH to yield HMPH (Fig. 1A). A
total of 240 mg (21% overall yield from curcumin) of HMPH was
obtained (Fig. S1). The
1H and 13C NMR (Figs. S2 and S3) and mass spectral data (data not
shown) were consistent with the reported values (24).
Cell line and culture
The mouse macrophage cell line, RAW264.7 was
purchased from American Type Culture Collection. Cells were
cultured in RPMI-1640 medium (Corning, Inc.) supplemented with 10%
foetal bovine serum (Biochrom, Ltd.), 1% penicillin/streptomycin
and 2 mM stable L-glutamine (Gibco; Thermo Fisher Scientific, Inc.)
in an atmosphere containing 95% humidity and 5% CO2 at
37°C. In all experiments, the cell line was cultured for 24 h and
cell passages 21–30 were used for experimentation.
Measurement of cytotoxicity
To measure cell viability of HMPH, the MTT assay
(Sigma-Aldrich; Merck KGaA) was performed. RAW264.7 cells were
plated into 96-well plates at a density of 3×105
cells/cm2. After 24 h, cells were pretreated with HMPH
or curcumin at different concentrations (0.63–20 µM) for 1 h at
37°C, then incubated with LPS (10 ng/ml) (Escherichia coli
0111:B4; Sigma-Aldrich; Merck KGaA) for 24 h. After a 3 h
incubation period with MTT (0.5 mg/ml) solution, the formazan
crystals were dissolved in DMSO and absorbance was measured at 560
nm.
Measurement of NO production
RAW264.7 cells (3.0×105
cells/cm2) were cultured in 96-well plates. To avoid the
direct interaction of the test compounds with LPS and the presence
of a high inflammatory background, cells were pretreated with the
test compounds for 1 h prior to LPS stimulation. Briefly, cells
were incubated with HMPH or curcumin (0.63–20 µM), an inhibitor of
IκBα phosphorylation (BAY 11-7082; 10 µM; Sigma-Aldrich; Merck
KGaA) and an anti-inflammatory drug (dexamethasone; 10 µM;
Sigma-Aldrich; Merck KGaA) for 1 h prior to LPS (10 ng/ml)
stimulation for 24 h. Nitrite concentration in the cultured medium
was measured using Griess reagent (Sigma-Aldrich; Merck KGaA)
(25). The optical density was
measured at 540 nm.
Measurement of inflammatory
cytokines
RAW264.7 cells were plated at a density of
3.0×105 cells/cm2 in 96-well plates for 24 h.
Cells were treated with HMPH and curcumin at different
concentrations (5–20 µM) and BAY 11–7082 (10 µM) for 1 h followed
by LPS (10 ng/ml) stimulation for 24 h. Subsequently, culture
medium was analysed using ELISA kits for TNF-α (cat. no. 430904),
IL-6 (cat. no. 431304) and IL-1β (cat. no. 432604), according to
the manufacturer's protocols (BioLegend, Inc.).
Preparation of nuclear and cytosolic
extracts
RAW264.7 cells were pretreated with HMPH and
curcumin (5–20 µM) and BAY 11-7082 (10 µM) for 1 h followed by LPS
(10 ng/ml) stimulation for 15 min. Nuclear and cytoplasmic extracts
were prepared using NE-PER™ Nuclear and Cytoplasmic Extraction
Reagents (cat. no. 78835; Thermo Fisher Scientific, Inc.) according
to the manufacturer's guidelines. The nuclear and cytoplasmic
proteins were then assessed by western blot analysis.
Western blot analysis
RAW264.7 cells were cultured at a concentration of
3.0×105 cells/cm2 on 6-well plates. Cells
were pretreated with HMPH and curcumin (5–20 µM), BAY 11-7082 (10
µM) and dexamethasone (10 µM) for 1 h, then cells were activated
with LPS (10 ng/ml) for 15 min for p65 NF-κB detection or 24 h for
iNOS and COX-2 detection. Cells were washed three times with
ice-cold PBS and lysed with lysis buffer containing protease
inhibitors (Cell Signaling Technology, Inc.). Cell lysates were
centrifuged at 12,000 × g for 15 min at 4°C. The supernatant was
collected and protein concentrations were determined using the
detergent compatible protein assay kit (BioRad Laboratories, Inc.).
Briefly, 30–50 µg protein was loaded, separated by SDS-PAGE on 7.5
and 10% gels, and then transferred to polyvinylidene fluoride
membranes. The membranes were blocked with 5% non-fat dry milk in
Tris-buffered saline and 0.1% Tween-20 (TBST) for 1 h at room
temperature, and incubated with primary antibodies against iNOS
(1:1,000; cat. no. 13120), COX-2 (1:1,000; cat. no. 4842), p65
NF-κB (1:1,000; cat. no. 8242), β-actin (1:5,000; cat. no. 4967)
and lamin B1 (1:1,000; cat. no. 13435) (Cell Signaling Technology,
Inc.) at 4°C overnight. Each membrane was then washed with TBST
followed by incubation with anti-rabbit HRP-conjugated secondary
antibody (1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.)
at room temperature for 1 h under agitation. Blots were visualized
using the chemiluminescent HRP detection reagent (EMD Millipore).
GeneSys software version 1.2.5.0 (Synoptics, Ltd.) was used to
acquire iNOS, COX-2 (HMPH treatment group) and associated β-actin
images, and Image Lab™ Touch Software (Bio-Rad Laboratories, Inc.)
was used to acquire the images of p65 NF-κB, lamin B1, COX-2
(curcumin treatment group) and associated β-actin. Signal
intensities were densitometrically semi-quantified using ImageJ
V1.8.0 software program (National Institute of Health).
Immunofluorescence staining
RAW264.7 cells (3.0×105
cells/cm2) were seeded on coverslips on 24-well plates
for 24 h. Cells were pretreated with HMPH (20 µM), curcumin (20 µM)
or BAY 11-7082 (10 µM) for 1 h, and were then treated with LPS (10
ng/ml) for 15 or 30 min. Cells were washed three times with
ice-cold PBS, fixed in pre-cooled methanol (−20°C) for 5 min and
immersed in cold acetone. The fixed cells were blocked in 1% bovine
serum albumin (Sigma-Aldrich; Merck KGaA) in PBS containing 0.1%
Tween-20 (PBST) for 30 min and then incubated with anti-p65 NF-κB
antibody (1:200; cat. no. 8242; Cell Signalling Technology, Inc.)
at room temperature for 1 h. After washing in PBST, the cells were
incubated with anti-rabbit secondary antibody conjugated with Alexa
Fluor-488 (1:400; cat. no. 4412S; Cell Signaling Technology, Inc.)
alongside propidium iodide (2.5 µg/ml) at room temperature for 1 h.
The coverslips were mounted on microscope slides and images were
captured using a Leica TCS SP5 II laser scanning confocal
microscope (Leica Microsystems GmbH).
Pretreatment with compounds before LPS
stimulation
RAW264.7 cells (3.0×105
cells/cm2) were seeded on 6-well plates and incubated at
37°C for 24 h in an incubator containing 5% CO2. Cells
were incubated in the presence or absence of various concentrations
of HMPH and curcumin (5–20 µM) and BAY 11-7082 (10 µM) for 1 h, and
were washed three times with serum-free medium before incubation
with LPS (10 ng/ml) for 24 h. The culture supernatant was collected
and the nitrite accumulated in the supernatant was determined by
the Griess assay.
Molecular docking analysis
The crystal structure of human MD2-lipid IVa complex
(PDB code 2E59) was used as the initial coordinates for the
molecular docking calculations using AutoDock4.2 (26). The docking input files and analysed
docking results were generated from AutoDockTools1.5.6 (26). A grid box dimension of 60×60×60
Å3 with a spacing of 0.375 Å between the grid points was
created and centred on the ligand. The Lamarckian genetic algorithm
was employed for the docking calculations to enable modification of
the gene population (27). The
docking parameters were as follows: A total of 250 independent
runs, a population size of 150, and a maximum of 25,000,000 energy
evaluations. MD2 protein was set rigid but all torsional bonds of
the ligand were freely rotated. Non-polar hydrogens of the molecule
were merged and each atom was assigned with Gasteiger partial
charges. The lowest docked energy of each conformation in the most
populated cluster was selected for the detailed analysis and
further study. Visualisation of the docking results was performed
using Biovia Discovery Studio Visualizer (Dassault Systèmes).
Molecular Mechanics/Poisson Boltzmann
Surface Area (MM/PBSA) free energy analysis
The energy minimisations of the docked MD2-HMPH and
MD2-curcumin complexes were performed using the SANDER module of
AMBER16 packages (28). To remove
bad contacts, the systems were minimised starting with the steepest
descent followed by the conjugate gradient. The minimized complexes
were solvated by water molecules. Then, the systems were heated
progressively from 0 to 300 K for 100 ps and equilibrated at 300 K
for 200 ps in the canonical (NVT) and isothermal-isobaric (NPT)
ensembles, respectively. The last snapshot obtained from the
equilibrated phase was used as the suitable structure of the
complex for calculating the binding free energy value using the
MM/PBSA module (∆GMM/PBSA) in the AMBER16 programme;
this methodology provides more reliable binding energy values.
Statistical analysis
Data are presented as the mean ± SEM of three
independent experiments. Statistical significance was evaluated by
one-way or two-way ANOVA followed by Bonferroni multiple comparison
test. P<0.05 was considered to indicate a statistically
significant difference. The statistical analyses were performed
using GraphPad Prism 6 (GraphPad Software, Inc.).
Results
Effect of HMPH on cell viability
The present study investigated the effect of HMPH on
the cell viability of RAW264.7 macrophages. Cells were treated with
the test compounds in increasing concentrations between 0.63 and 20
µM, followed by treatment with LPS (10 ng/ml) for 24 h. HMPH did
not affect the viability of RAW264.7 cells at concentrations up to
20 µM in combination with 10 ng/ml LPS; similar results were
observed for curcumin (Fig.
1B).
Effect of HMPH on NO production and
iNOS protein expression levels in LPS-activated RAW264.7
macrophages
The present study further explored the inhibitory
activity of HMPH on NO in LPS-activated RAW264.7 macrophages. The
inhibitory effect of HMPH on NO in LPS-activated RAW264.7 cells was
assessed by Griess assay. The results revealed that LPS
significantly induced NO production compared with in the control
cells (without LPS treatment) (Fig.
2A). Pretreatment with HMPH, at concentrations ranging between
0.63 and 20 µM, significantly inhibited LPS-induced NO secretion in
a dose-dependent manner, whereas curcumin suppressed NO production
at higher concentrations of 1.25–20 µM (Fig. 2A). These findings indicated that
HMPH was more effective than curcumin at inhibiting NO production.
The suppression of iNOS protein expression levels by HMPH were
further investigated in LPS-activated RAW264.7 cells. Upon LPS
stimulation, the protein expression levels of iNOS were
significantly increased (Fig. 2B and
C). Pretreatment with 5–20 µM HMPH and curcumin significantly
decreased the protein expression levels of iNOS in LPS-activated
macrophages compared with LPS treatment alone. The
anti-inflammatory drugs dexamethasone and BAY 11-7082 also
significantly inhibited NO production and iNOS protein expression
compared with LPS-treated cells.
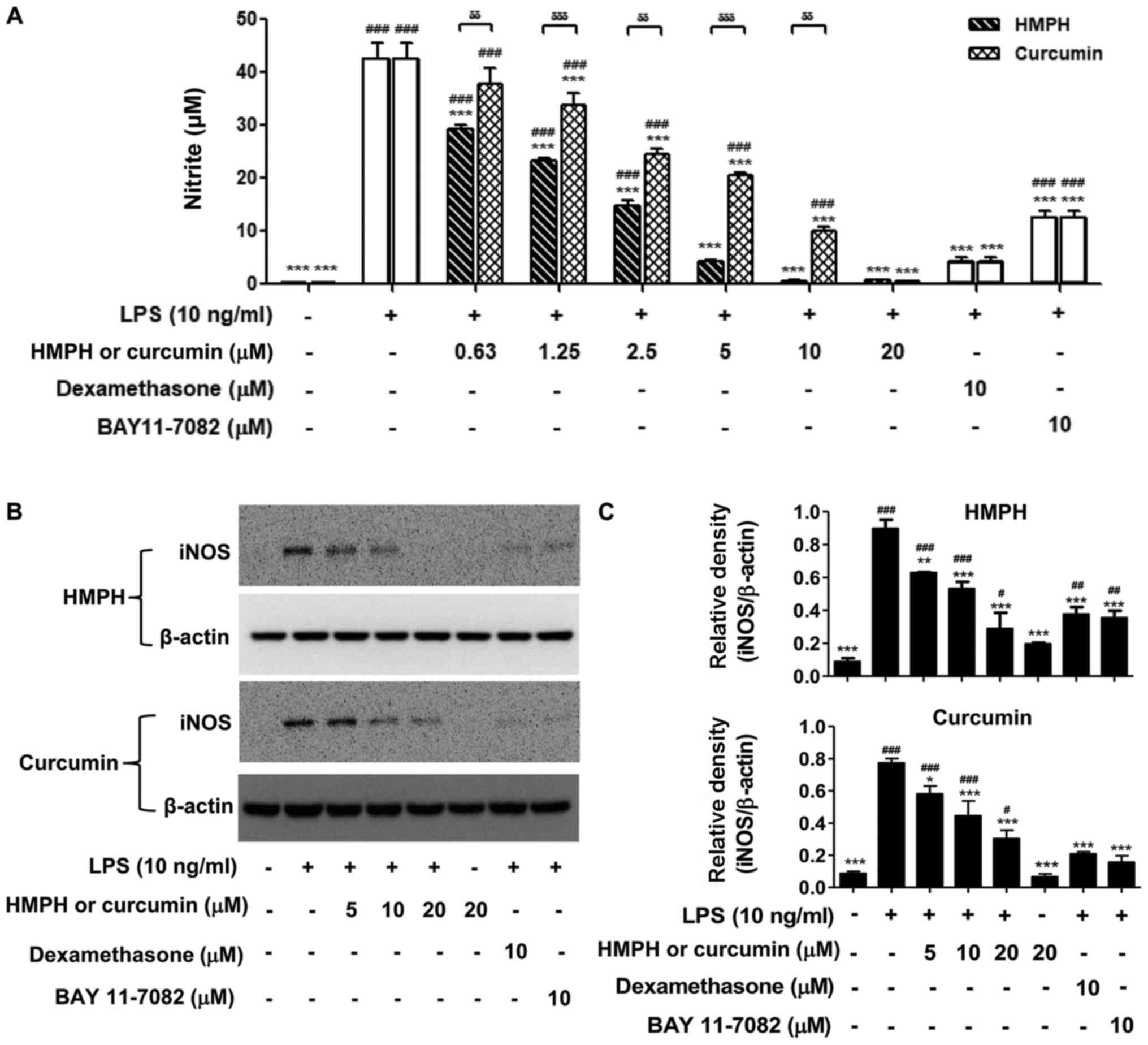 | Figure 2.HMPH suppresses NO production and
iNOS protein expression levels in LPS-activated RAW264.7
macrophages. (A) NO production upon stimulation with LPS (10 ng/ml)
with or without HMPH and curcumin (0.63–20 µM) was detected by
Griess assay. Cell lysates were prepared from RAW264.7 cells
pretreated with or without HMPH and curcumin for 1 h followed by
LPS treatment for 24 h. (B) Protein expression levels of iNOS were
determined by western blotting. The representative images of iNOS
and β-actin were from two different parts of the same blotted
membranes, with an exposure time of 15 min for iNOS and 10–25 sec
for β-actin. (C) Relative expression levels of iNOS were
semi-quantified by scanning densitometry and normalized to β-actin.
Data are presented as the mean ± SEM of three independent
experiments. #P<0.05, ##P<0.01,
###P<0.001 vs. control group; *P<0.05,
**P<0.01, ***P<0.00 vs. LPS-stimulated group;
δδP<0.01, δδδP<0.001 vs.
curcumin-treated group. HMPH,
1,7-bis(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one; LPS,
lipopolysaccharide; NO, nitric oxide; iNOS, inducible nitric oxide
synthase. |
Effect of HMPH on COX-2 protein
expression levels in LPS-activated RAW264.7 macrophages
As curcumin has been shown to inhibit COX-2 protein
expression (20), the present study
investigated the inhibition of COX-2 expression by HMPH. HMPH at 20
µM (Fig. 3A and B) significantly
inhibited the protein expression levels of COX-2 in LPS-activated
RAW264.7 macrophages; curcumin exhibited a similar effect at the
same concentration. Compared with HMPH and curcumin, dexamethasone
possessed a stronger inhibitory effect on COX-2 protein expression
(Fig. 3).
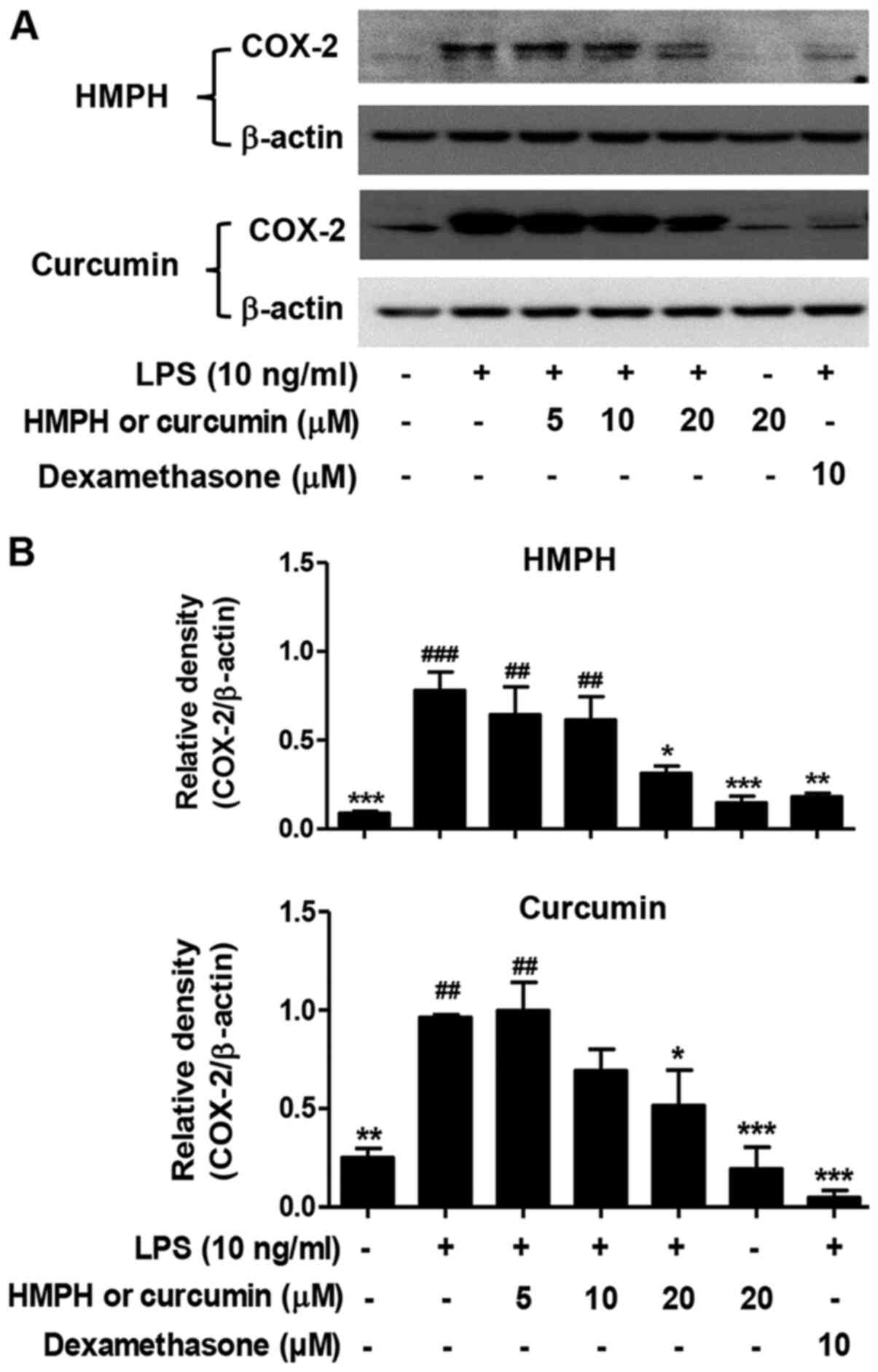 | Figure 3.HMPH suppresses COX-2 expression in
LPS-activated RAW264.7 macrophages. (A) Protein expression levels
of COX-2 were determined by western blotting. For HMPH treatment
group, the representative images of COX-2 (exposure time: 20 min)
and β-actin (exposure time: 1 min) were from two different parts of
the same blotted membranes. For the curcumin treatment group, the
two representative images of COX-2 (exposure time: 1.20 min) and
β-actin (exposure time: 1 min) were taken from different membranes.
The COX-2 blotting image from the curcumin treatment group was
acquired using a different imaging system. (B) Relative expression
levels of COX-2 were semi-quantified by scanning densitometry and
normalized to β-actin. Data are presented as the mean ± SEM of
three independent experiments. ##P<0.01,
###P<0.001 vs. control group; *P<0.05,
**P<0.01, ***P<0.001 vs. LPS-stimulated group. HMPH,
1,7-bis(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one; LPS,
lipopolysaccharide; COX-2, cyclooxygenase 2. |
Effect of HMPH on inflammatory
cytokine production in LPS-activated RAW264.7 macrophages
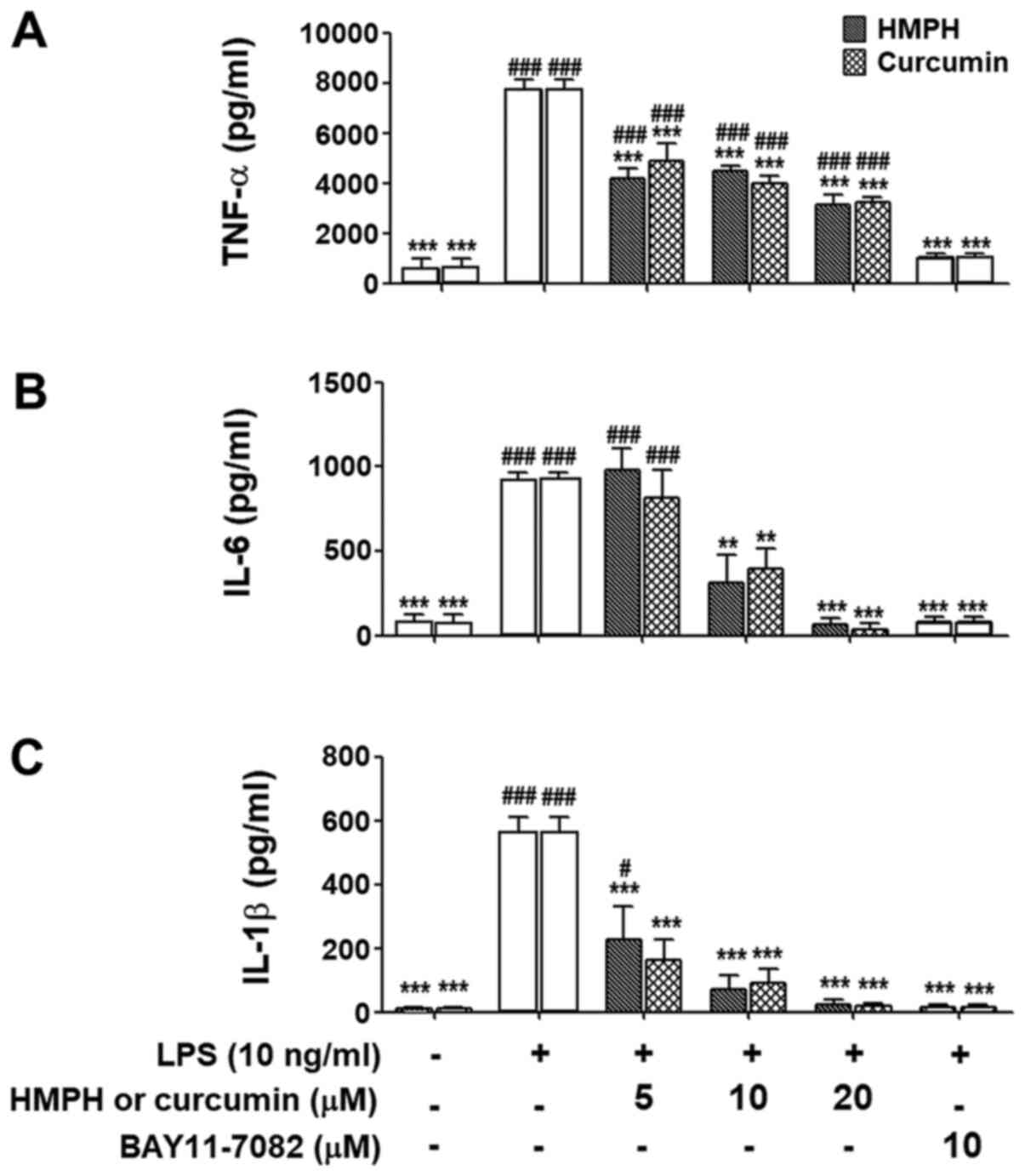
The present study also investigated the suppression
of cytokine production in LPS-activated macrophages using ELISA.
Upon stimulation with LPS for 24 h, the production of TNF-α
(Fig. 4A), IL-6 (Fig. 4B) and IL-1β (Fig. 4C) was significantly increased.
Pretreatment of cells with HMPH at 10–20 µM markedly inhibited
LPS-induced TNF-α, IL-6 and IL-1β production (Fig. 4A-C). Curcumin exhibited a similar
trend with regards to suppression of these inflammatory cytokines.
BAY 11-7082 (10 µM) also significantly inhibited the production of
all inflammatory cytokines measured.
Effect of HMPH on LPS-activated NF-κB
translocation in RAW264.7 macrophages
NF-κB is an important signalling protein involved in
the activation of inflammation (7,8). The
present study further investigated the effect of HMPH on the
suppression of p65 NF-κB nuclear translocation. Upon LPS
stimulation, p65 NF-κB translocated into the nucleus (Fig. 5). HMPH (10–20 µM) significantly
suppressed p65 NF-κB translocation into the nucleus; however, HMPH
had no effect on p65 NF-κB expression in the cytoplasm (Fig. 5A). The mechanism of action of HMPH
on the suppression of p65 NF-κB translocation was greater than that
of curcumin (Fig. 5B). BAY 11-7082
(10 µM) also significantly suppressed the translocation of p65
NF-κB into the nucleus. These findings are consistent with the
results obtained from immunofluorescence staining of p65 NF-κB in
RAW264.7 cells treated with LPS for 15 min (Fig. S4) and 30 min (Fig. S5). After treatment with HMPH, p65
NF-κB protein expression, as indicated by green fluorescence, was
retained mainly in the cytoplasm of LPS-activated macrophages,
similar to that observed in cells treated with BAY 11-7082
(Figs. S4 and S5). Cells pretreated with curcumin
exhibited both nuclear and cytoplasmic localisation of NF-κB
following LPS stimulation, indicating a less pronounced effect than
that observed in response to HMPH.
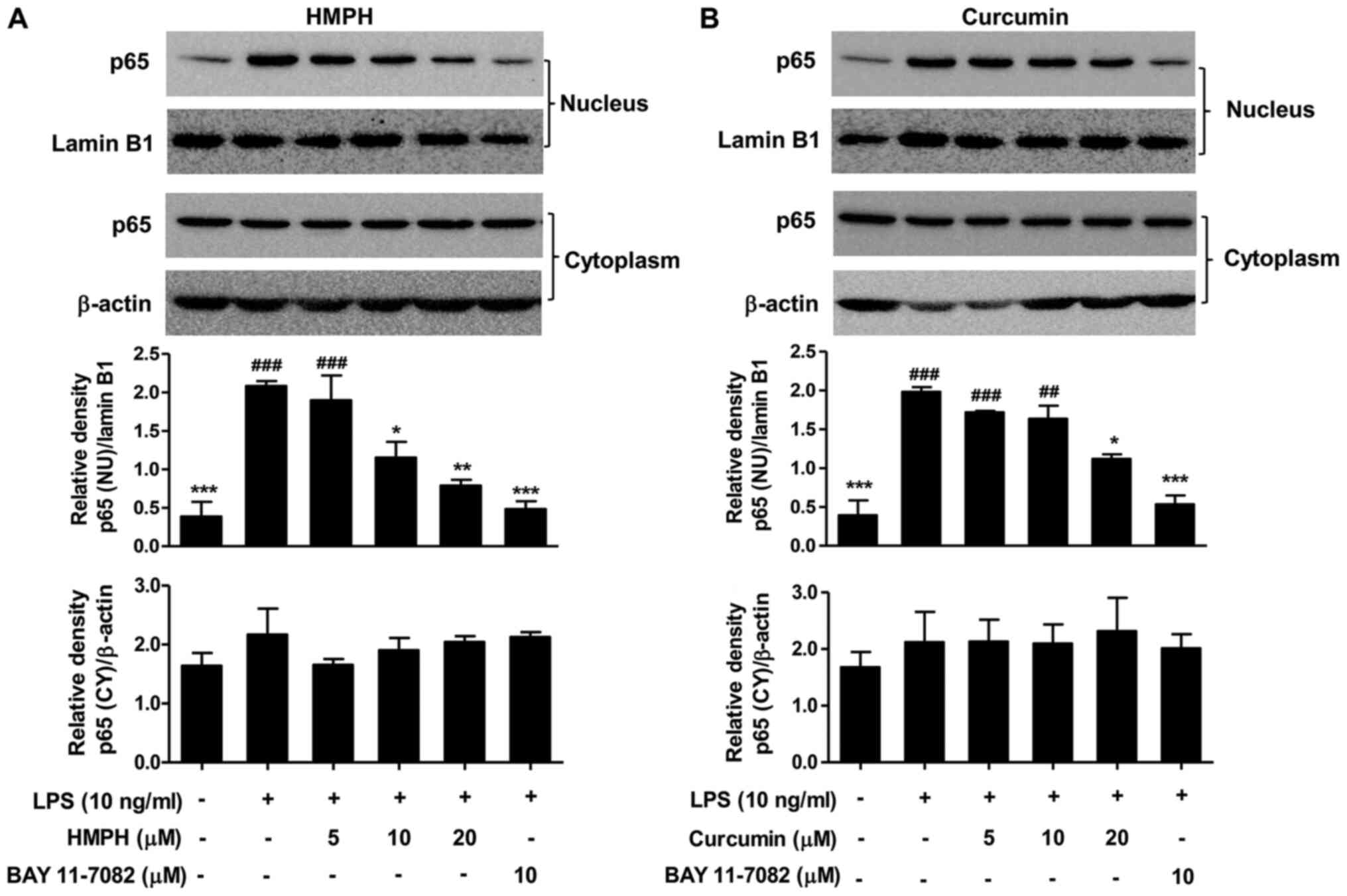 | Figure 5.HMPH suppresses LPS-activated p65
NF-κB translocation in RAW264.7 macrophages. Proteins were prepared
from RAW264.7 cells pretreated with or without different
concentrations (5–20 µM) of (A) HMPH and (B) curcumin for 1 h
followed by LPS treatment (10 ng/ml) for 15 min. The protein
expression levels of p65 NF-κB were determined by western blotting
(top panel) and relative expression levels were normalized to
β-actin or lamin B1 (bottom panel). For the NU fraction, the two
representative images of p65 NF-κB and lamin B1 were taken from
different blotted membranes. For the CY fraction, the two
representative images of p65 NF-κB and β-actin were from the same
membrane. Data are presented as the mean ± SEM of three independent
experiments. ##P<0.01, ###P<0.001 vs.
control group; *P<0.05, **P<0.01, ***P<0.001 vs.
LPS-stimulated group. HMPH,
1,7-bis(4-hydroxy-3-methoxyphenyl)-1,4,6-heptatrien-3-one; LPS,
lipopolysaccharide; NU, nuclear; CY, cytoplasmic; NF-κB, nuclear
factor-κB. |
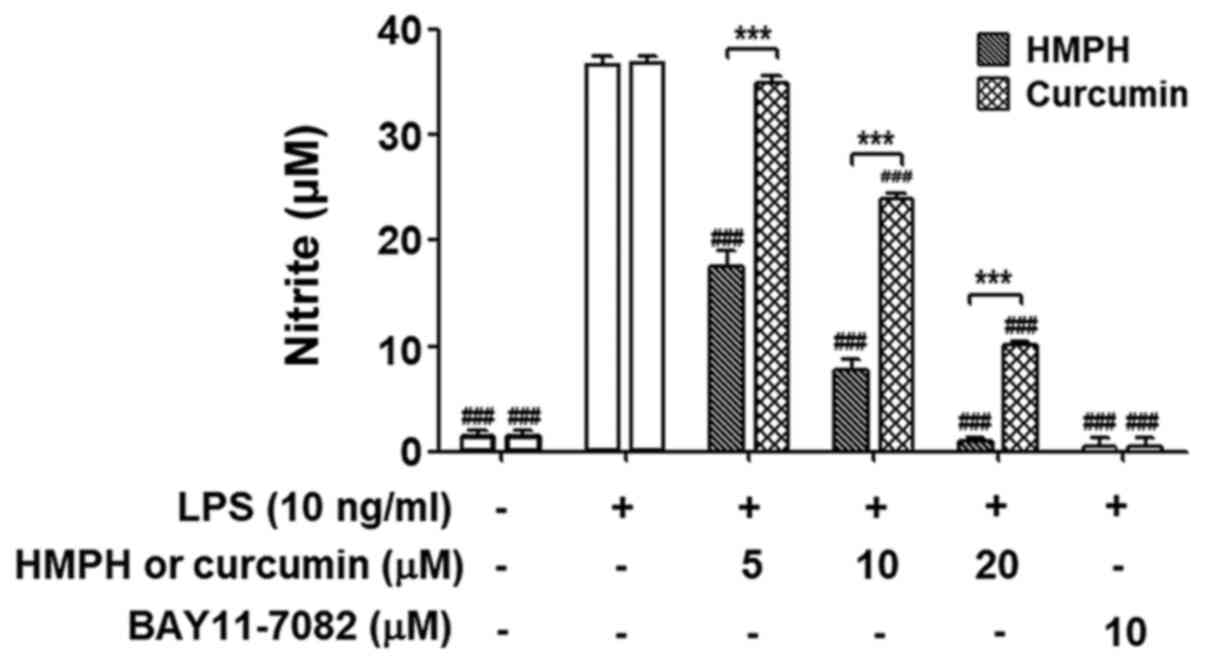
Effect of pretreatment with HMPH on
LPS stimulation in RAW264.7 macrophages
To investigate whether the anti-inflammatory effect
of HMPH on the LPS-induced inflammatory response was due to its
effect on the cells, RAW264.7 cells were pretreated with HMPH for 1
h and were then thoroughly washed to remove HMPH before LPS
stimulation for 24 h. Curcumin was used for comparison with HMPH
treatment. Notably, inhibition of NO production was still observed
after the removal of HMPH (5–20 µM) and curcumin (10–20 µM) prior
to LPS stimulation (Fig. 6). BAY
11–7082 (10 µM) completely inhibited NO production after the
washout, this finding was similar to that observed in cells treated
with 20 µM HMPH. These data suggested that HMPH may act on cells
and exert anti-inflammatory action prior to LPS stimulation.
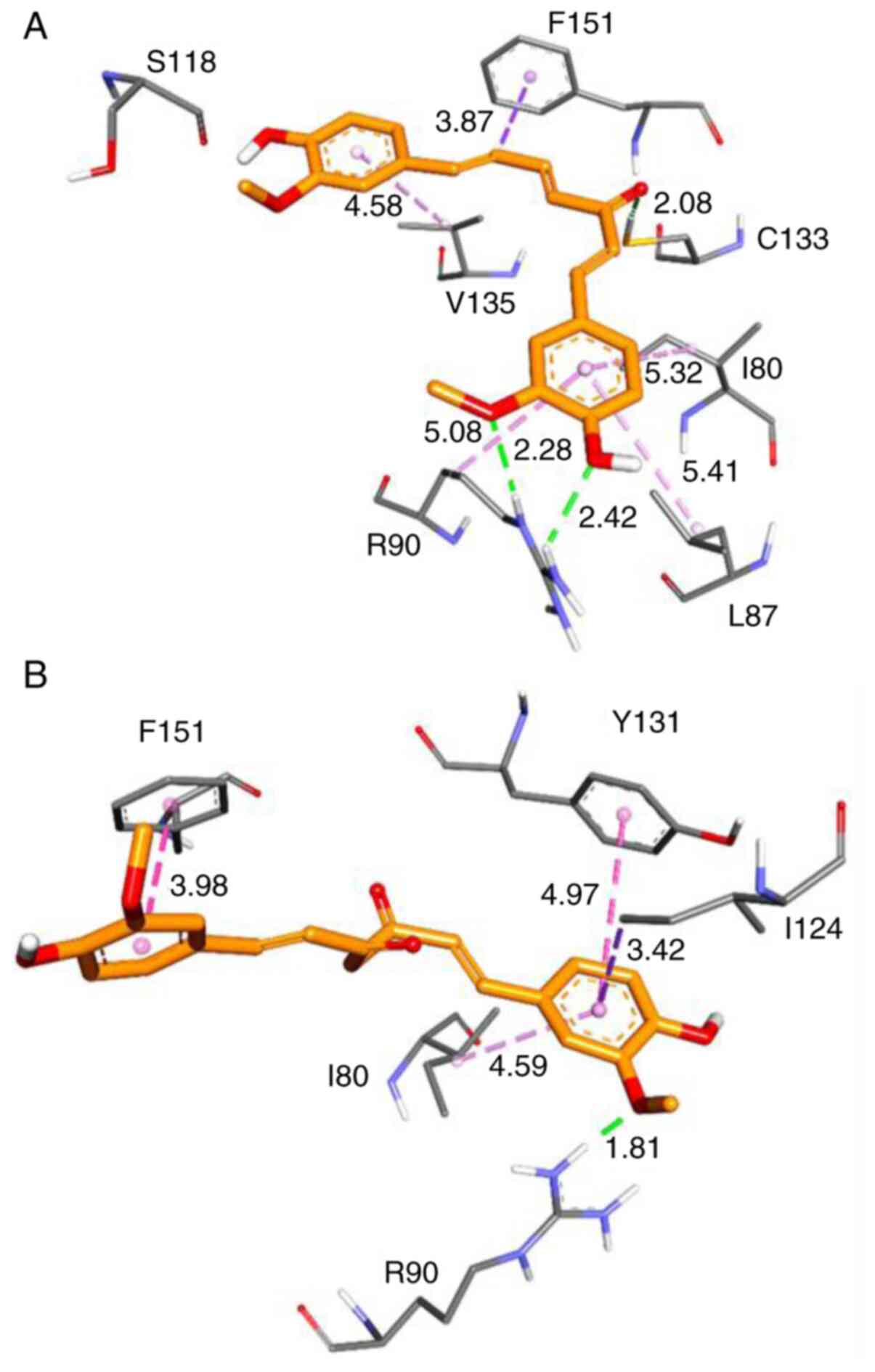
Effect of HMPH on MD2
Previous results demonstrated that HMPH could
inhibit inflammatory responses in activated macrophages and the
inhibition of LPS-induced inflammation may be through the complex
formation of HMPH with MD2 (10,21).
The binding of HMPH to the MD2 protein pocket was subsequently
assessed and compared with the binding of curcumin. Molecular
docking was applied to predict the possible interaction between
HMPH and the MD2 protein using AutoDock4.2. The lowest binding
energy represents the best binding conformation between the protein
and ligand. Table I summarises the
results of the docking study based on binding interactions and
energies. The minimised structures of the best conformation of
MD2-HMPH and MD2-curcumin docked complexes are shown in Fig. 7. The results revealed that HMPH can
form a stronger interaction with MD2 protein than curcumin. There
are two hydrogen bond interactions involving amino acids R90 and
C133, as well as the hydrophobic interactions with the residues
F151, I80, L87 and V135 that were observed in the MD2-HMPH complex
(Fig. 7A). Conversely, curcumin
interacted with MD2 by forming a hydrogen bond with R90, as well as
forming hydrophobic interactions with Y131, F151, I124 and I80
residues of the MD2 protein (Fig.
7B).
 | Table I.Summary of the binding interactions
and energies of MD2-HMPH and MD2-curcumin complexes. |
Table I.
Summary of the binding interactions
and energies of MD2-HMPH and MD2-curcumin complexes.
| Compound | Interaction | Residue | Distance (Å) |
ΔGdocking (kcal/mol) |
ΔGMM/PBSA (kcal/mol) |
|---|
| HMPH | Hydrogen bond | R90, C133 | 2.28/2.42,
2.08 | −6.12 | −29.56 |
|
| Pi-Sigma | F151 | 3.87 |
|
|
|
| Pi-Alkyl | I80, L87, V135 | 5.32, 5.41,
4.58 |
|
|
| Curcumin | Hydrogen bond | R90 | 1.81 | −5.64 | −25.63 |
|
| Pi-Pi | Y131, F151 | 4.97, 3.98 |
|
|
|
| Pi-Sigma | I124 | 3.42 |
|
|
|
| Pi-Alkyl | I80 | 4.59 |
|
|
MM/PBSA binding free energy of
MD2-HMPH complex
The present study further calculated the binding
free energy of MD2-HMPH compared with MD2-curcumin complexes using
a more reliable method, MM/PBSA. As shown in Table I, HMPH (∆GMM/PBSA, −29.56
kcal/mol) had a more energetically favourable value in the MD2
binding pocket than curcumin (∆GMM/PBSA, −25.63
kcal/mol), which indicated strong binding between MD2 and HMPH
compared with the MD2-curcumin complex. These results indicated
that the effect of HMPH on the inflammatory response in macrophages
may be achieved through interfering with MD2 protein.
Discussion
Numerous stable curcuminoid analogues have been
isolated and synthesised to investigate their potential biological
functions (23,29). The present study explored the
anti-inflammatory activity and the underlying molecular effect of
HMPH on LPS-activated RAW264.7 macrophages. To the best of our
knowledge, the present study was the first to reveal that HMPH
exhibited superior suppression of inflammatory mediators compared
with curcumin and its action was associated with the suppression of
NF-κB translocation. Furthermore, the inhibitory effect of HMPH on
the inflammatory response may be achieved through interfering with
MD2 binding, thereby interrupting the inflammatory cascade. In
previous studies, modification of the easily decomposed
α,β-unsaturated β-diketo moiety of curcumin could enhance the
stability and anti-inflammatory effect of curcuminoid analogues
(23,30), and thus, this may be the reason why
HMPH containing unsaturated trienone linker possessed superior
anti-inflammatory effects than curcumin.
Upon inflammation, LPS forms a complex with plasma
LPS-binding protein, which leads to the transfer of the complexes
to CD14 located at the cell membrane of macrophages (3). CD14 transfers LPS to the receptor
complex, which consists of TLR4 and MD2, leading to induction of
myeloid differentiation 88-dependent and TIR-domain-containing
adapter-inducing interferon-β-dependent signalling, thereby
directly activating the downstream signalling proteins of the NF-κB
pathway (31). The
anti-inflammatory action of curcumin in macrophages has also been
shown to be due to interfering with LPS binding to the TLR4/MD2
complex and blocking of signalling molecules in the inflammatory
pathway (21). It has been reported
that curcumin may form hydrogen bonds with residues R90 and Y102 of
the MD2 protein, and the binding affinity of these two interactions
was decreased in the MD2R90A/Y102A mutant system (32). In silico modelling revealed
that HMPH formed a stronger interaction with the MD2 protein than
curcumin. The present results are consistent with those of previous
reports, which reported that the amino acids R90, Y102 and C133
were possible key residues in the interaction between the MD2
protein and small molecules, such as curcumin and synthetic
chalcone L6H21 (21,32,33).
The three curcuminoid analogues L48H37, MAC17 and MAC28 have been
shown to attenuate the LPS-induced TLR4/MD2-downstream
proinflammatory NF-κB signalling pathway possibly through binding
with the R90 residue of the MD2 protein (10,34).
It was suggested that inhibition of MD2 by curcuminoid analogues at
the possible key residues may interfere with the upstream
components of the NF-κB signalling pathway, thus leading to the
attenuation of inflammation. However, further research is required
to confirm these findings.
Macrophages secrete large amounts of
pro-inflammatory mediators that can be harmful and result in a
deleterious effect during the inflammatory response (1). A previous study demonstrated that
curcumin markedly reduced inflammation via suppression of iNOS/NO,
COX-2, and the inflammatory cytokines IL-1β, IL-6 and TNF-α in
LPS-activated RAW264.7 cells by targeting multiple signalling
pathways (19,20,35).
It has also been reported that the mechanism of action of curcumin
may involve modulation of the NF-κB signalling pathway (20,36).
Furthermore, curcumin has been reported to inhibit LPS-induced IκBα
degradation, with concomitant inhibition of p65 and p50 nuclear
translocation in RAW264.7 cells (37). Similar to these findings with
curcumin, the present study demonstrated that HMPH could
significantly inhibit iNOS/NO, COX-2 and inflammatory cytokine
production via suppression of p65 NF-κB translocation into the
nucleus in a model of LPS-activated RAW264.7 cells. This evidence
may support the potential use of HMPH to attenuate macrophage
inflammatory activities.
The present study had some limitations. Typically,
the translocation of NF-κB into the nucleus after activation is
consistently related to its functions as a transcription factor. In
a previous study, the inhibition of nuclear p65 NF-κB translocation
by mycoepoxydiene in LPS-activated RAW264.7 cells was concomitant
with the suppression of NF-κB DNA binding and transcriptional
activities, which consequently attenuated the inflammatory response
(38). However, the lack of
electromobility shift assay (EMSA) detection was a limitation of
the present study. To further support the present findings, the
inhibition of nuclear NF-κB translocation by HMPH should be
conducted in parallel with EMSA detection to provide a better
explanation of the function of NF-κB in the transcriptional
regulation of target genes involved in the inflammatory response.
In addition, there are some challenges in assessing the interaction
of HMPH with MD2. The present study demonstrated a strong binding
between HMPH and MD2 using an in silico study; however, to
the best of our knowledge, there have been no reports on the
complex formation of HMPH with MD2, and its effects on the
suppression of NF-κB activation and inflammation. It would be
interesting to conduct further experiments based on the binding
affinity of HMPH with MD2 or mutant MD2, and the subsequent effects
on the inhibition of NF-κB activation and inflammation.
In conclusion, HMPH possessed greater
anti-inflammatory activity than curcumin via suppression of
inflammatory mediators in LPS-activated RAW264.7 macrophages. The
present finding provided an insight into the molecular mechanism by
which HMPH attenuates inflammation through the suppression of
iNOS/NO, COX-2, inflammatory cytokines and NF-κB translocation.
In silico modelling demonstrated that HMPH formed two stable
hydrogen bonds with the key residues R90 and C133 of the MD2
protein, and thus, may block inflammatory signalling pathways.
These findings indicated that HMPH may exert potential
anti-inflammatory effects and may be considered a promising novel
therapeutic agent for the treatment of inflammatory diseases.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Walailak University
Fund (grant nos. WU61201, 020101/02/2559 and 09/2561), The Royal
Golden Jubilee Ph.D. Program (grant no. RGJ PHD/0216/2561), The
Thailand Research Fund (grant no. DBG6180030) and the Center of
Excellence for Innovation in Chemistry (PERCH-CIC).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
CJ, WanC, WaralC, PY and KL performed experiments,
analysed data and reviewed the manuscript. WiC, ASa, PH, KK and TU
participated in study design and supervised the study. ASu and
WaranC aided in the experimental design, refined the manuscript,
and corresponded during the manuscript submission and revision. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
Glossary
Abbreviations
Abbreviations:
|
COX
|
cyclooxygenase
|
|
iNOS
|
inducible nitric oxide synthase
|
|
LPS
|
lipopolysaccharide
|
|
MD2
|
myeloid differentiation factor 2
|
|
NF-κB
|
nuclear factor-κB
|
|
NO
|
nitric oxide
|
|
TLR4
|
Toll-like receptor 4
|
References
|
1
|
Karakike E, Adami ME, Lada M, Gkavogianni
T, Koutelidakis IM, Bauer M, Giamarellos-Bourboulis EJ and
Tsangaris I: Late peaks of HMGB1 and sepsis outcome evidence for
synergy with chronic inflammatory disorders. Shock. 52:334–339.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stengel S, Quickert S, Lutz P, Ibidapo-Obe
O, Steube A, Köse-Vogel N, Yarbakht M, Reuken PA, Busch M, Brandt
A, et al: Peritoneal level of CD206 associates with mortality and
an inflammatory macrophage phenotype in patients with decompensated
cirrhosis and spontaneous bacterial peritonitis. Gastroenterology.
158:1745–1761. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rajaiah R, Perkins DJ, Ireland DD and
Vogel SN: CD14 dependence of TLR4 endocytosis and TRIF signaling
displays ligand specificity and is dissociable in endotoxin
tolerance. Proc Natl Acad Sci USA. 112:8391–8396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen L, Deng H, Cui H, Fang J, Zuo Z, Deng
J, Li Y, Wang X and Zhao L: Inflammatory responses and
inflammation-associated diseases in organs. Oncotarget.
9:7204–7218. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pizzuto M, Lonez C, Baroja-Mazo A,
Martínez-Banaclocha H, Tourlomousis P, Gangloff M, Pelegrin P,
Ruysschaert JM, Gay NJ and Bryant CE: Saturation of acyl chains
converts cardiolipin from an antagonist to an activator of
Toll-like receptor-4. Cell Mol Life Sci. 76:3667–3678. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taniguchi K and Karin M: NF-κB,
inflammation, immunity and cancer: Coming of age. Nat Rev Immunol.
18:309–324. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Obaid M, Udden SMN, Deb P, Shihabeddin N,
Zaki MH and Mandal SS: LncRNA HOTAIR regulates
lipopolysaccharide-induced cytokine expression and inflammatory
response in macrophages. Sci Rep. 8:156702018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Aoki T, Frȍsen J, Fukuda M, Bando K, Shioi
G, Tsuji K, Ollikainen E, Nozaki K, Laakkonen J and Narumiya S:
Prostaglandin E2-EP2-NF-κB signaling in macrophages as a potential
therapeutic target for intracranial aneurysms. Sci Signal.
10:eaah60372017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Manček-Keber M, Gradišar H, Iñigo Pestaña
M, Martinez de Tejada G and Jerala R: Free thiol group of MD-2 as
the target for inhibition of the lipopolysaccharide-induced cell
activation. J Biol Chem. 284:19493–19500. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Shan X, Dai Y, Jiang L, Chen G,
Zhang Y, Wang Z, Dong L, Wu J, Guo G, et al: Curcumin analog L48H37
prevents lipopolysaccharide-induced TLR4 signaling pathway
activation and sepsis via targeting MD2. J Pharmacol Exp Ther.
353:539–550. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kocaadam B and Şanlier N: Curcumin, an
active component of turmeric (Curcuma longa), and its
effects on health. Crit Rev Food Sci Nutr. 57:2889–2895. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akter J, Hossain MA, Takara K, Islam MZ
and Hou DX: Antioxidant activity of different species and varieties
of turmeric (Curcuma spp): Isolation of active compounds.
Comp Biochem Physiol C Toxicol Pharmacol. 215:9–17. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Park SY and Kim DS: Discovery of natural
products from Curcuma longa that protect cells from
beta-amyloid insult: A drug discovery effort against Alzheimer's
disease. J Nat Prod. 65:1227–1231. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chuprajob T, Changtam C, Chokchaisiri R,
Chunglok W, Sornkaew N and Suksamrarn A: Synthesis, cytotoxicity
against human oral cancer KB cells and structure-activity
relationship studies of trienone analogues of curcuminoids. Bioorg
Med Chem Lett. 24:2839–2844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang R, Zhang X, Chen C, Chen G, Zhong Q,
Zhang Q, Zheng S, Wang G and Chen QH: Synthesis and evaluation of
1,7-diheteroarylhepta-1,4,6-trien-3-ones as curcumin-based
anticancer agents. Eur J Med Chem. 110:164–180. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Utaipan T, Boonyanuphong P, Chuprajob T,
Suksamrarn A and Chunglok W: A trienone analog of curcumin, 1,7-bis
(3-hydroxyphenyl)-1,4,6-heptatrien-3-one, possesses ROS- and
caspase-mediated apoptosis in human oral squamous cell carcinoma
cells in vitro. Appl Biol Chem. 63:72020. View Article : Google Scholar
|
|
17
|
Srivilai J, Rabgay K, Khorana N, Waranuch
N, Nuengchamnong N, Wisuitiprot W, Chuprajob T, Changtam C,
Suksamrarn A, Chavasiri W, et al: Anti-androgenic curcumin
analogues as steroid 5-alpha reductase inhibitors. Med Chem Res.
26:1550–1556. 2017. View Article : Google Scholar
|
|
18
|
Jang MK, Sohn DH and Ryu JH: A curcuminoid
and sesquiterpenes as inhibitors of macrophage TNF-α release from
Curcuma zedoaria. Planta Med. 67:550–552. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tan RZ, Liu J, Zhang YY, Wang HL, Li JC,
Liu YH, Zhong X, Zhang YW, Yan Y, Lan HY, et al: Curcumin relieved
cisplatin-induced kidney inflammation through inhibiting
Mincle-maintained M1 macrophage phenotype. Phytomedicine.
52:284–294. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhao F, Gong Y, Hu Y, Lu M, Wang J, Dong
J, Chen D, Chen L, Fu F and Qiu F: Curcumin and its major
metabolites inhibit the inflammatory response induced by
lipopolysaccharide: Translocation of nuclear factor-κB as potential
target. Mol Med Rep. 11:3087–3093. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gradišar H, Keber MM, Pristovšek P and
Jerala R: MD-2 as the target of curcumin in the inhibition of
response to LPS. J Leukoc Biol. 82:968–974. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mirzaei H, Shakeri A, Rashidi B, Jalili A,
Banikazemi Z and Sahebkar A: Phytosomal curcumin: A review of
pharmacokinetic, experimental and clinical studies. Biomed
Pharmacother. 85:102–112. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koeberle A, Muñoz E, Appendino GB, Minassi
A, Pace S, Rossi A, Weinigel C, Barz D, Sautebin L, Caprioglio D,
et al: SAR studies on curcumin's pro-inflammatory targets:
Discovery of prenylated pyrazolocurcuminoids as potent and
selective novel inhibitors of 5-lipoxygenase. J Med Chem.
57:5638–5648. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Changtam C, de Koning HP, Ibrahim H, Sajid
MS, Gould MK and Suksamrarn A: Curcuminoid analogs with potent
activity against Trypanosoma and Leishmania species.
Eur J Med Chem. 45:941–956. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun J, Zhang X, Broderick M and Fein H:
Measurement of nitric oxide production in biological systems by
using Griess reaction assay. Sensors (Basel). 3:276–284. 2003.
View Article : Google Scholar
|
|
26
|
Morris GM, Huey R, Lindstrom W, Sanner MF,
Belew RK, Goodsell DS and Olson AJ: AutoDock4 and AutoDockTools4:
Automated docking with selective receptor flexibility. J Comput
Chem. 30:2785–2791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morris G, Goodsell D, Halliday R, Huey R,
Hart W, Belew R and Olson A: Automated docking using a lamarckian
genetic algorithm and an empirical binding free energy function. J
Comput Chem. 19:1639–1662. 1998. View Article : Google Scholar
|
|
28
|
Case DA, Betz RM, Cerutti DS, Cheatham TE,
Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, et al:
AMBER 2016. University of California; San Francisco: 2016
|
|
29
|
Liang G, Li X, Chen L, Yang S, Wu X,
Studer E, Gurley E, Hylemon PB, Ye F, Li Y, et al: Synthesis and
anti-inflammatory activities of mono-carbonyl analogues of
curcumin. Bioorg Med Chem Lett. 18:1525–1529. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao C, Zhang Y, Zou P, Wang J, He W, Shi
D, Li H, Liang G and Yang S: Synthesis and biological evaluation of
a novel class of curcumin analogs as anti-inflammatory agents for
prevention and treatment of sepsis in mouse model. Drug Des Devel
Ther. 9:1663–1678. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ullah MO, Sweet MJ, Mansell A, Kellie S
and Kobe B: TRIF-dependent TLR signaling, its functions in host
defense and inflammation, and its potential as a therapeutic
target. J Leukoc Biol. 100:27–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang Z, Chen G, Chen L, Liu X, Fu W, Zhang
Y, Li C, Liang G and Cai Y: Insights into the binding mode of
curcumin to MD-2: Studies from molecular docking, molecular
dynamics simulations and experimental assessments. Mol Biosyst.
11:1933–1938. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang Y, Shan X, Chen G, Jiang L, Wang Z,
Fang Q, Liu X, Wang J, Zhang Y, Wu W, et al: MD-2 as the target of
a novel small molecule, L6H21, in the attenuation of LPS-induced
inflammatory response and sepsis. Br J Pharmacol. 172:4391–4405.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Y, Liu Z, Wu J, Bai B, Chen H, Xiao
Z, Chen L, Zhao Y, Lum H, Wang Y, et al: New MD2 inhibitors derived
from curcumin with improved anti-inflammatory activity. Eur J Med
Chem. 148:291–305. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li B, Hu Y, Zhao Y, Cheng M, Qin H, Cheng
T, Wang Q, Peng X and Zhang X: Curcumin attenuates titanium
particle-induced inflammation by regulating macrophage polarization
in vitro and in vivo. Front Immunol. 8:552017.PubMed/NCBI
|
|
36
|
Zhang J, Zheng Y, Luo Y, Du Y, Zhang X and
Fu J: Curcumin inhibits LPS-induced neuroinflammation by promoting
microglial M2 polarization via TREM2/TLR4/NF-κB pathways in BV2
cells. Mol Immunol. 116:29–37. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oh SW, Cha JY, Jung JE, Chang BC, Kwon HJ,
Lee BR and Kim DY: Curcumin attenuates allergic airway inflammation
and hyper-responsiveness in mice through NF-κB inhibition. J
Ethnopharmacol. 136:414–421. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen Q, Chen T, Li W, Zhang W, Zhu J, Li
Y, Huang Y, Shen Y and Yu C: Mycoepoxydiene inhibits
lipopolysaccharide-induced inflammatory responses through the of
TRAF6 polyubiquitination. PLoS One. 7:e448902012. View Article : Google Scholar : PubMed/NCBI
|