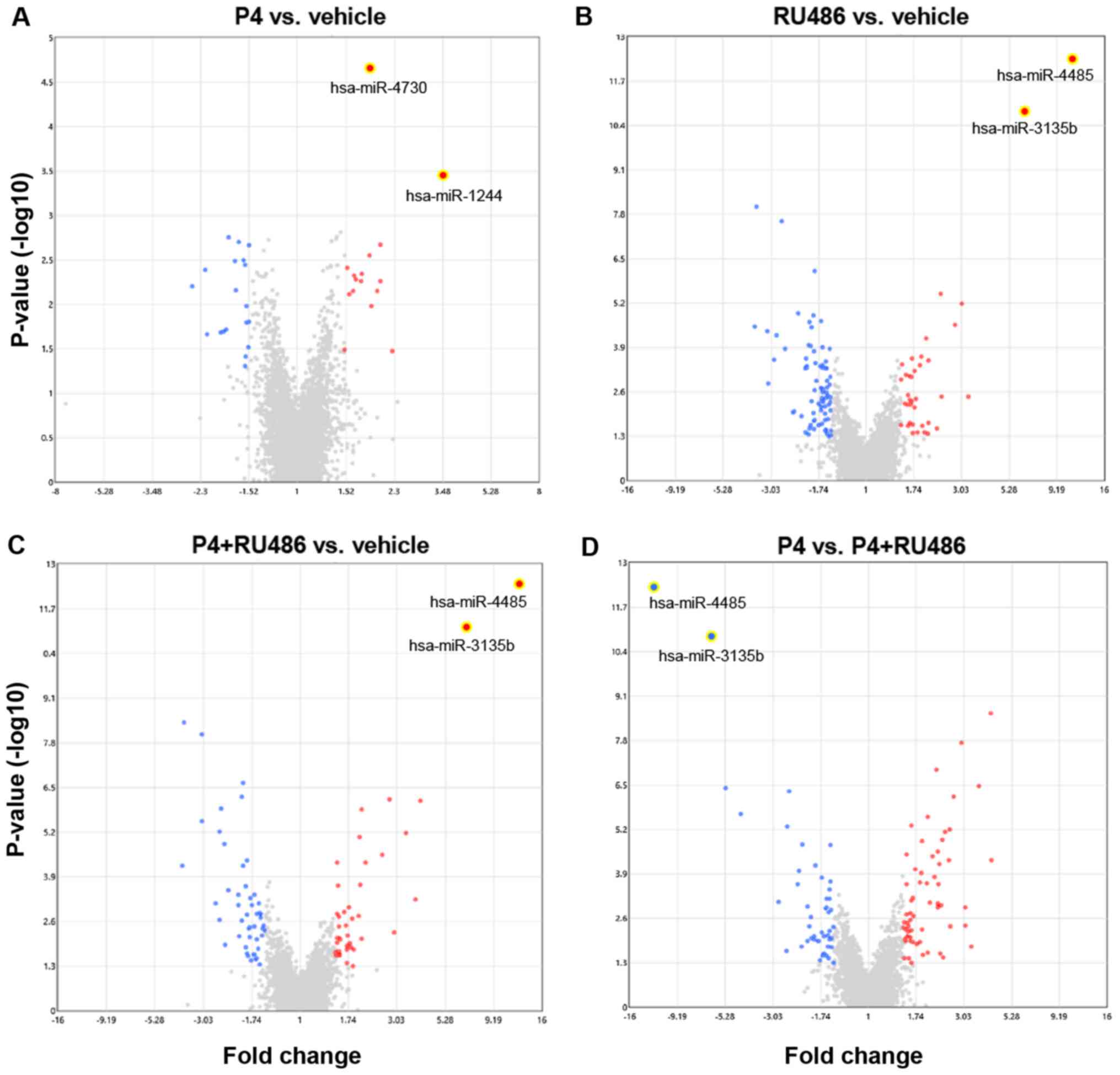
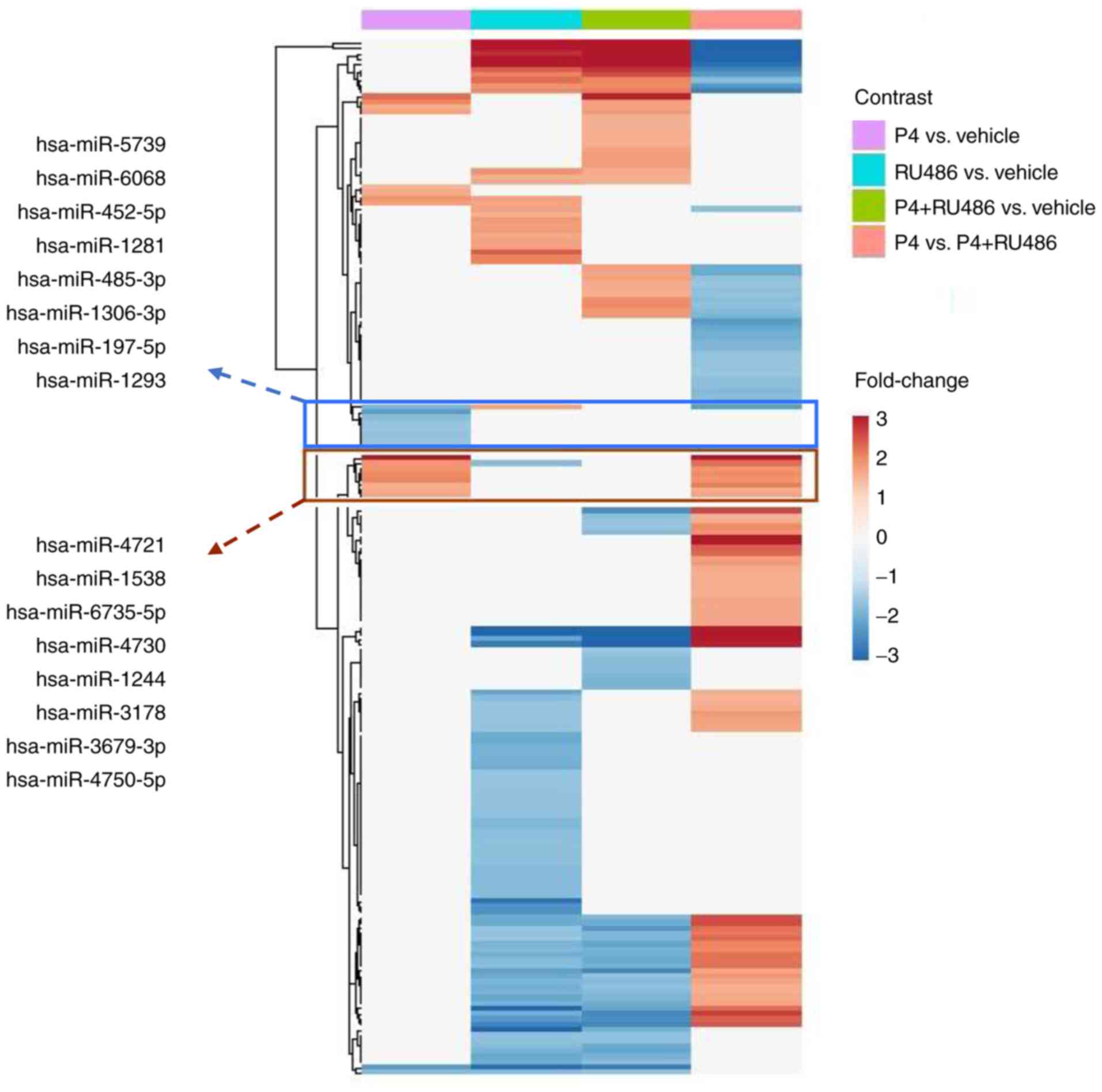
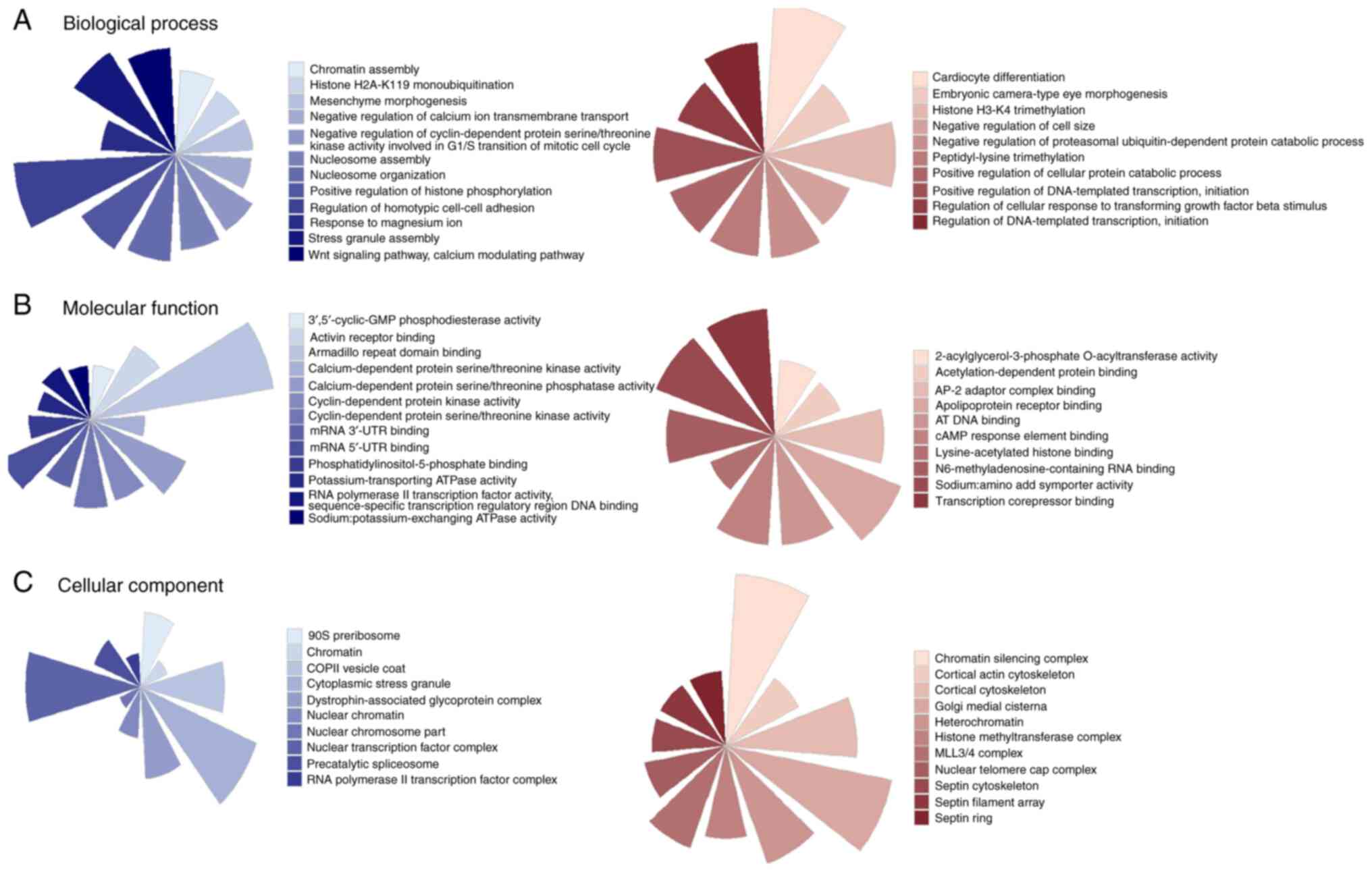
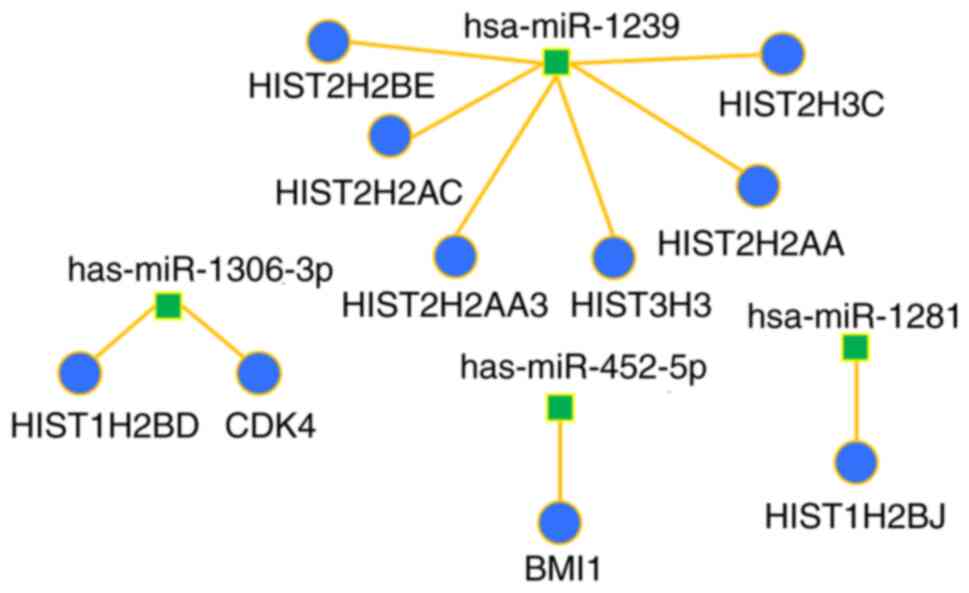
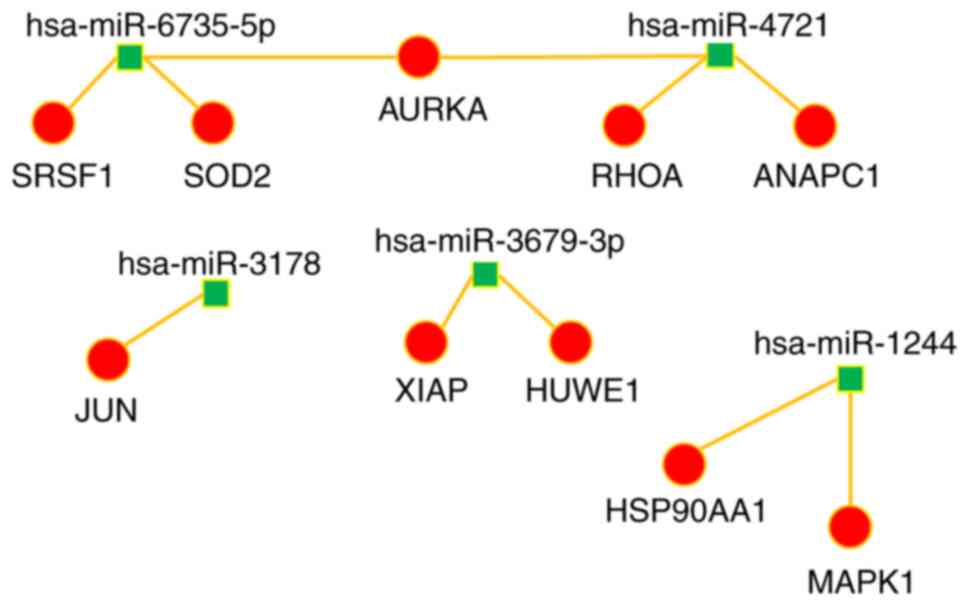
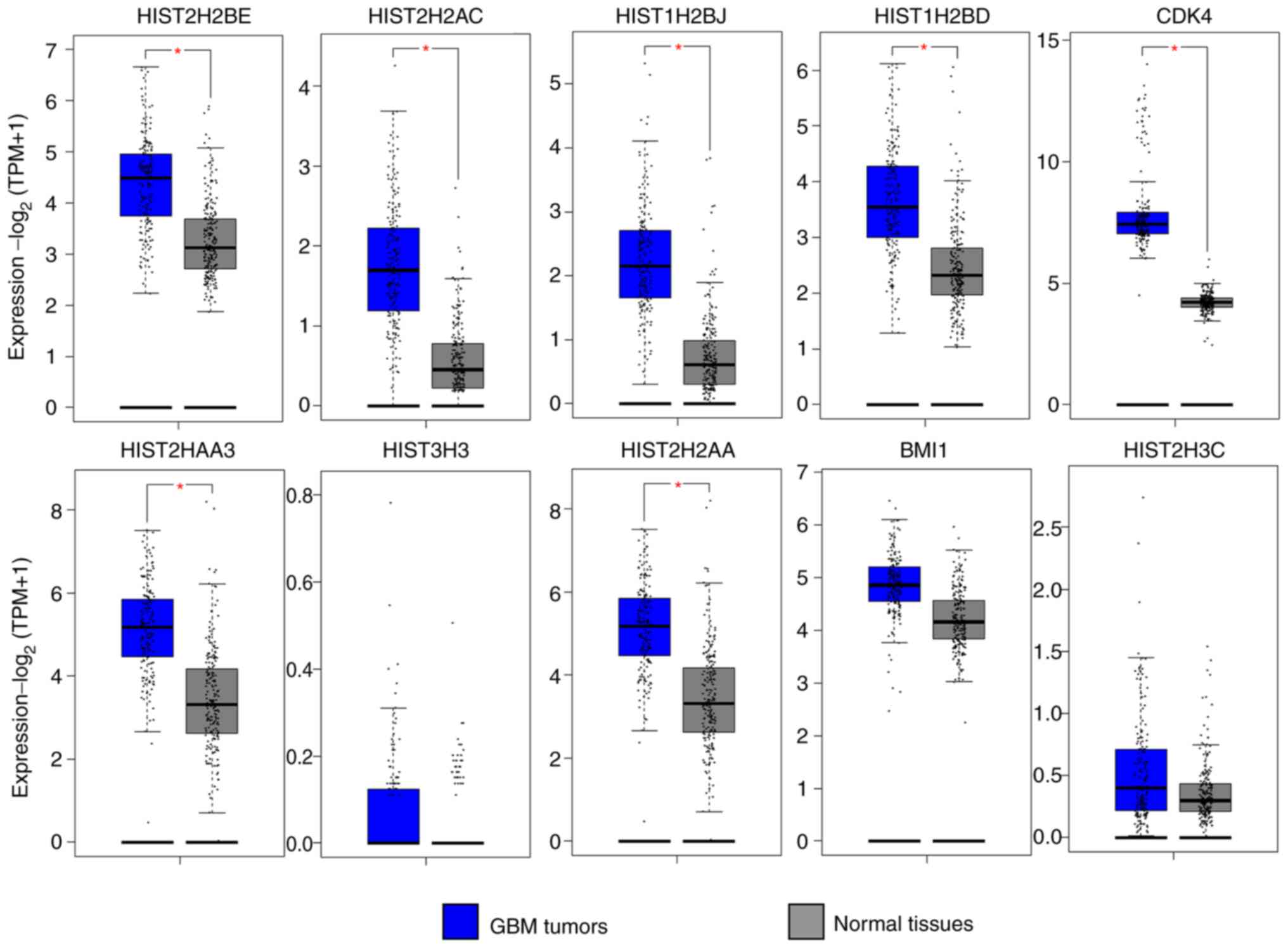
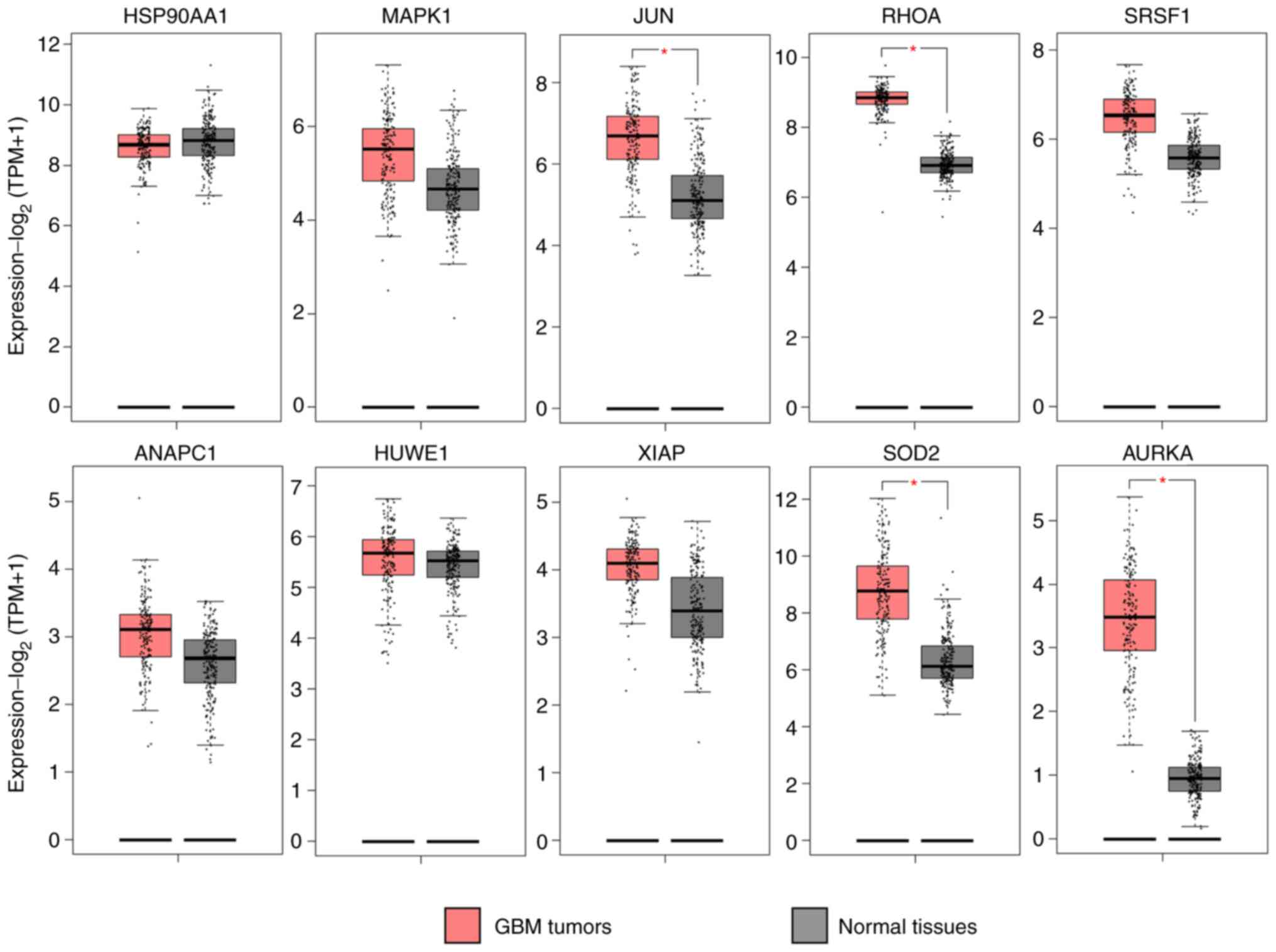
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Urbanska K, Sokolowska J, Szmidt M and
Sysa P: Glioblastoma multiforme-An overview. Contemp Oncol (Pozn).
18:307–312. 2014.PubMed/NCBI
|
|
3
|
Weller M, Wick W, Aldape K, Brada M,
Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R and
Reifenberger G: Glioma. Nat Rev Dis Primers. 1:150172015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ostrom QT, Gittleman H, Liao P,
Vecchione-Koval T, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and other central nervous
system tumors diagnosed in the United States in 2010–2014. Neuro
Oncol. 19 (Suppl 5):v1–v88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ciafrè SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar
|
|
6
|
Novakova J, Slaby O, Vyzula R and Michalek
J: MicroRNA involvement in glioblastoma pathogenesis. Biochem
Biophys Res Commun. 386:1–5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aldaz B, Sagardoy A, Nogueira L, Guruceaga
E, Grande L, Huse JT, Aznar MA, Díez-Valle R, Tejada-Solís S,
Alonso MM, et al: Involvement of miRNAs in the differentiation of
human glioblastoma multiforme stem-like cells. PLoS One.
8:e770982013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alfardus H, Mcintyre A and Smith S:
MicroRNA regulation of glycolytic metabolism in glioblastoma.
Biomed Res Int. 2017:91573702017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercatelli N, Galardi S and Ciafrè SA:
MicroRNAs as multifaceted players in glioblastoma multiforme. Int
Rev Cell Mol Biol. 333:269–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sana J, Busek P, Fadrus P, Besse A, Radova
L, Vecera M, Reguli S, Stollinova Sromova L, Hilser M, Lipina R, et
al: Identification of microRNAs differentially expressed in
glioblastoma stem-like cells and their association with patient
survival. Sci Rep. 8:28362018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ceballos-Chávez M, Subtil-Rodríguez A,
Giannopoulou EG, Soronellas D, Vázquez-Chávez E, Vicent GP,
Elemento O, Beato M and Reyes JC: The chromatin remodeler CHD8 is
required for activation of progesterone receptor-dependent
enhancers. PLoS Genet. 11:e10051742015. View Article : Google Scholar
|
|
12
|
Helsen C and Claessens F: Looking at
nuclear receptors from a new angle. Mol Cell Endocrinol.
382:97–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jacobsen BM and Horwitz KB: Progesterone
receptors, their isoforms and progesterone regulated transcription.
Mol Cell Endocrinol. 357:18–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
González-Agüero G, Gutiérrez AA,
González-Espinosa D, Solano JD, Morales R, González-Arenas A,
Cabrera-Muñoz E and Camacho-Arroyo I: Progesterone effects on cell
growth of U373 and D54 human astrocytoma cell lines. Endocrine.
32:129–135. 2007. View Article : Google Scholar
|
|
15
|
Piña-Medina AG, Hansberg-Pastor V,
González-Arenas A, Cerbón M and Camacho-Arroyo I: Progesterone
promotes cell migration, invasion and cofilin activation in human
astrocytoma cells. Steroids. 105:19–25. 2016. View Article : Google Scholar
|
|
16
|
Germán-Castelán L, Manjarrez-Marmolejo J,
González-Arenas A, González-Morán MG and Camacho-Arroyo I:
Progesterone induces the growth and infiltration of human
astrocytoma cells implanted in the cerebral cortex of the rat.
Biomed Res Int. 2014:3931742014. View Article : Google Scholar
|
|
17
|
González-Arenas A, Cabrera-Wrooman A, Díaz
NF, González- García TK, Salido-Guadarrama I, Rodríguez-Dorantes M
and Camacho-Arroyo I: Progesterone receptor subcellular
localization and gene expression profile in human astrocytoma cells
are modified by progesterone. Nucl Recept Res. 1:1–10. 2014.
View Article : Google Scholar
|
|
18
|
Zamora-Sánchez CJ, Hansberg-Pastor V,
Salido-Guadarrama I, Rodríguez-Dorantes M and Camacho-Arroyo I:
Allopregnanolone promotes proliferation and differential gene
expression in human glioblastoma cells. Steroids. 119:36–42. 2017.
View Article : Google Scholar
|
|
19
|
Zamora-Sánchez CJ, Del Moral-Morales A,
Hernández-Vega AM, Hansberg-Pastor V, Salido-Guadarrama I,
Rodríguez-Dorantes M and Camacho-Arroyo I: Allopregnanolone alters
the gene expression profile of human glioblastoma cells. Int J Mol
Sci. 19:8642018. View Article : Google Scholar
|
|
20
|
Hernández-Hernández OT, González-García TK
and Camacho-Arroyo I: Progesterone receptor and SRC-1 participate
in the regulation of VEGF, EGFR and cyclin D1 expression in human
astrocytoma cell lines. J Steroid Biochem Mol Biol. 132:127–134.
2012. View Article : Google Scholar
|
|
21
|
Wendler A, Keller D, Albrecht C, Peluso JJ
and Wehiling M: Involvement of let-7/miR-98 microRNAs in the
regulation of progesterone receptor membrane component 1 expression
in ovarian cancer cells. Oncol Rep. 25:273–279. 2011.PubMed/NCBI
|
|
22
|
Cittelly DM, Finlay-Schultz J, Howe EN,
Spoelstra NS, Axlund SD, Hendricks P, Jacobsen BM, Sartorius CA and
Richer JK: Progestin suppression of miR-29 potentiates
dedifferentiation of breast cancer cells via KLF4. Oncogene.
32:2555–2564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rivas MA, Venturutti L, Huang YW,
Schillaci R, Huang TH and Elizalde PV: Downregulation of the
tumor-suppressor miR-16 via progestin-mediated oncogenic signaling
contributes to breast cancer development. Breast Cancer Res.
14:R772012. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez
G, Bautista-Piña V, Arellano-Llamas R and Hidalgo-Miranda A:
Identification and pathway analysis of microRNAs with no previous
involvement in breast cancer. PLoS One. 7:e319042012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cochrane DR, Jacobsen BM, Connaghan KD,
Howe EN, Bain DL and Richer JK: Progestin regulated miRNAs that
mediate progesterone receptor action in breast cancer. Mol Cell
Endocrinol. 355:15–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Finlay-Schultz J, Cittelly DM, Hendricks
P, Patel P, Kabos P, Jacobsen BM, Richer JK and Sartorius CA:
Progesterone downregulation of miR-141 contributes to expansion of
stem-like breast cancer cells through maintenance of progesterone
receptor and Stat5a. Oncogene. 34:3676–3687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fletcher CE, Dart DA and Bevan CL:
Interplay between steroid signalling and microRNAs: Implications
for hormone-dependent cancers. Endocr Relat Cancer. 21:R409–R429.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Godbole M, Chandrani P, Gardi N, Dhamne H,
Patel K, Yadav N, Gupta S, Badwe R and Dutt A: miR-129-2 mediates
down-regulation of progesterone receptor in response to
progesterone in breast cancer cells. Cancer Biol Ther. 18:801–805.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McFall T, McKnight B, Rosati R, Kim S,
Huang Y, Viola-Villegas N and Ratnam M: Progesterone receptor A
promotes invasiveness and metastasis of luminal breast cancer by
suppressing regulation of critical microRNAs by estrogen. J Biol
Chem. 293:1163–1177. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nie L, Zhao YB, Pan JL, Lei Y, Liu M, Long
Y, Zhang JH, Hu Y, Xu MQ, Yuan DZ and Yue LM: Progesterone-Induced
miR-152 inhibits the proliferation of endometrial epithelial cells
by downregulating WNT-1. Reprod Sci. 24:1444–1453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Smyth GK: limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Springer; New York, NY: pp. 397–420. 2005,
View Article : Google Scholar
|
|
32
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: 2018, http://www.R-project.org/
|
|
33
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46(D1): D239–D245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dweep H, Gretz N and Sticht C: MiRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41((Web Server Issue)): W169–W73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z,
Meirelles GV, Clark NR and Ma'ayan A: Enrichr: Interactive and
collaborative HTML5 gene list enrichment analysis tool. BMC
Bioinformatics. 14:1282013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gene Ontology Consortium, . Gene ontology
consortium: Going forward. Nucleic Acids Res. 43((Database issue)):
D1049–D1056. 2015.PubMed/NCBI
|
|
39
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1): D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G and
Kerin MJ: MicroRNA signatures predict oestrogen receptor,
progesterone receptor and HER2/neu receptor status in breast
cancer. Breast Cancer Res. 11:R272009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Maillot G, Lacroix-Triki M, Pierredon S,
Gratadou L, Schmidt S, Bénès V, Roché H, Dalenc F, Auboeuf D,
Millevoi S and Vagner S: Widespread estrogen-dependent repression
of microRNAs involved in breast tumor cell growth. Cancer Res.
69:8332–8340. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun R, Fu X, Li Y, Xie Y and Mao Y: Global
gene expression analysis reveals reduced abundance of putative
microRNA targets in human prostate tumours. BMC Genomics.
10:932009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ribas J, Ni X, Haffner M, Wentzel EA,
Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J,
Rodriguez R, et al: miR-21: An androgen receptor-regulated microRNA
that promotes hormone-dependent and hormone-independent prostate
cancer growth. Cancer Res. 69:7165–7169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cadepond F, Ulmann A and Baulieu EE: RU486
(MIFEPRISTONE): Mechanisms of action and clinical uses. Annu Rev
Med. 48:129–156. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Catalano RD, Yanaihara A, Evans AL, Rocha
D, Prentice A, Saidi S, Print CG, Charnock-Jones DS, Sharkey AM and
Smith SK: The effect of RU486 on the gene expression profile in an
endometrial explant model. Mol Hum Reprod. 9:465–473. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Y, Sui R, Chen Y, Liang H, Shi J and
Piao H: Downregulation of miR-485-3p promotes glioblastoma cell
proliferation and migration via targeting RNF135. Exp Ther Med.
18:475–482. 2019.PubMed/NCBI
|
|
51
|
Yang W, Warrington NM, Taylor SJ, Whitmire
P, Carrasco E, Singleton KW, Wu N, Lathia JD, Berens ME, Kim AH, et
al: Sex differences in GBM revealed by analysis of patient imaging,
transcriptome and survival data. Sci Transl Med. 11:eaao52532019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang P, Kang W, Pan Y, Zhao X and Duan L:
Overexpression of HOXC6 promotes cell proliferation and migration
via MAPK signaling and predicts a poor prognosis in glioblastoma.
Cancer Manag Res. 11:8167–8179. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou L, Tang H, Wang F, Chen L, Ou S, Wu
T, Xu J and Guo K: Bioinformatics analyses of significant genes,
related pathways and candidate prognostic biomarkers in
glioblastoma. Mol Med Rep. 18:4185–4196. 2018.PubMed/NCBI
|
|
54
|
Zhang S, Wan Y, Pan T, Gu X, Qian C, Sun
G, Sun L, Xiang Y, Wang Z and Shi L: MicroRNA-21 inhibitor
sensitizes human glioblastoma U251 stem cells to chemotherapeutic
drug temozolomide. J Mol Neurosci. 47:346–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nicolas S, Abdellatef S, Haddad MA,
Fakhoury I and El-Sibai M: Hypoxia and EGF stimulation regulate
VEGF expression in human glioblastoma multiforme (GBM) cells by
differential regulation of the PI3K/Rho-GTPase and MAPK pathways.
Cells. 8:13972019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shan ZN, Tian R, Zhang M, Gui ZH, Wu J,
Ding M, Zhou XF and He J: MiR128-1 inhibits the growth of
glioblastoma multiforme and glioma stem-like cells via targeting
BMI1 and E2F3. Oncotarget. 7:78813–78826. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wen L, Tan Y, Dai S, Zhu Y, Meng T, Yang
X, Liu Y, Liu X, Yuan H and Hu F: VEGF-mediated tight junctions
pathological fenestration enhances doxorubicin-loaded
glycolipid-like nanoparticles traversing BBB for
glioblastoma-targeting therapy. Drug Deliv. 24:1843–1855. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bao S, Wu Q, Sathornsumetee S, Hao Y, Li
Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD and Rich JN: Stem
cell-like glioma cells promote tumor angiogenesis through vascular
endothelial growth factor. Cancer Res. 66:7843–7848. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cui X, Xu Z, Zhao Z, Sui D, Ren X, Huang
Q, Qin J, Hao L, Wang Z, Shen L and Lin S: Analysis of CD137l and
IL-17 expression in tumor tissue as prognostic indicators for
gliblastoma. Int J Biol Sci. 9:134–141. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Berenguer-Daizé C, Astorgues-Xerri L,
Odore E, Cayol M, Cvitkovic E, Noel K, Bekradda M, MacKenzie S,
Rezai K, Lokiec F, et al: OTX015 (MK-8628), a novel BET inhibitor,
displays in vitro and in vivo antitumor effects alone and in
combination with conventional therapies in glioblastoma models. Int
J Cancer. 139:2047–2055. 2016. View Article : Google Scholar
|
|
61
|
Jin X, Kim LJY, Wu Q, Wallace LC, Prager
BC, Sanvoranart T, Gimple RC, Wang X, Mack SC, Miller TE, et al:
Targeting glioma stem cells through combined BMI1 and EZH2
inhibition. Nat Med. 23:1352–1361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Elango R, Vishnubalaji R, Manikandan M,
Binhamdan SI, Siyal AA, Alshawakir YA, Alfayez M, Aldahmash A and
Alajez NM: Concurrent targeting of BMI1 and CDK4/6 abrogates tumor
growth in vitro and in vivo. Sci Rep. 9:136962019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Peng G, Liao Y and Shen C: miRNA-429
inhibits astrocytoma proliferation and invasion by targeting BMI1.
Pathol Oncol Res. 23:369–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Antonyak MA, Kenyon LC, Godwin AK, James
DC, Emlet DR, Okamoto I, Tnani M, Holgado-Madruga M, Moscatello DK
and Wong AJ: Elevated JNK activation contributes to the
pathogenesis of human brain tumors. Oncogene. 21:5038–5046. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tsuiki H, Tnani M, Okamoto I, Kenyon LC,
Emlet DR, Holgado-Madruga M, Lanham IS, Joynes CJ, Vo KT and Wong
AJ: Constitutively active forms of c-Jun NH2-terminal kinase are
expressed in primary glial tumors. Cancer Res. 63:250–255.
2003.PubMed/NCBI
|
|
66
|
Cui J, Han SY, Wang C, Su W, Harshyne L,
Holgado-Madruga M and Wong AJ: c-Jun NH(2)-terminal kinase 2alpha2
promotes the tumorigenicity of human glioblastoma cells. Cancer
Res. 66:10024–10031. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yu OM, Benitez JA, Plouffe SW, Ryback D,
Klein A, Smith J, Greenbaum J, Delatte B, Rao A, Guan KL, et al:
YAP and MRTF-A, transcriptional co-activators of RhoA-mediated gene
expression, are critical for glioblastoma tumorigenicity. Oncogene.
37:5492–5507. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu G, Yan T, Li X, Sun J, Zhang B, Wang H
and Zhu Y: Daam1 activates RhoA to regulate Wnt5a-induced
glioblastoma cell invasion. Oncol Rep. 39:465–472. 2018.PubMed/NCBI
|
|
69
|
Kusaczuk M, Krętowski R, Naumowicz M,
Stypułkowska A and Cechowska-Pasko M: Silica nanoparticle-induced
oxidative stress and mitochondrial damage is followed by activation
of intrinsic apoptosis pathway in glioblastoma cells. Int J
Nanomedicine. 13:2279–2294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cholia RP, Kumari S, Kumar S, Kaur M, Kaur
M, Kumar R, Dhiman M and Mantha AK: An in vitro study ascertaining
the role of H2O2 and glucose oxidase in modulation of antioxidant
potential and cancer cell survival mechanisms in glioblastoma U-87
MG cells. Metab Brain Dis. 32:1705–1716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tanaka H, Mizuno M, Katsumata Y, Ishikawa
K, Kondo H, Hashizume H, Okazaki Y, Toyokuni S, Nakamura K,
Yoshikawa N, et al: Oxidative stress-dependent and -independent
death of glioblastoma cells induced by non-thermal plasma-exposed
solutions. Sci Rep. 9:136572019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: EIF4A3-induced circular RNA MMP9 (circMMP9)
acts as a sponge of miR-124 and promotes glioblastoma multiforme
cell tumorigenesis. Mol Cancer. 17:1662018. View Article : Google Scholar : PubMed/NCBI
|