Introduction
Glioblastoma multiforme (GBM), a grade IV
astrocytoma, is the most common and aggressive brain tumor in
adults. It is characterized by being highly infiltrative,
angiogenic and resistant to chemotherapy and radiotherapy. The
medical history of patients with GBM is short as few of them
survive more than one year (1–3). GBM
is mainly diagnosed in adults >50 years old, but it can occur at
any age and the incidence is higher in men than in women (3:2)
(4).
Studies have focused on the identification of new
biomarkers and therapeutic agents in GBM. Of particular interest
are the microRNAs (miRNAs), which are single-stranded, short,
non-coding RNA sequences with a length between 18 and 25
nucleotides that regulate gene expression at the
post-transcriptional level by silencing their mRNA targets through
binding to their 3′-untranslated region (3′-UTR). Compelling
evidence continues to accumulate that miRNAs are involved in
cancer-related signaling pathways associated with gliomagenesis,
proliferation, apoptosis, invasion and malignancy of GBM (5–10).
Progesterone (P4), a sex steroid hormone, can exert
its effects through a classical mechanism by binding the
intracellular progesterone receptor (PR), a ligand-dependent
transcription factor. Upon activation, PRs are dimerized and
translocated to the nucleus where they recruit coactivators and
chromatin-remodeling complexes to activate transcription of
progesterone responsive genes (11–13).
The participation of PR in P4 actions in GBMs has been documented.
In two human astrocytoma cell lines (U373, grade III and D54, grade
IV), González-Agüero et al (14) studied the effects of progesterone
and RU486 on cell growth; at a concentration of 10 nM, P4 increased
the number of D54 and U373 cells. Also, it was observed that P4
increased S phase of the cell cycle in U373 cells (14). In scratch-wound and Transwell
assays, P4 increases the number of D54 and U-251MG cells migrating
and the number of invasive cells, respectively (15). In an in vivo study,
Germán-Castelán et al (16),
implanted U87 GBM xenografts into the cerebral cortex of male rats
and observed that P4 significantly increases GBM tumor area and
infiltration length. RU486, a PR antagonist and an oligonucleotide
antisense against PR, blocked the effects of P4.
It has been reported that P4, through PR activation,
increases proliferation, migration and invasion of cells derived
from human GBMs by regulating the expression of genes involved in
these processes including TGFβ, COF1, EGFR, VEGF and cyclin D1
(17–20). Although the role of P4 over the
expression of miRNAs in GBMs remains to be elucidated, it is known
that in breast cancer, P4 regulates the expression of miRNAs with
tumor suppressor or oncogenic action, via the classic PR (21–30).
The present study characterized the expression profile of
P4-regulated miRNAs in the U-251MG cell line, derived from a human
GBM and the biological processes that could be regulated by the
differentially expressed (DE) miRNAs.
Materials and methods
Cell culture and treatments
The U-251MG cell line derived from a human GBM was
acquired from the American Type Culture Collection (ATCC). It was
maintained in Dulbecco's modified Eagle's medium (DMEM; Biowest)
with high glucose, phenol red, supplemented with 10% fetal bovine
serum (FBS; Biowest), 1 mM sodium pyruvate (In Vitro S.A.),
0.1 mM of non-essential amino acids (In Vitro S.A.) and 1 mM
antibiotic (Streptomycin 10 g/l, Penicillin G 6.028 g/l and
Amphotericin B 0.025 g/l; In Vitro S.A.), at 37°C in a humid
atmosphere with 5% CO2. The media was replaced 12 h
before the treatments by DMEM-no phenol red (cat. no. ME-019; In
Vitro, MEX) supplemented with 10% charcoal/dextran treated FBS
(cat. no. SH30068.03; Thermo Fisher Scientific, Inc.). The
treatments consisted of P4 (10 nM), RU486 (10 µM), P4 (10 nM) +
RU486 (10 µM; all from Sigma-Aldrich; Merck KGaA) and vehicle
(cyclodextrin, 0.02%) for 6 h at 37°C.
RNA isolation
Following treatments, total RNA was extracted using
TRIzol reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The concentration and purity of the
extracted RNA were determined by the Agilent 2100 Bioanalyzer
(Agilent Technologies, Inc.) and the integrity of the RNA was
checked by electrophoresis on a 1.5% agarose gel. Only the samples
with an RNA Integrity Number of 9–10 were used.
Microarrays analysis
For miRNA expression analysis, a standard protocol
was followed. Briefly, for each sample, 250 ng of total RNA were
labelled with the FlashTag™ Biotin RNA Labeling kit
(Affymetrix®; Thermo Fisher Scientific, Inc.). The
labelled RNA was hybridized with the GeneChip miRNA 4.0 Array
(Affymetrix®; Thermo Fisher Scientific, Inc.) and then
the miRNA microarray chips were washed twice with 1X PBST (0.02%
Tween) and stained with FlashTag™ Biotin HSR (Affymetrix; Thermo
Fisher Scientific, Inc.) for 5 min at 35°C. The image was digitized
and the CEL files were generated. The fluorescence intensity values
in CEL format were loaded into Expression Console™ v1.4.1.46
software (Affymetrix; Thermo Fisher Scientific, Inc.), where they
were pre-processed with Robust Multiarray Analysis and normalized
by quartiles. The CHP files generated after normalization were
loaded into the Transcriptomic Analysis Console™ v4.0.1 software
(Affymetrix; Thermo Fisher Scientific, Inc.) for expression
analysis through the functions of the limma package (31) and for graphics generation. Heatmaps
were generated with pheatmap v1.0.12
(CRAN.R-project.org/package=pheatmap) package for R version 3.5.2
(32).
Prediction of target genes of
DE-miRNAs
A search of the target genes of DE-miRNAs was
performed in four different open access databases: DIANA-Tarbase v8
(33), miRWalk v2.0 (34), Diana-microT-CDS v5.0 (35) and TargetScan v7.2 (36). From these databases, DIANA-Tarbase
v8 and miRWalk reported validated miRNA-mRNA interactions, while
Diana-microT-CDS v5.0 and TargetScan v7.2 report only predicted
miRNA-mRNA interactions. False positives were avoided by taking the
intersections of at least 3 of these databases.
Gene Ontology (GO) enrichment analysis
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis
The target genes found in at least 3 databases were
used as input for Enrichr 3.0 platform (https://maayanlab.cloud/Enrichr/) (37) to perform a GO functional gene
annotation (38) and a KEGG pathway
enrichment analysis (39). The
resulting GO terms with P<0.05 were considered significantly
enriched.
Analysis of the protein-protein
interaction network (PPI)
The PPI network was established using the STRING
database (https://string-db.org/) (40). Hub genes were determined with the
help of Cytoscape software (v3.7.1) (41). Finally, the expression levels of the
hub genes were determined in Gene Expression Profiling Interactive
Analysis (GEPIA; v1.0; http://gepia.cancer-pku.cn/) (42), an interactive web server that was
developed to perform RNA-seq expression data analysis of 9,736
tumors and 8,587 normal samples of the Cancer Genome Atlas and
Genotype-Tissue Expression projects.
Statistical analysis
For the selection of differentially expressed miRNAs
between treatments in the microarrays, ANOVA was performed with the
t-moderated method of eBayes. The miRNAs with a fold-change (FC)
≥±1.5 and a P-value <0.05 were selected as differentially
expressed in the contrasts of treatments. All data were analyzed
and plotted by using GraphPad Prism v5.00 for Windows (GraphPad
Software, Inc.).
Results
Identification of miRNAs with
differential expression (DE-miRNAs)
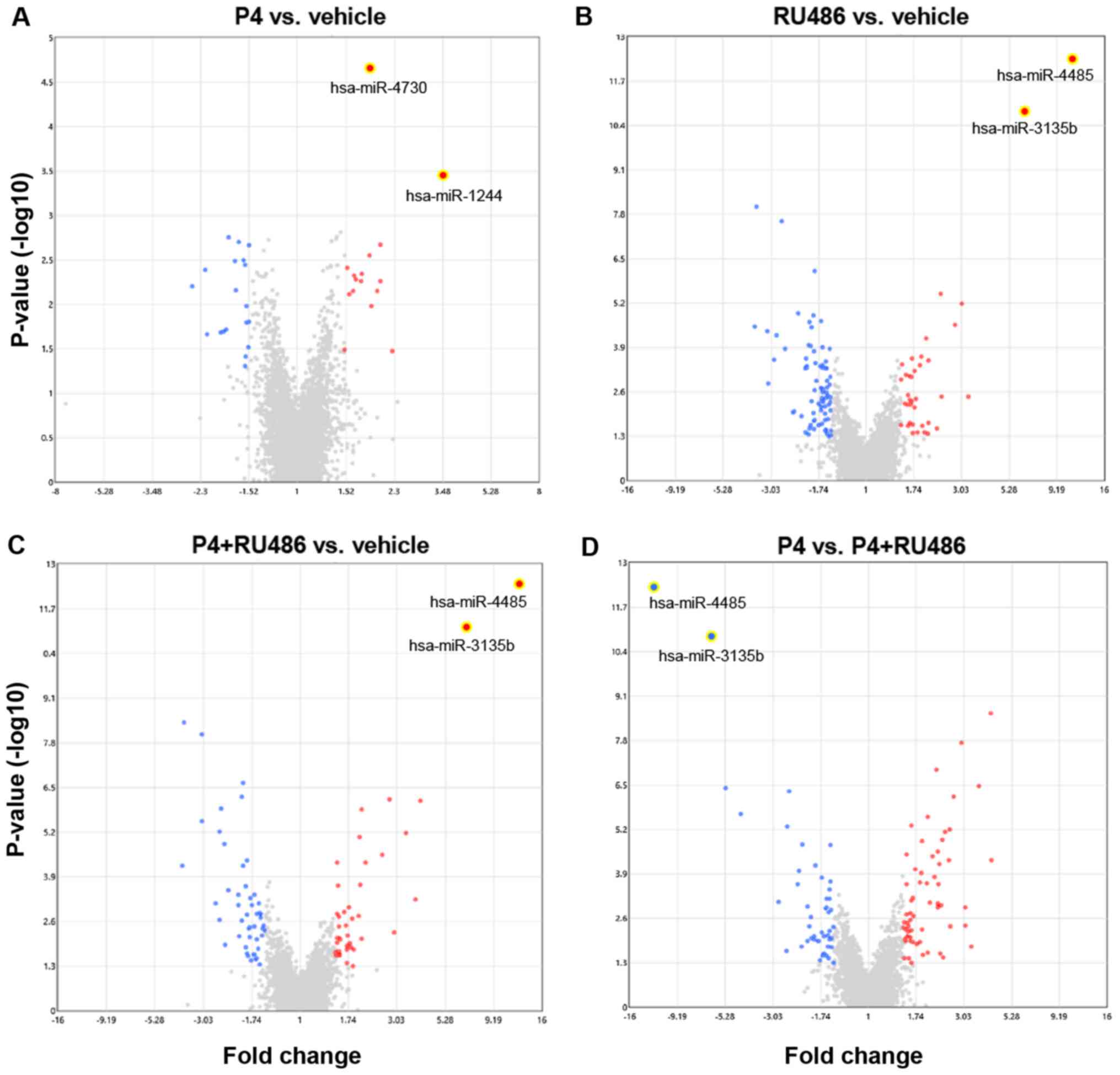
Global miRNA expression changes in the U-251MG cells
were evaluated after 6 h of treatment with vehicle (V), P4, RU486,
or P4 + RU486. A total of 190 DE-miRNAs were found after comparing
the different treatments. DE-miRNAs were evaluated in the following
comparisons (Table I): P4 vs.
vehicle (Fig. 1A), RU486 vs.
vehicle (Fig. 1B), P4+RU486 vs.
vehicle (Fig. 1C) and P4 vs.
P4+RU486 (Fig. 1D). The number of
upregulated and downregulated DE-miRNAs in each contrast is shown
in Table I. The data presented in
the present study have been deposited in NCBI's Gene Expression
Omnibus (43) and are accessible
through GEO Series accession number GSE144204.
 | Table I.Count of DE-miRNAs by comparison. |
Table I.
Count of DE-miRNAs by comparison.
| Comparison | Upregulated | Downregulated | Total |
|---|
| P4 vs. vehicle | 16 | 10 | 26 |
| RU486 vs.
vehicle | 27 | 77 | 104 |
| P4+RU486 vs.
vehicle | 38 | 47 | 85 |
| P4 vs.
P4+RU486 | 63 | 39 | 102 |
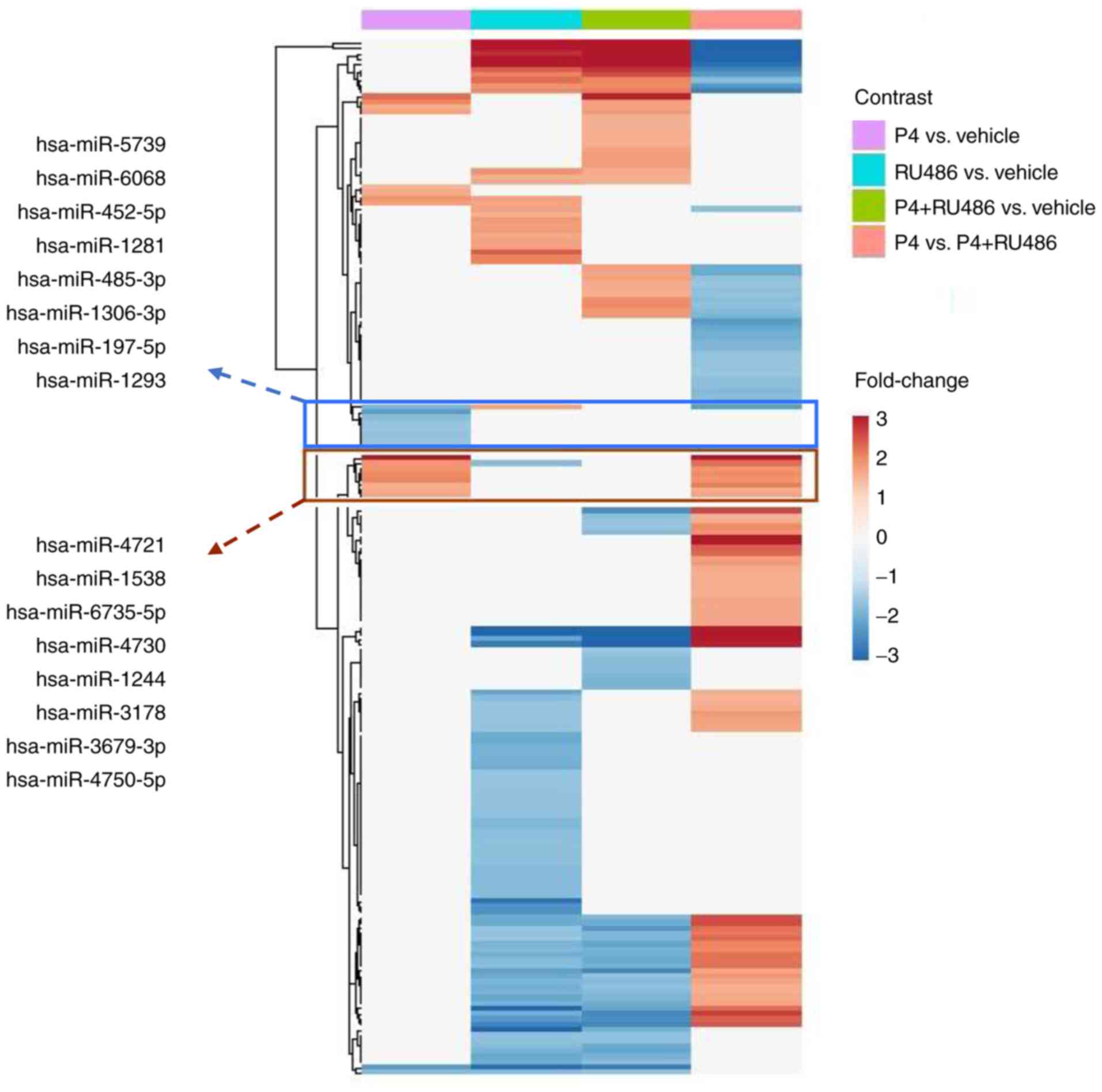
To determine the differences among treatments, a
heatmap with the DE-miRNAs was generated (Fig. 2). It was identified that P4
treatment decreased the expression of 8 miRNAs; these effects were
blocked by RU486 since none of these miRNAs exhibited a significant
differential expression in the comparisons RU486 vs. V or P4 vs.
P4+RU486; the present study also identified 8 miRNAs upregulated by
P4 compared with vehicle, however in this case, the effect of P4
was not blocked by RU486, which can be seen in the comparison of P4
vs. P4+RU486.
Prediction of molecular pathways
associated with putative targets of the DE-miRNAs
Bioinformatic analysis for the identification of
putative targets of the DE-miRNAs was performed as described in
Materials and methods. A total of 367 and 434 putative genes were
determined for downregulated and upregulated miRNAs, respectively
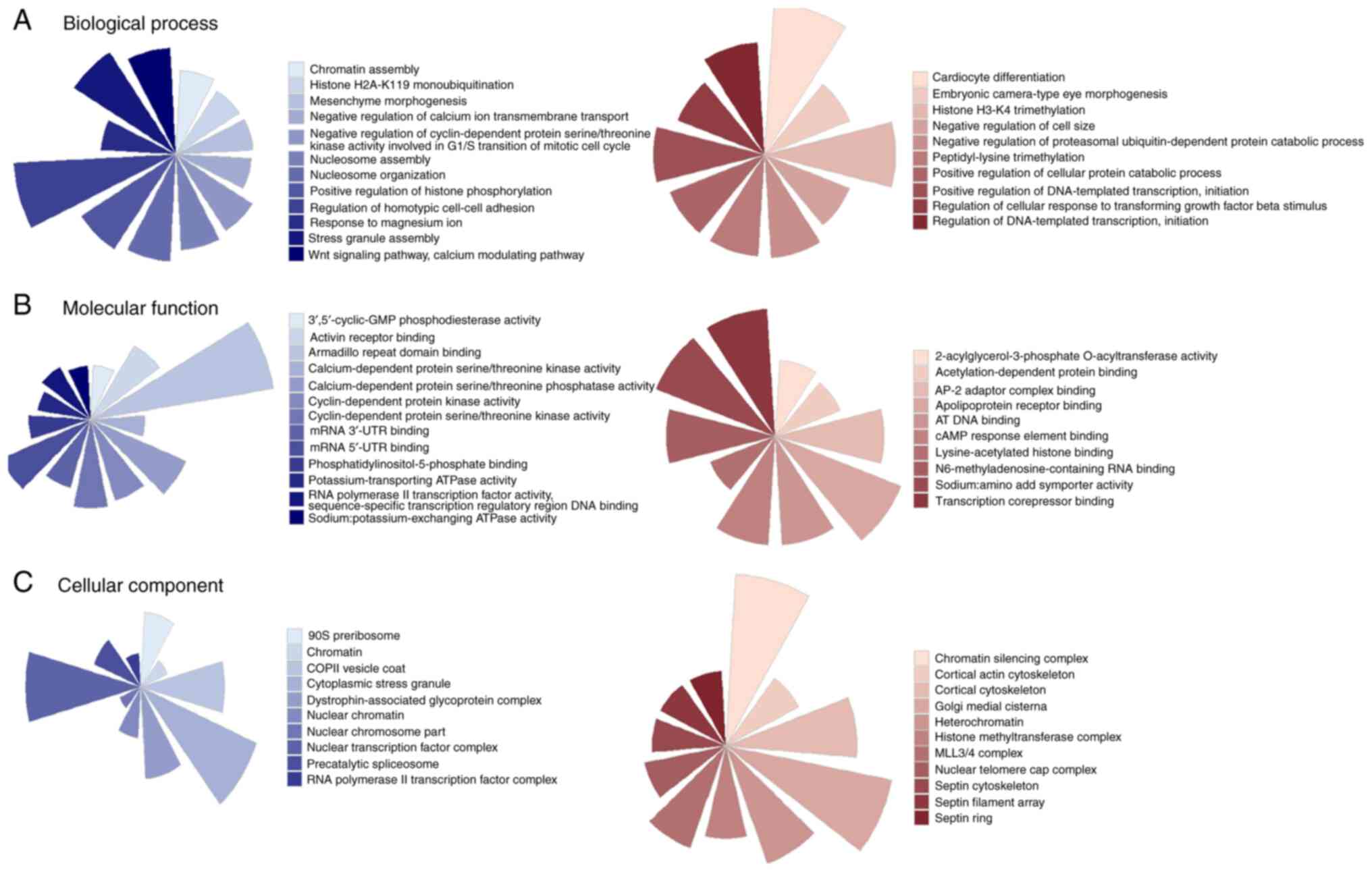
(Table SI). It was identified that
target genes of miRNAs downregulated by P4 participated in
cell-cell adhesion, regulation of histone posttranslational
modifications, cell cycle progression, mRNA binding, transcription
regulation, nucleosome assembly (Fig.
3). By contrast, the miRNAs upregulated by P4 participated in
the negative regulation of genes involved in transcription, histone
H3K4 modifications, regulation of TFGβ pathway, chromatin structure
and cytoskeleton conformation (Fig.
3).
Pathway enrichment analysis showed that the target
genes of the miRNAs downregulated by P4 regulate signaling pathways
participating in diseases such as systemic lupus erythematosus and
alcoholism, but also in long-term potentiation of synaptic
activity, cell cycle and maintenance of stem cells pluripotency
(Table II). The target genes of
the miRNAs upregulated by P4 participated in synaptic vesicle
cycle, fluid shear stress and atherosclerosis, in addition to in
TGF-β and IL-17 signaling pathways (Table III).
 | Table II.Pathway enrichment analysis of the
target genes of miRNAs downregulated by P4. |
Table II.
Pathway enrichment analysis of the
target genes of miRNAs downregulated by P4.
| Term | Overlap | P-value |
|---|
| Systemic lupus
erythematous | 14/133 | 0.0000001245 |
| Alcoholism | 16/180 | 0.0000001687 |
| Viral
carcinogenesis | 12/201 | 0.000291301 |
| Cushing
syndrome | 10/155 | 0.000503219 |
| Bladder cancer | 5/41 | 0.000798635 |
| Long-term
potentiation | 6/67 | 0.001274163 |
| Cell cycle | 8/124 | 0.001804709 |
| Mineral
absorption | 5/51 | 0.002163061 |
| MAPK signaling
pathway | 13/295 | 0.002737286 |
| Signaling pathways
regulating pluripotency of stem cells | 8/139 | 0.003665340 |
 | Table III.Pathway enrichment analysis of the
target genes of miRNAs upregulated by P4. |
Table III.
Pathway enrichment analysis of the
target genes of miRNAs upregulated by P4.
| Term | Overlap | P-value |
|---|
| Synaptic vesicle
cycle | 6/78 | 0.0028211 |
| Fluid shear stress
and atherosclerosis | 8/139 | 0.0037271 |
| Tight junction | 8/170 | 0.0120564 |
| Colorectal
cancer | 5/86 | 0.0195424 |
| TGFβ signaling
pathway | 5/90 | 0.0232857 |
| IL-17 signaling
pathway | 5/93 | 0.0263776 |
| Ferroptosis | 3/40 | 0.0349887 |
| Renal cell
carcinoma | 4/69 | 0.0357799 |
| Adherent
junction | 4/72 | 0.0408592 |
| Bacterial invasion
of epithelial cells | 4/74 | 0.0444609 |
Screening of hub Genes and
protein-protein interaction network (PPI)
The target genes of the miRNAs regulated by P4 were
mapped in the STRING database to obtain a clearer idea of their
interactions and their global action. The analysis of PPI networks
in Cytoscape, resulted in 259 and 267 pairs of nodes for the target
genes of the miRNAs downregulated and upregulated by P4,
respectively. The genes with a high degree of interactions in the
PPI network were defined as hub genes since they could serve a
critical role in the module (Table
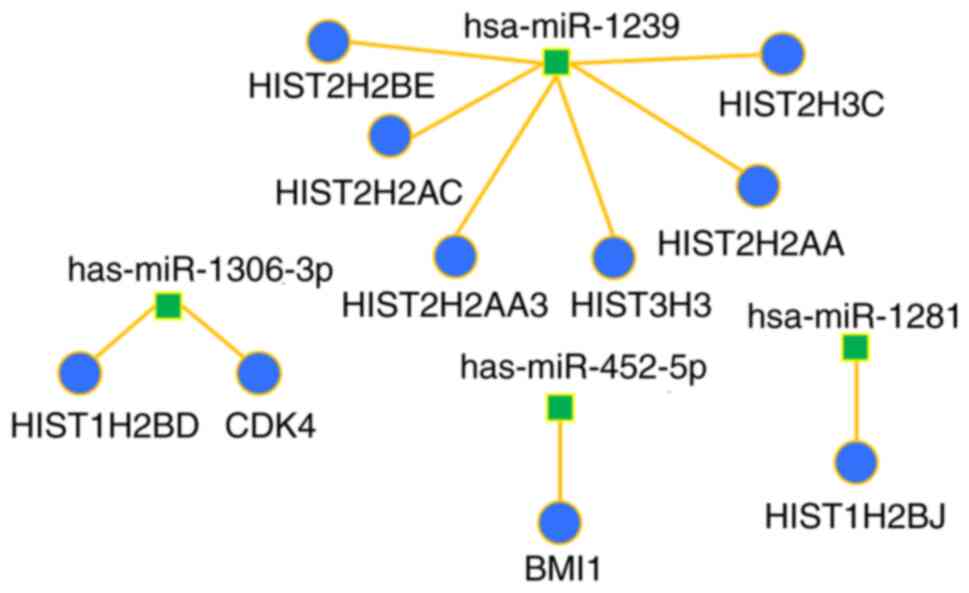
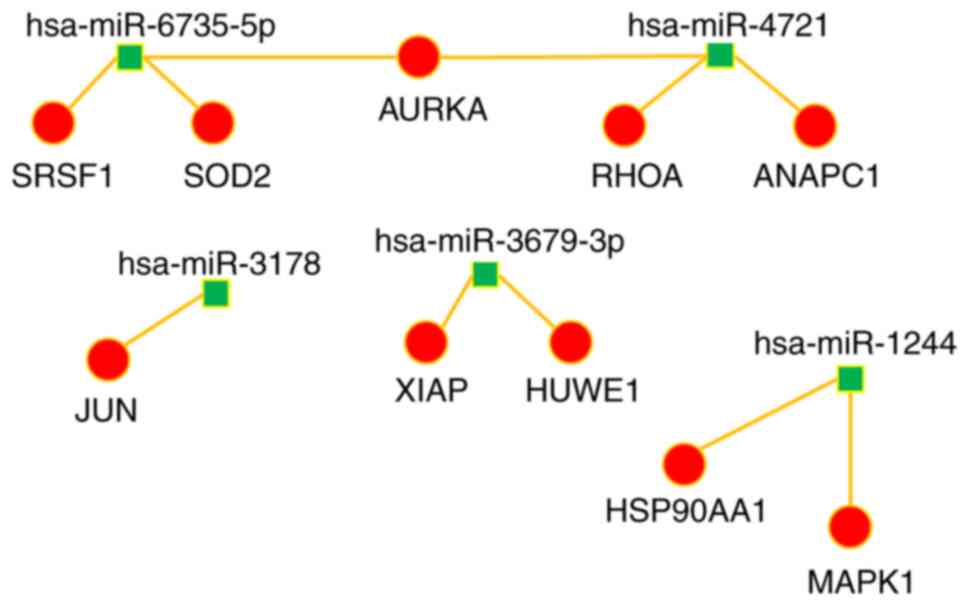
IV). A miRNA-target interaction network was performed to
visualize the interaction between the calculated hub genes and
their corresponding miRNA (Figs. 4
and 5). The majority of the hub
genes of miRNAs downregulated by P4 were histones; whereas the
principal hub gene of the miRNAs upregulated by P4 was HSP90AA1, a
gene that encodes an inducible molecular chaperone.
 | Table IV.Hub genes of the miRNAs regulated by
P4. |
Table IV.
Hub genes of the miRNAs regulated by
P4.
| Target genes of the
miRNAs downregulated by P4 |
|---|
|
|---|
| miRNA | Gene | Degree |
|---|
| hsa-miR-1239 | HIST2H2BE | 34 |
| hsa-miR-1239 | HIST2H2AC | 32 |
| hsa-miR-1281 | HIST1H2BJ | 24 |
|
hsa-miR-1306-3p | HIST1H2BD | 24 |
|
hsa-miR-1306-3p | CDK4 | 20 |
| hsa-miR-1239 | HIST2H2AA3 | 20 |
| hsa-miR-1239 | HIST3H3 | 20 |
| hsa-miR-1239 | HIST2H2AA | 20 |
| hsa-miR-452-5p | BMI1 | 20 |
| hsa-miR-1239 | HIST2H3C | 17 |
|
| Target genes of
the miRNAs upregulated by P4 |
|
| hsa-miR-1244 | HSP90AA1 | 30 |
| hsa-miR-1244 | MAPK1 | 29 |
| hsa-miR-3178 | JUN | 27 |
| hsa-miR-4721 | RHOA | 25 |
|
hsa-miR-6735-5p | SRSF1 | 15 |
| hsa-miR-4721 | ANAPC1 | 15 |
|
hsa-miR-3679-3p | HUWE1 | 14 |
|
hsa-miR-3679-3p | XIAP | 14 |
|
hsa-miR-6735-5p | SOD2 | 14 |
| hsa-miR-4721 | AURKA | 13 |
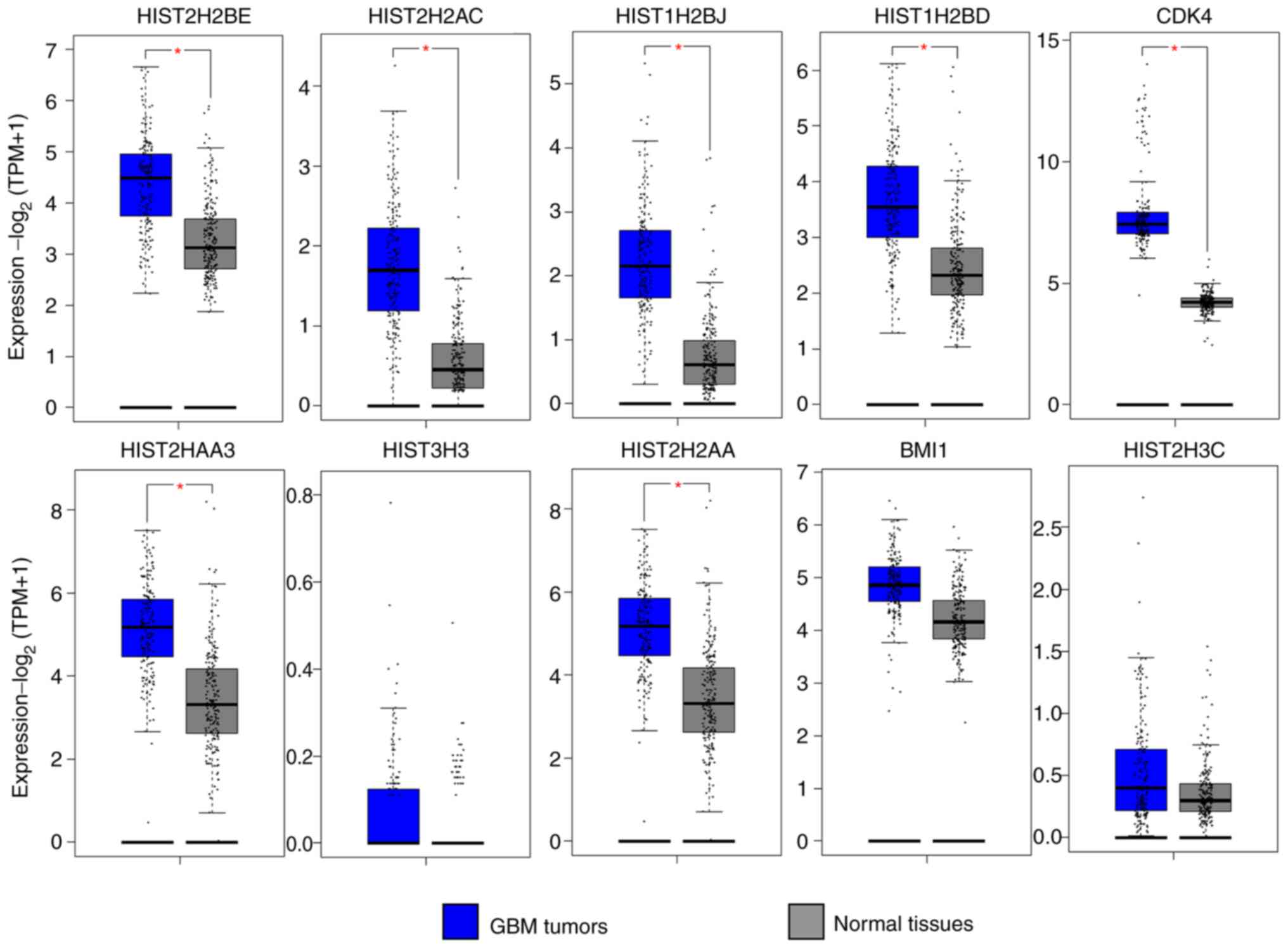
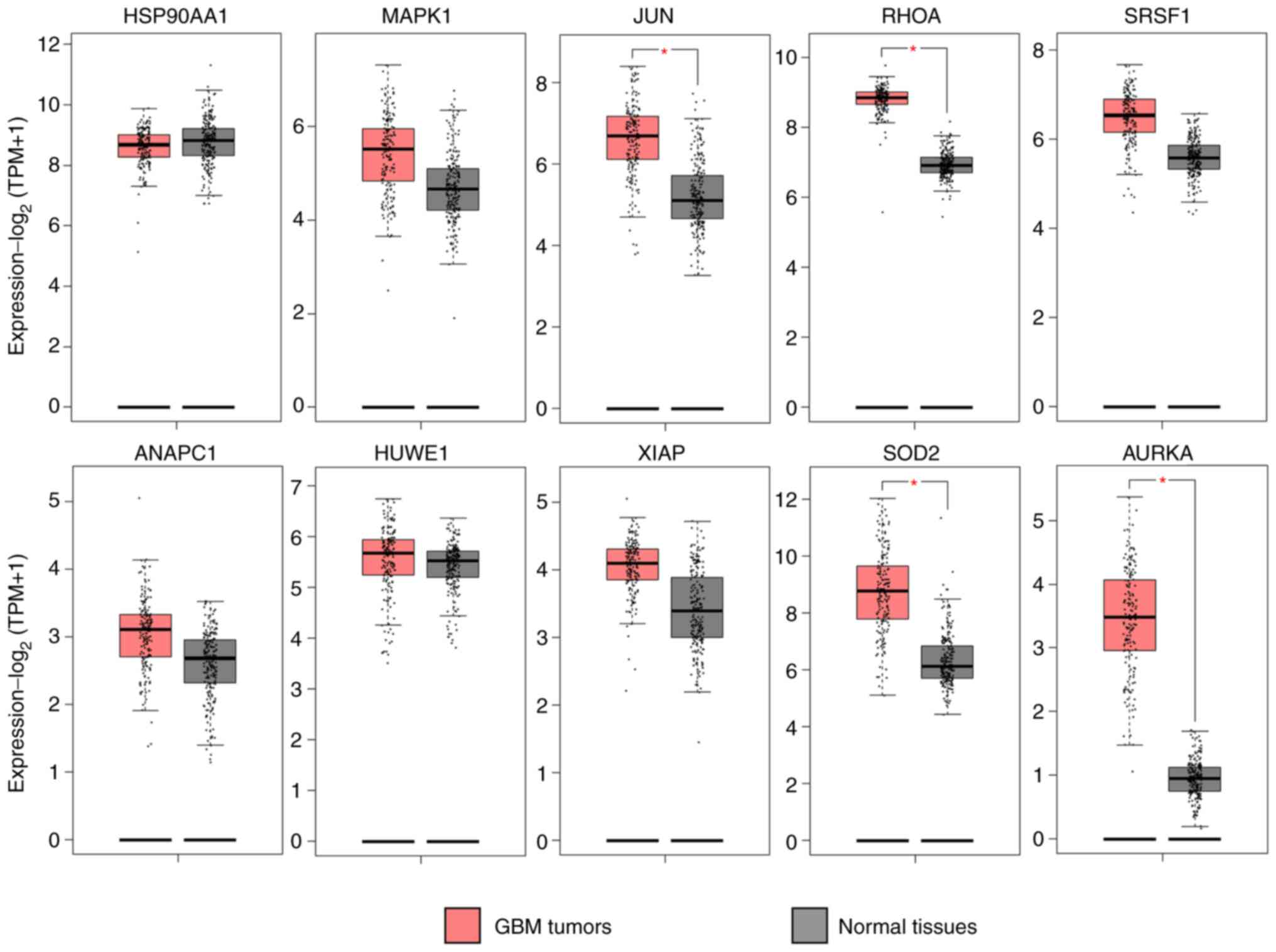
The GEPIA database was used to compare the
expression levels of the hub genes in biopsies of GBM compared with
normal tissue. The expression of 7 hub genes (HIST2H2BE, HIST2H2AC,
HIST1H2BJ, HIST1H2BD, CDK4, HIST2H2AA3 and HIST2H2AA) modulated by
the DE-miRNAs downregulated by P4 was significantly higher in GBM
compared with normal tissue (Fig.
6), as was expected given the negative association between a
miRNA and its target genes. Notably, the target genes of the
upregulated miRNAs, JUN, RHOA, SOD2 and AURKA exhibited a
significantly higher expression compared with normal healthy
tissue, contrary to the expected result; the remaining target genes
did not present significant differences (Fig. 7).
Discussion
GBM, a grade IV astrocytoma, is the commonest and
most aggressive brain tumor in adults (1–3).
Previous studies have suggested that alterations in miRNAs
expression are involved in GMB progression (5–10).
Changes in the expression profile of miRNAs by steroid hormones
have been noted in hormone-responsive cancers (44–47)
but not in GBM. Since it has been suggested that sex hormones, such
as P4, are involved in the several processes that contribute to GBM
progression (14–20), the objective of the present study
was to determine the expression profile of miRNAs regulated by P4
in GBM cells and identify the pathways regulated by them. The
effect of P4 on increased cell proliferation of GBMs was observed
at a concentration of 10 nM (14),
the same concentration that increases migration and invasion
(15). In all previous cases, 10 µM
of RU486 was determined to be the concentration that significantly
blocked the effects of P4 on GBM malignancy (14,15).
In the present study, the expression of 190 new
miRNAs was altered by P4, RU486 and P4+RU486 in U-251MG cells. Of
the 190 miRNAs, only 16 were regulated by P4 and this effect was
blocked by RU486 (an antagonist of PR). However, some miRNAs
exhibited changes in their expression upon treatment with of RU486
compared with the vehicle, suggesting that RU486 alone should exert
an effect on miRNA expression, probably due to the affinity that
RU486 also has for the glucocorticoid receptor (48,49).
It was also found that P4+RU486 treatment had several DE-miRNAs
respect to the vehicle, which suggests that RU486 does not entirely
block P4 actions, and that P4 and RU486 may exert their effects via
different mechanisms.
Of the 16 miRNAs reported in the present study, only
the human (has)-miR-485-3p miRNA, downregulated by P4, has been
previously evaluated in GBM cells. Agreeing with the present study,
Zhang et al (50) found that
the expression of hsa-miR-485-3p is downregulated in biopsies from
patients with compared with healthy brain tissue; these authors
determined that its target gene is the ring finger protein 35
(RNF135). By silencing RNF135, the hsa-miR-485-3p inactivated the
MAPK/ERK1/2 pathway in GBM cells, while functional assays showed
that hsa-miR-485-3p inhibited proliferation and migration of GBM
cells, which was reversed by the overexpression of RNF135; all
suggesting that the hsa-miR-485-3p miRNA has a tumor suppressor
function in GBMs (50).
To evaluate the possible targets of the DE-miRNAs,
the present study conducted functional annotation and pathway
analysis. A set of target genes for the miRNAs downregulated by P4
and another set for the ones upregulated were defined. The
functional annotation demonstrated that the target genes of the
downregulated miRNAs could be involved in post-transcriptional
histone modification, chromatin assembly, nucleosome assembly, cell
cycle progression, RNA binding and regulation of transcription. The
pathway analysis revealed that these genes could participate in
some pathways not related to cancer (Systemic lupus erythematosus
and alcoholism) as well as with pathways that are well
characterized in cancer and specifically in GBM, including cell
cycle (51), MAPK signaling pathway
(52,53) and regulation of stem cells
pluripotency (54–56).
Regarding the target genes of the upregulated
miRNAs, the results of the functional notation matched the results
of the pathway analysis as the regulation of transcription,
modifications of histone H3K4, regulation of the TGFβ pathway,
chromatin structure and conformation of the cytoskeleton are
related to the pathways of synaptic vesicles formation, maintenance
of tight junctions, TGFβ and the IL signaling pathways.
Particularly in GBMs, the secretion of VEGF by cancer cells
inhibits the formation of tight junctions (57,58)
and notably, P4 increases the expression of VEGF in cell lines
derived from GBMs (20). Based on
these data, it could be hypothesized that P4 increases the
malignancy of GBMs through the regulation of miRNAs that affect the
availability of VEGF and the maintenance of tight junctions.
Regarding the TGFβ pathway, it is known that TGFβ expression is
directly upregulated by P4 through PR (17). This suggests that the increase of
TGFβ expression should be due to the silencing of repressors
pathways, due to the action of miRNAs upregulated by P4. Finally,
when looking for an association between the IL-17 pathway and GBMs,
it was noted that IL-17 expression has been positively related to
the survival rates of patients with GBM (59); in this case, it is possible that P4
decreases the expression of IL-17 by the upregulation of miRNAs
directed to activators of this pathway.
Analysis of the protein-protein interaction network,
carried out with the target genes of those miRNAs regulated by P4,
determined a list of hub genes that could be important in the
regulation of the malignancy of GBMs. The results indicated that
eight of the 10 hub target genes of miRNAs downregulated by P4
correspond to histones; therefore, these histones would be expected
to be upregulated in GBMs. One way to confirm this deduction was
through the analysis of the expression data from the GEPIA
database. Based on the analysis of these data, 7 of the 8 histones
matched their miRNA regulators; markedly, only the expression of
histone HIST2H2BE has been observed in GBMs under the treatment of
OTX015, an inhibitor of bromodomain and extraterminal bromodomain
proteins (60). In addition to
histones, BMI1 and CDK4 were chosen as hub genes in the present
study's analysis of the target genes for the miRNAs downregulated
by P4 and, according to the results of their expression in GEPIA,
only CDK4 was significantly upregulated in GBMs biopsies; however,
according to literature (56,61–63),
BMI1 is also highly expressed in GBM. There is evidence that CDK4
and BMI1 are oncogenes involved in the regulation of the cell cycle
and the increase in proliferation and invasion of GBMs cells
(56,61–63).
In addition, the function of these hub genes coincided with the
results of functional gene notation and pathway enrichment
analysis.
In the case of the expression of hub genes of the
miRNAs upregulated by P4, genes are involved in the cell cycle,
proliferation, invasion, migration and inhibition of apoptosis and
genes that participate in antioxidant defence. To date, the
expression of these genes has not been evaluated after P4
treatment, however, when verifying their expression in the GEPIA
database and in published papers, the present study noted that
these genes are key for GBM biology and characterized as oncogenes.
As aforementioned, P4 promotes the malignancy of GBMs and according
to the results of the present study, it upregulated miRNAs whose
targets are these oncogenes. This was unexpected and therefore
further research in this field is required.
The JUN gene encodes the transcription factor AP-1,
which is constitutively activated in gliomas, contributing to their
malignancy (64–66) and RHOA has been implicated in
invasion and migration of human GBM cells through its signalization
with YAP, MRTF-A, Daam1 and Wnt5a, also contributing to the
malignancy of GBM (67,68). In the case of SOD2, its expression
has been observed to be that significantly increased, at the mRNA
and protein levels, in LN-239 and U87 cell lines (both derived from
GBMs) exposed to oxidative stress (69–71).
Although JUN, RHOA and SOD2 have been described as oncogenes, their
regulation by some type of non-coding RNA has not been studied, in
contrast to what has been observed in AUKRA. It can be hypothesized
that the regulatory miRNAs of these hub genes, could have another
level of regulation by including circular (circ)RNAs (69) or long non-coding RNAs, which also
have response elements to miRNAs (70) For example AUKRA is a target gene of
circMMP9 and hsa-miR-124 axis (72).
Finally, the interaction network between hub genes
and their miRNAs allows the visualization of the complexity of the
gene regulation by miRNAs; the value of the network is the
demonstration of the interaction between miRNAs regulated by P4 and
their target genes, which could aid in understanding the epigenetic
alterations induced by P4 in GBM.
The present study described the global changes in
the expression profile of miRNAs induced by P4 in cells derived
from human GBM. Microarray expression analysis identified 8 miRNAs
downregulated by P4 and 8 miRNAs upregulated by P4. As a result of
bioinformatic analyses, it was found that P4 could regulate
processes such as proliferation, cell cycle progression and cell
migration of GBMs through the regulation of a network of
miRNAs-mRNAs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The financial support was provided by the projects
PAPIIT 2020 IN217120, DGAPA,UNAM to Ignacio Camacho-Arroyo, by
Consejo Nacional de Ciencia y Tecnología (CONACYT) project no.
258589 to Eduardo Martínez-Martínez and by CONACYT project no.
A1-S-26446 to MRD.
Availability of data and materials
The miRNA expression data used to support the
findings of the present study have been deposited in the NCBI's
Gene Expression Omnibus and are accessible through GEO Series
accession number GSE144204 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144204).
Authors' contributions
All authors contributed to the study conception and
design. The material preparation, data collection and analysis were
performed by DEVV and ADMM. The verification of the raw data was
performed by ICA, MRD and EMM. The first draft of the manuscript
was written by DEVV. All authors provided technical and
experimental advice and reviewed and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wen PY and Kesari S: Malignant gliomas in
adults. N Engl J Med. 359:492–507. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Urbanska K, Sokolowska J, Szmidt M and
Sysa P: Glioblastoma multiforme-An overview. Contemp Oncol (Pozn).
18:307–312. 2014.PubMed/NCBI
|
|
3
|
Weller M, Wick W, Aldape K, Brada M,
Berger M, Pfister SM, Nishikawa R, Rosenthal M, Wen PY, Stupp R and
Reifenberger G: Glioma. Nat Rev Dis Primers. 1:150172015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ostrom QT, Gittleman H, Liao P,
Vecchione-Koval T, Wolinsky Y, Kruchko C and Barnholtz-Sloan JS:
CBTRUS statistical report: Primary brain and other central nervous
system tumors diagnosed in the United States in 2010–2014. Neuro
Oncol. 19 (Suppl 5):v1–v88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ciafrè SA, Galardi S, Mangiola A, Ferracin
M, Liu CG, Sabatino G, Negrini M, Maira G, Croce CM and Farace MG:
Extensive modulation of a set of microRNAs in primary glioblastoma.
Biochem Biophys Res Commun. 334:1351–1358. 2005. View Article : Google Scholar
|
|
6
|
Novakova J, Slaby O, Vyzula R and Michalek
J: MicroRNA involvement in glioblastoma pathogenesis. Biochem
Biophys Res Commun. 386:1–5. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aldaz B, Sagardoy A, Nogueira L, Guruceaga
E, Grande L, Huse JT, Aznar MA, Díez-Valle R, Tejada-Solís S,
Alonso MM, et al: Involvement of miRNAs in the differentiation of
human glioblastoma multiforme stem-like cells. PLoS One.
8:e770982013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alfardus H, Mcintyre A and Smith S:
MicroRNA regulation of glycolytic metabolism in glioblastoma.
Biomed Res Int. 2017:91573702017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mercatelli N, Galardi S and Ciafrè SA:
MicroRNAs as multifaceted players in glioblastoma multiforme. Int
Rev Cell Mol Biol. 333:269–323. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sana J, Busek P, Fadrus P, Besse A, Radova
L, Vecera M, Reguli S, Stollinova Sromova L, Hilser M, Lipina R, et
al: Identification of microRNAs differentially expressed in
glioblastoma stem-like cells and their association with patient
survival. Sci Rep. 8:28362018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ceballos-Chávez M, Subtil-Rodríguez A,
Giannopoulou EG, Soronellas D, Vázquez-Chávez E, Vicent GP,
Elemento O, Beato M and Reyes JC: The chromatin remodeler CHD8 is
required for activation of progesterone receptor-dependent
enhancers. PLoS Genet. 11:e10051742015. View Article : Google Scholar
|
|
12
|
Helsen C and Claessens F: Looking at
nuclear receptors from a new angle. Mol Cell Endocrinol.
382:97–106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jacobsen BM and Horwitz KB: Progesterone
receptors, their isoforms and progesterone regulated transcription.
Mol Cell Endocrinol. 357:18–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
González-Agüero G, Gutiérrez AA,
González-Espinosa D, Solano JD, Morales R, González-Arenas A,
Cabrera-Muñoz E and Camacho-Arroyo I: Progesterone effects on cell
growth of U373 and D54 human astrocytoma cell lines. Endocrine.
32:129–135. 2007. View Article : Google Scholar
|
|
15
|
Piña-Medina AG, Hansberg-Pastor V,
González-Arenas A, Cerbón M and Camacho-Arroyo I: Progesterone
promotes cell migration, invasion and cofilin activation in human
astrocytoma cells. Steroids. 105:19–25. 2016. View Article : Google Scholar
|
|
16
|
Germán-Castelán L, Manjarrez-Marmolejo J,
González-Arenas A, González-Morán MG and Camacho-Arroyo I:
Progesterone induces the growth and infiltration of human
astrocytoma cells implanted in the cerebral cortex of the rat.
Biomed Res Int. 2014:3931742014. View Article : Google Scholar
|
|
17
|
González-Arenas A, Cabrera-Wrooman A, Díaz
NF, González- García TK, Salido-Guadarrama I, Rodríguez-Dorantes M
and Camacho-Arroyo I: Progesterone receptor subcellular
localization and gene expression profile in human astrocytoma cells
are modified by progesterone. Nucl Recept Res. 1:1–10. 2014.
View Article : Google Scholar
|
|
18
|
Zamora-Sánchez CJ, Hansberg-Pastor V,
Salido-Guadarrama I, Rodríguez-Dorantes M and Camacho-Arroyo I:
Allopregnanolone promotes proliferation and differential gene
expression in human glioblastoma cells. Steroids. 119:36–42. 2017.
View Article : Google Scholar
|
|
19
|
Zamora-Sánchez CJ, Del Moral-Morales A,
Hernández-Vega AM, Hansberg-Pastor V, Salido-Guadarrama I,
Rodríguez-Dorantes M and Camacho-Arroyo I: Allopregnanolone alters
the gene expression profile of human glioblastoma cells. Int J Mol
Sci. 19:8642018. View Article : Google Scholar
|
|
20
|
Hernández-Hernández OT, González-García TK
and Camacho-Arroyo I: Progesterone receptor and SRC-1 participate
in the regulation of VEGF, EGFR and cyclin D1 expression in human
astrocytoma cell lines. J Steroid Biochem Mol Biol. 132:127–134.
2012. View Article : Google Scholar
|
|
21
|
Wendler A, Keller D, Albrecht C, Peluso JJ
and Wehiling M: Involvement of let-7/miR-98 microRNAs in the
regulation of progesterone receptor membrane component 1 expression
in ovarian cancer cells. Oncol Rep. 25:273–279. 2011.PubMed/NCBI
|
|
22
|
Cittelly DM, Finlay-Schultz J, Howe EN,
Spoelstra NS, Axlund SD, Hendricks P, Jacobsen BM, Sartorius CA and
Richer JK: Progestin suppression of miR-29 potentiates
dedifferentiation of breast cancer cells via KLF4. Oncogene.
32:2555–2564. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rivas MA, Venturutti L, Huang YW,
Schillaci R, Huang TH and Elizalde PV: Downregulation of the
tumor-suppressor miR-16 via progestin-mediated oncogenic signaling
contributes to breast cancer development. Breast Cancer Res.
14:R772012. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Romero-Cordoba S, Rodriguez-Cuevas S,
Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez
G, Bautista-Piña V, Arellano-Llamas R and Hidalgo-Miranda A:
Identification and pathway analysis of microRNAs with no previous
involvement in breast cancer. PLoS One. 7:e319042012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cochrane DR, Jacobsen BM, Connaghan KD,
Howe EN, Bain DL and Richer JK: Progestin regulated miRNAs that
mediate progesterone receptor action in breast cancer. Mol Cell
Endocrinol. 355:15–24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Finlay-Schultz J, Cittelly DM, Hendricks
P, Patel P, Kabos P, Jacobsen BM, Richer JK and Sartorius CA:
Progesterone downregulation of miR-141 contributes to expansion of
stem-like breast cancer cells through maintenance of progesterone
receptor and Stat5a. Oncogene. 34:3676–3687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fletcher CE, Dart DA and Bevan CL:
Interplay between steroid signalling and microRNAs: Implications
for hormone-dependent cancers. Endocr Relat Cancer. 21:R409–R429.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Godbole M, Chandrani P, Gardi N, Dhamne H,
Patel K, Yadav N, Gupta S, Badwe R and Dutt A: miR-129-2 mediates
down-regulation of progesterone receptor in response to
progesterone in breast cancer cells. Cancer Biol Ther. 18:801–805.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
McFall T, McKnight B, Rosati R, Kim S,
Huang Y, Viola-Villegas N and Ratnam M: Progesterone receptor A
promotes invasiveness and metastasis of luminal breast cancer by
suppressing regulation of critical microRNAs by estrogen. J Biol
Chem. 293:1163–1177. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nie L, Zhao YB, Pan JL, Lei Y, Liu M, Long
Y, Zhang JH, Hu Y, Xu MQ, Yuan DZ and Yue LM: Progesterone-Induced
miR-152 inhibits the proliferation of endometrial epithelial cells
by downregulating WNT-1. Reprod Sci. 24:1444–1453. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Smyth GK: limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
Using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Springer; New York, NY: pp. 397–420. 2005,
View Article : Google Scholar
|
|
32
|
R Core Team: R: A language and environment
for statistical computing. R Foundation for Statistical Computing;
Vienna, Austria: 2018, http://www.R-project.org/
|
|
33
|
Karagkouni D, Paraskevopoulou MD,
Chatzopoulos S, Vlachos IS, Tastsoglou S, Kanellos I, Papadimitriou
D, Kavakiotis I, Maniou S, Skoufos G, et al: DIANA-TarBase v8: A
decade-long collection of experimentally supported miRNA-gene
interactions. Nucleic Acids Res. 46(D1): D239–D245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dweep H, Gretz N and Sticht C: MiRWalk
database for miRNA-target interactions. Methods Mol Biol.
1182:289–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Paraskevopoulou MD, Georgakilas G,
Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C,
Dalamagas T and Hatzigeorgiou AG: DIANA-microT web server v5.0:
Service integration into miRNA functional analysis workflows.
Nucleic Acids Res. 41((Web Server Issue)): W169–W73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z,
Meirelles GV, Clark NR and Ma'ayan A: Enrichr: Interactive and
collaborative HTML5 gene list enrichment analysis tool. BMC
Bioinformatics. 14:1282013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gene Ontology Consortium, . Gene ontology
consortium: Going forward. Nucleic Acids Res. 43((Database issue)):
D1049–D1056. 2015.PubMed/NCBI
|
|
39
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1): D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45((W1)):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G and
Kerin MJ: MicroRNA signatures predict oestrogen receptor,
progesterone receptor and HER2/neu receptor status in breast
cancer. Breast Cancer Res. 11:R272009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Maillot G, Lacroix-Triki M, Pierredon S,
Gratadou L, Schmidt S, Bénès V, Roché H, Dalenc F, Auboeuf D,
Millevoi S and Vagner S: Widespread estrogen-dependent repression
of microRNAs involved in breast tumor cell growth. Cancer Res.
69:8332–8340. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun R, Fu X, Li Y, Xie Y and Mao Y: Global
gene expression analysis reveals reduced abundance of putative
microRNA targets in human prostate tumours. BMC Genomics.
10:932009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ribas J, Ni X, Haffner M, Wentzel EA,
Salmasi AH, Chowdhury WH, Kudrolli TA, Yegnasubramanian S, Luo J,
Rodriguez R, et al: miR-21: An androgen receptor-regulated microRNA
that promotes hormone-dependent and hormone-independent prostate
cancer growth. Cancer Res. 69:7165–7169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cadepond F, Ulmann A and Baulieu EE: RU486
(MIFEPRISTONE): Mechanisms of action and clinical uses. Annu Rev
Med. 48:129–156. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Catalano RD, Yanaihara A, Evans AL, Rocha
D, Prentice A, Saidi S, Print CG, Charnock-Jones DS, Sharkey AM and
Smith SK: The effect of RU486 on the gene expression profile in an
endometrial explant model. Mol Hum Reprod. 9:465–473. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Y, Sui R, Chen Y, Liang H, Shi J and
Piao H: Downregulation of miR-485-3p promotes glioblastoma cell
proliferation and migration via targeting RNF135. Exp Ther Med.
18:475–482. 2019.PubMed/NCBI
|
|
51
|
Yang W, Warrington NM, Taylor SJ, Whitmire
P, Carrasco E, Singleton KW, Wu N, Lathia JD, Berens ME, Kim AH, et
al: Sex differences in GBM revealed by analysis of patient imaging,
transcriptome and survival data. Sci Transl Med. 11:eaao52532019.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang P, Kang W, Pan Y, Zhao X and Duan L:
Overexpression of HOXC6 promotes cell proliferation and migration
via MAPK signaling and predicts a poor prognosis in glioblastoma.
Cancer Manag Res. 11:8167–8179. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zhou L, Tang H, Wang F, Chen L, Ou S, Wu
T, Xu J and Guo K: Bioinformatics analyses of significant genes,
related pathways and candidate prognostic biomarkers in
glioblastoma. Mol Med Rep. 18:4185–4196. 2018.PubMed/NCBI
|
|
54
|
Zhang S, Wan Y, Pan T, Gu X, Qian C, Sun
G, Sun L, Xiang Y, Wang Z and Shi L: MicroRNA-21 inhibitor
sensitizes human glioblastoma U251 stem cells to chemotherapeutic
drug temozolomide. J Mol Neurosci. 47:346–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Nicolas S, Abdellatef S, Haddad MA,
Fakhoury I and El-Sibai M: Hypoxia and EGF stimulation regulate
VEGF expression in human glioblastoma multiforme (GBM) cells by
differential regulation of the PI3K/Rho-GTPase and MAPK pathways.
Cells. 8:13972019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shan ZN, Tian R, Zhang M, Gui ZH, Wu J,
Ding M, Zhou XF and He J: MiR128-1 inhibits the growth of
glioblastoma multiforme and glioma stem-like cells via targeting
BMI1 and E2F3. Oncotarget. 7:78813–78826. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wen L, Tan Y, Dai S, Zhu Y, Meng T, Yang
X, Liu Y, Liu X, Yuan H and Hu F: VEGF-mediated tight junctions
pathological fenestration enhances doxorubicin-loaded
glycolipid-like nanoparticles traversing BBB for
glioblastoma-targeting therapy. Drug Deliv. 24:1843–1855. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bao S, Wu Q, Sathornsumetee S, Hao Y, Li
Z, Hjelmeland AB, Shi Q, McLendon RE, Bigner DD and Rich JN: Stem
cell-like glioma cells promote tumor angiogenesis through vascular
endothelial growth factor. Cancer Res. 66:7843–7848. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cui X, Xu Z, Zhao Z, Sui D, Ren X, Huang
Q, Qin J, Hao L, Wang Z, Shen L and Lin S: Analysis of CD137l and
IL-17 expression in tumor tissue as prognostic indicators for
gliblastoma. Int J Biol Sci. 9:134–141. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Berenguer-Daizé C, Astorgues-Xerri L,
Odore E, Cayol M, Cvitkovic E, Noel K, Bekradda M, MacKenzie S,
Rezai K, Lokiec F, et al: OTX015 (MK-8628), a novel BET inhibitor,
displays in vitro and in vivo antitumor effects alone and in
combination with conventional therapies in glioblastoma models. Int
J Cancer. 139:2047–2055. 2016. View Article : Google Scholar
|
|
61
|
Jin X, Kim LJY, Wu Q, Wallace LC, Prager
BC, Sanvoranart T, Gimple RC, Wang X, Mack SC, Miller TE, et al:
Targeting glioma stem cells through combined BMI1 and EZH2
inhibition. Nat Med. 23:1352–1361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Elango R, Vishnubalaji R, Manikandan M,
Binhamdan SI, Siyal AA, Alshawakir YA, Alfayez M, Aldahmash A and
Alajez NM: Concurrent targeting of BMI1 and CDK4/6 abrogates tumor
growth in vitro and in vivo. Sci Rep. 9:136962019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Peng G, Liao Y and Shen C: miRNA-429
inhibits astrocytoma proliferation and invasion by targeting BMI1.
Pathol Oncol Res. 23:369–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Antonyak MA, Kenyon LC, Godwin AK, James
DC, Emlet DR, Okamoto I, Tnani M, Holgado-Madruga M, Moscatello DK
and Wong AJ: Elevated JNK activation contributes to the
pathogenesis of human brain tumors. Oncogene. 21:5038–5046. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tsuiki H, Tnani M, Okamoto I, Kenyon LC,
Emlet DR, Holgado-Madruga M, Lanham IS, Joynes CJ, Vo KT and Wong
AJ: Constitutively active forms of c-Jun NH2-terminal kinase are
expressed in primary glial tumors. Cancer Res. 63:250–255.
2003.PubMed/NCBI
|
|
66
|
Cui J, Han SY, Wang C, Su W, Harshyne L,
Holgado-Madruga M and Wong AJ: c-Jun NH(2)-terminal kinase 2alpha2
promotes the tumorigenicity of human glioblastoma cells. Cancer
Res. 66:10024–10031. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Yu OM, Benitez JA, Plouffe SW, Ryback D,
Klein A, Smith J, Greenbaum J, Delatte B, Rao A, Guan KL, et al:
YAP and MRTF-A, transcriptional co-activators of RhoA-mediated gene
expression, are critical for glioblastoma tumorigenicity. Oncogene.
37:5492–5507. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Liu G, Yan T, Li X, Sun J, Zhang B, Wang H
and Zhu Y: Daam1 activates RhoA to regulate Wnt5a-induced
glioblastoma cell invasion. Oncol Rep. 39:465–472. 2018.PubMed/NCBI
|
|
69
|
Kusaczuk M, Krętowski R, Naumowicz M,
Stypułkowska A and Cechowska-Pasko M: Silica nanoparticle-induced
oxidative stress and mitochondrial damage is followed by activation
of intrinsic apoptosis pathway in glioblastoma cells. Int J
Nanomedicine. 13:2279–2294. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Cholia RP, Kumari S, Kumar S, Kaur M, Kaur
M, Kumar R, Dhiman M and Mantha AK: An in vitro study ascertaining
the role of H2O2 and glucose oxidase in modulation of antioxidant
potential and cancer cell survival mechanisms in glioblastoma U-87
MG cells. Metab Brain Dis. 32:1705–1716. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tanaka H, Mizuno M, Katsumata Y, Ishikawa
K, Kondo H, Hashizume H, Okazaki Y, Toyokuni S, Nakamura K,
Yoshikawa N, et al: Oxidative stress-dependent and -independent
death of glioblastoma cells induced by non-thermal plasma-exposed
solutions. Sci Rep. 9:136572019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang R, Zhang S, Chen X, Li N, Li J, Jia
R, Pan Y and Liang H: EIF4A3-induced circular RNA MMP9 (circMMP9)
acts as a sponge of miR-124 and promotes glioblastoma multiforme
cell tumorigenesis. Mol Cancer. 17:1662018. View Article : Google Scholar : PubMed/NCBI
|