Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide and the second leading cause of cancer-associated
death (1). In China, the incidence
and mortality rates of CRC have been increasing gradually over the
past decade, with 190,000 people dying of CRC every year, despite
improved surgical and medical management (2). Numerous studies have revealed that
microsatellite instability, CpG island methylation, frequent gene
mutations, and the PI3K, WNT, MAPK, p53, transforming growth factor
(TGF)-β and DNA mismatch repair pathways are involved in the
initiation and progression of CRC (3,4).
However, the genetic and genomic changes associated with colorectal
tumorigenesis and their significance are still unclear.
MicroRNAs (miRNAs/miRs) are endogenous long
non-coding RNAs of 21–25 nucleotides that are generally transcribed
from non-coding regions of the genome. miRNAs are processed from a
60–70-nucleotide hairpin precursor. Over the past decade, >2,000
miRNAs have been found in the human genome (5); they participate in several
physiological and pathological processes by regulating the
expression of >60% of human genes (5). Mature miRNAs are the most important
members of the active RNA-induced silencing complex, and play a key
role in gene regulation. Mature miRNAs suppress the translation or
degradation of their target mRNAs by interacting with the
3′-untranslated region (UTR), open reading frame (ORF), or 5′-UTR
of the mRNA (6,7). miRNAs repress protein production by
post-transcriptional suppression of mRNA translation or promotion
of mRNA degradation, allowing miRNAs to regulate cell
differentiation, proliferation, apoptosis and homeostasis (5,8,9).
Dysregulation of miRNAs has been implicated in human diseases and
tumorigenesis, including tumor invasion, metastasis, staging and
prognosis (10–14). Accumulating evidence has shown that
miRNAs function as regulators in CRC (11,12,15).
Since the low expression of miR-143 and miR-145 in
CRC was first reported (16), the
dysregulation of miRNAs has been found to be a frequent event in
CRC, and is considered to be associated with CRC tumorigenesis and
progression (11,12,15).
Nagy et al (17) found that
miR-18a, −18b, −431, −503, −1246 and −4417 are upregulated during
the adenoma to carcinoma sequence in CRC, whereas miR-133a, −375,
−378, −422, and −479 are downregulated during this sequence. It was
also shown that the expression profiles of miRNAs in colorectal
adenoma and CRC are significantly different (17). Further studies have revealed that
aberrant miRNAs, including miR-142-3p, are involved in cell
proliferation, cell cycle transition, cell apoptosis, autophagy,
epithelial-to-mesenchymal transition (EMT), invasion and metastasis
in CRC by regulating the WNT/β-catenin, epidermal growth factor
receptor, TGF-β, Rac family small GTPase 1 (RAC1) and p53 signaling
pathways (11,12,15,18).
The aforementioned results suggest that some miRNAs may be
effective biomarkers for diagnosis and prognosis, and can be used
as drug targets for therapy in CRC. However, although the
dysregulation of miRNAs plays a key role in CRC progression, the
specific mechanism by which miRNAs regulate colon tumorigenesis is
still largely unknown.
In the present study, the Exiqon miRNA
oligonucleotide microarray was used to assess miRNA expression
profiles in CRC, and bioinformatics was employed to analyze the
biological function of differentially expressed miRNAs and their
role in CRC tumorigenesis and progression. The functions and
mechanisms of miRNA-142-3p were assessed in CRC cell lines.
Materials and methods
Tissue specimens
A total of 63 CRC tissues and their corresponding
adjacent non-cancer tissues were collected from January 2017 to
December 2018 in the Department of Pathology of the First
Affiliated Hospital of Hainan Medical University (Haikou, China).
All patients (age, 28–86 years; mean age, 58.3 years) included in
the study had undergone surgical resection but without previous
surgery, radiotherapy or chemotherapy. All cases were diagnosed
pathologically by two senior pathologists. Clinicopathological
characteristics, such as age, sex, tumor size and tumor TNM stage
according to the AJCC 8th Edition (19), were obtained from electronic medical
records and are summarized in Table
I. To maintain the consistency of the biological information,
three pairs of cases with similar clinical data were selected for
miRNA microarray analysis (Table
SI and Fig. S1). All the
patients provided written consent for their specimens to be used in
the study. The study was approved by the Ethics Committee of the
First Affiliated Hospital of Hainan Medical University.
 | Table I.Association between miR-142-3p
expression and pathological characteristics of patients with colon
cancer. |
Table I.
Association between miR-142-3p
expression and pathological characteristics of patients with colon
cancer.
|
|
| miR-142-3p |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
parameter | n | Low expression | High
expression | χ2 | P-value |
|---|
| Age, years |
|
|
| 0.226 | 0.464 |
|
≥60 | 42 | 8 | 34 |
|
|
|
<60 | 21 | 3 | 18 |
|
|
| Sex |
|
|
| 0.017 | >0.99 |
|
Male | 39 | 7 | 32 |
|
|
|
Female | 24 | 4 | 20 |
|
|
| Depth of
invasion |
|
|
| 2.56 | 0.16 |
|
T1+T2 | 22 | 7 | 15 |
|
|
|
T3+T4 | 41 | 4 | 37 |
|
|
| Clinical TNM
stage |
|
|
| 8.53 | 0.01 |
|
I+II | 27 | 9 | 18 |
|
|
|
III+IV | 36 | 2 | 34 |
|
|
| Tumor grade |
|
|
| 0.49 | 0.31 |
|
I+II | 41 | 6 | 35 |
|
|
|
III+IV | 22 | 5 | 17 |
|
|
| Lymph node
metastasis |
|
|
| 4.88 | 0.04 |
| N0 | 28 | 8 | 19 |
|
|
|
N1+N2 | 35 | 3 | 33 |
|
|
| Distant
metastasis |
|
|
| 0.15 | 0.55 |
|
Yes | 4 | 1 | 3 |
|
|
| No | 59 | 10 | 49 |
|
Microarray analysis
CRC and paired non-cancer mucosa tissues from three
sets of cases were homogenized using the TissueLyser II (Qiagen
GmbH), and total RNA was isolated using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.) and purified with the RNeasy mini kit
(Qiagen) according to the manufacturer's instructions. Microarray
hybridization and data analysis were performed by KangChen BioTech
Co., Ltd., according to the protocols for the miRCURY miRNA Array
(v.18.0, Exiqon) which comprised >3,100 probes for capturing
miRNAs. Samples were labeled with Hy3 using the miRCURY Array Power
Labeling kit (Exiqon). rRNA was removed using the rRNA removal kit,
and the remaining mRNA was hybridized to each array in a
hybridization oven at 55°C and 20 rpm for 20 h. The array was
washed with the Gene Expression Wash Buffer (Exiqon) and scanned
using the Axon Genepix 4000B Scanner (Exiqon). Then scanned images
were analyzed with the GenePix Pro 6.0 software (Axon Instruments;
Molecular Devices, LLC). Background subtraction was performed, and
the raw data were normalized using the quantile algorithm.
Aberrantly expressed miRNAs were classified as those with a 2-fold
change in expression and a P<0.05. The obtained microarray data
were deposited in the Gene Expression Omnibus database (accession
no. GSE101502).
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Total RNA and miRNAs were extracted from 63 pairs of
CRC and matched non-cancer mucosa tissues using the RNA and miRcute
miRNA isolation kits, respectively (cat. nos. DP439 and DP502;
Tiangen Biotech Co., Ltd.). Total RNA was reverse transcribed into
first-strand cDNA by incubating at 42°C for 15 min using the RNA
first-strand cDNA kit (cat. no. KR118; TIANGEN Biotech Co., Ltd.)
and the TaqMan MicroRNA Reverse Transcription kit (cat. no.
4366596; Thermo Fisher Scientific, Inc.). The specific stem-looped
RT primers were synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.) and are shown in Table II. The PCR primer sequences for the
miRNAs, small nuclear RNA U6 (U6), RAC1, and GAPDH were synthesized
by Invitrogen (Thermo Fisher Scientific, Inc.) and are listed in
Table III. Quantification of
miRNAs and RAC1 mRNA was performed using the ABI viia7 system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). qPCR was
performed using the FastFire qPCR kit and the miRcute miRNA qPCR
Detection kit (cat. nos. FP208 and FP401, TIANGEN Biotech Co.,
Ltd.), according to the manufacturer's instructions. The initial
denaturation step during qPCR was performed at 95°C for 120 sec,
followed by 40 amplification cycles at 95°C for 15 sec, and
annealing and extension at 60°C for 20 sec. The relative expression
levels of RAC1 mRNA and miRNAs were calculated using the
2−ΔΔCq method (20). U6
and GAPDH were used as internal references.
 | Table II.Stem-looped reverse transcription
primers of miRNAs. |
Table II.
Stem-looped reverse transcription
primers of miRNAs.
| miRNAs | Primer
sequence |
|---|
| miR-15b-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGTAAA-3′ |
| miR-31-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGCTAT-3′ |
| miR-139-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACTGGA-3′ |
| miR-142-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCCATAAA-3′ |
| miR-196b-3p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGAAGGC-3′ |
| miR-342-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCAATCAC-3′ |
| miR-378a-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACACAGG-3′ |
| miR-455-5p |
5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCGATGTA-3′ |
 | Table III.Primer sequences for miRNAs,
GAPDH and RAC1. |
Table III.
Primer sequences for miRNAs,
GAPDH and RAC1.
| Gene | Primer
sequences | Annealing
temperature, °C | Product length,
bp |
|---|
| miR-15b-5p | F:
5′-GGGTAGCAGCACATCATGG-3′; R: 5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
| miR-31-5p | F:
5′-GGGAGGCAAGATGCTGGC-3′; R: 5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 65 |
| miR-139-5p | F:
5′-GGGGTCTACAGTGCACGTGT-3′; R: 5′-CAGTGCGTGTCGTGGAGT-3′ | 60 | 65 |
| miR-142-3p | F:
5′-GGGGGTGTAGTGTTTCCTA-3′; R: 5′-CAGTGCGTGTCGTGGA-3′ | 60 | 68 |
| miR-196b-3p | F:
5′-GGCTCGACAGCACGACACT-3′; R: 5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 63 |
| miR-342-5p | F:
5′-GGGAGAGGGGTGCTATCTG-3′; R: 5′-GTGCGTGTCGTGGAGTCG-3′ | 60 | 64 |
| miR-378a-5p | F:
5′-GGCTCGTGACTCCAGGT-3′; R: 5′-CAGTGCGTGTCGTGGAG-3′ | 60 | 64 |
| miR-455-5p | F:
5′-GGGCAGTATGTGCCTTTGG-3′; R: 5′-CAGTGCGTGTCGTGGAGT-3′ | 60 | 68 |
| U6 | F:
5′-GCTTCGGCAGCACATATACTAAAAT-3′; | 60 | 89 |
|
| R:
5′-CGCTTCACGAATTTGCGTGTCAT-3′ |
| RAC1 | F:
5′-ATGTCCGTGCAAAGTGGTATC-3′; | 60 | 86 |
|
| R:
5′-CTCGGATCGCTTCGTCAAACA-3′ |
| GAPDH | F:
5′-GCCAAAAGGGTCATCATCTC-3′; | 60 | 124 |
|
| R:
5′-GTAGAGGCAGGGATGATGTTC-3′ |
Bioinformatics analysis
CRC-associated miRNAs were identified in the dbDEMC
2.0 (21) and miRCancer (22) databases. These databases were also
used to find the tripartite overlapping data of differentially
expressed miRNAs and miRNAs associated with CRC using Biovenn
(23). The target genes of
differentially expressed miRNAs were predicted by the PicTar
(24), Targetscan (25) and miRanda (26) databases. The cut-off value
determined using mirSVR was −0.49. The regulatory network of
miR-142-3p and its targeted genes were visualized using Cytoscape
(27). Tripartite overlapping genes
were identified, and enriched pathways and cellular functions were
identified by Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) analyses (david.ncifcrf.gov/tools.jsp) using the Database for
Annotation, Visualization, and Integrated Discovery (DAVID) v6.8
(28).
Cell culture
The human colon cancer cell lines, SW620 and 293T,
were purchased from the China Center for Type Culture Collection.
Cells were incubated in Dulbecco's modified Eagle's supplemented
with 10% fetal bovine serum (both Gibco; Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin and 100 U/ml streptomycin in a 5%
CO2 incubator at 37°C. When the cells reached 70%
confluence at the logarithmic growth phase, they were used for the
experimental study.
Dual-luciferase assay
A dual-luciferase assay was performed using a
Nano-Glo® Dual-luciferase® Reporter Assay
System (cat. no. N1630; Promega Corporation) according to the
manufacturer's instructions. Briefly, a wild-type sequence of the
RAC1 3′-UTR (WT-RAC1,
AAGACAGTATTTTGACAAAATACGAAGTGGAGATTTACACTACATTGTACAAGGAATGAA) and a
mutant sequence of the RAC1 3′-UTR (MUT-RAC1,
AAGACAGTATTTTGACAAAATACGAAGTGGAGATTTTGTGATGTTTGTACAAGGAATGAA) were
inserted into the luc2 site of the pmirGLO Dual-Luciferase miRNA
Target Expression Vector (Promega Corporation) using SacI
and XhoI. The 293T cells were seeded at 5×104
cells per well into 24-well plates, co-transfected with 5 ng
reporter vectors and 40 pmol miR-142-3p mimics
(UGUAGUGUUUCCUACUUUAUGGA) or miR-142-3p negative control (NC)
mimics (UUUGUACUACACAAAAGUACUG; both Guangzhou RiboBio Co., Ltd.)
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 12 h, the transfection medium was replaced
with a complete culture medium, and cells were further cultured for
48 h. At that point, the cells were lysed, and dual-luciferase
activity was measured. All assays were performed in triplicate.
Renilla luciferase was used for normalization.
Cell viability and colony-formation
assays
SW620 cells at the logarithmic growth phase were
seeded into 96-well plates or 6-well plates at a density of 1,000
cells/well. The cells were transfected with 40 pmol miR-142-3p
mimics or miR-142-3p NC mimics using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) or treated with RAC1
inhibitor (NSC23766 trihydrochloride; cat. no. ab142161; Abcam).
Cell proliferation was assessed using Cell Counting Kit-8 (CCK-8)
(Dojindo Molecular Technologies, Inc.) according to the
manufacturer's instructions. The transfection medium was replaced
after 12 h, and cells were cultured for 24, 48, 72 or 96 h. At the
appropriate time point, 10 µl CCK-8 reagent and 90 µl DMEM were
added into each well for 2 h. Absorbance was measured at a
wavelength of 450 nm using a multifunctional enzyme-labeling
instrument.
For the colony-formation assay, SW620 cells were
cultured until the number of colonies in the control group was
>50, which took ~15 days. The cell colonies were fixed in 10%
formalin at 25°C for 1 h and stained with 0.1% crystal violet
(Beyotime Institute of Biotechnology) at 25°C for 5 min. The number
and average size of colonies were counted using an inverted light
microscope at ×100 magnification (Olympus Corporation). All assays
were performed in triplicate.
Cell migration and invasion
assays
The cell invasion and migration assays were
performed using Transwell chambers (Corning, Inc.), with or without
Matrigel (BD Biosciences), respectively. Briefly, 5×104
or 1×105 transfected cells were suspended in serum-free
medium and plated in the upper chamber for the migration and
invasion assays, respectively. The medium in the lower chamber was
supplemented with 10% FBS. After 16 or 24 h, cells in the lower
chamber were fixed in 10% formalin at 25°C for 10 min and stained
with 0.1% crystal violet at 25°C for 5 min and counted using an
inverted light microscope at ×100 magnification (Olympus
Corporation).
Western blotting
SW620 cells were lysed in lysis buffer [1% SDS, 50
mM Tris-HCl (pH 8.0), 5 mM DTT, 1 mM EDTA, 1 mM NaF, 10 mM PMSF, 1
mM Na3VO4 and protease inhibitor cocktail],
denatured, and centrifuged at 10,000 × g and 4°C for 10 min, and
the supernatant was collected. The protein concentration was
measured using the BCA method (Beyotime Institute of
Biotechnology). Protein (30 µg) was used for sodium dodecyl sulfate
polyacrylamide gel electrophoresis using 12% acrylamide gels. The
proteins were transferred and fixed on the polyvinylidene fluoride
membranes, blocked with 5% skimmed milk supplemented with 95% PBS
at 25°C for 2 h, and incubated with the primary antibodies at 4°C
overnight. All primary and secondary antibodies were purchased from
Abcam. The primary antibodies used were: RAC1 (cat. no. ab155938;
1:200), GAPDH (cat. no. ab9485; 1:2,000), MMP9 (cat. no. ab76003;
1:500), extracellular signal-regulated kinase (ERK)1/2 (cat. no.
ab184699; 1:500), pERK1/2 (cat. no. ab201015; 1:500), vimentin
(cat. no. ab8978, 1:1,000), E-cadherin (cat. no. ab1416, 1:500) and
N-cadherin (cat. no. ab18203, 1:500; all Abcam). After washing the
membranes three times, horseradish peroxidase-conjugated secondary
antibodies (goat anti-Rabbit, cat. no. ab150077, 1:2,000 and
anti-mouse IgG, cat. no. ab150113, 1:2,000) were added, and the
membranes were incubated at 25°C for 0.5 h. The membranes were
washed and developed using BeyoECL Plus (Beyotime Institute of
Biotechnology, Suzhou, China). Bands were imaged using Tanon 4600SF
Automatic Chemiluminescence System (Tanon Science and Technology
Co., Ltd.) and intensities were quantified using ImageJ v1.8.0
(imagej.nih.gov/ij/).
Statistical analysis
IBM SPSS Statistics for Windows, version 22.0 (IBM
Corp.) was used for the statistical analysis. Student's t-test
(paired or unpaired depending on the data) was used for the
analysis of continuous variables (mean ± standard deviation).
One-way analysis of variance with Tukey's post hoc test was used to
evaluate the differences between multiple groups. Associations
among pathological features (categorical variables) were evaluated
using the χ2 and Fisher's exact tests. Correlations were
assessed using the Pearson correlation coefficient. P<0.05 was
considered to indicate a statistically significant difference.
Results
Aberrant expression of miRNAs in CRC
tissues
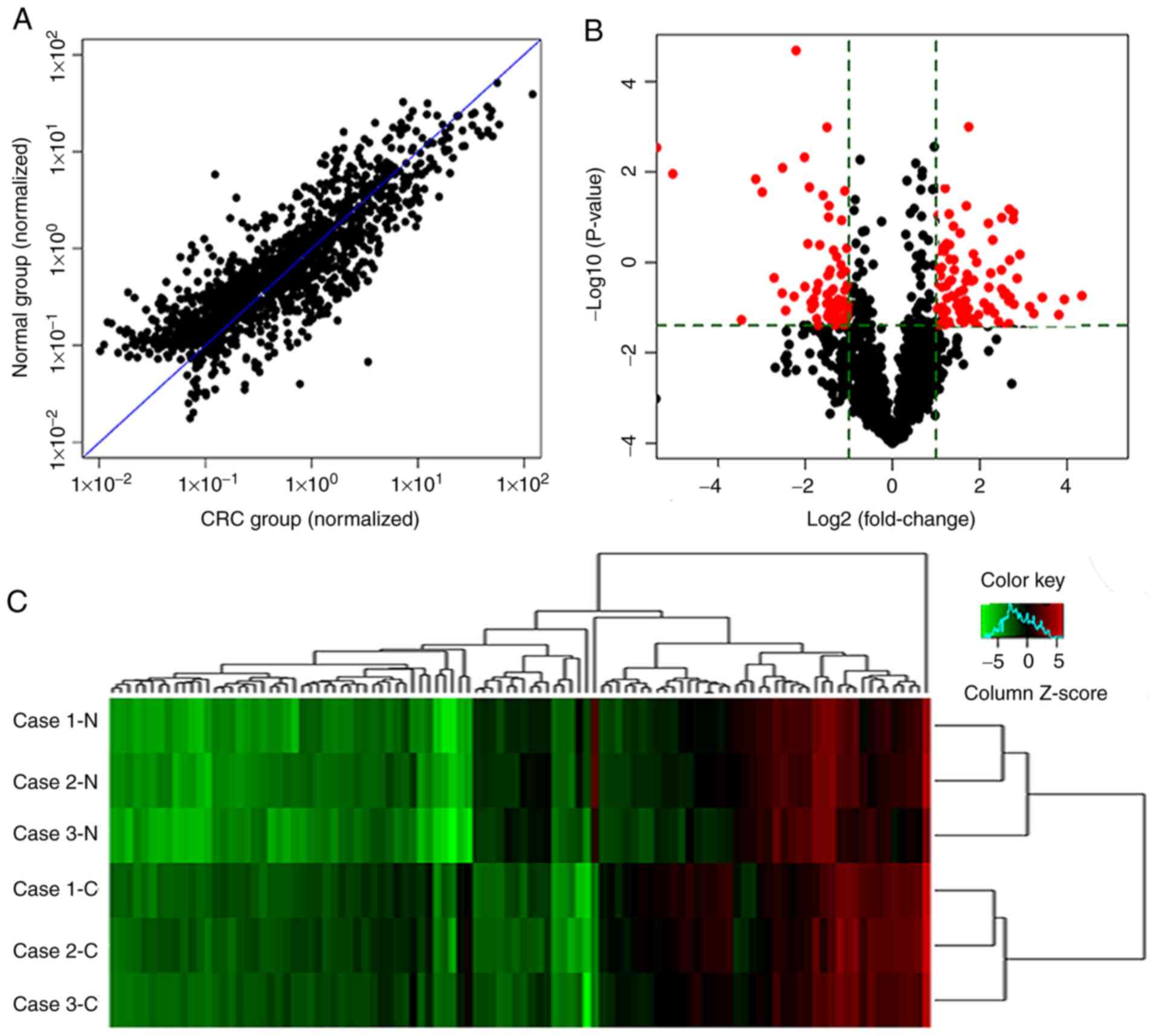
A microarray analysis was performed to determine the
differentially expressed miRNAs between CRC and paired non-cancer
tissue. In total, >2,000 miRNAs were detected, and 178 miRNAs
were differentially expressed between cancer and non-cancer mucosal
tissues. Of the, 178 miRNAs, 76 were upregulated in CRC tissues and
102 were downregulated (Fig. 1A and
B). Unsupervised hierarchical clustering of 3 CRC and paired
non-cancer tissue samples analyzed by miRNAs microarray using the
178 differentially expressed miRNAs showed that the expression
profile of miRNAs was clearly divided into two groups,
corresponding to CRC and paired non-cancer tissues (Fig. 1C). Therefore, the expression of
miRNAs in CRC was different from that in non-cancer mucosa
tissue.
Analysis and validation of miRNAs
dysregulated in CRC
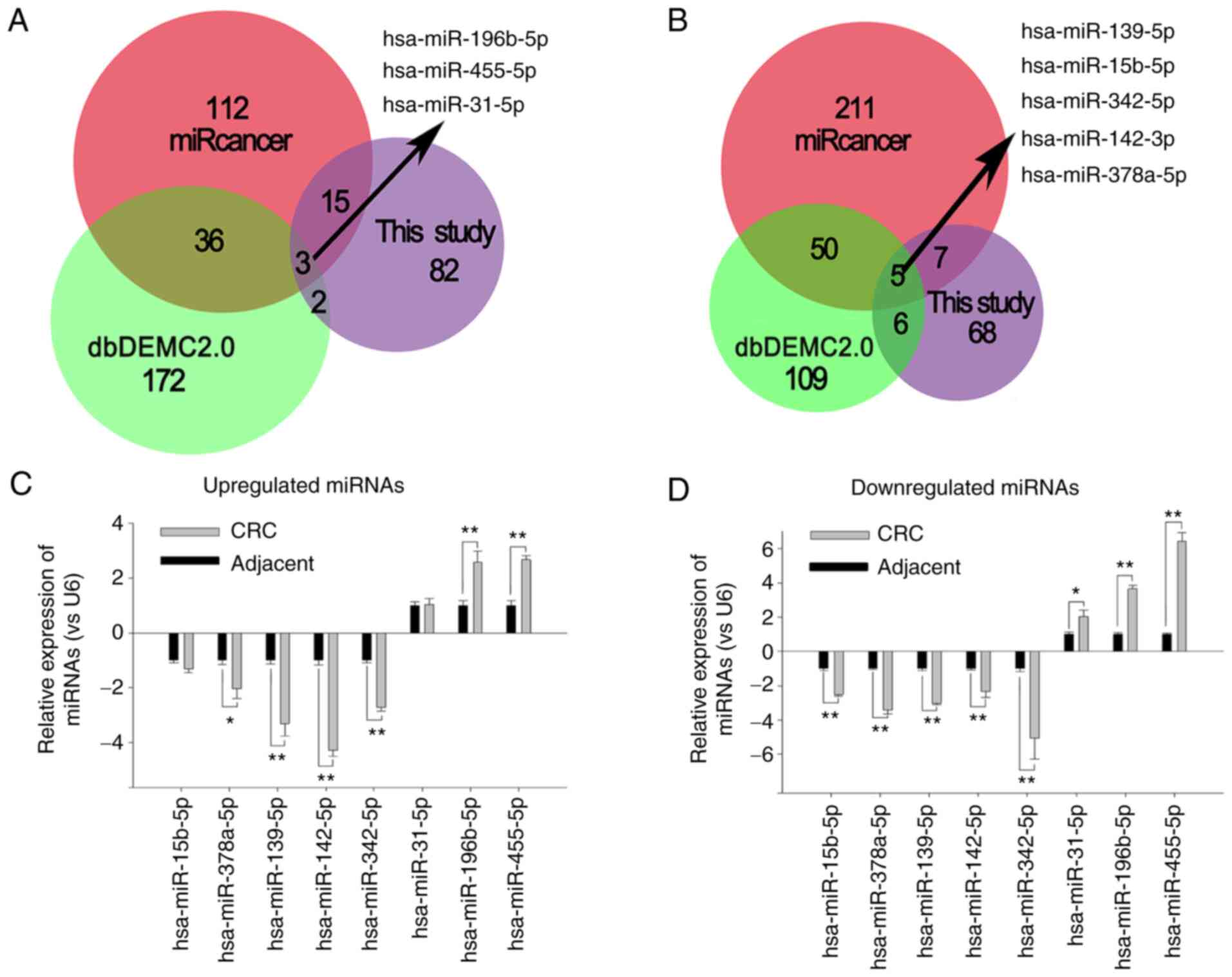
In order to identify the miRNAs that were most
closely associated with CRC, the dbDEMC2 and miRcancer databases
were utilized to search for the dysregulated miRNAs in CRC. A total
of 359 and 371 miRNAs that were dysregulated in CRC were obtained
from the dbDEMC2 and miRcancer databases, respectively. The
differentially expressed miRNAs identified from the microarray
analysis were compared with the miRNAs obtained from the two
databases (Fig. 2A and B), and 8
miRNAs found in all databases were identified. The downregulated
miRNAs included miR-139-5p, miR-15b-5p, miR-342-5p, miR-142-3p and
miR-378a-5p; and the upregulated miRNAs included miR-196b-5p,
miR-455-5p and miR-31-5p. In the present study, qPCR (Fig. 2C) showed that the expression of
these miRNAs was consistent with the microarray analysis results
(Fig. 2D) (r=0.903; P=0.02). The
expression of miR-142-3p was downregulated 4.3-fold in CRC tissue
compared with that in adjacent non-cancer mucosa tissue (t=12.473;
P=0.006).
Bioinformatics analysis of
miR-142-3p
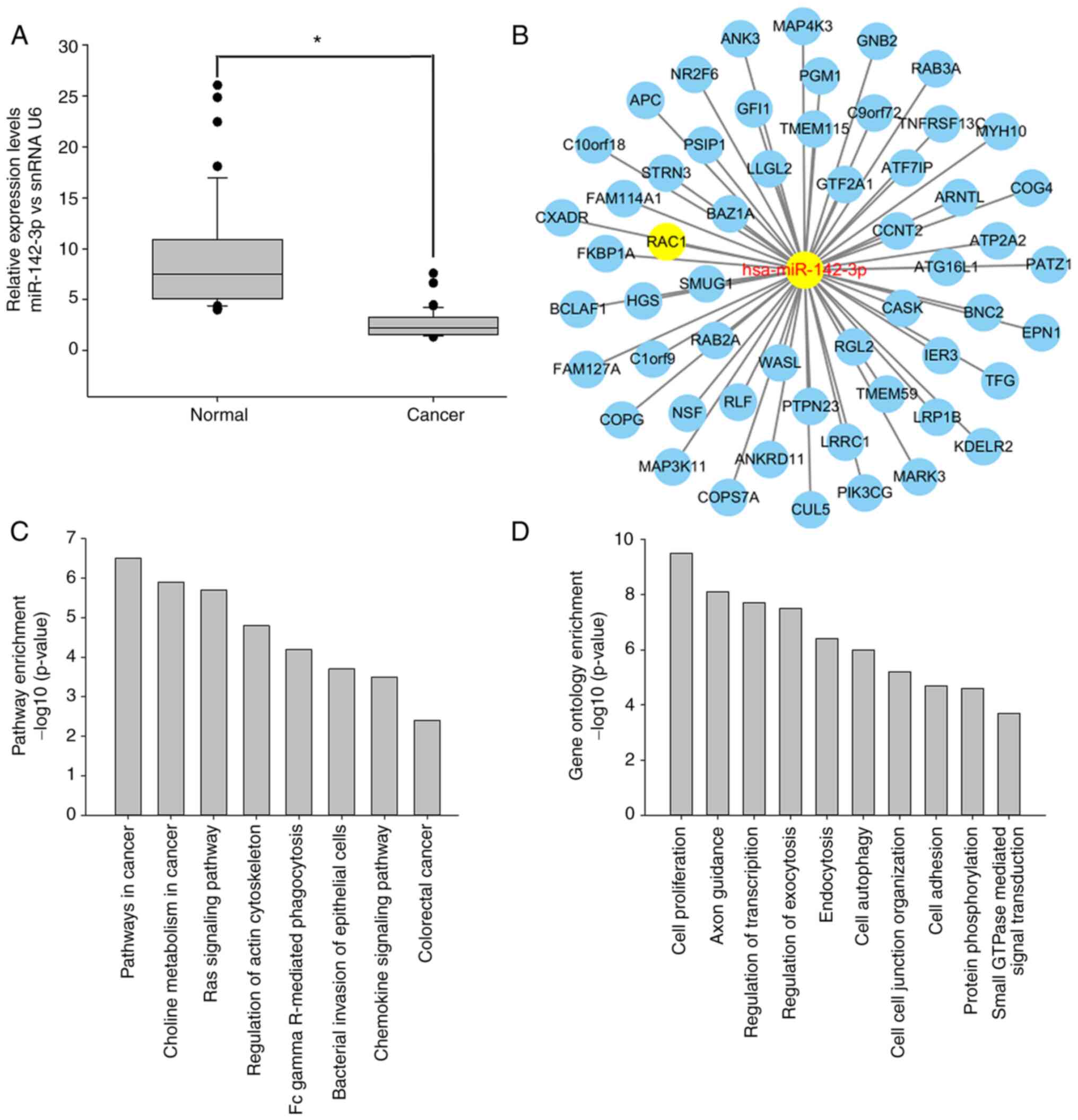
The expression level of miR-142-3p was further
examined by qPCR in 63 cases of CRC and matched non-cancer tissues.
The results showed that miR-142-3p was downregulated in 81.0% of
primary CRC tissues compared with that in matched non-cancer
colorectal mucosa tissues (t=2.595; P=0.012; Fig. 3A). Additionally, miR-142-3p
expression was associated with lymph node metastasis and
Tumor-Node-Metastasis (TNM) stage (Table I). In order to understand the
biological roles of miR-142-3p, the PicTar, Targetscan and miRanda
databases were used to predict its possible targets. A total of 56
potential targets were identified after examining genes that were
present in all three databases (Fig.
3B). Next, those genes were imported into DAVID to perform GO
and KEGG pathway enrichment analyses. The results revealed that
these targets were associated with ‘cell adhesion’, ‘cell
autophagy’, ‘small GTPase-mediated signal transduction’, ‘cell
proliferation’ and ‘regulation of transcription’, among others, and
the involved pathways included ‘pathways in cancer’, ‘regulation of
actin cytoskeleton’, the ‘Ras signaling pathway’, ‘colorectal
cancer’ and other associated signaling pathways (Fig. 3C).
miR-142-3p has the function of
regulating RAC1 expression
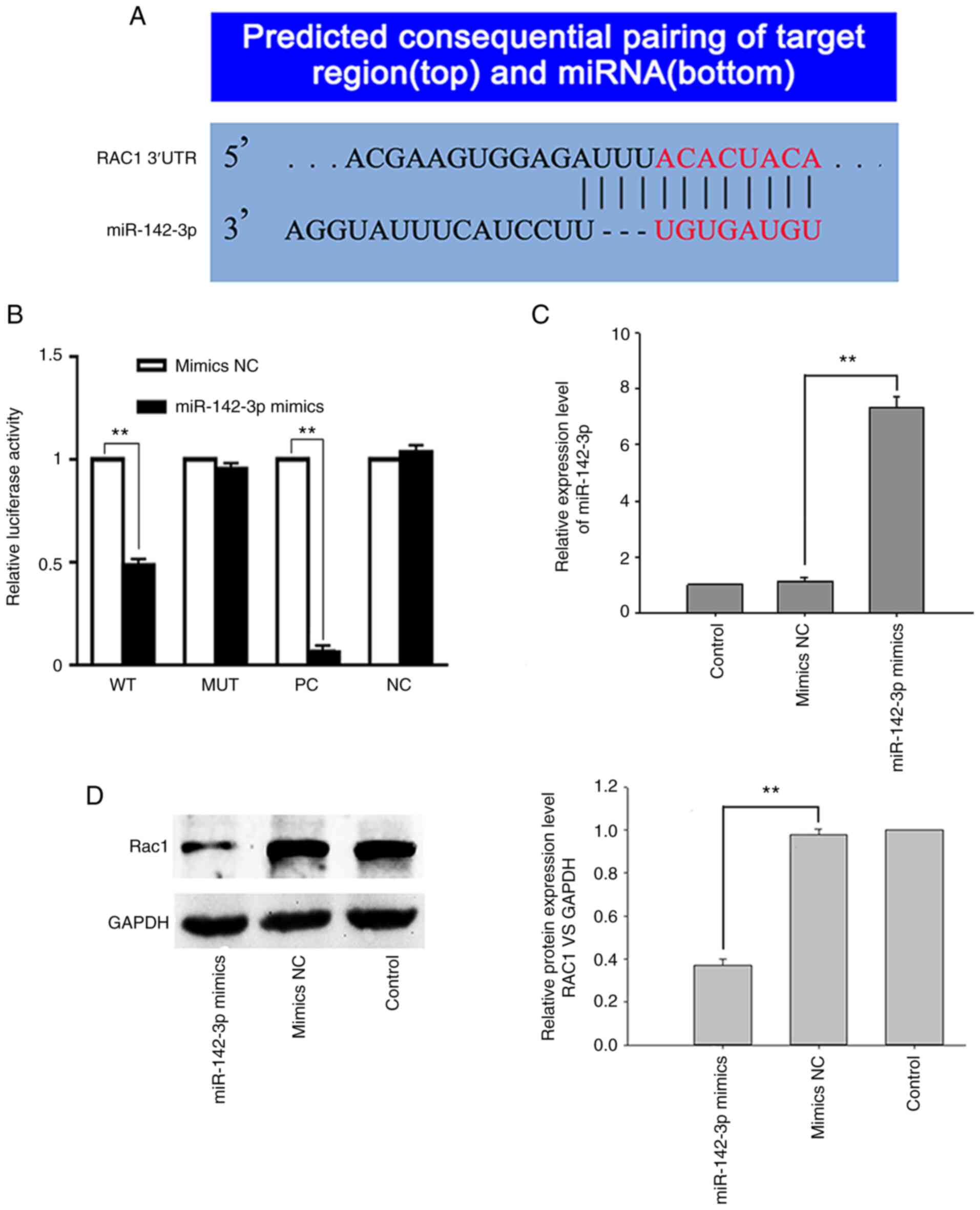
RAC1 had the highest score as a predicted target of
miR-142-3p and had a putative binding site within its 3′-UTR
(Fig. 4A). In order to investigate
whether miR-142-3p regulates the expression of RAC1, a
dual-luciferase assay was performed to determine whether miR-142-3p
can bind to the 3′-UTR of RAC1 and repress RAC1 expression. The
results showed that luciferase activity in 293T cells
co-transfected with miR-142-3p mimics and RAC1-WT was decreased by
>40% compared with that in control cells co-transfected with
miR-142-3p NC mimics and RAC1-WT (t=35.64; P=0.001). However, 293T
cells co-transfected with miR-142-3p mimics and the mutant 3′-UTR
of RAC1 had no significant decrease in luciferase activity (t=3.64;
P=0.68; Fig. 4B). In order to
further verify that RAC1 is a target of miR-142-3p, SW620 cells
were transfected with miR-142-3p mimics (KD cells), which increased
miR-142-3p expression by 732.1% (t=11.47; P=0.008; Fig. 4C), while decreasing RAC1 expression
by 63.8%, compared with the SW620 cells transfected with miR-142-3p
NC mimics (NC cells) (t=23.99; P=0.002; Fig. 4D). The aforementioned results
suggest that RAC1 may be a target of miR-142-3p.
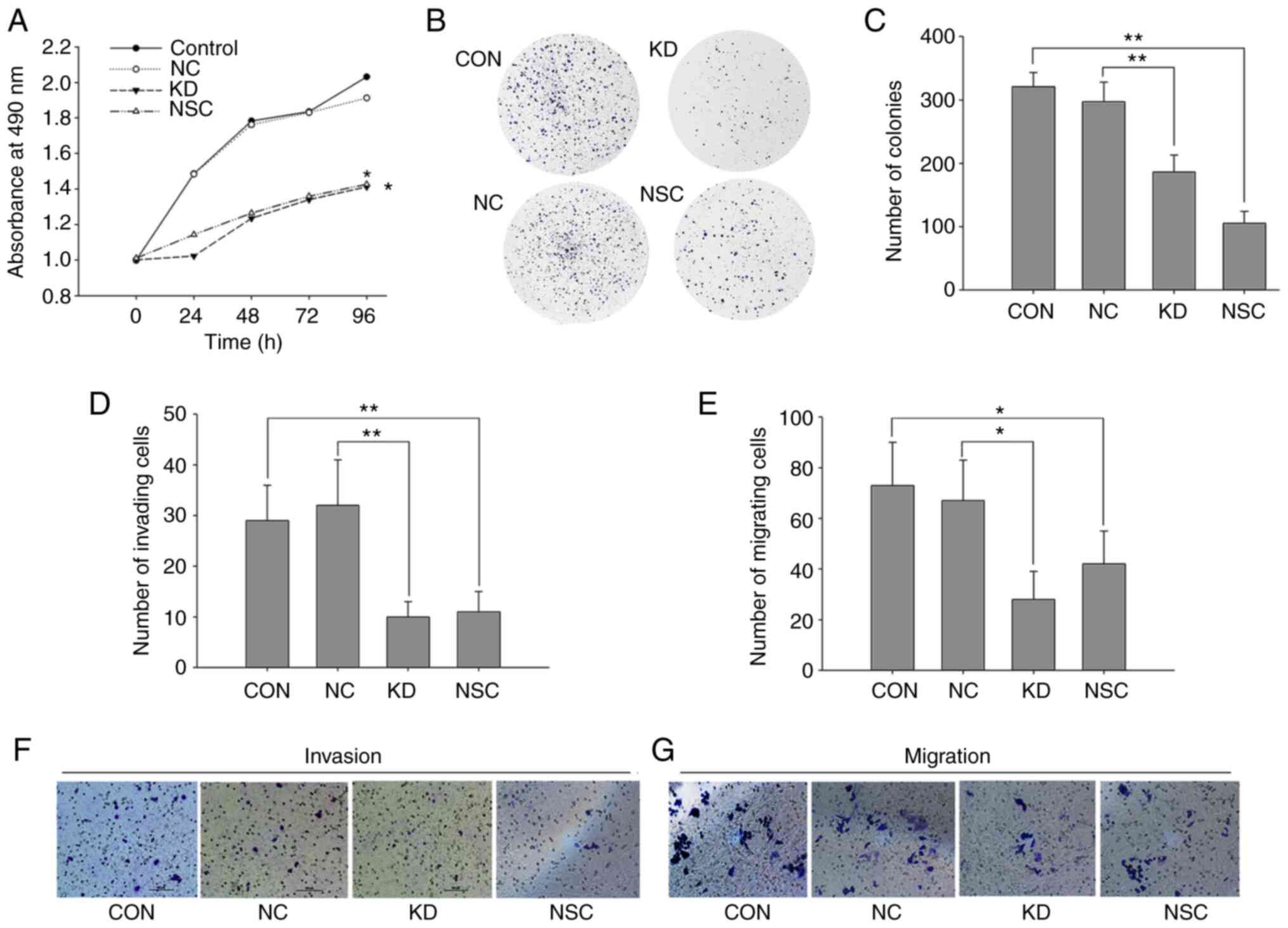
Increased miR-142-3p inhibits the
viability, invasion and migration of SW620 cells
In order to analyze the role of miR-142-3p in CRC,
miR-142-3p mimics were transfected into SW620 cells and cell
viability was assessed by the CCK-8 and colony-formation assays.
After transfection, the proliferation of SW620 cells was inhibited
in a time-dependent manner (t=12.32; P=0.001), whereas no
significant difference was detected between NC cells and control
cells (t=1.325; P=0.277). Cell proliferation decreased by 26.2% 4
days after transfection of miR-142-3p mimics, compared with NC
cells (t=4.94; P=0.039; Fig. 5A).
In addition, following miR-142-3p transfection, the colony-forming
capacity of SW620 cells was significantly decreased by 37.4%
(t=32.04; P=0.001; Fig. 5B and C).
Transwell assays were used to assess the effect of miR-142-3p on
invasion and migration in SW620 cells. Compared with NC cells, cell
invasion was decreased by 68.7% (t=10.39; P=0.009) after cells were
transfected with miR-142-3p mimics (Fig. 5D and F), and migration was decreased
by 58.2% (t=6.351; P=0.028; Fig. 5E and
G). When SW620 cells were treated with the RAC1 inhibitor
NSC23766, similar results were found. Compared with the control
cells, the NSC cells showed a significant decrease in
proliferation, cloning, invasion and migration abilities by 29.8%
(t=8.893; P=0.003), 67.3% (t=7.336; P=0.018), 62.1% (t=13.423;
P=0.006) and 42.5% (t=13.510; P=0.006), respectively (Fig. 5). These results suggest that
miR-142-3p may play a key role in blocking the viability, invasion
and migration of SW620 cells, which may be associated with aberrant
expression of RAC1.
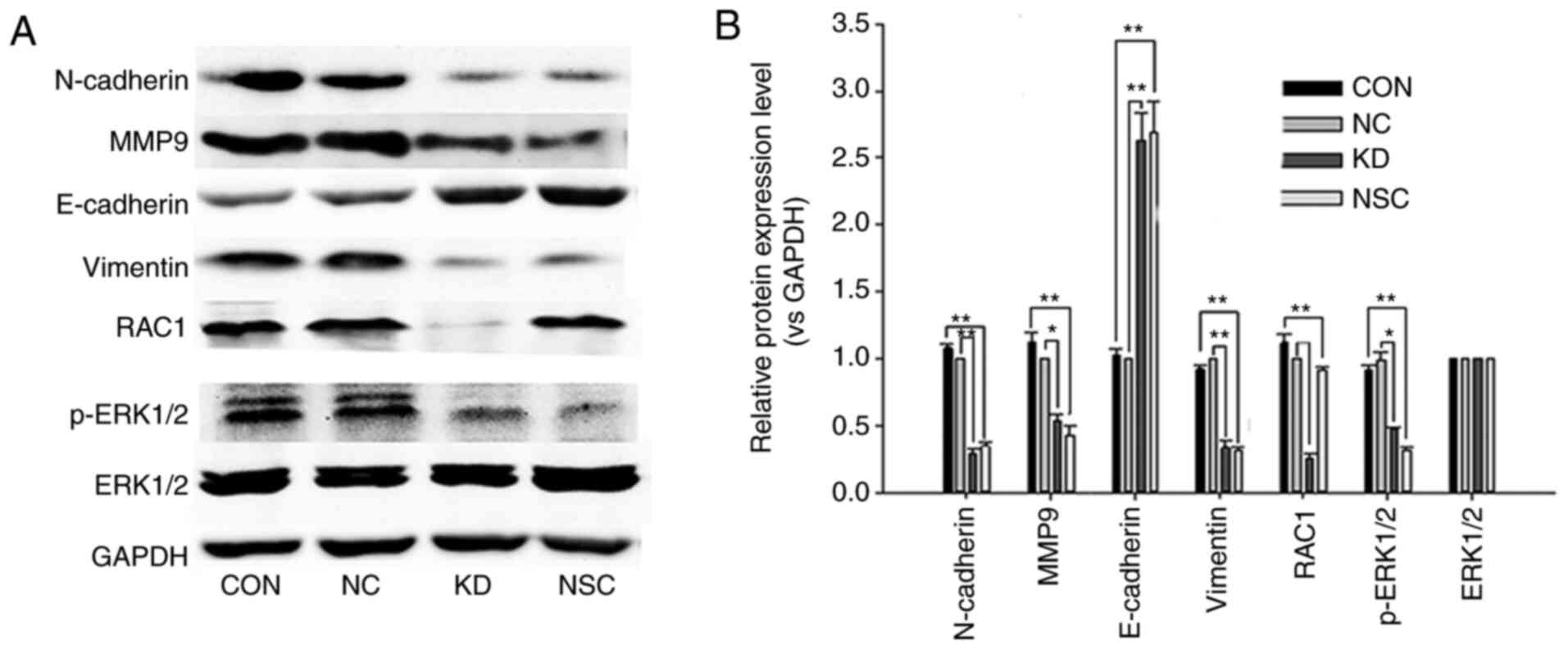
miR-142-3p inhibits EMT by targeting
RAC1 in SW620 cells
In order to investigate whether miR-142-3p affects
EMT in SW620 cells, western blotting was used to detect the
expression of EMT-associated markers and the phosphorylation of
ERK1/2. The expression of RAC1, vimentin, MMP9, N-cadherin and
phosphorylated (p)-ERK was decreased, but that of E-cadherin was
increased in SW620 cells transfected with miR-142-3p mimics,
compared with NC cells (Fig. 6).
Similar results were observed when cells were treated with the RAC1
inhibitor NSC23766 (Fig. 6).
Compared with the control cells, the expression of RAC1 did not
change in NSC cells (Fig. 6). The
aforementioned results indicate that miR-142-3p may also play an
important role in blocking EMT in CRC cells by regulating
RAC1/ERK1/2 signaling. Increasing miR-142-3p inhibited EMT of colon
cancer cells by decreasing RAC1 expression; this was consistent
with the effect of NSC23766 suppressing RAC1-GTP activation.
Discussion
The miRNA microarray is one of the most common
methods of screening disease-associated miRNAs, and several studies
using this technology have revealed aberrant miRNA expression in
human malignant tumors (10,29),
including CRC (17). Volinia et
al (29) used miRNA microarrays
to study the expression patterns of miRNAs in 540 tumor tissues and
found that miRNAs are differentially expressed in non-cancer and
cancer tissues. Further, these miRNAs are strongly tissue-specific.
Zhang et al (30) found that
35 miRNAs demonstrated aberrant expression in colon cancer tissues
using miRNA microarrays. In the present study, the Exiqon miRNA
microarray was used to assess differentially expressed miRNAs in
paired CRC and non-cancer tissues. Compared with paired non-cancer
tissues, 76 miRNAs were upregulated and 102 were downregulated in
CRC samples, and qPCR verified the microarray results.
Numerous recent reports have identified a
dysregulation of miRNAs in CRC (11,15,16,18).
Through analyzing data from miR2Disease (21) and miRcancer (22), it was found that there are >300
differentially expressed miRNAs in CRC. These miRNAs are involved
in cell proliferation, invasion, metastasis (31), drug resistance (32) and other biological functions
(11,15,16,18).
Therefore, these miRNAs can be used as markers for diagnosis,
prognosis, recurrence and drug efficacy in CRC (11,15,16,18).
The present study identified 8 shared miRNAs from microarray
analysis, using the dbDEMC2 and miRcancer databases; of these
miRNAs, miR-142-3p was the most significantly downregulated.
Furthermore, GO and KEGG pathway enrichment analysis showed that 56
predicted targets of miR-142-3p were associated with ‘cell
adhesion’, ‘cell autophagy’, ‘small GTPase-mediated signal
transduction’, ‘cell proliferation’, ‘regulation of transcription’
and other functional regulation, and the involved pathways included
‘pathways in cancer’, ‘regulation of actin cytoskeleton’, the
‘Ras-signaling pathway’, ‘colorectal cancer’ and other associated
signaling pathways. Therefore, miR-142-3p may play a key role in
the tumorigenesis and development of CRC.
Several studies have confirmed that downregulation
of miR-142-3p is important in the development and progression of
various tumors (33), such as lung
(34), breast (35) and gastric (36) cancer. The inhibition of miR-142-3p
can promote the growth, invasion and metastasis of tumor cells,
whereas increased expression of miR-142-3p inhibits cell migration
and proliferation (33–36). Recent studies revealed that
miR-142-3p was downregulated in CRC, which is associated with CRC
development and progression, and that miR-142-3p could be used as a
marker for the diagnosis of the disease (37–39).
It was also found that miR-142-3p was significantly downregulated
in 51 CRC tissues compared with that in matched non-cancer mucosa
tissues, and that miR-142-3p expression was associated with lymph
node metastasis and TNM stage. Transfection of miR-142-3p into
SW620 colon cancer cells decreased their proliferation and
colony-forming capacity, and decreased invasion and migration. The
aforementioned data suggest that miR-142-3p may play a key role in
regulating the behavior of CRC cells.
Some studies have reported that overexpression of
miR-142-3p leads to downregulation of RAC1 in multiple cancer
types, and that RAC1 is a target of miR-142-3p (40–42).
miR-142-3p downregulates RAC1 by binding to its 3′-UTR (40–43).
In the present study, the level of RAC1 was decreased in SW620
cells after transfection with miR-142-3p mimics. Moreover, when the
3′-UTR of wild-type RAC1 was fused to a luciferase construct, there
was a significant decrease in luciferase activity after
transfection of miR-142-3p mimics; however, this effect dwindled
when the luciferase construct was fused to a mutant RAC1 sequence.
The aforementioned results indicated that miR-142-3p can regulate
the expression of RAC1 to regulate tumorigenesis.
RAC1 is an important member of the Rho GTPase
family, which regulates various cell biological processes,
including cell survival, proliferation, adhesion, EMT, metastasis
and invasion. The Rho GTPase family is also involved in PAK,
nuclear factor-κB, MAPK, STAT3 and WNT/β-catenin signaling
(44–46). RAC1-GTP is the active conformation
of RAC1, and RAC1 hyperactivation is found in several human cancer
types (46,47). It has been reported that eliminating
RAC1 GTPase activity can disrupt EMT in CRC cells and significantly
inhibit the growth, invasion and metastasis of colon cancer cells
(48). Meanwhile, one study showed
that upregulated miR-142-3p leads to an increase in RAC1 expression
(49), which has a major conflict
with the function of most miRNAs. miRNA interacts with the 5′-UTR,
ORF or 3′-UTR of its target mRNA to suppress its translation or
induce its degradation, but usually does not upregulate its
expression; there are only a few cases where miRNAs upregulate
protein translation, primarily in mitochondria (6–8,50,51).
In addition, these views are inconsistent with the study results of
other authors (40–43), including that of the present study.
The present study results showed that RAC1 was significantly
downregulated after transfection with miR-142-3p mimics, and EMT,
proliferation, invasion and migration were also inhibited. Further
studies revealed that the expression of p-ERK decreased after
transfection with miR-142-3p mimics or inhibition of RAC1 activity.
Meanwhile the present results are consistent with those of other
studies (40–43); if increasing the expression of RAC1
protein in CRC cells can restore the phenotypic changes caused by
miR-142-3p upregulation, the result would be more reliable. Further
exploration of the effect of changes in RAC1 expression on
miR-142-3p function will be helpful to elucidate the molecular
mechanism of CRC metastasis.
In conclusion, the expression of miR-142-3p was
decreased in CRC; this may be associated with lymph node metastasis
and TNM stage. Transfection of colon cancer cells with miR-142-3p
mimics inhibited the expression of RAC1 and decreased the
proliferation, invasion and migration of cancer cells; these
effects may be associated with the inhibition of ERK1/2 signaling.
The results in the present study suggest that miR-142-3p may be a
potential target for the treatment of CRC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the Natural Science
Foundation of Hainan province, China (grant nos. 818MS144 and
819MS120).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The obtained microarray data
were deposited in the Gene Expression Omnibus database (accession
no. GSE101502).
Authors' contributions
NX analyzed and interpreted the data regarding the
miRNA microarray and CRC tissues. QM and YZ performed the
bioinformatics analysis and qPCR assay, ZL and YL performed the
statistical analysis. FX and SL collected the tissue specimens. YH
performed the experimental design, cell function experiments, and
was a major contributor in writing the manuscript. NX and YH
confirm the authenticity of all the raw data. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
the First Affiliated Hospital of Hainan Medical University
(approval no. HYFYKJ-2018-32). All the patients provided written
consent for their specimens to be used in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
MAPK
|
mitogen-activated protein kinase
|
|
TGF
|
transforming growth factor
|
|
miRNA
|
microRNA
|
|
UTR
|
untranslated region
|
|
EMT
|
epithelial-to-mesenchymal
transition
|
|
qPCR
|
quantitative polymerase chain
reaction
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
ERK
|
extraceullar signal-regulated
kinase
|
|
TNM
|
Tumor-Node-Metastasis
|
|
RAC1
|
Rac family small GTPase 1
|
|
CCK-8
|
Cell Counting Kit-8
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koveitypour Z, Panahi F, Vakilian M,
Peymani M, Seyed Forootan F, Nasr Esfahani MH and Ghaedi K:
Signaling pathways involved in colorectal cancer progression. Cell
Biosci. 9:972019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wongfieng W, Jumnainsong A, Chamgramol Y,
Sripa B and Leelayuwat C: 5′-UTR and 3′-UTR regulation of MICB
expression in human cancer cells by novel microRNAs. Genes (Basel).
8:2132017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gebert LF and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He B, Zhao Z, Cai Q, Zhang Y, Zhang P, Shi
S, Xie H, Peng X, Yin W, Tao Y and Wang X: miRNA-based biomarkers,
therapies, and resistance in cancer. Int J Biol Sci. 16:2628–2647.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu G and Li B: Role of miRNA in
transformation from normal tissue to colorectal adenoma and cancer.
J Cancer Res Ther. 15:278–285. 2019.PubMed/NCBI
|
|
12
|
Pidíkova P, Reis R and Herichova I: miRNA
clusters with down-regulated expression in human colorectal cancer
and their regulation. Int J Mol Sci. 21:46332020. View Article : Google Scholar
|
|
13
|
Bartels CL and Tsongalis GJ: MicroRNAs:
Novel biomarkers for human cancer. Ann Biol Clin (Paris).
68:263–272. 2010.(Artcle in French). PubMed/NCBI
|
|
14
|
Santos P and Almeida F: Role of Exosomal
miRNAs and the tumor microenvironment in drug resistance. Cells.
9:14502020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ren A, Dong Y, Tsoi H and Yu J: Detection
of miRNA as non-invasive biomarkers of colorectal cancer. Int J Mol
Sci. 16:2810–2823. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Michael MZ, Connor SM, van Holst Pellekaan
NG, Young GP and James RJ: Reduced accumulation of specific
microRNAs in colorectal neoplasia. Mol Cancer Res. 1:882–891.
2003.PubMed/NCBI
|
|
17
|
Nagy ZB, Wichmann B, Kalmár A, Galamb O,
Barták BK, Spisák S, Tulassay Z and Molnár B: Colorectal adenoma
and carcinoma specific miRNA profiles in biopsy and their
expression in plasma specimens. Clin Epigenetics. 9:222017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Long J, He Q, Yin Y, Lei X, Li Z and Zhu
W: The effect of miRNA and autophagy on colorectal cancer. Cell
Prolif. 53:e129002020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weiser MR: AJCC 8th edition: Colorectal
cancer. Ann Surg Oncol. 25:1454–1455. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang Z, Wu L, Wang A, Tang W, Zhao Y, Zhao
H and Teschendorff AE: dbDEMC 2.0: Updated database of
differentially expressed miRNAs in human cancers. Nucleic Acids
Res. 45:D812–D818. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xie B, Ding Q, Han H and Wu D: miRCancer:
A microRNA-cancer association database constructed by text mining
on literature. Bioinformatics. 29:638–644. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hulsen T, de Vlieg J and Alkema W:
BioVenn-a web application for the comparison and visualization of
biological lists using area-proportional Venn diagrams. BMC
Genomics. 9:4882008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Enright AJ, John B, Gaul U, Tuschl T,
Sander C and Marks DS: MicroRNA targets in drosophila. Genome Biol.
5:R12003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang JX, Song W, Chen ZH, Wei JH, Liao
YJ, Lei J, Hu M, Chen GZ, Liao B, Lu J, et al: Prognostic and
predictive value of a microRNA signature in stage II colon cancer:
A microRNA expression analysis. Lancet Oncol. 14:1295–1306. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mansoori B, Mohammadi A, Naghizadeh S,
Gjerstorff M, Shanehbandi D, Shirjang S, Najafi S, Holmskov U,
Khaze V, Duijf PH and Baradaran B: miR-330 suppresses EMT and
induces apoptosis by downregulating HMGA2 in human colorectal
cancer. J Cell Physiol. 235:920–931. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anandappa G, Lampis A, Cunningham D, Khan
KH, Kouvelakis K, Vlachogiannis G, Hedayat S, Tunariu N, Rao S,
Watkins D, et al: miR-31-3p expression and benefit from anti-egfr
inhibitors in metastatic colorectal cancer patients enrolled in the
prospective phase II PROSPECT-C Trial. Clin Cancer Res.
25:3830–3838. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shrestha A, Mukhametshina RT, Taghizadeh
S, Vásquez- Pacheco E, Cabrera-Fuentes H, Rizvanov A, Mari B,
Carraro G and Bellusci S: MicroRNA-142 is a multifaceted regulator
in organogenesis, homeostasis, and disease. Dev Dyn. 246:285–290.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lawson J, Dickman C, Towle R, Jabalee J,
Javer A and Garnis C: Extracellular vesicle secretion of miR-142-3p
from lung adenocarcinoma cells induces tumor promoting changes in
the stroma through cell-cell communication. Mol Carcinog.
58:376–387. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mansoori B, Mohammadi A, Ghasabi M,
Shirjang S, Dehghan R, Montazeri V, Holmskov U, Kazemi T, Duijf P,
Gjerstorff M and Baradaran B: miR-142-3p as tumor suppressor miRNA
in the regulation of tumorigenicity, invasion and migration of
human breast cancer by targeting Bach-1 expression. J Cell Physiol.
234:9816–9825. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Y, Cao Z, Wang L, Liu S and Cai J:
Downregulation of microRNA-142-3p and its tumor suppressor role in
gastric cancer. Oncol Lett. 15:8172–8180. 2018.PubMed/NCBI
|
|
37
|
Gao W, Pang D and Yu S: Serum level of
miR-142-3p predicts prognostic outcome for colorectal cancer
following curative resection. J Int Med Res. 47:2116–2125. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen WW, Zeng Z, Zhu WX and Fu GH:
MiR-142-3p functions as a tumor suppressor by targeting CD133,
ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl).
91:989–1000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ghanbari R, Mosakhani N, Asadi J, Nouraee
N, Mowla SJ, Yazdani Y, Mohamadkhani A, Poustchi H, Knuutila S and
Malekzadeh R: Downregulation of plasma MiR-142-3p and MiR-26a-5p in
patients With colorectal carcinoma. Iran J Cancer Prev.
8:e23292015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu T, He BS, Pan B, Pan YQ, Sun HL, Liu
XX, Xu XN, Chen XX, Zeng KX, Xu M and Wang SK: MiR-142-3p functions
as a tumor suppressor by targeting RAC1/PAK1 pathway in breast
cancer. J Cell Physiol. 235:4928–4940. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Peng D, Dong J, Zhao Y, Peng X, Tang J,
Chen X, Wang L, Hu DN, Reinach PS, Qu J and Yan K: miR-142-3p
suppresses uveal melanoma by targeting CDC25C, TGFβR1, GNAQ, WASL,
and RAC1. Cancer Manag Res. 11:4729–4742. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li WQ, Zhao WC, Xin J, Niu TL, Chao YF,
Zhou P, Zheng MH and Xu B: MicroRNA-142-3p suppresses cell
proliferation and migration in bladder cancer via Rac1. J Biol
Regul Homeost Agents. Feb 28–2020.(Epub ahead of print).
|
|
43
|
Liu J, Li W and Wang S, Wu Y, Li Z, Wang
W, Liu R, Ou J, Zhang C and Wang S: MiR-142-3p attenuates the
migration of CD4+ T cells through regulating actin
cytoskeleton via RAC1 and ROCK2 in arteriosclerosis obliterans.
PLoS One. 9:e955142014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sahajpal N, Kowluru A and Kowluru RA: The
regulatory role of rac1, a small molecular weight GTPase, in the
development of diabetic retinopathy. J Clin Med. 8:9652019.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nguyen LK, Kholodenko BN and von
Kriegsheim A: Rac1 and RhoA: Networks, loops and bistability. Small
GTPases. 9:316–321. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
De P, Aske JC and Dey N: RAC1 Takes the
lead in solid tumors. Cells. 8:3822019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kotelevets L and Chastre E: Rac1
signaling: From intestinal homeostasis to colorectal cancer
metastasis. Cancers (Basel). 12:6652020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou K, Rao J, Zhou ZH, Yao XH, Wu F, Yang
J, Yang L, Zhang X, Cui YH, Bian XW, et al: RAC1-GTP promotes
epithelial-mesenchymal transition and invasion of colorectal cancer
by activation of STAT3. Lab Invest. 98:989–998. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Gao X, Xu W, Lu T, Zhou J, Ge X and Hua D:
MicroRNA-142-3p promotes cellular invasion of colorectal cancer
cells by activation of RAC1. Technol Cancer Res Treat. Jan
1–2018.(Epub ahead of print). View Article : Google Scholar
|
|
50
|
Vasudevan S, Tong Y and Steitz JA:
Switching from repression to activation: MicroRNAs can up-regulate
translation. Science. 318:1931–1934. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang X, Zuo X, Yang B, Li Z, Xue Y, Zhou
Y, Huang J, Zhao X, Zhou J, Yan Y, et al: MicroRNA directly
enhances mitochondrial translation during muscle differentiation.
Cell. 158:607–619. 2014. View Article : Google Scholar : PubMed/NCBI
|