Introduction
Polycystic ovary syndrome (PCOS) is one of the most
common endocrinopathies; it is characterized by a number of
clinical symptoms, including menstrual disorders, hyperandrogenism,
infertility, insulin resistance, hirsutism, obesity and cystic
ovaries (1–3). The worldwide incidence of PCOS is
5–10% among women of reproductive age, and it accounted for 70% of
ovulation barrier sterility (4–6).
Early diagnosis and timely treatment can contribute to the
mitigation of the PCOS and may help in avoiding associated
metabolic complications (7).
However, due to the fact that many of the symptoms and signs of
PCOS are also normal in puberty, PCOS diagnose criteria needs to be
improved to prevent overdiagnosis of the syndrome (3,8).
The Rotterdam diagnostic criteria for PCOS is the most widely used
standard and includes hyperandrogenism, chronic anovulation
(oligomenorrhoea or amenorrhea) and polycystic ovary morphology
(volume of the ovary >10 cm3 or at least 12 follicles
measuring 2–9 mm) (9–11). Ruling out hyperandrogenism caused
by other endocrinopathies, a patient with any two of above three
criteria can be diagnosed with PCOS (12).
Confirmation of the etiology is key to determining
the treatment of PCOS. To date, environmental factors and genetic
factors are thought to be involved (13), including diet, unhealthy lifestyle
and certain diseases (such as thyroid dysfunction,
hyperprolactinemia and androgen-secreting tumors). Apart from
these, a large number of genes have been identified as serving a
role in the development of PCOS, such as GDF9 and androgen receptor
(AR). Androgenic effect, exerted through the AR, is regarded as one
of the most predominant mechanisms responsible for PCOS (14,15). As a steroid receptor and nuclear
transcription factor, AR can control the expression of downstream
target genes (16–18). In theory, hyperandrogenism is
influenced by the changes of testosterone (T) level and AR
activity. Previous studies have demonstrated that AR functions as a
dynamic heterocomplex with a number of coactivators and
corepressors (19–23). C-terminal-binding protein 1
antisense (CTBP1-AS) is a long non-coding RNA (lncRNA) that can
directly suppress CTBP1 expression through sense-antisense binding,
which subsequently promotes the transcriptional activity of AR
(24,25). In addition to promoting prostate
cancer progression (24),
CTBP1-AS was also positively correlated with PCOS (12). However, the molecular mechanism of
CTBP1-AS in PCOS remains largely unknown.
In the present study, the levels of CTBP1-AS were
compared between patients with PCOS and healthy controls. The
effects of CTBP1-AS on the proliferation and apoptosis of granulosa
cells was explored, and enhancer of zeste homolog 2 and embryonic
(EZH2) and ectoderm development protein (EED) were indicated to
interact with CTBP1-AS in primary granulosa cells and KGN cells.
Notably, it was demonstrated that the level of CTBP1-AS could be
significantly reduced by treatment of cryptotanshinone. The present
findings may reveal a new pathophysiological relevance of CTBP1-AS
in PCOS and may provide new potential target for PCOS
treatment.
Materials and methods
Isolation and culture of ovarian
granulosa cells
A total of 60 patients with PCOS and 60 healthy
controls were recruited to the present study. After ruling out
hyperandrogenism caused by other endocrinopathy, the Rotterdam
diagnostic criteria was used to diagnose PCOS, particularly those
with any two of the following features: Hyperandrogenism, chronic
anovulation (oligomenorrhoea or amenorrhea) and polycystic ovary
morphology. All women with PCOS included in the study fulfilled all
three criteria aforementioned; none of the healthy control women
recruited in this study met any of the Rotterdam diagnostic
criteria. All 120 patients were non-smokers, had no concurrent
illness and took no regular medications prior to the study; a
pregnancy test was performed prior to their inclusion in the study.
The average age of patient group and control group were 29.21±4.78
and 29.43±3.82 years, respectively. No significant differences were
identified for age or body mass index (BMI) between the wo groups
(Table I). The present study was
approved by the ethics committee of The First Affiliated Hospital
of Zhejiang Chinese Medical University (Hangzhou, China; approval
no. 2020-K-060-02); all subjects provided signed consent forms
prior to recruitment to the study.
 | Table I.Clinicopathological characteristics
of the patients with PCOS and the healthy control patients. |
Table I.
Clinicopathological characteristics
of the patients with PCOS and the healthy control patients.
| Clinicopathological
feature | Control
(n=60)a | PCOS
(n=60)a | P-value |
|---|
| Age, years | 29.43±3.82 | 29.21±4.78 | NS |
| BMI,
kg/m2 | 23.12±2.13 | 24.35±3.32 | NS |
| Total T,
nmol/l | 1.43±0.23 | 3.17±0.31 | <0.001 |
| Free T, pmol/l | 19.32±5.65 | 25.32±5.21 | 0.038 |
| E2, nmol/l | 0.31±0.021 | 0.26±0.029 | NS |
| SHBG, nmol/l | 65.21±6.57 | 62.56±4.28 | NS |
| DHEAS, nmol/l |
5,119.45±666.21 |
6,351.32±996.67 | 0.025 |
| FSH, IU/l | 3.92±0.63 | 6.13±0.32 | 0.031 |
| LH, IU/l | 4.99±.34 | 9.32±1.85 | 0.015 |
| Prolactin,
ng/ml | 11.27±1.29 | 13.98±3.65 | NS |
| Fasting insulin,
mIU/ml | 5.52± 0.41 | 10.07±0.82 | 0.01 |
| Fasting glucose,
mmol/l | 5.27±0.63 | 6.78±1.93 | NS |
Both healthy controls and patients with PCOS were
given clomiphene citrate at a dose of 50 mg/day on cycle days 5–9
to induce ovulation. Following ovulation induction, and 36 h after
human chorionic gonadotropin (10,000 U) injection, two eggs were
collected from each candidate using transvaginal ultrasound-guided
retrieval; eggs from patients with PCOS were aspirated by an
experienced operator before laparoscopic ovarian drilling (LOD)
surgery, whereas those from the healthy control patients were
immediately collected through 19-gauge single-lumen aspiration
needles (K-OPS-7035-REH-ET; Cook) to aspirate under a suction
pressure of 80 mmHg. Eggs from patients and healthy control were
both performed transvaginally under ultrasound guidance in
conscious sedation with fentanyl 1 µg/kg IV (Fentanyl®,
B. Braun Medical AB) and propofol 30–40 mg IV
(Propofol-®Lipuro, B. Braun Medical AB) was given when
needed. The granulosa cells were detached under a compact cell
culture light microscope (Olympus CKX53, Olympus)and kept in
follicular fluids. Granulosa cells were centrifuged at 395 × g for
15 min at 4°C, the precipitate was harvested and mixed with 50% v/v
Percoll solution (cat. no. 65455-52-9; Sigma-Aldrich; Merck KGaA)
and centrifuged again at 395 × g for 10 min to remove red blood
cells. Then, 0.2 g/l collagenase I solution (cat. no. 9001-12-1;
Sigma-Aldrich; Merck KGaA) was used to resuspend and digest the
granulosa cells for 30 min at 37°C followed by centrifugation as
aforementioned. Cells were harvested and seeded into a 12-well cell
culture plate at 1×105 cells/ml. Each well contained 1
ml RPMI-1640 medium (cat. no. SH30809.01; HyClone; Cytiva) with 10%
fetal bovine serum (cat. no. FB25015; Clark Bioscience) and 1%
penicillin-streptomycin (PS; cat. no. SV30010; Hyclone; Cytiva).
The cells were cultured in an incubator at 37°C with an atmosphere
of 5% CO2.
KGN cell culture
The KGN human ovarian granulosa tumor cell line
(cat. no. CL0544) was purchased from Hunan Fenghui Biotechnology
Co., Ltd. and cultured in DMEM/F-12 at 37°C with 5% CO2
(cat. no. SH30023.01; HyClone; Cytiva) supplemented with 10% FBS
and 1% PS.
RNA fluorescent in situ hybridization
(FISH)
To verify the subcellular localization of CTBP1-AS,
a DIG RNA Labeling Kit (cat. no. 11175025910; Roche Diagnostics)
was used to label CTBP1-AS probe (synthesized by RiboBio) according
to the manufacturer's protocol. The sequence of the CTBP1-AS probe,
complementary to CTBP1-AS, was 5′-TCATCATAATTTCTTATCCTAAG-3′. A
total of 50,000 primary granulosa cells were cultured on cover
glasses in 24-well culture dishes for 24 h at 37°C with 5%
CO2. The cells were subsequently rinsed with PBS, fixed
at room temperature in a solution of 3% formaldehyde and 10% acetic
acid in PBS for 30 min, and then permeabilized with 0.5% Triton
X-100 for 5 min at room temperature. After three washes with PBS,
hybridization was performed in a moist chamber at 42°C overnight.
Then, cells were washed by 4X saline sodium citrate) buffer with
0.1% Tween-20, and followed by 2X SSC, 1X SSC and PBS for 10 min at
42°C in dark. Cells were then blocked with 20% goat serum (cat. no.
SL038; Beijing Solarbio) for 1 h at room temperature and followed
by incubation with anti-DIG antibody (cat. no. 14682; Cell
Signaling Technology) which diluted with 10% goat serum at 1:200
overnight at 4°C. After three washes with PBS, FITC-conjugated goat
anti-rabbit secondary antibody (diluted with 10% goat serum at
1:1,000, cat. no. ab6717; Abcam) was added and incubated for 1 h at
room temperature. Finally, the nucleus was counterstained with DAPI
(cat. no. C0065; Beijing Solarbio Science and Biotechnology Co.,
Ltd.) for 20 min at room temperature. Results were visualized at
400 magnification using a Leica DMI4000B inverted fluorescence
microscope (Leica Microsystems GmbH).
CTBP1-AS knockdown
For CTBP1-AS knockdown, two small interfering
(si)RNAs were purchased from Sigma Genosys (Sigma-Aldrich; Merck
KGaA) and their sequences are as follows: siCTBP1-AS #1,
5′-CCAAUUAAUUAGACCACAAAA-3′; siCTBP1-AS#2,
5′-CAACUGUCAAGAAACAAUUAG-3′. Scrambled Stealth RNAi™ Med GC (Thermo
Fisher Scientific, Inc.) was used as a negative control
(siControl). Cells were seeded in 6-well plate and grown to ~40%
confluence at the time of transfection. 200 pmol (20 µM) siCTBP1-AS
#1, siCTBP1-AS #1 or siControl was respectively transfected at room
temperature by using the Lipofectamine RNAiMAX (cat. no. 13778-150;
Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The transfected cells were grown in
antibiotic free DMEM/F-12 (8119239, Gibco; Thermo Fisher
Scientific, Inc.) for 24 h and then re-seeded and cultured for an
additional 24 h for further experiments with the density of 200
cells/well for colony formation assay and 30,000 for
Cryptotanshinone treatment and immunofluorescence detection
assay.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated with the High Purity Total
RNA Extraction kit (cat. no. RP5611; BioTeke Corporation) and
reverse-transcribed into cDNA with the super M-MLV reverse
transcriptase (cat. no. PR6502; BioTeke Corporation) according to
the manufacturer's instructions. qPCR was conducted to analyze
CTBP1-AS and β-actin expression levels. qPCR was performed on an
Applied Biosystems 7300 cycler using SYBR Green PCR MasterMix
(Invitrogen; Thermo Fisher Scientific, Inc.). Thermocycling
parameters are as follows: 95°C for 3 min, 40 cycles of 95°C for 20
sec and 60°C for 30 sec with FAM acquisition. The primer sequences
used were as follows: CTBP1-AS forward, 5′-AACCTGGCAGCACGGAAGT-3′;
CTBP1-AS reverse, 5′-GAGCACAACCACCACCTCATC-3′; β-actin forward,
5′-CTGTGCCCATCTACGAGGGCTAT-3′; β-actin reverse,
5′-TTTGATGTCACGCACGATTTCC-3′. Relative expression levels were
calculated using the 2−ΔΔCq method (26) and normalized to β-actin.
Preparation of nuclear and cytoplasmic
RNA
A total of 5×106 cells were harvested,
washed with cold PBS and centrifuged at 1,000 × g for 10 min at
4°C. The supernatant was removed and the pellet was resuspended
with cold nuclear fractionation buffer [140 mM NaCl, 10 mM Tris-HCl
(pH 7.8), 1.5 mM MgCl2, 0.5% NP-40 and 3 U/ml RNaseOUT
(Thermo Fisher Scientific)]. After 5 min, the suspension was
centrifuged 500 × g for 5 min at 4°C. The supernatant was collected
as the cytoplasmic fraction, whereas the pellet was further washed
with fractionation buffer three additional time, centrifuged at
2,000 × g for 5 min at 4°C and used as the nuclear fraction. Total
cytoplasmic and nuclear RNAs were extracted using TRIzol
(Invitrogen; Thermo Fisher Scientific, Inc.) and subsequently
reverse transcribed to cDNA using super M-MLV reverse transcriptase
(cat. no. PR6502; BioTeke Corporation) according to the
manufacturer's instructions. The primer sequences used were as
follows: U48 forward, 5′-AGTGATGATGACCCCAGGTA-3′; U48 reverse,
5′-GGTCAGAGCGCTGCGGTGAT-3′; 7SL forward,
5′-GTAGCTTTTCGCAGCGTCTC-3′; 7SL reverse,
5′-GCACTACAGCCCAGAACTCC-3′. The relative expression of CTBP1-AS was
detected as aforementioned and calculated using the
2−ΔΔCq method (26)
and normalized to U48 and 7SL for nuclear and cytoplasmic
fractions, respectively.
RNA pulldown and mass spectrometry
analysis
Pierce Magnetic RNA-Protein Pull-Down Kit (cat. no.
20164; Thermo Fisher Scientific, Inc.) was used to perform RNA
pulldown assay according to the manufacturer's instructions.
Briefly, CTBP1-AS was synthesized by RiboBio Co. Ltd. and labeled
using desthiobiotinylated cytidine bisphosphate and T4 RNA ligase.
After labeling, CTBP1-AS was captured by streptavidin magnetic
beads (50 µl), which was contained in the kit, in RNA capture
buffer at room temperature for 30 min. 5×107 KGN cells
in logarithmic growth phase were harvested and lysed with Pierce IP
Lysis Buffer (cat. no. 87787, Thermo Fisher Scientific, Inc.) with
RNase inhibitor and protease/phosphatase inhibitor (cat. no. P0013;
Beyotime). Subsequently, 1,000 µg protein lysate were added to
Protein-RNA Binding Buffer equilibrated RNA-bound beads. The
mixture was incubated overnight with rotation at 4°C. The beads
were then rinsed with wash buffer three times followed by vortexing
and separation using a magnetic stand. 50 µl Elution Buffer was
added and incubated for 15–30 min at 37°C with agitation to elute
the RNA-binding protein complex. After separation by the magnetic
stand, the eluted samples were then harvested denatured for 10 min
at 100°C in loading buffer [50 mM Tris-HCl (pH 6.8), 2% SDS (w/v),
0.1% BPB (w/v), 10% glycerol (v/v), 1% β-mercaptoethanol (v/v)] for
SDS-PAGE electrophoresis followed closely by Coomassie brilliant
blue staining and mass spectrometry analysis or western blotting
detection. Mass spectrometry data was acquired by sending sample to
Shanghai Bioprofile Ltd for mass spectrometry assay.
Western blotting
Protein concentrations from RNA pulldown assays were
determined with a BCA Protein Assay Kit (cat. no. P0011; Beyotime
Institute of Biotechnology). Proteins were denatured for 10 min at
100°C in loading buffer [50 mM Tris-HCl (pH 6.8), 2% SDS (w/v),
0.1% BPB (w/v), 10% glycerol (v/v), 1% β-mercaptoethanol (v/v)],
separated by SDS-PAGE (10%), transferred into PVDF membranes and
blocked with 5% skim milk for 2 h at room temperature. Membranes
were incubated with anti-EZH2 (cat. no. 5246; Cell Signaling
Technology, Inc.) or anti-EED (cat. no. 85322; Cell Signaling
Technology, Inc.) antibodies, followed by incubation with
HRP-conjugated goat anti-rabbit secondary antibody (cat. no. 7074S,
Cell Signaling Technology). All antibodies were diluted with 5%
skim milk at 1:1,000 and incubated with the membranes for 2 h at
room temperature. Protein bands were visualized with the Tanon
High-sig ECL Western Blotting Substrate (cat. no. 180-501; Tanon
Science and Technology Co., Ltd.).
Cell Counting Kit-8 (CCK-8) assay
Cell viability was measured using CCK-8 assay (cat.
no. CK04; Dojindo Molecular Technologies, Inc.). Briefly, 5,000
primary granulosa cells were seeded into 96-well plate and
transfected for 24 h with 10 pmol siCTBP1-AS #1, siCTBP1-AS #2 or
siControl at room temperature. Then, transfected cells were
cultured at 37°C with 5% CO2 for ~24 h until the
confluence reached 30–40%. 0, 24, 48 and 72 h later, CCK-8 solution
was added and incubated for 2 h at 37°C. Absorbance at 450 nm was
measured using CMax Plus (Molecular Devices, LLC).
ELISA
Blood samples (5 ml) were collected from all
participants. The serum concentrations of luteinizing hormone (LH),
follicle stimulating hormone (FSH), prolactin, estradiol (E2),
total T, free T, sex hormone-binding globulin (SHBG) and
dehydroepiandrosterone-sulfate (DHEAS), as well as fasting glucose
and fasting insulin were determined by commercial ELISA kits for
Testosterone (cat. no. ab108666) and DHEAS (cat. no. ab108669)
purchased from Abcam. All detection were performed according to the
manufacturer's protocol.
Flow cytometry detection of
apoptosis
Cell apoptosis were detected using a FITC Annexin V
Apoptosis Detection Kit I (cat. no. 556547; BD Biosciences).
Primary granulosa cells were cultured to 30–50% confluence in
6-well plate, and then transfected with siRNAs for 48 h. Cells were
harvested, washed with cold PBS and stained with propidium iodide
(PI) alone or PI and Annexin V-FITC for 20 min at room temperature.
Data were obtained using CytoFLEX (Beckman Coulter, Inc.) and
analyzed by CytExpert software (Beckman Coulter, Inc.version
2.3).
Colony formation assay
SiCTBP1-AS #1, siCTBP1-AS #2 or siControl
transfected primary granulosa cells were plated at 200 cells/well
in 6-well dishes and cultured for 2–3 weeks in DMEM/F-12 (8119239,
Gibco) at 37°C with 5% CO2, supplemented with 10% FBS
(fetal bovine serum, FB25015, Clark) and 1% PS
(penicillin-streptomycin, SV30010, Hyclone) and washed with PBS.
The cells were fixed in cold methanol for 30 min. Fixed colonies
were visualized by incubating the cells with 0.5% (w/v) crystal
violet for 30 min. Residual crystal violet was rinsed away with
three washes with PBS. Visible colonies, consisting of ≥50 cells,
were counted. Each experiment was repeated three times.
Cryptotanshinone treatment and
immunofluorescence
A total of 30,000 KGN cells were cultured on cover
glasses in 24-well culture dishes. After 24 h incubation, different
concentrations cryptotanshinone (5 µM in Fig. 4 and 0, 2.5, 5, 10 µM for Fig. S3) was added and the cells were
incubated for another 24 h. The cells were rinsed with PBS, fixed
in a solution of 3% formaldehyde and 10% acetic acid in PBS for 30
min at room temperature and then permeabilized with 0.5% Triton
X-100 for 5 min at room temperature. After three washes with PBS,
cells were incubated with diluted (1:500) anti-5-methylcytosine
(5mC) antibody (cat. no. ab10805; Abcam) to label methylated DNA
overnight at 4°C and the nuclei were counterstained with DAPI (5
µg/ml) for 20 min at room temperature. Results were visualized and
images captured under a Leica DMI4000B inverted fluorescence
microscope and analyzed by Leica Application Suite V4 software
(Leica Microsystems GmbH).
Analyzing the promoter region of
CTBP1-AS
To analyze the promoter region of CTBP1-AS, we
recruited the UCSC Genome Browser database (genome.ucsc.edu).
CTBP1-AS (Homo sapiens, NR_104331) was used to analyze the CpG
island existed in the promoter region.
Statistical analysis
Data are presented as the mean ± SEM. Comparisons of
non-normally distributed variables between clinicopathological
characteristics of healthy control and patients with PCOS (Table I) and the comparisons of CTBP1-AS
level in healthy controls and patients with PCOS (Fig. 1) were determined using
Mann-Whitney U test or Kruskal-Wallis test followed by Dunn's post
hoc test. were performed using the Mann-Whitney U test. Student's t
test was used to determine the statistical significance when only
two groups were compared, and when three groups or more were
compared, ANOVA followed by Tukey's post hoc test was used.
P<0.05 was considered to indicate a statistically significant
difference. All data were analyzed using IBM SPSS Statistics 25.0
(IBM Corp.). All the experiments were repeated three times.
Results
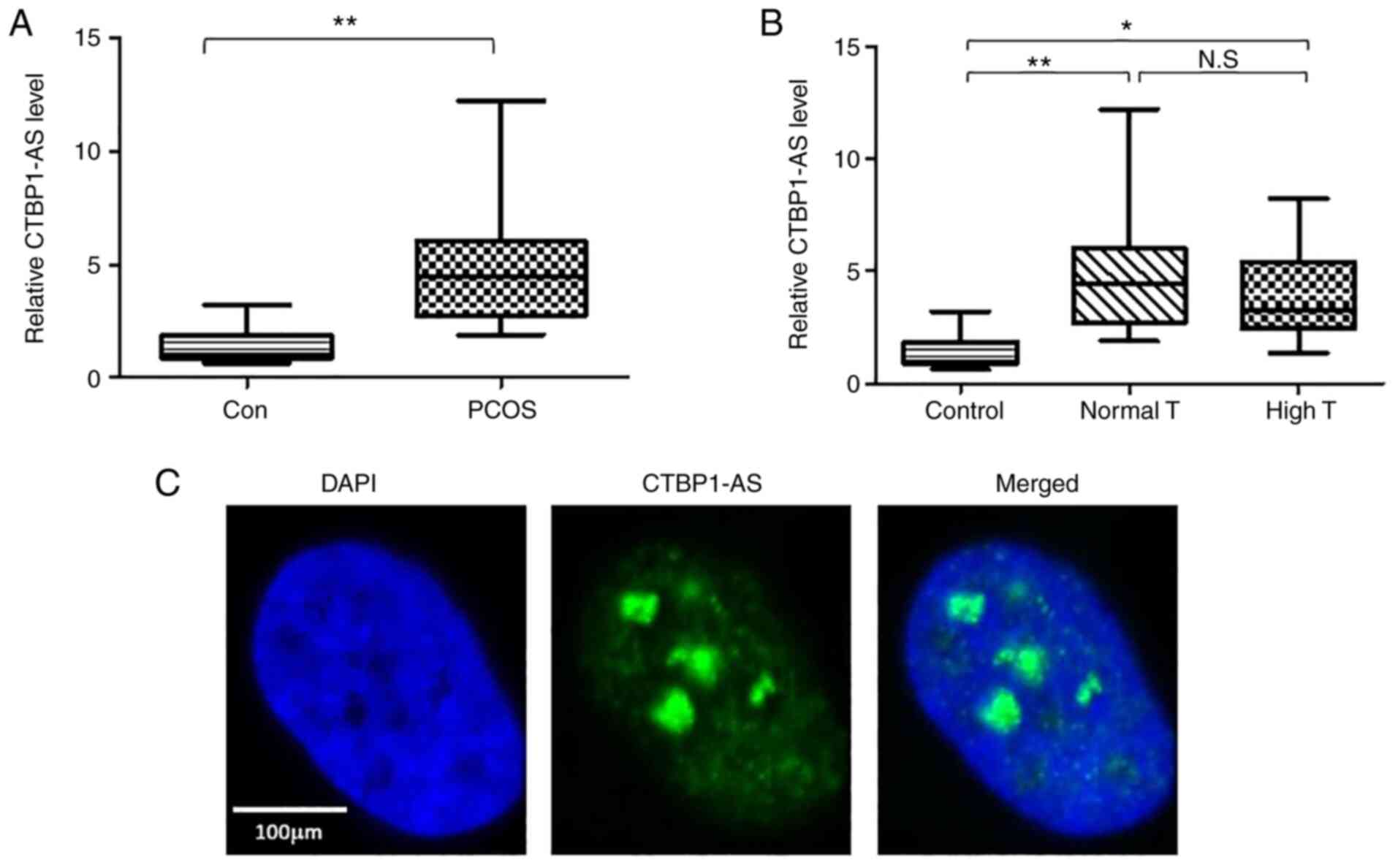
CTBP1-AS is upregulated in PCOS
To investigate whether CTBP1-AS served a role in
PCOS, 60 patients with PCOS and 60 healthy controls were recruited
to the present study; there were no significant differences in age
and BMI (Table I). ELISA serum
analysis results demonstrated that patients with PCOS had higher
levels of total T, free T, DHEAS, FSH, LH and fasting insulin than
controls (Table I). However, no
significant differences were identified for the concentrations E2,
SHBG, prolactin and fasting glucose between the two groups.
Two oocytes were harvested from each patient and the
ovarian granulosa cells were separated, cultured and subsequently
used to detect the expression level of CTBP1-AS by RT-qPCR.
Compared with healthy controls, the level of CTBP1-AS was
significantly higher in patients with PCOS (Fig. 1A). Total T levels were notably
different among patients with PCOS; therefore, the patients with
PCOS were divided into high T and normal T group. Both groups
exhibited higher levels of CTBP1-AS compared with the control group
(Fig. 1B). These data suggested
that CTBP1-AS may participate in PCOS process. Next, the
localization of CTBP1-AS in granulosa cells was examined using RNA
FISH. Consistent with a previous report (24), CTBP1-AS was expressed throughout
the nucleus (Fig. 1C). To further
evaluate the distribution of CTBP1-AS in granulosa cells, the
expression levels in cytoplasmic and nuclear fractions were
analyzed by RT-qPCR. The results demonstrated that the CTBP1-AS
levels in the nucleus were ~150 times than that of the expression
in cytoplasm (Fig. S1).
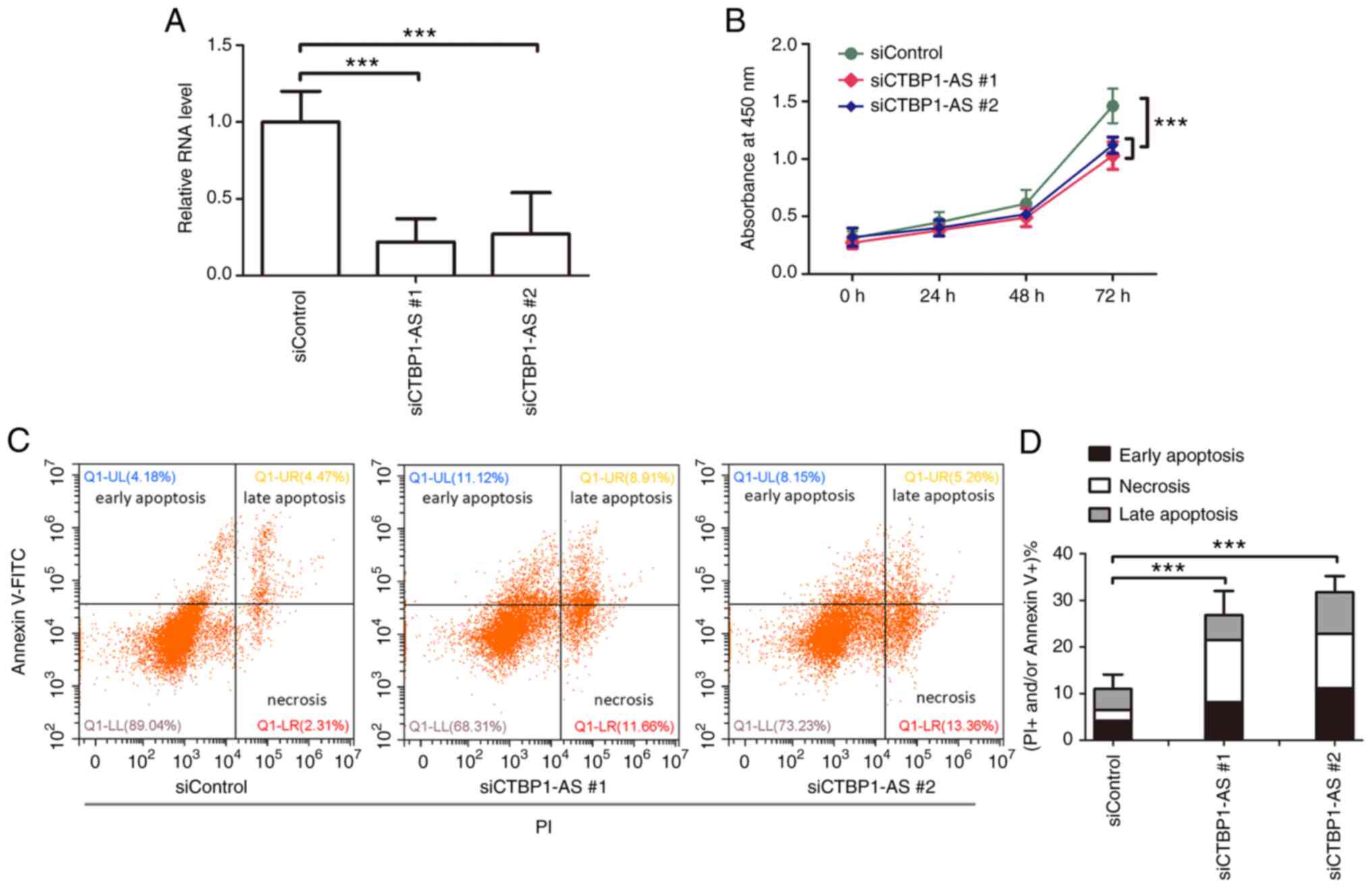
CTBP1-AS balances proliferation and
apoptosis of granulosa cells
To further investigate the biological significance
of CTBP1-AS in PCOS, CTBP1-AS expression was knocked down using
siRNA. As shown in Fig. 2A, the
expression levels of CTBP1-AS were significantly decreased in
primary granulosa cells transfected with siCTBP1-AS #1 and siCTBP1
#2 compared with siControl transfected cells. The proliferative
ability of granulosa cells was examined through CCK-8 assays, and
the results demonstrated that proliferation was significantly
inhibited by the siCTBP1-AS transfections compared with the control
(Fig. 2B). Colony formation
assays further demonstrated that CTBP1-AS knockdown suppressed the
colony formation ability of primary granulosa cells compared with
the control group (Fig. S2). In
addition, the effect of CTBP1-AS on granulosa cell apoptosis was
examined by flow cytometry. The results showed that the number of
both apoptotic and necrotic cells significantly increased following
CTBP1-AS knockdown (Fig. 2C and
D). Taken together, these results indicated that CTBP1-AS may
facilitate PCOS through balancing the proliferative and apoptotic
rates of granulosa cells.
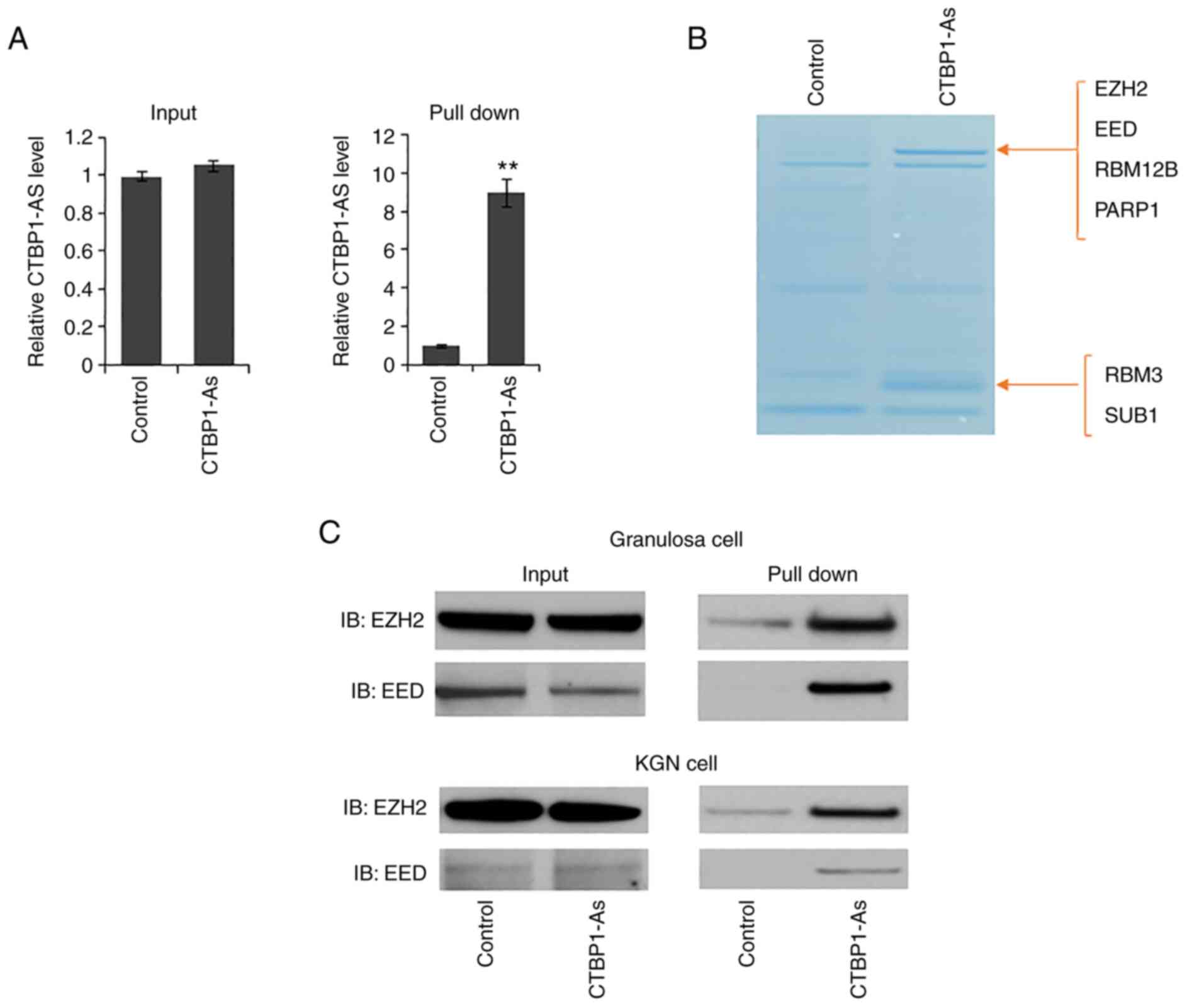
CTBP1 directly interacts with EZH2 and
EED in granulosa cells
As CTBP1-AS could recruit and bind PTB-associated
splicing factor in prostate cancer cells (24), the present study attempted to
identify the interacting proteins of CTBP1-AS in granulosa cells
using RNA pulldown and mass spectrometry. As shown in Fig. 3A, the enrichment efficiency of
CTBP1-AS was ~9 times higher compared with the control group. After
incubation with cell lysate, the mixture was collected and examined
by SDS-PAGE, followed closely by Coomassie brilliant blue staining.
The specific bands were further analyzed by mass spectrometry, and
two Polycomb-group proteins, EZH2 and EED, were of particular
interest (Fig. 3B). Both proteins
make up the core catalytic subunits of Polycomb repressive complex
2 (PRC2) and PRC2 is responsible for catalyzing histone 3, lysine
27 trimethylation (H3K27me3) on chromatin (26,27). The interaction was further
confirmed in primary granulosa cells and KGN cells by RNA pulldown
(Fig. 3C). It was inferred from
these results that CTBP1-AS may regulate PCOS through EZH2- and
EED-dependent H3K27me3 methylation.
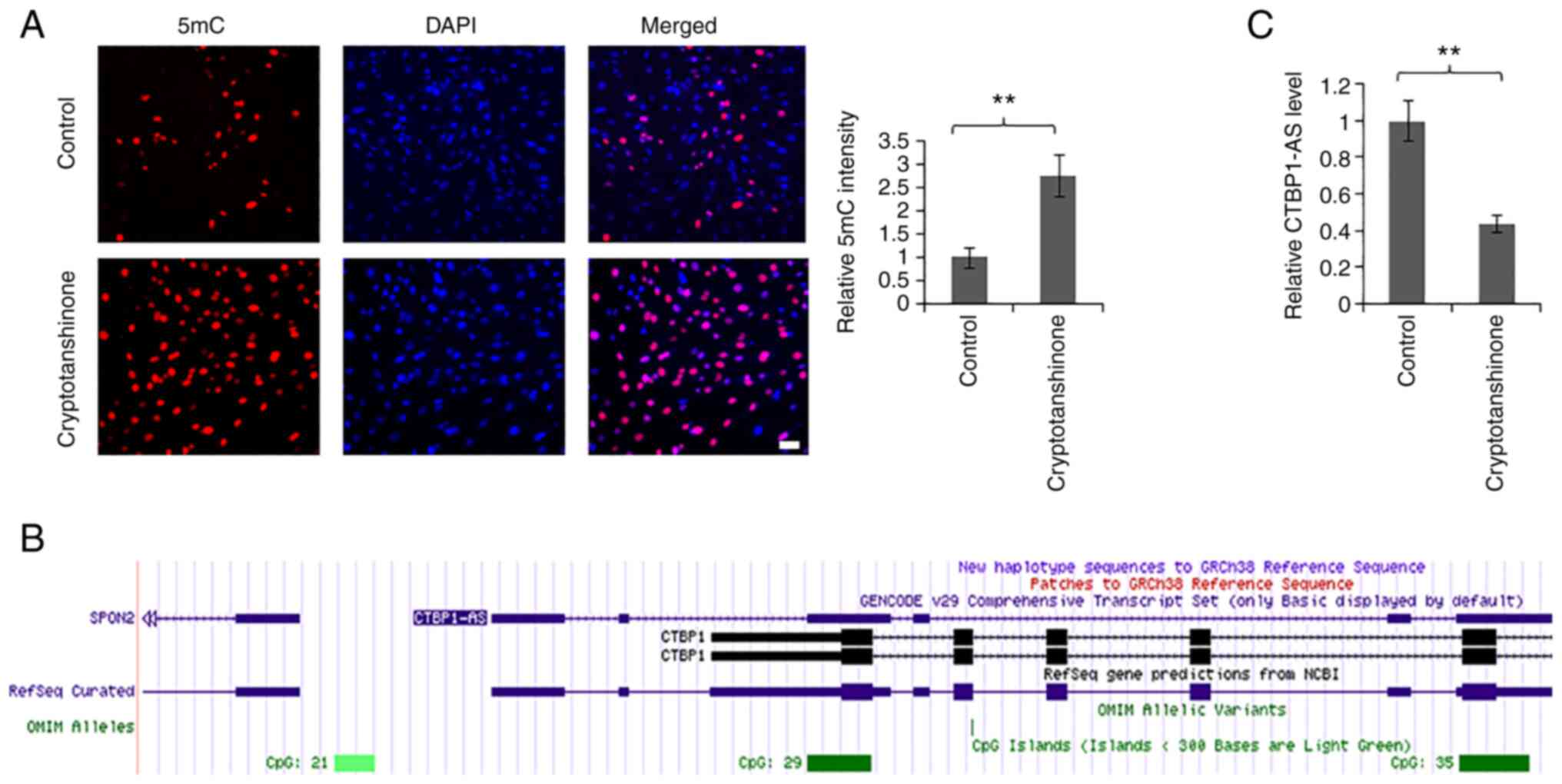
Cryptotanshinone effectively reduces
the level of CTBP1-AS in KGN cells
Cryptotanshinone can ameliorate insulin resistance
and androgen excess in a PCOS rat model (28,29), but the molecular mechanism is
still not fully understood. To better elucidate this mechanism, KGN
cells were treated with 5 µM cryptotanshinone for 24 h (Fig. 4A). The results of 5mC
immunofluorescence staining suggested that the methylation level of
the whole genome had a positive association with the concentration
of cryptotanshinone (Fig. S3).
Immunofluorescence intensity induced by 10 µM cryptotanshinone was
weaker compared with that induced by 5 µM (Fig. S3); it was speculated that this
may be caused by the cytotoxicity of cryptotanshinone. Furthermore,
UCSC Genome Browser analysis showed that there existed CpG island
in the promoter region of CTBP1-AS (Fig. 4B); therefore, we hypothesized that
cryptotanshinone may regulate the level of CTBP1-AS through
methylation. Consistent with this, cryptotanshinone treatment
significantly inhibited the level of CTBP1-AS (Fig. 4C). These results indicated that
CTBPA1-AS may be a potential target of cryptotanshinone for PCOS
treatment.
Discussion
As the main functional cells in the ovaries,
granulosa cells participate in the development of follicles and
steroid secretion (30). Atresia
is usually triggered by oocyte apoptosis followed by granulosa cell
apoptosis in the early stage of follicular development; however,
granulosa cell apoptosis initiates atresia in mature follicles
(31). Communication between
oocytes and granulosa cells lasts throughout the oocyte cell
development process. Granulosa cells can provide nutrients and
growth factors for oocyte cells, and oocytes promote the growth and
differentiation of granulosa cells (32). Previous reports have demonstrated
that dysfunction of these cells induces abnormal follicle and
related to PCOS (33,34); however, the molecular mechanism is
still unknown. AR is generally thought to be closely related to
PCOS, and lncRNA CTBP1-AS has been demonstrated to serve a role in
PCOS regulation through AR (12).
In the present study, the expression levels of CTBP1-AS were
compared in the granulosa cells from 60 patients with PCOS and 60
healthy control patients. The data revealed that CTBP1-AS is
upregulated in patients with PCOS, and knockdown of CTBP1-AS could
significantly inhibit the proliferation and promote the apoptosis
of granulosa cells in vitro.
Polycomb group proteins are considered paradigmatic
epigenetic modulators that regulate cell proliferation and
differentiation, and organ development, as well as initiation and
progression of certain diseases, such as prostate cancer and
diffuse large B-cell lymphoma (DLBCL), through remodeling the
chromatin structure and subsequent transcriptional repression
(35). EZH2 and EED are the two
core catalytic subunits of PRC2 and are required for histone H3
methylation (35). In prostate
cancer, AR directly binds to the promoter and enhancer of EZH2 to
regulate its expression (36).
However, in castration-resistant prostate cancer cells, EZH2 can
directly bind to and regulate AR (37). A recent study showed that EZH2 and
EED could bind AR to regulate its expression level and the
expression of downstream target genes (35). A previous study by Takayama et
al (24) demonstrated that
CTBP1-AS could promote AR transcriptional activity in prostate
cancer. However, whether there exists interaction between EZH2, EED
and CTBP1-AS is still not clear. The present study identified an
interaction between these components in granulosa cells in patients
with PCOS, which may provide a potential target for PCOS
treatment.
Results from the present study demonstrated that the
traditional Chinese medicine, cryptotanshinone, could effectively
decrease the expression level of CTBP1-AS. CpG islands can be found
in the promoter region of CTBP1-AS, and the methylation of this
region could inhibit its transcription. Therefore, drugs that
target the methylation of CTBP1-AS CpG islands may provide a new
direction for PCOS treatment.
In conclusion, the present study results
demonstrated an upregulated expression level of CTBP1-AS in PCOS
granulosa cells and identified the interaction between CTBP1-AS and
the Polycomb group proteins, EZH2 and EED. Most importantly,
results demonstrated the efficiency of cryptotanshinone in reducing
the level of CTBP1-AS expression. However, there are limitations to
the present study. As PCOS is a complex pathological process caused
by many factors, the effects of up- or downregulation of CTBP1-AS
in a PCOS model was not comprehensively verified by in vitro
and in vivo experiments. Therefore, additional studies are
needed to further clarify the role of CTBP1-AS in PCOS.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was funded by the Natural Science Foundation of
Zhejiang Province (grant no. LY20H290007).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MW contributed to experimental data analyses and
wrote the manuscript. SZ, BW, JX, WZ and FW contributed
experimental data. XD designed the experiments and supervised the
research. MW and XD confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of The First Affiliated Hospital of Zhejiang Chinese
Medical University (Hangzhou, China; approval no. 2020-K-060-02);
all subjects provided signed consent forms prior to recruitment to
the study.
Patient consent for publication
All the patients in this project signed the consent
forms.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mortada R and Williams T: Metabolic
syndrome: Polycystic ovary syndrome. FP Essent. 435:30–42.
2015.PubMed/NCBI
|
|
2
|
Hardiman P, Pillay OS and Atiomo W:
Polycystic ovary syndrome and endometrial carcinoma. Lancet.
361:1810–1812. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Akgül S, Düzçeker Y, Kanbur N and Derman
O: Do different diagnostic criteria impact polycystic ovary
syndrome diagnosis for adolescents? J Pediatr Adolesc Gynecol.
31:258–262. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Goodarzi MO, Dumesic DA, Chazenbalk G and
Azziz R: Polycystic ovary syndrome: Etiology, pathogenesis and
diagnosis. Nat Rev Endocrinol. 7:219–231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Asunción M, Calvo RM, Millán JL, Sancho
J, Avila S and Escobar-Morreale HF: A prospective study of the
prevalence of the polycystic ovary syndrome in unselected Caucasian
women from Spain. J Clin Endocrinol Metab. 85:2434–2438. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Amiri M, Tehrani FR, Bidhendi-Yarandi R,
Behboudi-Gandevani S, Azizi F and Carmina E: Relationships between
biochemical markers of hyperandrogenism and metabolic parameters in
women with polycystic ovary syndrome: A systematic review and
meta-analysis. Horm Metab Res. 51:22–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fitzgerald S, DiVasta A and Gooding H: An
update on PCOS in adolescents. Curr Opin Pediatr. 30:459–465. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Szydlarska D, Machaj M and Jakimiuk A:
History of discovery of polycystic ovary syndrome. Adv Clin Exp
Med. 26:555–558. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rotterdam ESHRE/ASRM-Sponsored PCOS
Consensus Workshop Group, : Revised 2003 consensus on diagnostic
criteria and long-term health risks related to polycystic ovary
syndrome. Fertil Steril. 81:19–25. 2004. View Article : Google Scholar
|
|
10
|
Neven ACH, Laven J, Teede HJ and Boyle JA:
A summary on polycystic ovary syndrome: Diagnostic criteria,
prevalence, clinical manifestations, and management according to
the latest international guidelines. Semin Reprod Med. 36:5–12.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang R and Mol BWJ: The Rotterdam criteria
for polycystic ovary syndrome: Evidence-based criteria? Hum Reprod.
32:261–264. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu Z, Hao C, Song D, Zhang N, Bao H and
Qu Q: Androgen receptor coregulator CTBP1-AS is associated with
polycystic ovary syndrome in Chinese women: A preliminary study.
Reprod Sci. 22:829–837. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Patel S: Polycystic ovary syndrome (PCOS),
an inflammatory, systemic, lifestyle endocrinopathy. J Steroid
Biochem Mol Biol. 182:27–36. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sung YA, Oh JY, Chung H and Lee H:
Hyperandrogenemia is implicated in both the metabolic and
reproductive morbidities of polycystic ovary syndrome. Fertil
Steril. 101:840–845. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Norman RJ, Dewailly D, Legro RS and Hickey
TE: Polycystic ovary syndrome. Lancet. 370:685–697. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gelmann EP: Molecular biology of the
androgen receptor. J Clin Oncol. 20:3001–3015. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Meng D, Yang S, Wan X, Zhang Y, Huang W,
Zhao P, Li T, Wang L, Huang Y, Li T and Li Y: A transcriptional
target of androgen receptor, miR-421 regulates proliferation and
metabolism of prostate cancer cells. Int J Biochem Cell Biol.
73:30–40. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Takayama KI: Splicing factors have an
essential role in prostate cancer progression and androgen receptor
signaling. Biomolecules. 9:1312019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baculescu N: The role of androgen receptor
activity mediated by the CAG repeat polymorphism in the
pathogenesis of PCOS. J Med Life. 6:18–25. 2013.PubMed/NCBI
|
|
20
|
Bevan CL, Hoare S, Claessens F, Heery DM
and Parker MG: The AF1 and AF2 domains of the androgen receptor
interact with distinct regions of SRC1. Mol Cell Biol.
19:8383–8392. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu J, Qiu Y, DeMayo FJ, Tsai SY, Tsai MJ
and O'Malley BW: Partial hormone resistance in mice with disruption
of the steroid receptor coactivator-1 (SRC-1) gene. Science.
279:1922–1925. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cheng S, Brzostek S, Lee SR, Hollenberg AN
and Balk SP: Inhibition of the dihydrotestosterone-activated
androgen receptor by nuclear receptor corepressor. Mol Endocrinol.
16:1492–1501. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Berrevoets CA, Umar A, Trapman J and
Brinkmann AO: Differential modulation of androgen receptor
transcriptional activity by the nuclear receptor co-repressor
(N-CoR). Biochem J. 379:731–738. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Takayama KI, Horie-Inoue K, Katayama S,
Suzuki T, Tsutsumi S, Ikeda K, Urano T, Fujimura T, Takagi K,
Takahashi S, et al: Androgen-responsive long noncoding RNA CTBP1-AS
promotes prostate cancer. EMBO J. 32:1665–1680. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Takayama K, Tsutsumi S, Katayama S,
Okayama T, Horie-Inoue K, Ikeda K, Urano T, Kawazu C, Hasegawa A,
Ikeo K, et al: Integration of cap analysis of gene expression and
chromatin immunoprecipitation analysis on array reveals genome-wide
androgen receptor signaling in prostate cancer cells. Oncogene.
30:619–630. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Czermin B, Melfi R, McCabe D, Seitz V,
Imhof A and Pirrotta V: Drosophila enhancer of Zeste/ESC complexes
have a histone H3 methyltransferase activity that marks chromosomal
Polycomb sites. Cell. 111:185–196. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Montgomery ND, Yee D, Chen A, Kalantry S,
Chamberlain SJ, Otte AP and Magnuson T: The murine polycomb group
protein Eed is required for global histone H3 lysine-27
methylation. Curr Biol. 15:942–947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji XY, Tan B and Zhu YZ: Salvia
miltiorrhiza and ischemic diseases. Acta Pharmacol Sin.
21:1089–1094. 2000.PubMed/NCBI
|
|
30
|
Wang X, Morris-Natschke SL and Lee KH: New
developments in the chemistry and biology of the bioactive
constituents of Tanshen. Med Res Rev. 27:133–148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Havelock JC, Rainey WE and Carr BR:
Ovarian granulosa cell lines. Mol Cell Endocrinol. 228:67–78. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Morita Y and Tilly JL: Oocyte apoptosis:
Like sand through an hourglass. Dev Biol. 213:1–17. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Buccione R, Schroeder AC and Eppig JJ:
Interactions between somatic cells and germ cells throughout
mammalian oogenesis. Biol Reprod. 43:543–547. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Erickson GF, Magoffin DA, Garzo VG, Cheung
AP and Chang RJ: Granulosa cells of polycystic ovaries: Are they
normal or abnormal? Hum Reprod. 7:293–299. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jakimiuk AJ, Weitsman SR, Navab A and
Magoffin DA: Luteinizing hormone receptor, steroidogenesis acute
regulatory protein, and steroidogenic enzyme messenger ribonucleic
acids are overexpressed in thecal and granulosa cells from
polycystic ovaries. J Clin Endocrinol Metab. 86:1318–1323. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Q, Wang G, Li Q, Jiang W, Kim JS, Wang
R, Zhu S, Wang X, Yan L, Yi Y, et al: Polycomb group proteins EZH2
and EED directly regulate androgen receptor in advanced prostate
cancer. Int J Cancer. 145:415–426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu J, Yu J, Mani RS, Cao Q, Brenner CJ,
Cao X, Wang W, Wu L, Li J, Hu M, et al: An integrated network of
androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in
prostate cancer progression. Cancer Cell. 17:443–454. 2010.
View Article : Google Scholar : PubMed/NCBI
|