Infectious diseases, which can be caused by a
variety of bacteria, viruses, parasites and fungi, are one of the
leading causes of morbidity and mortality worldwide (1,2).
According to recent statistics, there are ~60 million deaths
worldwide annually, at least 25% of which are caused by infectious
diseases (3). The emergence of
infectious diseases has created great challenges to public health
and socioeconomic stability. Although the use of vaccines and
antimicrobial and antiviral drugs can help combat infections to a
certain extent, their efficacy can decrease with the emergence of
new infectious agents and unknown drug-resistant pathogens
(4,5). In addition, the development of
vaccines and medicines requires lengthy clinical trials, which does
not allow for the rapid and effective control of infectious
diseases (6). In the absence of
specific vaccines and drugs, the selection of appropriate detection
techniques for the rapid and accurate identification of pathogens
is the most effective means of dealing with infectious diseases,
which can improve the efficiency of treatment for infectious
diseases and reduce their spread, leading to a rapid response to
serious public health events (7).
Traditional diagnostic techniques include microbial
culture, hemagglutination inhibition tests and enzyme-linked
immunosorbent assays (ELISAs). Of these, the culture of pathogenic
microorganisms is the most time-consuming and their identification
is mainly based on morphological characteristics, with low
specificity and sensitivity. In addition, immunological methods
such as hemagglutination inhibition assays and ELISAs, used for the
detection of pathogen-specific antibodies or antigens, are simple
to perform; however, they have disadvantages such as high
false-positives, high cost and poor thermal stability (8,9).
With the continuous development of genetic and genomic research,
molecular diagnostic techniques focused on nucleic acid detection
have provided new methods for the diagnosis of infectious diseases,
with a short turnaround time and high sensitivity. Molecular
diagnostic techniques can not only detect multiple pathogens, but
can also analyze drug resistance genes of pathogens and pathogen
homology analysis, and have gradually become an important tool in
the early diagnosis of infectious diseases (10–12).
At present, the commonly used molecular diagnostic techniques for
infectious diseases include PCR, isothermal amplification reaction,
gene chip technology and high-throughput sequencing technology.
Since 1985, PCR has become the most widely used
nucleic acid amplification method for pathogen detection (13). With the development of PCR
technology, molecular diagnostic techniques such as quantitative
PCR (qPCR), digital PCR (dPCR) and high-resolution melting (HRM)
based on the principle of conventional PCR (cPCR) have been widely
used for the rapid and straightforward identification and drug
resistance detection of known infectious disease pathogens
(14–16). PCR performs an important role in
the early diagnosis of infectious diseases.
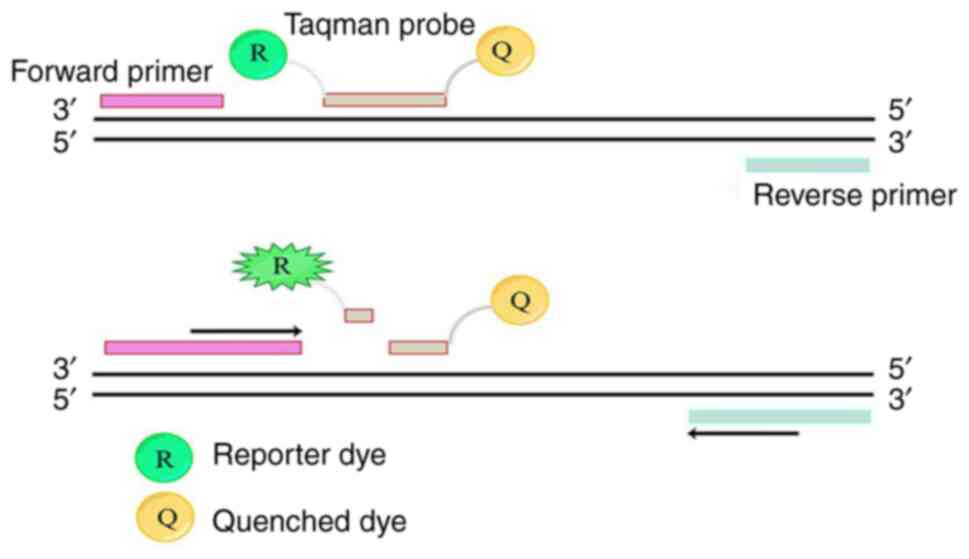
qPCR uses fluorescently labeled probes or
double-stranded DNA-specific fluorescent dye to qualitatively and
quantitatively analyze the fluorescence signal of amplification
products in real time without the need to detect PCR products
through complex electrophoresis steps (Fig. 1). This method is more automated and
has a lower risk of contamination compared with cPCR (17). qPCR has been widely used for the
early diagnosis and drug resistance detection of common clinical
pathogens (18) and has the
advantages of higher sensitivity, specificity, simplicity and
rapidity compared with traditional diagnostic methods (19). A study comparing the detection rate
of pneumococci using culture-based methods and direct detection by
qPCR showed that the detection rate of conventional culture was
41.2%, while the positive colonization rate using qPCR was 43.7%,
indicating a higher overall colonization rate of pneumococci using
qPCR methods compared with conventional culture methods (20). Ingalagi et al (21) detected 200 subgingival plaque
samples from patients with chronic periodontitis using qPCR and
cell culture simultaneously, and results showed that the positive
rate of qPCR and cell culture was 91.5 and 89.5%, respectively,
suggesting that the qPCR method had a higher detection rate and
clinical application value in the diagnosis of Porphyromonas
gingivalis compared with the traditional bacterial culture
method. Rolon Marrero et al (22) developed a qPCR assay for the
simultaneous detection of Helicobacter pylori and genotypic
markers of clarithromycin resistance directly from stool specimens
by designing primers and TaqMan probes targeting the 23S rRNA gene
of Helicobacter pylori. The assay can quickly, accurately
and non-invasively diagnose Helicobacter pylori, and provide
information on genotype susceptibility. This markedly shortens the
detection time and helps reduce the use of invasive diagnostic
processes, such as endoscopy and biopsy. Although qPCR is faster,
more sensitive and more specific than traditional diagnostic
methods, it can only detect one pathogen in a single amplification
reaction and is regarded as low-throughput; therefore, to meet the
requirements for high-throughput detection, researchers developed
multiplex qPCR (MqPCR). MqPCR can simultaneously detect multiple
pathogenic infections in a single sample using various sets of
primers and probes, which reduces detection time, labor and reagent
costs, and sample consumption. MqPCR also has sensitivity and
specificity rates comparable to those of qPCR. Bennett and Gunson
(23) developed an MqPCR that
could simultaneously detect adenovirus, astrovirus, rotavirus and
sapovirus in stool samples; this had a reduced turnaround time and
overall cost compared with qPCR. Recently, Jiang et al
(24) developed an MqPCR assay
capable of concurrently detecting nine respiratory pathogens with
no cross-reactivity and a limit of detection (LoD) of 250–500
copies/ml (1.25–2.5 copies/reaction), which is a promising
alternative for the early screening of acute respiratory tract
infections; however, qPCR is the preferred method for the
quantitative detection of common pathogens in general laboratories.
Despite the low cost and mature nature of the technology (Table I), qPCR is prone to nucleic acid
contamination, primer dimer formation, improper baseline setting
and a number of other issues which can lead to false-positives.
Furthermore, sample inhibitors, enzyme inactivation, insufficient
enzyme concentration, a low template amount and/or an inappropriate
annealing temperature may lead to false-negatives (25). In addition, qPCR is time-consuming,
requires relatively complex and expensive instruments to achieve
accurate (±0.5°C) and rapid (>10°C/s) thermal cycles, and
requires knowledgeable operators, making this technology difficult
to utilize in areas and hospitals with limited access to precision
instruments (Table II) (26–28).
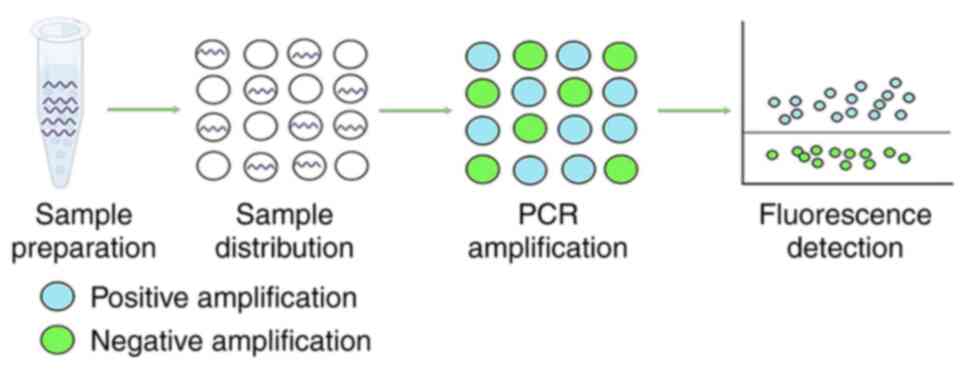
dPCR performs absolute quantification of target
genes in samples by dividing the amplification reaction into
thousands of independent sections using microplates, capillaries,
oil emulsions or microarrays, amplifying each target gene in
separate compartments, distinguishing the generated droplets as
negative or positive based on the setting of the fluorescence
threshold, and calculating the target gene content through the
ratio of negative and positive droplets (Fig. 2) (29). This partitioned amplification
reduces template competition, increases the sensitivity of the
reaction and allows dPCR to detect low levels of pathogens, minor
mutations and rare allele targets (30–33).
Therefore, it is particularly suitable for quantifying viruses with
sequence diversity and samples with low microorganism content
(34), such as BK polyomavirus,
human rhinovirus (HRV) and human immunodeficiency virus (HIV).
Studies have shown that dPCR can accurately monitor HRV serum viral
load with sequence diversity (35)
and latent HIV reservoirs (36),
and accurately quantify small amounts of human papillomavirus (HPV)
in the blood circulation (37) and
Mycobacterium tuberculosis (MTB) in whole blood samples from
infants not exhibiting early respiratory symptoms (38). In addition, dPCR can accurately
quantify minor mutations in DNA; for example, it can quantify the
frequency of drug resistance gene mutations in influenza viruses
and identify mutations in the hepatitis C virus core protein gene.
The method is able to detect <0.1% of rare variants in the
wild-type viral background, which is difficult to achieve through
the use of other molecular diagnostic techniques (39). Compared with qPCR, the main
advantage of dPCR is that it achieves absolute quantification
without relying on a standard curve. As a result, dPCR is
particularly well suited for quantitative monitoring of low
pathogen content during disease incubation or following the
administration of medication (Table
II). However, the accurate quantification of dPCR relies on the
correct threshold setting to distinguish between positive and
negative droplets, while the discrimination itself is influenced by
a number of factors, such as the quality and quantity of the
sample, the melting temperature, and primer and probe length
(40). In addition, the
instruments and reagents of dPCR technology are expensive,
resulting in high detection costs, while the exposed nature of the
droplet preparation system leads to an increased risk of
contamination and makes it easy to cause false-positives (41) (Table
I) (42–44).
Isothermal amplification is a simple nucleic acid
amplification technique performed at constant temperature, which
does not require a thermal cycler and is therefore simple in terms
of temperature control (54). The
amplified products can often be monitored by turbidity, color
change, lateral flow test strips and fluorescence curves, making it
suitable for resource-poor areas and primary healthcare units. To
date, loop-mediated isothermal amplification (LAMP), recombinase
polymerase amplification (RPA) and nucleic acid sequence-based
amplification (NASBA) have been used for rapid field detection of
infectious disease pathogens (55). However, the target product of the
isothermal amplification reaction is short and susceptible to
contamination by exogenous genetic material, which can result in
false-positives. In addition, isothermal amplification primer
design is complex, and to date, there is no dedicated primer design
software, which may limit the widespread use of isothermal
amplification for the diagnosis of clinical pathogens to some
extent (56).
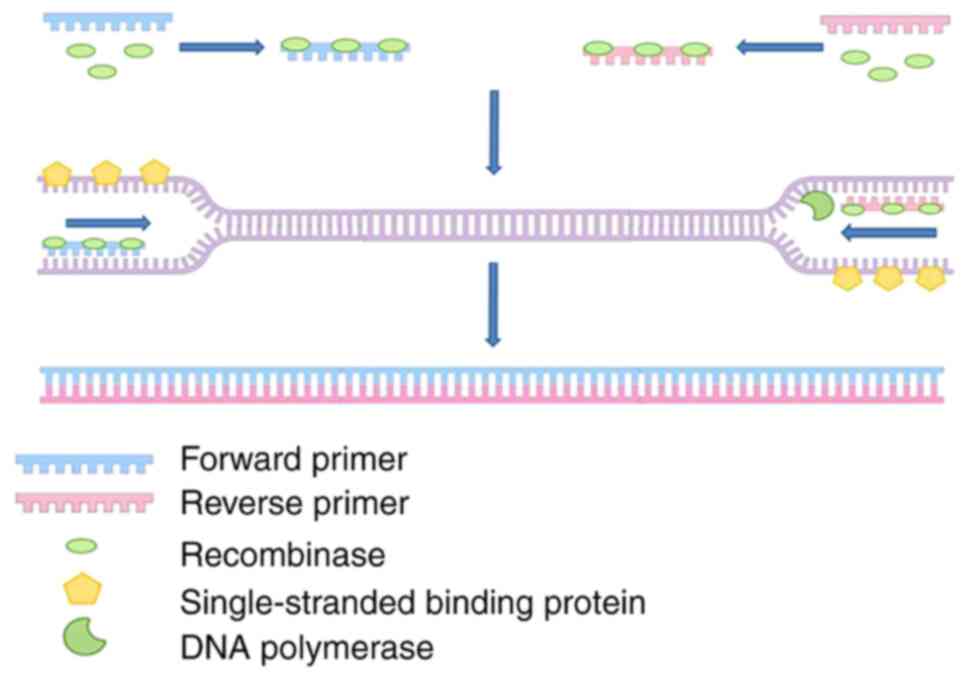
RPA is a sensitive, specific and rapid isothermal
amplification method that was first reported in 2006 (63). RPA, whose core components include
DNA polymerases and DNA-binding proteins and recombinases, expands
target DNA from as low as 1–10 copies copies of target DNA in a
single reaction to detectable levels within 30 min at 37–42°C
(Fig. 5). This method is
advantageous since it does not require amplification apparatus, has
a short detection time and high sample tolerance, and has been
successfully combined with different assays for rapid field
detection of infectious disease pathogens. The most commonly used
endpoint detection method for RPA is LFA (64–67).
Qi et al (68) successfully
established a universal typing method for the detection of human
adenovirus by combining RPA technology with lateral flow test
strips, which allows for detection within 25 min using only basic
constant temperature equipment. This method has good detection
performance and is suitable for the rapid detection of human
adenovirus in resource-limited areas. Mayran et al (69) combined rapid DNA extraction with
isothermal RPA and monitored the results by LFA, developing a new
RPA-LFA screening method to detect Hepatitis B virus (HBV) DNA in
pregnant women with hypervolemia. Achieving a sensitivity and
specificity of 98.6 and 88.2%, respectively, this new RPA-LFA
method allowed for the rapid detection of HBV. To further optimize
and improve the throughput of RPA-LFA, Li et al (70) developed a seven-fold assay using
different markers, which further reduced detection time and cost,
making it suitable for the rapid field detection of infectious
diseases (Table I) (71). To promote the application of RPA
for in-field detection of infectious diseases, it is necessary to
develop sample preparation methods and portable or fully automatic
RPA diagnostic equipment that are also suitable for field detection
in areas with poor medical infrastructure (Table II) (72).
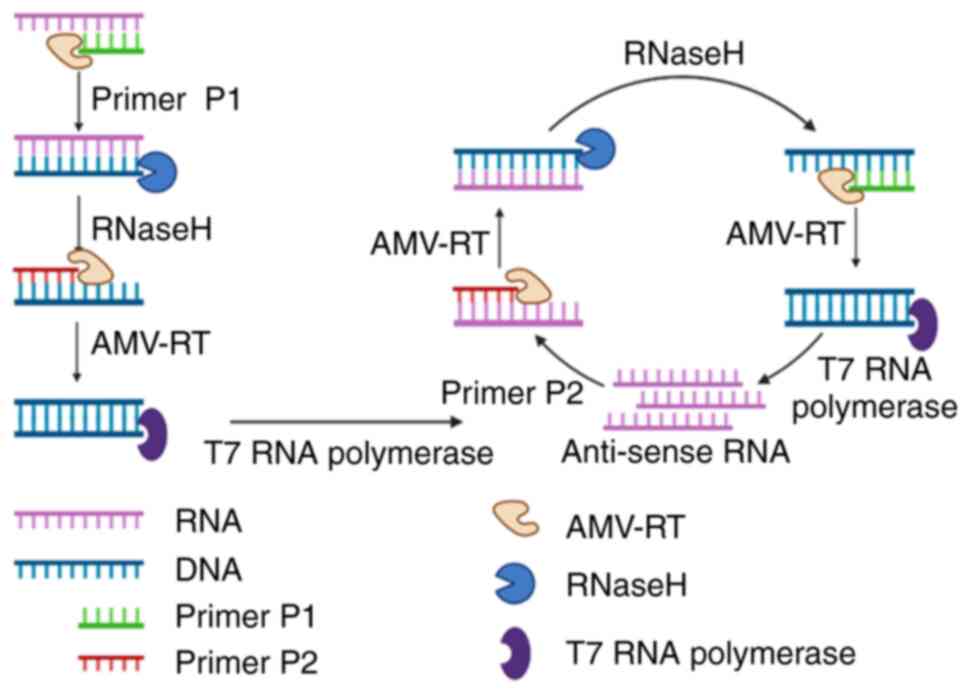
NASBA is an efficient, isothermal amplification
method developed by Compton in 1991 (73). The amplification reaction was
performed at 41°C, and the 100- to 250-bp nucleic acid target
sequence was amplified ~1012 times within 90 min
(74). The NASBA reaction mixture
involves three enzymes, T7 RNA polymerase, RNase H and avian
myeloblast virus reverse transcriptase, which selectively and
rapidly amplify RNA in the presence of background DNA, with good
sensitivity, making NASBA most suited for the analysis of RNA
samples (Fig. 6). During the
COVID-19 pandemic, Kia et al (75) developed a reverse
transcription-NASBA (RT-NASBA) assay for detecting severe acute
respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA using molecular
beacon probes based on nucleocapsid and RNA-dependent RNA
polymerase genes, with an LoD of 200 copies/ml (Table I). Compared with the Sansure
RT-qPCR US Food and Drug Administration (FDA)-approved kit (Sansure
Biotech, Inc.), its clinical sensitivity was 97.64%, which renders
it a simpler and faster detection method for SARS-CoV-2. Yrad et
al (76) developed an
NASBA-based LFA device that can detect dengue virus RNA at
concentrations of as low as 0.01 µM within 20 min, with an LoD of
1.2×104 PFU/ml in the sera of patients with a dengue
virus infection. This method has broad applications and will be
especially useful for dengue virus detection in resource-limited
areas. However, it should be noted that the NASBA amplification
efficiency is low when the nucleotide sequence is >250 or
<120 bp (Table II) (77). In addition, due to the low
temperature requirements of the NASBA reaction, it is easy for
primer dimerization and non-specific amplification to occur, which
markedly increases the false-positive rate (78).
Gene chip technology mainly includes traditional
solid- and liquid-phase chips; it is based on the principle of
nucleic acid molecular hybridization. Gene sequences in samples are
detected by hybridization between target sequences and probes fixed
on different materials. Multiple pathogens can be simultaneously
detected and identified, and clinicians can be quickly provided
with multiple pathogen information (79). However, this technology requires a
large amount of known pathogen genetic information as a basis and
can only be used for the intentional screening of known pathogen
genomes; it cannot detect novel pathogens.
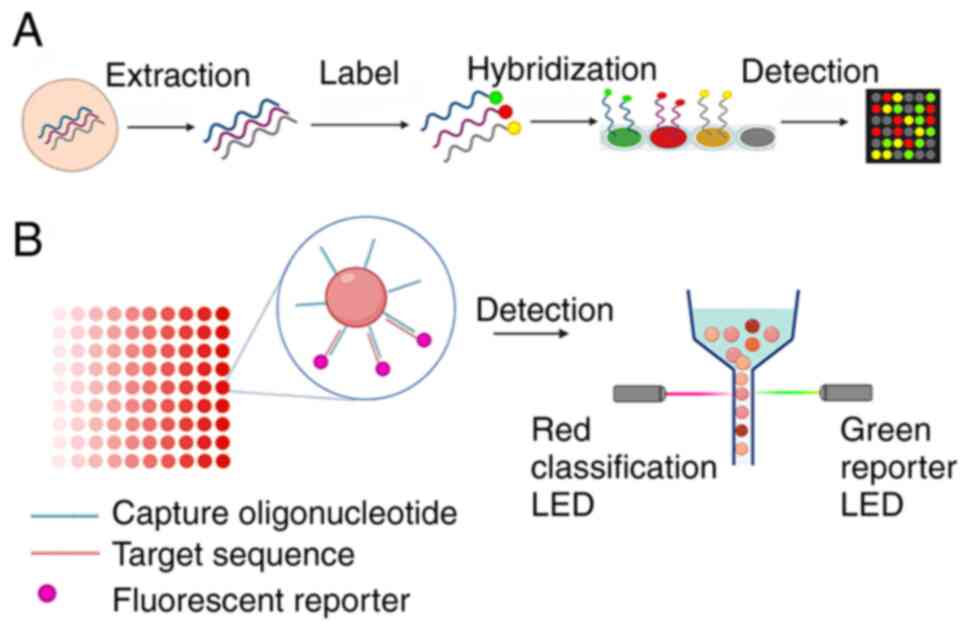
Solid-phase microarrays use specific probes attached
to solid supports to detect labeled target molecules in solution,
and can detect and analyze a large number of pathogens
simultaneously, which effectively shortens detection time (Fig. 7A). It is a high-throughput
molecular diagnostic tool suitable for the detection of multiple
infectious disease pathogens and drug resistance gene analysis
(80). Nasrabadi et al
(81) developed 16S and 23S
rDNA-based probes able to simultaneously detect and identify eight
food-borne bacterial pathogens. Ma et al (82) developed and evaluated a solid-phase
chip for the simultaneous detection of 15 types of bacteria
directly from respiratory tract specimens of patients with
pneumonia, thus reducing detection time, facilitating the early
administration of antimicrobial drugs and preventing bacterial
resistance caused by empirical antibiotic treatment. Recent studies
have shown that the CapitalBio DNA chip (CapitalBio Corporation)
can detect the resistance of MTB to rifampicin and isoniazid, and
the corresponding gene mutations within 6 h, which is quicker and
more accurate than traditional bacterial culture and drug
susceptibility tests (83–85). Currently, a number of automatic
detection platforms based on solid-phase chips are entering the
market. For example, FilmArray (BioFire Diagnostics) is an
integrated platform that combines fully automated sample
preparation, nucleic acid extraction, PCR amplification and
automatic detection, which can detect >100 different nucleic
acid targets at a time and can identify a variety of common
respiratory pathogens from a single sample within 1 h (86). This technique is often used for the
qualitative detection of common pathogens such as influenza virus,
adenovirus, Streptococcus pneumoniae and other common
respiratory tract psathogens, as well as common pathogens such as
Norovirus, Rotavirus, Salmonella, Shigella and other common
intestinal infection-causing pathogens (Table II). The technology is especially
useful in a clinical setting for determining the pathogen
composition of mixed infections and for large-scale screening of
infectious diseases (87).
However, solid-phase chip technology has certain limitations.
First, the use of fluorescent oligonucleotides represents a
significant cost (Table I). In
addition, the reference sequence information of the pathogen is
usually used to design the array, so it may lead to false-negatives
for highly variable pathogens (88).
Liquid-phase suspension chip technology couples
oligonucleotide probes to fluorescent microspheres with different
proportions of color configurations and classifies them according
to their internal colors using laser detection and flow cytometry
to achieve high-resolution automated detection (Fig. 7B) (89). Liquid-phase chip technology is
commonly used in gene expression analysis, microRNA analysis,
single nucleotide polymorphism analysis, specific sequence analysis
and microbial detection. For pathogen detection, liquid chip
technology can simultaneously identify and genotype a variety of
pathogens in a single complex sample, and the technology exhibits
high sensitivity, high throughput and high automation (90); it is suitable for large-scale
screening of infectious diseases at entry and exit ports (Table II) (91). Currently, its representative
technology, xMAP® technology (Luminex Corporation), has
been approved by the FDA for the multiplexed detection of pathogens
such as viruses, bacteria, parasites and fungi, allowing for the
rapid diagnosis of a single sample with good detection performance.
One of these systems, the xTAG® Respiratory Viral Panel
(Luminex Corporation), is a commercial kit for detecting multiple
respiratory virus nucleic acids in human nasopharyngeal samples
simultaneously. In addition to respiratory viruses,
microsphere-based multiplex nucleic acid detection has been
successfully used for the detection of various bacteria, viruses
and parasites in human fecal samples. One of these systems, the
xTAG Gastrointestinal Pathogen Panel (Luminex Corporation), is a
multiplex nucleic acid detection kit designed to rapidly detect
various bacterial, viral and parasitic nucleic acids in human fecal
samples, with an overall sensitivity and specificity of 96.3 and
99.8%, respectively (92).
Gene sequencing, including first-, second- and
third-generation sequencing (TGS), is the most accurate method of
identifying infectious pathogens; it has been successfully used for
the diagnosis of known pathogens and the identification of unknown
pathogens. First generation sequencing, represented by Sanger
sequencing, is mainly used for targeted sequencing to reveal the
sequences of several specific low-throughput sites, which is only
suitable for small-scale analysis (93). With the completion of the Human
Genome Project in the early 21st century and the rapid development
of sequencing technology, high-throughput and low-cost
next-generation sequencing (NGS) and TGS technologies have emerged
(94,95).
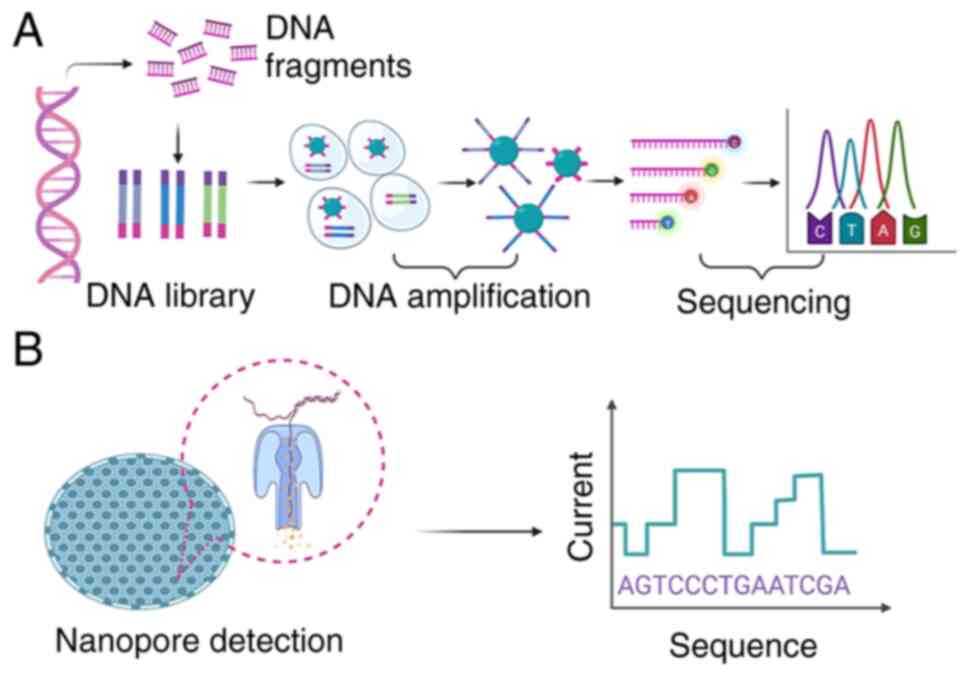
NGS can read billions of nucleotide sequences in a
single assay, it does not rely on cell culture and it retrieves all
DNA (Fig. 8A); it can also
comprehensively detect microbial species and sequences (96), which makes it suitable as a
complementary means of pathogen detection when no clear etiological
evidence is available from routine laboratory testing (Table II). For the identification and
genotyping of known pathogens, NGS is significantly more sensitive
than traditional methods. Zhang et al (97) used NGS technology to diagnose
bacterial meningitis in patients. By comparing bacterial culture
with the Alere BinaxNow® Streptococcus pneumoniae
Antigen test (Abbott Rapid Diagnostics), it was found that the
sensitivity and specificity of NGS for the diagnosis of bacterial
meningitis was 70.3 and 93.9%, respectively. The positive and
negative predictive values were 81.4 and 91.3%, respectively, which
revealed high sensitivity and specificity for the identification of
Streptococcus pneumoniae. A study found that in the
detection of pulmonary infectious disease pathogens, the positive
rates of bacterial culture and NGS (a measure of the sensitivity of
detection between culture and NGS) were 17.54 and 42.11%,
respectively. In addition, 94.49% of other pathogens associated
with human infectious diseases were detected by NGS in samples from
patients with pulmonary infection who tested negative for
traditional pathogens, suggesting that NGS could detect and
identify multiple pathogens simultaneously with higher accuracy
(98). According to NGS results,
not only pathogen identification and typing, but also drug
resistance gene detection, virulence gene detection and host immune
response analysis, can be performed (94), thus guiding clinical diagnosis,
disease treatment, and vaccine research and development. In
addition, NGS can detect pathogens and identify rare unknown
pathogens from different types of biological samples that cannot be
detected by conventional assays, such as Chlamydia psittaci
in bronchoalveolar lavage fluid (99), Naegleria flexneri and
Brucella in cerebrospinal fluid (100), and the Chikungunya and mumps
viruses in the blood (101),
which allows for the timely diagnosis of rare pathogens, and
promotes early and accurate treatment. NGS is suitable for the
rapid identification of emerging and re-emerging pathogens,
whole-genome sequencing, genomic variation and evolution, and
epidemiological investigation and tracking. NGS is a powerful tool
for tracking the source and chain of transmission of epidemics and
for monitoring the evolution of pathogens (102); it allows comprehensive access to
pathogen genome information in a short period of time, enabling
researchers and healthcare providers to rapidly respond to
infectious diseases. However, there are several challenges in the
practical application of clinical pathogen diagnosis, such as high
purity and high concentrations of nucleic acids for nucleic acid
processing in the preparation stages, the need for PCR
amplification, the inability to directly detect RNA, the short read
length and the need to use special bioinformatics tools for complex
data analysis (103). The biggest
obstacle, however, is interpreting complex sequencing data for
pathogen determination (104,105).
TGS, represented by the Oxford Nanopore Technologies
(ONT) MinION (ONT), was launched in 2015 (106); it combines genetic engineering
and computer-aided technology to determine the base sequence by
detecting the changes in electrical current as DNA or RNA passes
through the nanopore (Fig. 8B).
The high-throughput, rapid DNA sequencing technology produces
ultralong reads without the need for labeling, which simplifies the
detection process and reduces costs (107). TGS has a number of advantages
over NGS. NGS cannot directly detect RNA and has a short read
length in the field of pathogen diagnosis. Furthermore, TGS can
directly carry out RNA sequencing without a reverse transcription
process, which shortens detection time. Keller et al
(108) described direct RNA
sequencing of five influenza A virus genomes, with 100% nucleotide
coverage and 99% agreement between Nanopore and Illumina Inc.-based
sequencing results by modifying the RNA sequencing method published
by Oxford Nanopore Technologies, Ltd. Recently, TGS has been used
for the RNA sequencing of SARS-CoV-2, where it can detect
SARS-CoV-2 and other respiratory viruses simultaneously within 6–10
h (109), markedly reducing
detection time compared with NGS. In addition, TGS sequencing has
significantly increased read length capabilities, enabling complete
gene sequencing and identifying novel full-length transcript
variants and gene fusions that cannot be detected by NGS (110), thus improving the likelihood of
detecting and identifying pathogenic species. At present, portable
sequencing devices based on the Nanopore system have been used for
real-time on-site analysis and out-of-hospital bedside detection of
novel coronavirus, Ebola virus, adenovirus and a number of other
viruses (111–113). They are suitable for epidemic
surveillance and virus mutation monitoring in resource-limited
situations and also for the identification of rare and
difficult-to-culture bacteria (Table
II) (105). However, the high
error rate of TGS is the biggest obstacle to its application in the
field of microbial detection. Nonetheless, with continued
optimization of nanopore structures and base-calling algorithms,
and with improvements in sequence acquisition speed and base
recognition accuracy, TGS is expected to become an ideal tool for
the detection of known pathogens and the discovery of rare and
unknown pathogens (114).
Biosensing technology uses a combination of target
biomarkers and ionic conductive materials to generate signals,
which are detected and analyzed by sensors (optical,
electrochemical or piezoelectric) and reading devices (115). Currently, the use of
photoelectric biosensing of nucleic acids is increasing in
popularity due to its sensitivity and speed for the early diagnosis
and quantitative analysis of infectious diseases. Sheng et
al (116) developed a
label-free biosensor with an RNA aptamer for the sensitive, rapid
quantitative detection of food pathogens without the isolation,
purification and enrichment processes. A further study found that
optical label-free biosensors can detect and quantify MTB,
mycobacterial proteins and interferons quickly and efficiently,
making them beneficial for the early detection of tuberculosis
(117). Fluorescence in
situ hybridization (FISH) is a molecular diagnostic technique
used to detect and localize specific nucleic acid sequences in
cells. To improve throughput, FISH can be used in combination with
flow cytometry to detect target nucleic acid sequences in thousands
of individual cells. Flow cytometry-based FISH (flow-FISH) uses
fluorescent probes that target DNA or RNA to detect specific genes
or pathogens; it can also be multiplexed so that multiple gene
targets or pathogens can be measured simultaneously (118). Flow-FISH has been used for
bacterial identification and detecting gene expression, for
monitoring viral multiplication in infected cells, and for colony
analysis and counting. Recently, the use of in vivo
bacterial sorting technology assisted by flow-FISH has made it
easier to isolate, classify and purify live bacteria based on
target genes, and to study the role of target genes in the growth,
substance metabolism, bacterial virulence and antibiotic resistance
of bacteria (119). Mass
spectrometry analysis of the molecular mass and charge of
biomarkers will improve the quality of the model compared with the
reference spectra and can be used for the identification of
pathogenic microorganisms at the species and genus levels; with its
high accuracy and signal acquisition speed, it is expected to
become a routine tool for rapid clinical analysis of multiple
pathogenic microorganisms in a single sample (120,121).
In conclusion, molecular diagnostic technology
provides an improved choice for the diagnosis of infectious
diseases compared to traditional diagnostic techniques, such as
microbial culture, hemagglutination inhibition tests, and ELISA.
The careful selection and combination of different molecular
diagnostic technologies according to a user's needs can provide a
timely and accurate diagnosis of infectious disease pathogens and
facilitate precision treatment, to effectively control diseases.
qPCR technology is mature, low-cost and suitable for the
qualitative and quantitative analysis of common pathogens in
standard laboratories. dPCR can be used for the absolute
quantification of target genes in samples, and is particularly
suitable for the analysis of samples containing low pathogen
levels, and the detection of small mutations and rare allele
targets. As a fast, high-throughput and cost-effective technique,
HRM is often used for mutation detection and large-scale analysis
of single nucleotide polymorphisms. Isothermal PCR can be used for
nucleic acid amplification at a constant temperature, which does
not require a thermocycler, and is more suitable for the rapid
detection of pathogens in resource-limited areas and primary
medical units. Gene chip technology has the ability to detect and
identify multiple pathogens simultaneously, which is particularly
useful in clinical settings for the pathogenic composition
determination of mixed infections. However, this technology can
only screen for the genomes of known pathogens and cannot detect
new, unknown pathogens, unlike gene sequencing technology, which
can comprehensively detect the types and sequences of pathogens.
The molecular diagnostic techniques outlined in the present review
need further improvement. First, nucleic acid extraction and
purification steps in molecular diagnostic techniques are
cumbersome. Therefore, it is essential to streamline the existing
nucleic acid extraction procedures or develop molecular techniques
to avoid nucleic acid extraction. Second, most molecular diagnostic
reagents require low-temperature transport and storage, which
increases the cost of molecular diagnostics and hinders their
application in remote or resource-limited areas, so ready-to-use,
room temperature-stable reaction mixtures need to be studied to
reduce costs and increase their applicability in these areas.
Finally, molecular diagnostic techniques such as qPCR, dPCR and
sequencing are instrument-dependent, meaning that rapid on-site
detection of pathogens in resource-limited conditions can prove
difficult. Continuous improvement of molecular diagnostic
technology will help to create more high-throughput, automated and
portable instruments with high sensitivity and specificity to aid
in the rapid diagnosis and treatment of infectious diseases
worldwide.
Not applicable.
This study was supported by the Natural Science Foundation of
Zhejiang Province (grant. no. LGF20H200009), the Hangzhou Medical
and Health Technology Planning Project (grant. no. OO20190415) and
the Zhejiang Medical and Health Technology Planning Project (grant.
no. 2019331539).
Not applicable.
YZD, YJZ, QQL and XJJ developed the manuscript
concept and produced the initial draft. JC and HJZ provided
valuable comments on this first draft. YZD, YJZ, QQL, XJJ, JC and
HJZ critically revised the manuscript for intellectual content. All
authors read and approved the final version of the manuscript. Data
authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Zhu L, Ling J, Zhu Z, Tian T, Song Y and
Yang C: Selection and applications of functional nucleic acids for
infectious disease detection and prevention. Anal Bioanal Chem.
413:4563–4579. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ling Z, Xiao H and Chen W: Gut microbiome:
The cornerstone of life and health. Adv Gut Microbiome Res.
2022:1–3. 2022. View Article : Google Scholar
|
|
3
|
Vengesai A, Kasambala M, Mutandadzi H,
Mduluza-Jokonya TL, Mduluza T and Naicker T: Scoping review of the
applications of peptide microarrays on the fight against human
infections. PLoS One. 17:e02486662022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Casanova JL and Abel L: Lethal Infectious
diseases as inborn errors of immunity: Toward a synthesis of the
germ and genetic theories. Annu Rev Pathol. 16:23–50. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang L, Jianying L and Pei-Yong S:
SARS-CoV-2 variants and vaccination. Zoonoses (Burlingt).
2:62022.
|
|
6
|
Micoli F, Bagnoli F, Rappuoli R and
Serruto D: The role of vaccines in combatting antimicrobial
resistance. Nat Rev Microbiol. 19:287–302. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mercer A: Protection against severe
infectious disease in the past. Pathog Glob Health. 115:151–167.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L and Moore MD: A survey of analytical
techniques for noroviruses. Foods. 9:3182020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiang Z, Jiang B, Li W, Zhai G, Zhou H,
Wang Y and Wu J: The diagnostic and prognostic value of serum
exosome-derived carbamoyl phosphate synthase 1 in HEV-related acute
liver failure patients. J Med Virol. 94:5015–5025. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang HS, Tsai CL, Chang J, Hsu TC, Lin S
and Lee CC: Multiplex PCR system for the rapid diagnosis of
respiratory virus infection: Systematic review and meta-analysis.
Clin Microbiol Infect. 24:1055–1063. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Bortolanza M, Zhai G, Shang A, Ling
Z, Jiang B, Shen X, Yao Y, Yu J, Li L and Cao H: Gut microbiota
dysbiosis associated with plasma levels of Interferon-γ and viral
load in patients with acute hepatitis E infection. J Med Virol.
94:692–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu J, Xu Y, Cui Y, Bortolanza M, Wang M,
Jiang B, Yan M, Liang W, Yao Y, Pan Q, et al: Dynamic changes of
serum metabolites associated with infection and severity of
patients with acute hepatitis E infection. J Med Virol.
94:2714–2726. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang B, Zhou J, Li M, Wei Y, Wang J, Wang
Y, Shi P, Li X, Huang Z, Tang H and Song Z: Evaluation of
CRISPR/Cas9 site-specific function and validation of sgRNA sequence
by a Cas9/sgRNA-assisted reverse PCR technique. Anal Bioanal Chem.
413:2447–2456. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sidstedt M, Rådström P and Hedman J: PCR
inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal
Bioanal Chem. 412:2009–2023. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
García-Bernalt Diego J, Fernández-Soto P,
Crego-Vicente B, Alonso-Castrillejo S, Febrer-Sendra B,
Gómez-Sánchez A, Vicente B, López-Abán J and Muro A: Progress in
loop-mediated isothermal amplification assay for detection of
Schistosoma mansoni DNA: Towards a ready-to-use test. Sci Rep.
9:147442019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang W, Ji W, Zhang Y, Xie Y, Chen S, Jin
Y and Duan G: An update on detection technologies for SARS-CoV-2
variants of concern. Viruses. 14:23242022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv C, Deng W, Wang L, Qin Z, Zhou X and Xu
J: Molecular techniques as alternatives of diagnostic tools in
china as schistosomiasis moving towards elimination. Pathogens.
11:2872022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mackay IM, Arden KE and Nitsche A:
Real-time PCR in virology. Nucleic Acids Re. 30:1292–1305. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castelli G, Bruno F, Reale S, Catanzaro S,
Valenza V and Vitale F: Molecular diagnosis of leishmaniasis:
Quantification of parasite load by a Real-Time PCR assay with high
sensitivity. Pathogens. 10:8652021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vidanapathirana G, Angulmaduwa ALSK,
Munasinghe TS, Ekanayake EWMA, Harasgama P, Kudagammana ST,
Dissanayake BN and Liyanapathirana LVC: Comparison of pneumococcal
colonization density among healthy children and children with
respiratory symptoms using real time PCR (RT-PCR). BMC Microbiol.
22:312022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ingalagi P, Bhat KG, Kulkarni RD,
Kotrashetti VS, Kumbar V and Kugaji M: Detection and comparison of
prevalence of Porphyromonas gingivalis through culture and
Real Time-polymerase chain reaction in subgingival plaque samples
of chronic periodontitis and healthy individuals. J Oral Maxillofac
Pathol. 26:2882022.PubMed/NCBI
|
|
22
|
Marrero Rolon R, Cunningham SA, Mandrekar
JN, Polo ET and Patel R: Erratum for Marrero Rolon et al.,
‘Clinical evaluation of a real-time pCR assay for simultaneous
detection of helicobacter pylori and genotypic markers of
clarithromycin resistance directly from stool’. J Clin Microbiol.
60:e02452212022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bennett S and Gunson RN: The development
of a multiplex real-time RT-PCR for the detection of adenovirus,
astrovirus, rotavirus and sapovirus from stool samples. J Virol
Methods. 242:30–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang XW, Huang TS, Xie L, Chen SZ, Wang
SD, Huang ZW, Li XY and Ling WP: Development of a diagnostic assay
by three-tube multiplex real-time PCR for simultaneous detection of
nine microorganisms causing acute respiratory infections. Sci Rep.
12:133062022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu L, Zhang Y, Cui P, Wang C, Zeng X,
Deng G and Wang X: Development of a duplex TaqMan real-time RT-PCR
assay for simultaneous detection of newly emerged H5N6 influenza
viruses. Virol J. 16:1192019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Das Mukhopadhyay C, Sharma P, Sinha K and
Rajarshi K: Recent trends in analytical and digital techniques for
the detection of the SARS-Cov-2. Biophys Chem. 270:1065382021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu CY, Chan KG, Yean CY and Ang GY:
Nucleic acid-based diagnostic tests for the detection SARS-CoV-2:
An Update. Diagnostics (Basel). 11:532021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z,
Jian M, Ding Z, Dai X, Bao F and Liu A: Application of droplet
digital PCR to detect the pathogens of infectious diseases. Biosci
Rep. 38:BSR201811702018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lei S, Chen S and Zhong Q: Digital PCR for
accurate quantification of pathogens: Principles, applications,
challenges and future prospects. Int J Biol Macromol. 184:750–759.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Das S, Hammond-McKibben D, Guralski D,
Lobo S and Fiedler PN: Development of a sensitive molecular
diagnostic assay for detecting Borrelia burgdorferi DNA from the
blood of Lyme disease patients by digital PCR. PLoS One.
15:e02353722020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao Y, Yu M, Dong G, Chen B and Zhang B:
Digital PCR as an emerging tool for monitoring of microbial
biodegradation. Molecules. 25:7062020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Parvin R, Fan Q and Ye F:
Emerging digital PCR technology in precision medicine. Biosens
Bioelectron. 211:1143442020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Košir AB, Spilsberg B, Holst-Jensen A, Žel
J and Dobnik D: Development and inter-laboratory assessment of
droplet digital PCR assays for multiplex quantification of 15
genetically modified soybean lines. Sci Rep. 9:37352019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu L, Qu H, Alonso DG, Yu Z, Yu Y, Shi Y,
Hu C, Zhu T, Wu N and Shen F: Portable integrated digital PCR
system for the point-of-care quantification of BK virus from urine
samples. Biosens Bioelectron. 175:1129082021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sedlak RH, Nguyen T, Palileo I, Jerome KR
and Kuypers J: Superiority of Digital Reverse Transcription-PCR
(RT-PCR) over Real-Time RT-PCR for Quantitation of Highly Divergent
Human Rhinoviruses. J Clin Microbiol. 55:442–449. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Snippenberg W, Gleerup D, Rutsaert S,
Vandekerckhove L, De Spiegelaere W and Trypsteen W: Triplex digital
PCR assays for the quantification of intact proviral HIV-1 DNA.
Methods. 201:41–48. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bønløkke S, Stougaard M, Sorensen BS,
Booth BB, Høgdall E, Nyvang GB, Lindegaard JC, Blaakær J, Bertelsen
J, Fuglsang K, et al: The diagnostic value of circulating Cell-Free
HPV DNA in plasma from cervical cancer patients. Cells.
11:21702022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lyu L, Li Z, Pan L, Jia H, Sun Q, Liu Q
and Zhang Z: Evaluation of digital PCR assay in detection of M.
tuberculosis IS6110 and IS1081 in tuberculosis patients plasma.
BMC Infect Dis. 20:6572020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Salipante SJ and Jerome KR: Digital PCR-An
emerging technology with broad applications in microbiology. Clin
Chem. 66:117–123. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rutsaert S, Bosman K, Trypsteen W, Nijhuis
M and Vandekerckhove L: Digital PCR as a tool to measure HIV
persistence. Retrovirology. 15:162018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kojabad AA, Farzanehpour M, Galeh HEG,
Dorostkar R, Jafarpour A, Bolandian M and Nodooshan MM: Droplet
digital PCR of viral DNA/RNA, current progress, challenges, and
future perspectives. J Med Virol. 93:4182–4197. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dingle TC, Sedlak RH, Cook L and Jerome
KR: Tolerance of droplet-digital PCR vs real-time quantitative PCR
to inhibitory substances. Clin Chem. 59:1670–1672. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pan SW, Su WJ, Chan YJ, Chuang FY, Feng JY
and Chen YM: Mycobacterium tuberculosis-derived circulating
cell-free DNA in patients with pulmonary tuberculosis and persons
with latent tuberculosis infection. PLoS One. 16:e02538792021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang D, Liu E, Liu H, Jin X, Niu C, Gao Y
and Su X: A droplet digital PCR assay for detection and
quantification of Verticillium nonalfalfae and V.
albo-atrum. Front Cell Infect Microbiol. 12:11106842023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gundry CN, Vandersteen JG, Reed GH, Pryor
RJ, Chen J and Wittwer CT: Amplicon melting analysis with labeled
primers: A closed-tube method for differentiating homozygotes and
heterozygotes. Clin Chem. 49:396–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tamburro M and Ripabelli G: High
Resolution Melting as a rapid, reliable, accurate and
cost-effective emerging tool for genotyping pathogenic bacteria and
enhancing molecular epidemiological surveillance: A comprehensive
review of the literature. Ann Ig. 29:293–316. 2017.PubMed/NCBI
|
|
47
|
Hu M, Yang D, Wu X, Luo M and Xu F: A
novel high-resolution melting analysis-based method for Salmonella
genotyping. J Microbiol Methods. 172:1058062020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wen X, Chen Q, Yin H, Wu S and Wang X:
Rapid identification of clinical common invasive fungi via a
multi-channel real-time fluorescent polymerase chain reaction
melting curve analysis. Medicine (Baltimore). 99:e191942020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Banowary B, Dang VT, Sarker S, Connolly
JH, Chenu J, Groves P, Ayton M, Raidal S, Devi A, Vanniasinkam T
and Ghorashi SA: Differentiation of Campylobacter jejuni and
campylobacter coli using Multiplex-PCR and high resolution melt
curve analysis. PLoS One. 10:e01388082015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tong SY, Dakh F, Hurt AC, Deng YM, Freeman
K, Fagan PK, Barr IG and Giffard PM: Rapid detection of the H275Y
oseltamivir resistance mutation in influenza A/H1N1 2009 by single
base pair RT-PCR and high-resolution melting. PLoS One.
6:e214462020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kafi H, Emaneini M, Halimi S, Rahdar HA,
Jabalameli F and Beigverdi R: Multiplex high-resolution melting
assay for simultaneous detection of five key bacterial pathogens in
urinary tract infections: A pilot study. Front Microbiol.
13:10491782022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tong SY and Giffard PM: Microbiological
applications of high-resolution melting analysis. J Clin Microbiol.
50:3418–3421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ghorbani J, Hashemi FB, Jabalameli F,
Emaneini M and Beigverdi R: Multiplex detection of five common
respiratory pathogens from bronchoalveolar lavages using high
resolution melting curve analysis. BMC Microbiol. 22:1412020.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zamani M, Furst AL and Klapperich CM:
Strategies for engineering affordable technologies for
point-of-Care diagnostics of infectious diseases. Acc Chem Res.
54:3772–3779. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Du J, Ma B, Li J, Wang Y, Dou T, Xu S and
Zhang M: Rapid detection and differentiation of legionella
pneumophila and Non-legionella pneumophila Species by using
recombinase polymerase amplification combined with EuNPs-based
lateral flow immunochromatography. Front Chem. 9:8151892022.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Soroka M, Wasowicz B and Rymaszewska A:
Loop-Mediated isothermal amplification (LAMP): The better sibling
of PCR? Cells. 10:19312021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Notomi T, Okayama H, Masubuchi H, Yonekawa
T, Watanabe K, Amino N and Hase T: Loop-mediated isothermal
amplification of DNA. Nucleic Acids Res. 28:E632000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Parija SC and Poddar A: Molecular
diagnosis of infectious parasites in the post-COVID-19 era. Trop
Parasitol. 11:3–10. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vo DT and Story MD: Facile and direct
detection of human papillomavirus (HPV) DNA in cells using
loop-mediated isothermal amplification (LAMP). Mol Cell Probes.
59:1017602021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen N, Si Y, Li G, Zong M, Zhang W, Ye Y
and Fan L: Development of a loop-mediated isothermal amplification
assay for the rapid detection of six common respiratory viruses.
Eur J Clin Microbiol Infect Dis. 40:2525–2532. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim J, Park BG, Lim DH, Jang WS, Nam J,
Mihn DC and Lim CS: Development and evaluation of a multiplex
loop-mediated isothermal amplification (LAMP) assay for
differentiation of Mycobacterium tuberculosis and non-tuberculosis
mycobacterium in clinical samples. PLoS One. 16:e02447532021.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Phillips EA, Moehling TJ, Ejendal KFK,
Hoilett OS, Byers KM, Basing LA, Jankowski LA, Bennett JB, Lin LK,
Stanciu LA and Linnes JC: Microfluidic rapid and autonomous
analytical device (microRAAD) to detect HIV from whole blood
samples. Lab Chip. 19:3375–3386. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen X, Zhang J, Pan M, Qin Y, Zhao H, Qin
P, Yang Q, Li X, Zeng W, Xiang Z, et al: Loop-mediated isothermal
amplification (LAMP) assays targeting 18S ribosomal RNA genes for
identifying P. vivax and P. ovale species and
mitochondrial DNA for detecting the genus Plasmodium. Parasit
Vectors. 14:2782021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Trinh KTL and Lee NY: Fabrication of
wearable PDMS device for rapid detection of nucleic acids via
recombinase polymerase amplification operated by human body heat.
Biosensors (Basel). 12:722022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Islam MN, Moriam S, Umer M, Phan HP,
Salomon C, Kline R, Nguyen NT and Shiddiky MJA: Naked-eye and
electrochemical detection of isothermally amplified HOTAIR long
non-coding RNA. Analyst. 143:3021–3028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mota DS, Guimarães JM, Gandarilla AMD,
Filho JCBS, Brito WR and Mariúba LAM: Recombinase polymerase
amplification in the molecular diagnosis of microbiological targets
and its applications. Can J Microbiol. 68:383–402. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li J, Macdonald J and von Stetten F:
Review: A comprehensive summary of a decade development of the
recombinase polymerase amplification. Analyst. 145:1950–1960. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qi Y, Li W, Li X, Shen W, Zhang J, Li J,
Lv R, Lu N, Zong L, Zhuang S, et al: Development of rapid and
visual nucleic acid detection methods towards four serotypes of
human adenovirus species B based on RPA-LF test. Biomed Res Int.
2021:99577472021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mayran C, Foulongne V, Van de Perre P,
Fournier-Wirth C, Molès JP and Cantaloube JF: Rapid diagnostic test
for hepatitis B virus viral load based on recombinase polymerase
amplification combined with a lateral flow read-out. Diagnostics
(Basel). 12:6212022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li J, Pollak NM and Macdonald J: Multiplex
detection of nucleic acids using recombinase polymerase
amplification and a molecular colorimetric 7-Segment display. ACS
Omega. 4:11388–11396. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Munawar MA: Critical insight into
recombinase polymerase amplification technology. Expert Rev Mol
Diagn. 22:725–737. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xu L, Duan J, Chen J, Ding S and Cheng W:
Recent advances in rolling circle amplification-based biosensing
strategies-A review. Anal Chim Acta. 1148:2381872021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Compton J: Nucleic acid sequence-based
amplification. Nature. 350:91–92. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Glökler J, Lim TS, Ida J and Frohme M:
Isothermal amplifications-a comprehensive review on current
methods. Crit Rev Biochem Mol Biol. 56:543–586. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kia V, Tafti A, Paryan M and
Mohammadi-Yeganeh S: Evaluation of real-time NASBA assay for the
detection of SARS-CoV-2 compared with real-time PCR. Ir J Med Sci.
6:1–7. 2022.
|
|
76
|
Yrad FM, Castañares JM and Alocilja EC:
Visual detection of Dengue-1 RNA using gold nanoparticle-based
lateral flow biosensor. Diagnostics (Basel). 9:742019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mohammadi-Yeganeh S, Paryan M, Mirab
Samiee S, Kia V and Rezvan H: Molecular beacon probes-base
multiplex NASBA Real-time for detection of HIV-1 and HCV. Iran J
Microbiol. 4:47–54. 2012.PubMed/NCBI
|
|
78
|
Gao YP, Huang KJ, Wang FT, Hou YY, Xu J
and Li G: Recent advances in biological detection with rolling
circle amplification: Design strategy, biosensing mechanism, and
practical applications. Analyst. 147:3396–3414. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wöhrle J, Krämer SD, Meyer PA, Rath C,
Hügle M, Urban GA and Roth G: Digital DNA microarray generation on
glass substrates. Sci Rep. 10:57702020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xie C, Hu X, Liu Y and Shu C: Performance
comparison of GeneXpert MTB/RIF, gene chip technology, and modified
roche culture method in detecting mycobacterium tuberculosis and
drug susceptibility in sputum. Contrast Media Mol Imaging.
2022:29954642022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nasrabadi Z, Ranjbar R, Poorali F and
Sarshar M: Detection of eight foodborne bacterial pathogens by
oligonucleotide array hybridization. Electron Physician.
9:4405–4411. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ma X, Li Y, Liang Y, Liu Y, Yu L, Li C,
Liu Q and Chen L: Development of a DNA microarray assay for rapid
detection of fifteen bacterial pathogens in pneumonia. BMC
Microbiol. 20:1772020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Feng G, Han W, Shi J, Xia R and Xu J:
Analysis of the application of a gene chip method for detecting
Mycobacterium tuberculosis drug resistance in clinical specimens: A
retrospective study. Sci Rep. 11:179512021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhu L, Liu Q, Martinez L, Shi J, Chen C,
Shao Y, Zhong C, Song H, Li G, Ding X, et al: Diagnostic value of
GeneChip for detection of resistant Mycobacterium tuberculosis in
patients with differing treatment histories. J Clin Microbiol.
53:131–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun B and Sun Y: Diagnostic performance of
DNA microarray for detecting rifampicin and isoniazid resistance in
Mycobacterium tuberculosis. J Thorac Dis. 13:4448–4454. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chandran S, Arjun R, Sasidharan A, Niyas
VK and Chandran S: Clinical performance of FilmArray
Meningitis/Encephalitis multiplex polymerase chain reaction panel
in central nervous system infections. Indian J Crit Care Med.
26:67–70. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Senescau A, Kempowsky T, Bernard E,
Messier S, Besse P, Fabre R and François JM: Innovative
DendrisChips® Technology for a syndromic approach of in
vitro diagnosis: Application to the respiratory infectious
diseases. Diagnostics (Basel). 8:772018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dien Bard J and McElvania E: Panels and
syndromic testing in clinical microbiology. Clin Lab Med.
40:393–420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gonsalves S, Mahony J, Rao A, Dunbar S and
Juretschko S: Multiplexed detection and identification of
respiratory pathogens using the NxTAG® respiratory
pathogen panel. Methods. 158:61–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ma ZY, Deng H, Hua LD, Lei W, Zhang CB,
Dai QQ, Tao WJ and Zhang L: Suspension microarray-based comparison
of oropharyngeal swab and bronchoalveolar lavage fluid for pathogen
identification in young children hospitalized with respiratory
tract infection. BMC Infect Dis. 20:1682020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Dunbar SA: Applications of Luminex xMAP
technology for rapid, high-throughput multiplexed nucleic acid
detection. Clin Chim Acta. 363:71–82. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Reslova N, Michna V, Kasny M, Mikel P and
Kralik P: xMAP technology: Applications in detection of pathogens.
Front Microbiol. 8:552017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dai Z, Li T, Li J, Han Z, Pan Y, Tang S,
Diao X and Luo M: High-throughput long paired-end sequencing of a
Fosmid library by PacBio. Plant Methods. 15:1422019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Duan H, Li X, Mei A, Li P, Liu Y, Li X, Li
W, Wang C and Xie S: The diagnostic value of metagenomic
next-generation sequencing in infectious diseases. BMC Infect Dis.
21:622021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Grumaz S, Stevens P, Grumaz C, Decker SO,
Weigand MA, Hofer S, Brenner T, von Haeseler A and Sohn K:
Next-generation sequencing diagnostics of bacteremia in septic
patients. Genome Med. 8:732016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lyimo BM, Popkin-Hall ZR, Giesbrecht DJ,
Mandara CI, Madebe RA, Bakari C, Pereus D, Seth MD, Ngamba RM,
Mbwambo RB, et al: Potential opportunities and challenges of
deploying next generation sequencing and CRISPR-Cas systems to
support diagnostics and surveillance towards malaria control and
elimination in africa. Front Cell Infect Microbiol. 12:7578442022.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang XX, Guo LY, Liu LL, Shen A, Feng WY,
Huang WH, Hu HL, Hu B, Guo X, Chen TM, et al: The diagnostic value
of metagenomic next-generation sequencing for identifying
Streptococcus pneumoniae in paediatric bacterial meningitis.
BMC Infect Dis. 19:4952019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Huang J, Jiang E, Yang D, Wei J, Zhao M,
Feng J and Cao J: Metagenomic Next-generation sequencing versus
traditional pathogen detection in the diagnosis of peripheral
pulmonary infectious lesions. Infect Drug Resist. 13:567–576. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Dong Y, Gao Y, Chai Y and Shou S: Use of
quantitative metagenomics next-generation sequencing to confirm
fever of unknown origin and infectious disease. Front Microbio.
13:9310582022. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Gu L, Liu W, Ru M, Lin J, Yu G, Ye J, Zhu
ZA, Liu Y, Chen J, Lai G and Wen W: The application of metagenomic
next-generation sequencing in diagnosing Chlamydia psittaci
pneumonia: A report of five cases. BMC Pulm Med. 20:652020.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Jerome H, Taylor C, Sreenu VB, Klymenko T,
Filipe ADS, Jackson C, Davis C, Ashraf S, Wilson-Davies E,
Jesudason N, et al: Metagenomic next-generation sequencing aids the
diagnosis of viral infections in febrile returning travellers. J
Infect. 79:383–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Simner PJ, Miller S and Carroll KC:
Understanding the promises and hurdles of metagenomic
next-generation sequencing as a diagnostic tool for infectious
diseases. Clin Infect Dis. 66:778–788. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Yu X, Jiang W, Shi Y, Ye H and Lin J:
Applications of sequencing technology in clinical microbial
infection. J Cell Mol Med. 23:7143–7150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gu W, Miller S and Chiu CY: Clinical
metagenomic next-generation sequencing for pathogen detection. Annu
Rev Patho. 14:319–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang L, Chen F, Zeng Z, Xu M, Sun F, Yang
L, Bi X, Lin Y, Gao Y, Hao H, et al: Advances in metagenomics and
its application in environmental microorganisms. Front Microbiol.
12:7663642021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang X, Liu Y, Liu H, Pan W, Ren J, Zheng
X, Tan Y, Chen Z, Deng Y, He N, et al: Recent advances and
application of whole genome amplification in molecular diagnosis
and medicine. Med Comm. 3:e1162022.
|
|
107
|
Athanasopoulou K, Boti MA, Adamopoulos PG,
Skourou PC and Scorilas A: Third-Generation sequencing: The
spearhead towards the radical transformation of modern genomics.
Life (Basel). 12:302021.PubMed/NCBI
|
|
108
|
Keller MW, Rambo-Martin BL, Wilson MM,
Ridenour CA, Shepard SS, Stark TJ, Neuhaus EB, Dugan VG, Wentworth
DE and Barnes JR: Author Correction: Direct RNA Sequencing of the
coding complete influenza A virus genome. Sci Rep. 8:157462018.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z,
Gu J, Xiang B, Liu J, Jiang W, et al: Nanopore targeted sequencing
for the accurate and comprehensive detection of SARS-CoV-2 and
other respiratory viruses. Small. 16:e20021692020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mongan AE, Tuda JSB and Runtuwene LR:
Portable sequencer in the fight against infectious disease. J Hum
Genet. 65:35–40. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Holshue ML, DeBolt C, Lindquist S, Lofy
KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural
A, et al: First case of 2019 novel coronavirus in the united
states. N Engl J Med. 382:929–936. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wongsurawat T, Jenjaroenpun P, Taylor MK,
Lee J, Tolardo AL, Parvathareddy J, Kandel S, Wadley TD, Kaewnapan
B, Athipanyasilp N, et al: Rapid Sequencing of Multiple RNA Viruses
in Their Native Form. Front Microbiol. 10:2602019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Fang Y, Changavi A, Yang M, Sun L, Zhang
A, Sun D, Sun Z, Zhang B and Xu MQ: Nanopore Whole Transcriptome
Analysis and Pathogen Surveillance by a Novel Solid-Phase Catalysis
Approach. Adv Sci (Weinh). 9:e21033732022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Akaçin İ, Ersoy Ş, Doluca O and
Güngörmüşler M: Comparing the significance of the utilization of
next generation and third generation sequencing technologies in
microbial metagenomics. Microbiol Res. 264:1271542022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Gradisteanu Pircalabioru G, Iliescu FS,
Mihaescu G, Cucu AI, Ionescu ON, Popescu M, Simion M, Burlibasa L,
Tica M, Chifiriuc MC and Iliescu C: Advances in the rapid
diagnostic of viral respiratory tract infections. Front Cell Infect
Microbiol. 12:8072532022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sheng L, Lu Y, Deng S, Liao X, Zhang K,
Ding T, Gao H, Liu D, Deng R and Li J: A transcription aptasensor:
Amplified, label-free and culture-independent detection of
foodborne pathogens via light-up RNA aptamers. Chem Commun (Camb).
55:10096–10099. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Andryukov BG, Lyapun IN, Matosova EV and
Somova LM: Biosensor technologies in medicine: From detection of
biochemical markers to research into molecular targets (review).
Sovrem Tekhnologii Med. 12:70–83. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Robertson KL and Vora GJ: Locked nucleic
acid flow cytometry-fluorescence in situ hybridization (LNA
flow-FISH): A method for bacterial small RNA detection. J Vis Exp.
10:e36552012.PubMed/NCBI
|
|
119
|
Freen-van Heeren JJ: Flow-FISH as a tool
for studying bacteria, fungi and viruses. BioTech (Basel).
10:212021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Israr MZ, Bernieh D, Salzano A, Cassambai
S, Yazaki Y and Suzuki T: Matrix-assisted laser desorption
ionisation (MALDI) mass spectrometry (MS): basics and clinical
applications. Clin Chem Lab Med. 58:883–896. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Kailasa SK, Koduru JR, Park TJ, Wu HF and
Lin YC: Progress of electrospray ionization and rapid evaporative
ionization mass spectrometric techniques for the broad-range
identification of microorganisms. Analyst. 145:70722020. View Article : Google Scholar : PubMed/NCBI
|